File size: 2,950 Bytes
8917a83 4a2ad50 8917a83 4a2ad50 8917a83 27a9414 8917a83 4a2ad50 5810f7d 4a2ad50 5810f7d 4a2ad50 5810f7d 8917a83 5810f7d 8917a83 4a2ad50 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 |
---
license: apache-2.0
tags:
- vision
- image-classification
datasets:
- sartajbhuvaji/Brain-Tumor-Classification
language:
- en
pipeline_tag: image-classification
---
# MRI-Reader(small-sized model)
MRI-Reader is a fine-tuned version of [swin-base](https://huggingface.co/microsoft/swin-base-patch4-window7-224-in22k). It was introduced in this [paper](https://arxiv.org/abs/2103.14030) by Liu et al. and first released in this [repository](https://github.com/microsoft/Swin-Transformer).
## Model description
The Swin Transformer is a type of Vision Transformer. It builds hierarchical feature maps by merging image patches (shown in gray) in deeper layers and has linear computation complexity to input image size due to computation of self-attention only within each local window (shown in red). It can thus serve as a general-purpose backbone for both image classification and dense recognition tasks. In contrast, previous vision Transformers produce feature maps of a single low resolution and have quadratic computation complexity to input image size due to computation of self-attention globally.
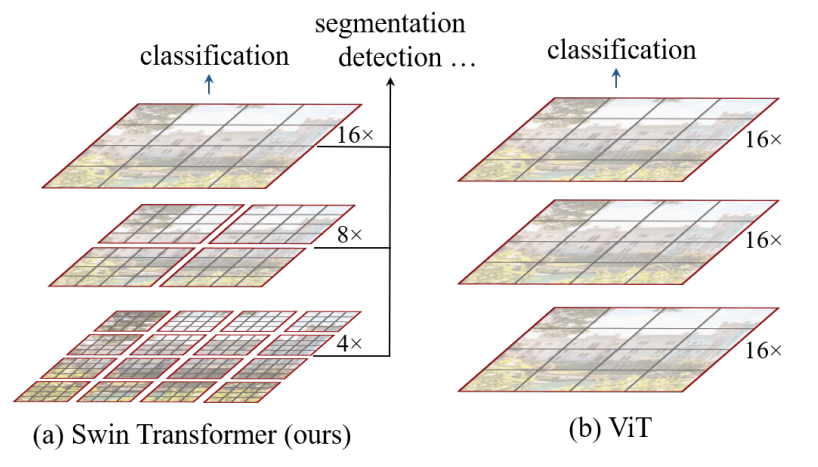
[Source](https://paperswithcode.com/method/swin-transformer)
### How to use
Here is how to use this model to identify meningioma tumor from a MRI scan:
```python
from transformers import AutoImageProcessor, AutoModelForImageClassification
from PIL import Image
import requests
processor = AutoImageProcessor.from_pretrained("NeuronZero/MRI-Reader")
model = AutoModelForImageClassification.from_pretrained("NeuronZero/MRI-Reader")
# Dataset url: https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri
image_url = "https://storage.googleapis.com/kagglesdsdata/datasets/672377/1183165/Testing/meningioma_tumor/image%28112%29.jpg?X-Goog-Algorithm=GOOG4-RSA-SHA256&X-Goog-Credential=databundle-worker-v2%40kaggle-161607.iam.gserviceaccount.com%2F20240326%2Fauto%2Fstorage%2Fgoog4_request&X-Goog-Date=20240326T125018Z&X-Goog-Expires=345600&X-Goog-SignedHeaders=host&X-Goog-Signature=32461d8d00888de5030d0dac653ecf5301c79a9445320a29c515713611fc8ec5bd6de1f1be490041f0dd937d7165f2bd3176ca926f2f33787a6ca7dbae1db2ce0b3a482a27a6258d4fe64c92ef7004c81488bfede784e50f22742e214cc303e8e9a52c6b4bc1db20e8aafba80589e87028e2f3212436c45fd7bc0a6978af3c2a2a5cbc25dcddf1489aecacaeebc75b93b2e111d391cf82c50a38906f88eec30e928285f043527972eed6d0dd2cd53b7e61c1be82bbefd6f8f38ffe438155e0dcf386425693a61c8c5857d6f4dbea7a8351e496160da261778c5f26d5496243f863ca65caf2b630701a998e79ce0bfa32291b19410a0f72d3399cea86b695c7dd"
image = Image.open(requests.get(image_url, stream=True).raw)
inputs = processor(images=image, return_tensors="pt")
outputs = model(**inputs)
logits = outputs.logits
predicted_class_idx = logits.argmax(-1).item()
print("Predicted class:", model.config.id2label[predicted_class_idx])
``` |