Commit
•
83a006f
1
Parent(s):
3f72d00
Upload 6 files
Browse files- M3D_Seg.jpg +3 -0
- data_load_demo.py +22 -0
- data_process.py +175 -0
- dataset_info.json +383 -0
- dataset_info.txt +153 -0
- term_dictionary.json +0 -0
M3D_Seg.jpg
ADDED
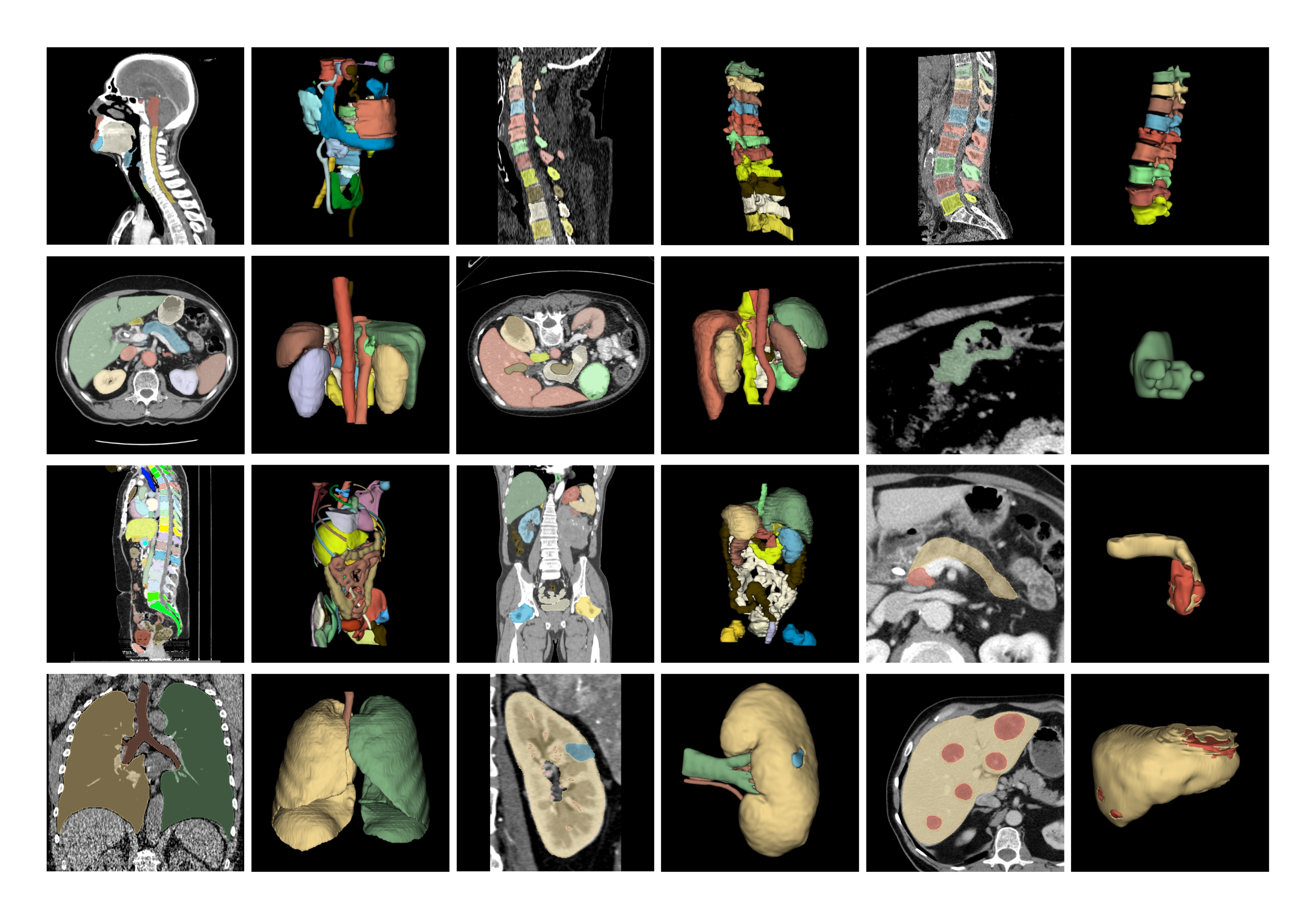
|
Git LFS Details
|
data_load_demo.py
ADDED
|
@@ -0,0 +1,22 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
from scipy import sparse
|
| 3 |
+
import ast
|
| 4 |
+
import os
|
| 5 |
+
import json
|
| 6 |
+
|
| 7 |
+
uniseg_path = '/PATH/M3D_Seg' # your path
|
| 8 |
+
dataset_code = '0001'
|
| 9 |
+
json_path = os.path.join('./', dataset_code, dataset_code + '.json')
|
| 10 |
+
with open(json_path, 'r') as f:
|
| 11 |
+
dataset_dict = json.load(f)
|
| 12 |
+
|
| 13 |
+
ct_file_path = os.path.join(uniseg_path, dataset_dict['train'][0]['image'])
|
| 14 |
+
gt_file_path = os.path.join(uniseg_path, dataset_dict['train'][0]['label'])
|
| 15 |
+
|
| 16 |
+
img_array = np.load(ct_file_path)[0]
|
| 17 |
+
print('img_array.shape ', img_array.shape)
|
| 18 |
+
|
| 19 |
+
allmatrix_sp= sparse.load_npz(gt_file_path)
|
| 20 |
+
gt_shape = ast.literal_eval(gt_file_path.split('.')[-2].split('_')[-1])
|
| 21 |
+
gt_array=allmatrix_sp.toarray().reshape(gt_shape)
|
| 22 |
+
print('gt_array.shape ', gt_array.shape)
|
data_process.py
ADDED
|
@@ -0,0 +1,175 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import numpy as np
|
| 3 |
+
import multiprocessing
|
| 4 |
+
import argparse
|
| 5 |
+
from scipy import sparse
|
| 6 |
+
from sklearn.model_selection import train_test_split
|
| 7 |
+
import json
|
| 8 |
+
|
| 9 |
+
from monai.transforms import (
|
| 10 |
+
AddChanneld,
|
| 11 |
+
Compose,
|
| 12 |
+
LoadImaged,
|
| 13 |
+
Orientationd,
|
| 14 |
+
)
|
| 15 |
+
|
| 16 |
+
def set_parse():
|
| 17 |
+
# %% set up parser
|
| 18 |
+
parser = argparse.ArgumentParser()
|
| 19 |
+
parser.add_argument("-category", default=['liver', 'right kidney', 'spleen', 'pancreas', 'aorta', 'inferior vena cava', 'right adrenal gland', 'left adrenal gland', 'gallbladder', 'esophagus', 'stomach', 'duodenum', 'left kidney'], type=list)
|
| 20 |
+
parser.add_argument("-image_dir", type=str, required=True)
|
| 21 |
+
parser.add_argument("-label_dir", type=str, required=True)
|
| 22 |
+
parser.add_argument("-dataset_code", type=str, required=True)
|
| 23 |
+
parser.add_argument("-save_root", type=str, required=True)
|
| 24 |
+
parser.add_argument("-test_ratio", type=float, required=True)
|
| 25 |
+
|
| 26 |
+
args = parser.parse_args()
|
| 27 |
+
return args
|
| 28 |
+
|
| 29 |
+
args = set_parse()
|
| 30 |
+
|
| 31 |
+
# get ct> dir
|
| 32 |
+
image_list_all = [item for item in sorted(os.listdir(args.image_dir))]
|
| 33 |
+
label_list_all = [item for item in sorted(os.listdir(args.label_dir))]
|
| 34 |
+
assert len(image_list_all) == len(label_list_all)
|
| 35 |
+
print('dataset size ', len(image_list_all))
|
| 36 |
+
|
| 37 |
+
# build dataset
|
| 38 |
+
data_path_list_all = []
|
| 39 |
+
for idx in range(len(image_list_all)):
|
| 40 |
+
img_path = os.path.join(args.image_dir, image_list_all[idx])
|
| 41 |
+
label_path = os.path.join(args.label_dir, label_list_all[idx])
|
| 42 |
+
name = image_list_all[idx].split('.')[0]
|
| 43 |
+
info = (idx, name, img_path, label_path)
|
| 44 |
+
data_path_list_all.append(info)
|
| 45 |
+
|
| 46 |
+
img_loader = Compose(
|
| 47 |
+
[
|
| 48 |
+
LoadImaged(keys=['image', 'label']),
|
| 49 |
+
AddChanneld(keys=['image', 'label']),
|
| 50 |
+
Orientationd(keys=['image', 'label'], axcodes="RAS"),
|
| 51 |
+
]
|
| 52 |
+
)
|
| 53 |
+
|
| 54 |
+
# save
|
| 55 |
+
save_path = os.path.join(args.save_root, args.dataset_code)
|
| 56 |
+
ct_save_path = os.path.join(save_path, 'ct')
|
| 57 |
+
gt_save_path = os.path.join(save_path, 'gt')
|
| 58 |
+
if not os.path.exists(ct_save_path):
|
| 59 |
+
os.makedirs(ct_save_path)
|
| 60 |
+
if not os.path.exists(gt_save_path):
|
| 61 |
+
os.makedirs(gt_save_path)
|
| 62 |
+
|
| 63 |
+
# exist file:
|
| 64 |
+
exist_file_list = os.listdir(ct_save_path)
|
| 65 |
+
print('exist_file_list ', exist_file_list)
|
| 66 |
+
|
| 67 |
+
def normalize(ct_narray):
|
| 68 |
+
ct_voxel_ndarray = ct_narray.copy()
|
| 69 |
+
ct_voxel_ndarray = ct_voxel_ndarray.flatten()
|
| 70 |
+
# for all data
|
| 71 |
+
thred = np.mean(ct_voxel_ndarray)
|
| 72 |
+
voxel_filtered = ct_voxel_ndarray[(ct_voxel_ndarray > thred)]
|
| 73 |
+
# for foreground data
|
| 74 |
+
upper_bound = np.percentile(voxel_filtered, 99.95)
|
| 75 |
+
lower_bound = np.percentile(voxel_filtered, 00.05)
|
| 76 |
+
mean = np.mean(voxel_filtered)
|
| 77 |
+
std = np.std(voxel_filtered)
|
| 78 |
+
### transform ###
|
| 79 |
+
ct_narray = np.clip(ct_narray, lower_bound, upper_bound)
|
| 80 |
+
ct_narray = (ct_narray - mean) / max(std, 1e-8)
|
| 81 |
+
return ct_narray
|
| 82 |
+
|
| 83 |
+
def run(info):
|
| 84 |
+
idx, file_name, case_path, label_path = info
|
| 85 |
+
item = {}
|
| 86 |
+
if file_name + '.npy' in exist_file_list:
|
| 87 |
+
print(file_name + '.npy exist, skip')
|
| 88 |
+
return
|
| 89 |
+
print('process ', idx, '---' ,file_name)
|
| 90 |
+
# generate ct_voxel_ndarray
|
| 91 |
+
item_load = {
|
| 92 |
+
'image' : case_path,
|
| 93 |
+
'label' : label_path,
|
| 94 |
+
}
|
| 95 |
+
item_load = img_loader(item_load)
|
| 96 |
+
ct_voxel_ndarray = item_load['image']
|
| 97 |
+
gt_voxel_ndarray = item_load['label']
|
| 98 |
+
|
| 99 |
+
ct_shape = ct_voxel_ndarray.shape
|
| 100 |
+
item['image'] = ct_voxel_ndarray
|
| 101 |
+
|
| 102 |
+
# generate gt_voxel_ndarray
|
| 103 |
+
gt_voxel_ndarray = np.array(gt_voxel_ndarray).squeeze()
|
| 104 |
+
present_categories = np.unique(gt_voxel_ndarray)
|
| 105 |
+
gt_masks = []
|
| 106 |
+
for cls_idx in range(len(args.category)):
|
| 107 |
+
cls = cls_idx + 1
|
| 108 |
+
if cls not in present_categories:
|
| 109 |
+
gt_voxel_ndarray_category = np.zeros(ct_shape)
|
| 110 |
+
gt_masks.append(gt_voxel_ndarray_category)
|
| 111 |
+
print('case {} ==> zero category '.format(idx) + args.category[cls_idx])
|
| 112 |
+
print(gt_voxel_ndarray_category.shape)
|
| 113 |
+
else:
|
| 114 |
+
gt_voxel_ndarray_category = gt_voxel_ndarray.copy()
|
| 115 |
+
gt_voxel_ndarray_category[gt_voxel_ndarray != cls] = 0
|
| 116 |
+
gt_voxel_ndarray_category[gt_voxel_ndarray == cls] = 1
|
| 117 |
+
gt_masks.append(gt_voxel_ndarray_category)
|
| 118 |
+
gt_voxel_ndarray = np.stack(gt_masks, axis=0)
|
| 119 |
+
|
| 120 |
+
assert gt_voxel_ndarray.shape[0] == len(args.category), str(gt_voxel_ndarray.shape[0])
|
| 121 |
+
assert gt_voxel_ndarray.shape[1:] == ct_voxel_ndarray.shape[1:]
|
| 122 |
+
item['label'] = gt_voxel_ndarray.astype(np.int32)
|
| 123 |
+
print(idx, ' load done!')
|
| 124 |
+
|
| 125 |
+
#############################
|
| 126 |
+
item['image'] = normalize(item['image'])
|
| 127 |
+
print(idx, ' transform done')
|
| 128 |
+
|
| 129 |
+
############################
|
| 130 |
+
print(file_name + ' ct gt <--> ', item['image'].shape, item['label'].shape)
|
| 131 |
+
np.save(os.path.join(ct_save_path, file_name + '.npy'), item['image'])
|
| 132 |
+
allmatrix_sp=sparse.csr_matrix(item['label'].reshape(item['label'].shape[0], -1))
|
| 133 |
+
sparse.save_npz(os.path.join(gt_save_path, file_name + '.' + str(item['label'].shape)), allmatrix_sp)
|
| 134 |
+
print(file_name + ' save done!')
|
| 135 |
+
|
| 136 |
+
def generate_dataset_json(root_dir, output_file, test_ratio=0.2):
|
| 137 |
+
ct_dir = os.path.join(root_dir, 'ct')
|
| 138 |
+
gt_dir = os.path.join(root_dir, 'gt')
|
| 139 |
+
ct_paths = sorted([os.path.join(ct_dir, f) for f in sorted(os.listdir(ct_dir))])
|
| 140 |
+
gt_paths = sorted([os.path.join(gt_dir, f) for f in sorted(os.listdir(gt_dir))])
|
| 141 |
+
|
| 142 |
+
data = list(zip(ct_paths, gt_paths))
|
| 143 |
+
train_data, val_data = train_test_split(data, test_size=test_ratio)
|
| 144 |
+
labels = {}
|
| 145 |
+
labels['0'] = 'background'
|
| 146 |
+
for idx in range(len(args.category)):
|
| 147 |
+
label_name = args.category[idx]
|
| 148 |
+
label_id = idx + 1
|
| 149 |
+
labels[str(label_id)] = label_name
|
| 150 |
+
dataset = {
|
| 151 |
+
'name': f'{args.dataset_code} Dataset',
|
| 152 |
+
'description': f'{args.dataset_code} Dataset',
|
| 153 |
+
'tensorImageSize': '4D',
|
| 154 |
+
'modality': {
|
| 155 |
+
'0': 'CT',
|
| 156 |
+
},
|
| 157 |
+
'labels': labels,
|
| 158 |
+
'numTraining': len(train_data),
|
| 159 |
+
'numTest': len(val_data),
|
| 160 |
+
'training': [{'image': ct_path, 'label': gt_path} for ct_path, gt_path in train_data],
|
| 161 |
+
'validation': [{'image': ct_path, 'label': gt_path} for ct_path, gt_path in val_data]
|
| 162 |
+
}
|
| 163 |
+
with open(output_file, 'w') as f:
|
| 164 |
+
print(f'{output_file} dump')
|
| 165 |
+
json.dump(dataset, f, indent=2)
|
| 166 |
+
|
| 167 |
+
if __name__ == "__main__":
|
| 168 |
+
with multiprocessing.Pool(processes=10) as pool:
|
| 169 |
+
pool.map(run, data_path_list_all)
|
| 170 |
+
print('Process Finished!')
|
| 171 |
+
|
| 172 |
+
generate_dataset_json(root_dir=save_path,
|
| 173 |
+
output_file=os.path.join(save_path, f'{args.dataset_code}.json'),
|
| 174 |
+
test_ratio=args.test_ratio)
|
| 175 |
+
print('Json Split Done!')
|
dataset_info.json
ADDED
|
@@ -0,0 +1,383 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"0000": [
|
| 3 |
+
"liver"
|
| 4 |
+
],
|
| 5 |
+
"0001": [
|
| 6 |
+
"A_Carotid_L",
|
| 7 |
+
"A_Carotid_R",
|
| 8 |
+
"Arytenoid",
|
| 9 |
+
"Bone_Mandible",
|
| 10 |
+
"Brainstem",
|
| 11 |
+
"BuccalMucosa",
|
| 12 |
+
"Cavity_Oral",
|
| 13 |
+
"Cochlea_L",
|
| 14 |
+
"Cochlea_R",
|
| 15 |
+
"Cricopharyngeus",
|
| 16 |
+
"Esophagus_S",
|
| 17 |
+
"Eye_AL",
|
| 18 |
+
"Eye_AR",
|
| 19 |
+
"Eye_PL",
|
| 20 |
+
"Eye_PR",
|
| 21 |
+
"Glnd_Lacrimal_L",
|
| 22 |
+
"Glnd_Lacrimal_R",
|
| 23 |
+
"Glnd_Submand_L",
|
| 24 |
+
"Glnd_Submand_R",
|
| 25 |
+
"Glnd_Thyroid",
|
| 26 |
+
"Glottis",
|
| 27 |
+
"Larynx_SG",
|
| 28 |
+
"Lips",
|
| 29 |
+
"OpticChiasm",
|
| 30 |
+
"OpticNrv_L",
|
| 31 |
+
"OpticNrv_R",
|
| 32 |
+
"Parotid_L",
|
| 33 |
+
"Parotid_R",
|
| 34 |
+
"Pituitary",
|
| 35 |
+
"SpinalCord"
|
| 36 |
+
],
|
| 37 |
+
"0002": [
|
| 38 |
+
"spleen",
|
| 39 |
+
"right kidney",
|
| 40 |
+
"left kidney",
|
| 41 |
+
"gall bladder",
|
| 42 |
+
"esophagus",
|
| 43 |
+
"liver",
|
| 44 |
+
"stomach",
|
| 45 |
+
"arota",
|
| 46 |
+
"postcava",
|
| 47 |
+
"pancreas",
|
| 48 |
+
"right adrenal gland",
|
| 49 |
+
"left adrenal gland",
|
| 50 |
+
"duodenum",
|
| 51 |
+
"bladder",
|
| 52 |
+
"prostate/uterus"
|
| 53 |
+
],
|
| 54 |
+
"0003": [
|
| 55 |
+
"liver",
|
| 56 |
+
"kidney",
|
| 57 |
+
"spleen",
|
| 58 |
+
"pancreas"
|
| 59 |
+
],
|
| 60 |
+
"0004": [
|
| 61 |
+
"kidney",
|
| 62 |
+
"kidney tumor",
|
| 63 |
+
"kidney cyst"
|
| 64 |
+
],
|
| 65 |
+
"0005": [
|
| 66 |
+
"Renal vein",
|
| 67 |
+
"Kidney",
|
| 68 |
+
"Renal artery",
|
| 69 |
+
"Tumor"
|
| 70 |
+
],
|
| 71 |
+
"0006": [
|
| 72 |
+
"kidney",
|
| 73 |
+
"kidney tumor"
|
| 74 |
+
],
|
| 75 |
+
"0007": [
|
| 76 |
+
"spleen",
|
| 77 |
+
"right kidney",
|
| 78 |
+
"left kidney",
|
| 79 |
+
"gallbladder",
|
| 80 |
+
"esophagus",
|
| 81 |
+
"liver",
|
| 82 |
+
"stomach",
|
| 83 |
+
"aorta",
|
| 84 |
+
"inferior vena cava",
|
| 85 |
+
"portal vein and splenic vein",
|
| 86 |
+
"pancreas",
|
| 87 |
+
"right adrenal gland",
|
| 88 |
+
"left adrenal gland"
|
| 89 |
+
],
|
| 90 |
+
"0008": [
|
| 91 |
+
"pancreas"
|
| 92 |
+
],
|
| 93 |
+
"0009": [
|
| 94 |
+
"Stones",
|
| 95 |
+
"artery",
|
| 96 |
+
"biliarysystem",
|
| 97 |
+
"bladder",
|
| 98 |
+
"bone",
|
| 99 |
+
"colon",
|
| 100 |
+
"gallbladder",
|
| 101 |
+
"heart",
|
| 102 |
+
"kidneys",
|
| 103 |
+
"leftkidney",
|
| 104 |
+
"leftlung",
|
| 105 |
+
"leftsurrenalgland",
|
| 106 |
+
"leftsurretumor",
|
| 107 |
+
"liver",
|
| 108 |
+
"livercyst",
|
| 109 |
+
"liverkyst",
|
| 110 |
+
"liverkyste",
|
| 111 |
+
"livertumor",
|
| 112 |
+
"livertumor01",
|
| 113 |
+
"livertumor02",
|
| 114 |
+
"livertumor03",
|
| 115 |
+
"livertumor04",
|
| 116 |
+
"livertumor05",
|
| 117 |
+
"livertumor06",
|
| 118 |
+
"livertumor07",
|
| 119 |
+
"livertumor1",
|
| 120 |
+
"livertumor2",
|
| 121 |
+
"livertumors",
|
| 122 |
+
"lungs",
|
| 123 |
+
"metal",
|
| 124 |
+
"metastasectomie",
|
| 125 |
+
"pancreas",
|
| 126 |
+
"portalvein",
|
| 127 |
+
"portalvein1",
|
| 128 |
+
"rightkidney",
|
| 129 |
+
"rightlung",
|
| 130 |
+
"rightsurrenalgland",
|
| 131 |
+
"rightsurretumor",
|
| 132 |
+
"skin",
|
| 133 |
+
"smallintestin",
|
| 134 |
+
"spleen",
|
| 135 |
+
"stomach",
|
| 136 |
+
"surrenalgland",
|
| 137 |
+
"tumor",
|
| 138 |
+
"uterus",
|
| 139 |
+
"venacava",
|
| 140 |
+
"venoussystem"
|
| 141 |
+
],
|
| 142 |
+
"0010": [
|
| 143 |
+
"liver",
|
| 144 |
+
"right kidney",
|
| 145 |
+
"spleen",
|
| 146 |
+
"pancreas",
|
| 147 |
+
"aorta",
|
| 148 |
+
"inferior vena cava",
|
| 149 |
+
"right adrenal gland",
|
| 150 |
+
"left adrenal gland",
|
| 151 |
+
"gallbladder",
|
| 152 |
+
"esophagus",
|
| 153 |
+
"stomach",
|
| 154 |
+
"duodenum",
|
| 155 |
+
"left kidney"
|
| 156 |
+
],
|
| 157 |
+
"0011": [
|
| 158 |
+
"adrenal_gland_left",
|
| 159 |
+
"adrenal_gland_right",
|
| 160 |
+
"aorta",
|
| 161 |
+
"autochthon_left",
|
| 162 |
+
"autochthon_right",
|
| 163 |
+
"brain",
|
| 164 |
+
"clavicula_left",
|
| 165 |
+
"clavicula_right",
|
| 166 |
+
"colon",
|
| 167 |
+
"duodenum",
|
| 168 |
+
"esophagus",
|
| 169 |
+
"face",
|
| 170 |
+
"femur_left",
|
| 171 |
+
"femur_right",
|
| 172 |
+
"gallbladder",
|
| 173 |
+
"gluteus_maximus_left",
|
| 174 |
+
"gluteus_maximus_right",
|
| 175 |
+
"gluteus_medius_left",
|
| 176 |
+
"gluteus_medius_right",
|
| 177 |
+
"gluteus_minimus_left",
|
| 178 |
+
"gluteus_minimus_right",
|
| 179 |
+
"heart_atrium_left",
|
| 180 |
+
"heart_atrium_right",
|
| 181 |
+
"heart_myocardium",
|
| 182 |
+
"heart_ventricle_left",
|
| 183 |
+
"heart_ventricle_right",
|
| 184 |
+
"hip_left",
|
| 185 |
+
"hip_right",
|
| 186 |
+
"humerus_left",
|
| 187 |
+
"humerus_right",
|
| 188 |
+
"iliac_artery_left",
|
| 189 |
+
"iliac_artery_right",
|
| 190 |
+
"iliac_vena_left",
|
| 191 |
+
"iliac_vena_right",
|
| 192 |
+
"iliopsoas_left",
|
| 193 |
+
"iliopsoas_right",
|
| 194 |
+
"inferior_vena_cava",
|
| 195 |
+
"kidney_left",
|
| 196 |
+
"kidney_right",
|
| 197 |
+
"liver",
|
| 198 |
+
"lung_lower_lobe_left",
|
| 199 |
+
"lung_lower_lobe_right",
|
| 200 |
+
"lung_middle_lobe_right",
|
| 201 |
+
"lung_upper_lobe_left",
|
| 202 |
+
"lung_upper_lobe_right",
|
| 203 |
+
"pancreas",
|
| 204 |
+
"portal_vein_and_splenic_vein",
|
| 205 |
+
"pulmonary_artery",
|
| 206 |
+
"rib_left_1",
|
| 207 |
+
"rib_left_10",
|
| 208 |
+
"rib_left_11",
|
| 209 |
+
"rib_left_12",
|
| 210 |
+
"rib_left_2",
|
| 211 |
+
"rib_left_3",
|
| 212 |
+
"rib_left_4",
|
| 213 |
+
"rib_left_5",
|
| 214 |
+
"rib_left_6",
|
| 215 |
+
"rib_left_7",
|
| 216 |
+
"rib_left_8",
|
| 217 |
+
"rib_left_9",
|
| 218 |
+
"rib_right_1",
|
| 219 |
+
"rib_right_10",
|
| 220 |
+
"rib_right_11",
|
| 221 |
+
"rib_right_12",
|
| 222 |
+
"rib_right_2",
|
| 223 |
+
"rib_right_3",
|
| 224 |
+
"rib_right_4",
|
| 225 |
+
"rib_right_5",
|
| 226 |
+
"rib_right_6",
|
| 227 |
+
"rib_right_7",
|
| 228 |
+
"rib_right_8",
|
| 229 |
+
"rib_right_9",
|
| 230 |
+
"sacrum",
|
| 231 |
+
"scapula_left",
|
| 232 |
+
"scapula_right",
|
| 233 |
+
"small_bowel",
|
| 234 |
+
"spleen",
|
| 235 |
+
"stomach",
|
| 236 |
+
"trachea",
|
| 237 |
+
"urinary_bladder",
|
| 238 |
+
"vertebrae_C1",
|
| 239 |
+
"vertebrae_C2",
|
| 240 |
+
"vertebrae_C3",
|
| 241 |
+
"vertebrae_C4",
|
| 242 |
+
"vertebrae_C5",
|
| 243 |
+
"vertebrae_C6",
|
| 244 |
+
"vertebrae_C7",
|
| 245 |
+
"vertebrae_L1",
|
| 246 |
+
"vertebrae_L2",
|
| 247 |
+
"vertebrae_L3",
|
| 248 |
+
"vertebrae_L4",
|
| 249 |
+
"vertebrae_L5",
|
| 250 |
+
"vertebrae_T1",
|
| 251 |
+
"vertebrae_T10",
|
| 252 |
+
"vertebrae_T11",
|
| 253 |
+
"vertebrae_T12",
|
| 254 |
+
"vertebrae_T2",
|
| 255 |
+
"vertebrae_T3",
|
| 256 |
+
"vertebrae_T4",
|
| 257 |
+
"vertebrae_T5",
|
| 258 |
+
"vertebrae_T6",
|
| 259 |
+
"vertebrae_T7",
|
| 260 |
+
"vertebrae_T8",
|
| 261 |
+
"vertebrae_T9"
|
| 262 |
+
],
|
| 263 |
+
"0012": [
|
| 264 |
+
"Liver",
|
| 265 |
+
"Bladder",
|
| 266 |
+
"Lungs",
|
| 267 |
+
"Kidneys",
|
| 268 |
+
"Bone",
|
| 269 |
+
"Brain"
|
| 270 |
+
],
|
| 271 |
+
"0013": [
|
| 272 |
+
"Liver",
|
| 273 |
+
"Spleen",
|
| 274 |
+
"Kidney (L)",
|
| 275 |
+
"Kidney (R)",
|
| 276 |
+
"Stomach",
|
| 277 |
+
"Gallbladder",
|
| 278 |
+
"Esophagus",
|
| 279 |
+
"Pancreas",
|
| 280 |
+
"Duodenum",
|
| 281 |
+
"Colon",
|
| 282 |
+
"Intestine",
|
| 283 |
+
"Adrenal",
|
| 284 |
+
"Rectum",
|
| 285 |
+
"Bladder",
|
| 286 |
+
"Head of femur (L)",
|
| 287 |
+
"Head of femur (R)"
|
| 288 |
+
],
|
| 289 |
+
"0014": [
|
| 290 |
+
"cervical spine C1",
|
| 291 |
+
"cervical spine C2",
|
| 292 |
+
"cervical spine C3",
|
| 293 |
+
"cervical spine C4",
|
| 294 |
+
"cervical spine C5",
|
| 295 |
+
"cervical spine C6",
|
| 296 |
+
"cervical spine C7",
|
| 297 |
+
"thoracic spine T1",
|
| 298 |
+
"thoracic spine T2",
|
| 299 |
+
"thoracic spine T3",
|
| 300 |
+
"thoracic spine T4",
|
| 301 |
+
"thoracic spine T5",
|
| 302 |
+
"thoracic spine T6",
|
| 303 |
+
"thoracic spine T7",
|
| 304 |
+
"thoracic spine T8",
|
| 305 |
+
"thoracic spine T9",
|
| 306 |
+
"thoracic spine T10",
|
| 307 |
+
"thoracic spine T11",
|
| 308 |
+
"thoracic spine T12",
|
| 309 |
+
"lumbar spine L1",
|
| 310 |
+
"lumbar spine L2",
|
| 311 |
+
"lumbar spine L3",
|
| 312 |
+
"lumbar spine L4",
|
| 313 |
+
"lumbar spine L5",
|
| 314 |
+
"lumbar spine L6",
|
| 315 |
+
"sacrum",
|
| 316 |
+
"cocygis",
|
| 317 |
+
"additional 13th thoracic vertebra, T13"
|
| 318 |
+
],
|
| 319 |
+
"0015": [
|
| 320 |
+
"cervical spine C1",
|
| 321 |
+
"cervical spine C2",
|
| 322 |
+
"cervical spine C3",
|
| 323 |
+
"cervical spine C4",
|
| 324 |
+
"cervical spine C5",
|
| 325 |
+
"cervical spine C6",
|
| 326 |
+
"cervical spine C7",
|
| 327 |
+
"thoracic spine T1",
|
| 328 |
+
"thoracic spine T2",
|
| 329 |
+
"thoracic spine T3",
|
| 330 |
+
"thoracic spine T4",
|
| 331 |
+
"thoracic spine T5",
|
| 332 |
+
"thoracic spine T6",
|
| 333 |
+
"thoracic spine T7",
|
| 334 |
+
"thoracic spine T8",
|
| 335 |
+
"thoracic spine T9",
|
| 336 |
+
"thoracic spine T10",
|
| 337 |
+
"thoracic spine T11",
|
| 338 |
+
"thoracic spine T12",
|
| 339 |
+
"lumbar spine L1",
|
| 340 |
+
"lumbar spine L2",
|
| 341 |
+
"lumbar spine L3",
|
| 342 |
+
"lumbar spine L4",
|
| 343 |
+
"lumbar spine L5",
|
| 344 |
+
"lumbar spine L6",
|
| 345 |
+
"sacrum",
|
| 346 |
+
"cocygis",
|
| 347 |
+
"additional 13th thoracic vertebra, T13"
|
| 348 |
+
],
|
| 349 |
+
"0016": [
|
| 350 |
+
"liver"
|
| 351 |
+
],
|
| 352 |
+
"0017": [
|
| 353 |
+
"kidney",
|
| 354 |
+
"pancreas",
|
| 355 |
+
"pancreatic-lesion"
|
| 356 |
+
],
|
| 357 |
+
"0018": [
|
| 358 |
+
"colon cancer"
|
| 359 |
+
],
|
| 360 |
+
"0019": [
|
| 361 |
+
"hepatic vessels",
|
| 362 |
+
"tumour"
|
| 363 |
+
],
|
| 364 |
+
"0020": [
|
| 365 |
+
"liver",
|
| 366 |
+
"tumour"
|
| 367 |
+
],
|
| 368 |
+
"0021": [
|
| 369 |
+
"lung tumours"
|
| 370 |
+
],
|
| 371 |
+
"0022": [
|
| 372 |
+
"pancreas",
|
| 373 |
+
"tumour"
|
| 374 |
+
],
|
| 375 |
+
"0023": [
|
| 376 |
+
"spleen"
|
| 377 |
+
],
|
| 378 |
+
"0024": [
|
| 379 |
+
"left lung",
|
| 380 |
+
"right lung",
|
| 381 |
+
"trachea"
|
| 382 |
+
]
|
| 383 |
+
}
|
dataset_info.txt
ADDED
|
@@ -0,0 +1,153 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
dataset_name = {
|
| 2 |
+
'0000': 'CHAOS',
|
| 3 |
+
'0001': 'HaN-Seg',
|
| 4 |
+
'0002': 'AMOS22',
|
| 5 |
+
'0003': 'AbdomenCT-1k',
|
| 6 |
+
'0004': 'KiTS23',
|
| 7 |
+
'0005': 'KiPA22',
|
| 8 |
+
'0006': 'KiTS19',
|
| 9 |
+
'0007': 'BCTV',
|
| 10 |
+
'0008': 'Pancreas-CT',
|
| 11 |
+
'0009': '3D-IRCADb',
|
| 12 |
+
'0010': 'AbdomenCT-12organ',
|
| 13 |
+
'0011': 'TotalSegmentator',
|
| 14 |
+
'0012': 'CT-ORG',
|
| 15 |
+
'0013': 'WORD',
|
| 16 |
+
'0014': 'VerSe19',
|
| 17 |
+
'0015': 'VerSe20',
|
| 18 |
+
'0016': 'SLIVER07',
|
| 19 |
+
'0017': 'QUBIQ',
|
| 20 |
+
'0018': 'MSD-colon',
|
| 21 |
+
'0019': 'MSD-hepatic_vessel',
|
| 22 |
+
'0020': 'MSD-liver',
|
| 23 |
+
'0021': 'MSD-lung',
|
| 24 |
+
'0022': 'MSD-pancreas',
|
| 25 |
+
'0023': 'MSD-spleen',
|
| 26 |
+
'0024': 'LUNA16',
|
| 27 |
+
}
|
| 28 |
+
|
| 29 |
+
dataset_info_raw = {
|
| 30 |
+
'0000': ['liver'],
|
| 31 |
+
'0001': ['A_Carotid_L','A_Carotid_R','Arytenoid','Bone_Mandible','Brainstem','BuccalMucosa','Cavity_Oral','Cochlea_L','Cochlea_R','Cricopharyngeus','Esophagus_S','Eye_AL','Eye_AR','Eye_PL','Eye_PR','Glnd_Lacrimal_L','Glnd_Lacrimal_R','Glnd_Submand_L','Glnd_Submand_R','Glnd_Thyroid','Glottis','Larynx_SG','Lips','OpticChiasm','OpticNrv_L','OpticNrv_R','Parotid_L','Parotid_R','Pituitary','SpinalCord'],
|
| 32 |
+
'0002': ["spleen", "right kidney", "left kidney", "gall bladder", "esophagus", "liver", "stomach", "arota", "postcava", "pancreas", "right adrenal gland", "left adrenal gland", "duodenum", "bladder", "prostate/uterus"],
|
| 33 |
+
'0003': ['liver', 'kidney', 'spleen', 'pancreas'],
|
| 34 |
+
'0004': ['kidney', 'kidney tumor', 'kidney cyst'],
|
| 35 |
+
'0005': ['Renal vein', 'Kidney', 'Renal artery', 'Tumor'],
|
| 36 |
+
'0006': ['kidney', 'kidney tumor'],
|
| 37 |
+
'0007': ['spleen','right kidney','left kidney','gallbladder','esophagus','liver','stomach','aorta','inferior vena cava','portal vein and splenic vein','pancreas','right adrenal gland','left adrenal gland'],
|
| 38 |
+
'0008': ['pancreas'],
|
| 39 |
+
'0009': ['Stones', 'artery', 'biliarysystem', 'bladder', 'bone', 'colon', 'gallbladder', 'heart', 'kidneys', 'leftkidney', 'leftlung', 'leftsurrenalgland', 'leftsurretumor', 'liver', 'livercyst', 'liverkyst', 'liverkyste', 'livertumor', 'livertumor01', 'livertumor02', 'livertumor03', 'livertumor04', 'livertumor05', 'livertumor06', 'livertumor07', 'livertumor1', 'livertumor2', 'livertumors', 'lungs', 'metal', 'metastasectomie', 'pancreas', 'portalvein', 'portalvein1', 'rightkidney', 'rightlung', 'rightsurrenalgland', 'rightsurretumor', 'skin', 'smallintestin', 'spleen', 'stomach', 'surrenalgland', 'tumor', 'uterus', 'venacava', 'venoussystem'],
|
| 40 |
+
'0010': ['liver', 'right kidney', 'spleen', 'pancreas', 'aorta', 'inferior vena cava', 'right adrenal gland', 'left adrenal gland', 'gallbladder', 'esophagus', 'stomach', 'duodenum', 'left kidney'],
|
| 41 |
+
'0011': ['adrenal_gland_left', 'adrenal_gland_right', 'aorta', 'autochthon_left', 'autochthon_right', 'brain', 'clavicula_left', 'clavicula_right', 'colon', 'duodenum', 'esophagus', 'face', 'femur_left', 'femur_right', 'gallbladder', 'gluteus_maximus_left', 'gluteus_maximus_right', 'gluteus_medius_left', 'gluteus_medius_right', 'gluteus_minimus_left', 'gluteus_minimus_right', 'heart_atrium_left', 'heart_atrium_right', 'heart_myocardium', 'heart_ventricle_left', 'heart_ventricle_right', 'hip_left', 'hip_right', 'humerus_left', 'humerus_right', 'iliac_artery_left', 'iliac_artery_right', 'iliac_vena_left', 'iliac_vena_right', 'iliopsoas_left', 'iliopsoas_right', 'inferior_vena_cava', 'kidney_left', 'kidney_right', 'liver', 'lung_lower_lobe_left', 'lung_lower_lobe_right', 'lung_middle_lobe_right', 'lung_upper_lobe_left', 'lung_upper_lobe_right', 'pancreas', 'portal_vein_and_splenic_vein', 'pulmonary_artery', 'rib_left_1', 'rib_left_10', 'rib_left_11', 'rib_left_12', 'rib_left_2', 'rib_left_3', 'rib_left_4', 'rib_left_5', 'rib_left_6', 'rib_left_7', 'rib_left_8', 'rib_left_9', 'rib_right_1', 'rib_right_10', 'rib_right_11', 'rib_right_12', 'rib_right_2', 'rib_right_3', 'rib_right_4', 'rib_right_5', 'rib_right_6', 'rib_right_7', 'rib_right_8', 'rib_right_9', 'sacrum', 'scapula_left', 'scapula_right', 'small_bowel', 'spleen', 'stomach', 'trachea', 'urinary_bladder', 'vertebrae_C1', 'vertebrae_C2', 'vertebrae_C3', 'vertebrae_C4', 'vertebrae_C5', 'vertebrae_C6', 'vertebrae_C7', 'vertebrae_L1', 'vertebrae_L2', 'vertebrae_L3', 'vertebrae_L4', 'vertebrae_L5', 'vertebrae_T1', 'vertebrae_T10', 'vertebrae_T11', 'vertebrae_T12', 'vertebrae_T2', 'vertebrae_T3', 'vertebrae_T4', 'vertebrae_T5', 'vertebrae_T6', 'vertebrae_T7', 'vertebrae_T8', 'vertebrae_T9'],
|
| 42 |
+
'0012': ['Liver','Bladder','Lungs','Kidneys','Bone','Brain'],
|
| 43 |
+
'0013': ['Liver','Spleen','Kidney (L)','Kidney (R)','Stomach','Gallbladder','Esophagus','Pancreas','Duodenum','Colon','Intestine','Adrenal','Rectum','Bladder','Head of femur (L)','Head of femur (R)'],
|
| 44 |
+
'0014': ['cervical spine C1', 'cervical spine C2', 'cervical spine C3', 'cervical spine C4', 'cervical spine C5', 'cervical spine C6', 'cervical spine C7', 'thoracic spine T1', 'thoracic spine T2', 'thoracic spine T3', 'thoracic spine T4', 'thoracic spine T5', 'thoracic spine T6', 'thoracic spine T7', 'thoracic spine T8', 'thoracic spine T9', 'thoracic spine T10', 'thoracic spine T11', 'thoracic spine T12', 'lumbar spine L1', 'lumbar spine L1', 'lumbar spine L3', 'lumbar spine L4', 'lumbar spine L5', 'lumbar spine L6', 'sacrum','cocygis','additional 13th thoracic vertebra, T13',],
|
| 45 |
+
'0015': ['cervical spine C1', 'cervical spine C2', 'cervical spine C3', 'cervical spine C4', 'cervical spine C5', 'cervical spine C6', 'cervical spine C7', 'thoracic spine T1', 'thoracic spine T2', 'thoracic spine T3', 'thoracic spine T4', 'thoracic spine T5', 'thoracic spine T6', 'thoracic spine T7', 'thoracic spine T8', 'thoracic spine T9', 'thoracic spine T10', 'thoracic spine T11', 'thoracic spine T12', 'lumbar spine L1', 'lumbar spine L1', 'lumbar spine L3', 'lumbar spine L4', 'lumbar spine L5', 'lumbar spine L6', 'sacrum','cocygis','additional 13th thoracic vertebra, T13',],
|
| 46 |
+
'0016': ['liver'],
|
| 47 |
+
'0017': ['kidney', 'pancreas', 'pancreatic-lesion'],
|
| 48 |
+
'0018': ['colon cancer'],
|
| 49 |
+
'0019': ['hepatic vessels', 'tumour'],
|
| 50 |
+
'0020': ['liver', 'tumour'],
|
| 51 |
+
'0021': ['lung tumours'],
|
| 52 |
+
'0022': ['pancreas', 'tumour'],
|
| 53 |
+
'0023': ['spleen'],
|
| 54 |
+
'0024': ['left lung', 'right lung', 'trachea'],
|
| 55 |
+
}
|
| 56 |
+
|
| 57 |
+
|
| 58 |
+
|
| 59 |
+
dataset_info = {
|
| 60 |
+
'0000': ['liver'],
|
| 61 |
+
'0001': ['carotid artery left', 'carotid artery right', 'arytenoid', 'bone mandible', 'brainstem',
|
| 62 |
+
'buccal mucosa',
|
| 63 |
+
'oral cavity', 'cochlea left', 'cochlea right', 'cricopharyngeal inlet', 'cervical esophagus',
|
| 64 |
+
'anterior eyeball left', 'anterior eyeball right',
|
| 65 |
+
'posterior eyeball left', 'posterior eyeball right', 'lacrimal gland left', 'lacrimal gland right',
|
| 66 |
+
'submandibular gland left', 'submandibular gland right',
|
| 67 |
+
'thyroid', 'larynx glottis', 'larynx supraglottic', 'lips', 'optic chiasm', 'optic nerve left',
|
| 68 |
+
'optic nerve right',
|
| 69 |
+
'parotid gland left', 'parotid gland right', 'pituitary gland', 'spinal cord'],
|
| 70 |
+
'0002': ["spleen", "right kidney", "left kidney", "gall bladder", "esophagus", "liver", "stomach", "aorta",
|
| 71 |
+
"postcava", "pancreas", "right adrenal gland", "left adrenal gland", "duodenum", "bladder",
|
| 72 |
+
"prostate or uterus"],
|
| 73 |
+
'0003': ['liver', 'kidney', 'spleen', 'pancreas'],
|
| 74 |
+
'0004': ['kidney', 'kidney tumor', 'kidney cyst'],
|
| 75 |
+
'0005': ['renal vein', 'kidney', 'renal artery', 'tumor'],
|
| 76 |
+
'0006': ['kidney', 'kidney tumor'],
|
| 77 |
+
'0007': ['spleen', 'right kidney', 'left kidney', 'gallbladder', 'esophagus', 'liver', 'stomach', 'aorta',
|
| 78 |
+
'inferior vena cava', 'portal vein and splenic vein', 'pancreas', 'right adrenal gland',
|
| 79 |
+
'left adrenal gland'],
|
| 80 |
+
'0008': ['pancreas'],
|
| 81 |
+
'0009': ['stones', 'artery', 'biliary system', 'bladder', 'bone', 'colon', 'gallbladder', 'heart',
|
| 82 |
+
'kidneys',
|
| 83 |
+
'left kidney', 'left lung', 'left suprarenal gland', 'left suprarenal tumor', 'liver',
|
| 84 |
+
'liver cyst', 'liver kyst',
|
| 85 |
+
|
| 86 |
+
'liver kyste', 'liver tumor', 'liver tumor 01', 'liver tumor 02', 'liver tumor 03',
|
| 87 |
+
'liver tumor 04',
|
| 88 |
+
'liver tumor 05', 'liver tumor 06', 'liver tumor 07', 'liver tumor 1', 'liver tumor 2',
|
| 89 |
+
'liver tumors',
|
| 90 |
+
|
| 91 |
+
'lungs', 'metal', 'metastasectomie', 'pancreas', 'portal vein', 'portal vein 1', 'right kidney',
|
| 92 |
+
'right lung', 'right suprarenal gland', 'right suprarenal tumor', 'skin', 'small intestin',
|
| 93 |
+
'spleen', 'stomach',
|
| 94 |
+
'suprarenal gland', 'tumor', 'uterus', 'vena cava', 'venous system'],
|
| 95 |
+
'0010': ['liver', 'right kidney', 'spleen', 'pancreas', 'aorta', 'inferior vena cava',
|
| 96 |
+
'right adrenal gland', 'left adrenal gland', 'gallbladder', 'esophagus', 'stomach', 'duodenum',
|
| 97 |
+
'left kidney'],
|
| 98 |
+
'0011': ['adrenal gland left', 'adrenal gland right', 'aorta', 'autochthon left', 'autochthon right',
|
| 99 |
+
'brain', 'clavicula left', 'clavicula right', 'colon', 'duodenum', 'esophagus', 'face',
|
| 100 |
+
'femur left', 'femur right', 'gallbladder', 'gluteus maximus left', 'gluteus maximus right',
|
| 101 |
+
'gluteus medius left', 'gluteus medius right', 'gluteus minimus left', 'gluteus minimus right',
|
| 102 |
+
|
| 103 |
+
'heart atrium left', 'heart atrium right', 'heart myocardium', 'heart ventricle left',
|
| 104 |
+
'heart ventricle right', 'hip left', 'hip right', 'humerus left', 'humerus right',
|
| 105 |
+
'iliac artery left', 'iliac artery right', 'iliac vena left', 'iliac vena right', 'iliopsoas left',
|
| 106 |
+
|
| 107 |
+
'iliopsoas right', 'inferior vena cava', 'kidney left', 'kidney right', 'liver',
|
| 108 |
+
'lung lower lobe left', 'lung lower lobe right', 'lung middle lobe right', 'lung upper lobe left',
|
| 109 |
+
'lung upper lobe right', 'pancreas', 'portal vein and splenic vein', 'pulmonary artery',
|
| 110 |
+
|
| 111 |
+
'rib left 1', 'rib left 10', 'rib left 11', 'rib left 12', 'rib left 2', 'rib left 3',
|
| 112 |
+
'rib left 4', 'rib left 5', 'rib left 6', 'rib left 7', 'rib left 8', 'rib left 9', 'rib right 1',
|
| 113 |
+
|
| 114 |
+
'rib right 10', 'rib right 11', 'rib right 12', 'rib right 2', 'rib right 3', 'rib right 4',
|
| 115 |
+
'rib right 5', 'rib right 6', 'rib right 7', 'rib right 8', 'rib right 9', 'sacrum',
|
| 116 |
+
|
| 117 |
+
'scapula left', 'scapula right', 'small bowel', 'spleen', 'stomach', 'trachea', 'urinary bladder',
|
| 118 |
+
'vertebrae C1', 'vertebrae C2', 'vertebrae C3', 'vertebrae C4', 'vertebrae C5', 'vertebrae C6',
|
| 119 |
+
|
| 120 |
+
'vertebrae C7', 'vertebrae L1', 'vertebrae L2', 'vertebrae L3', 'vertebrae L4', 'vertebrae L5',
|
| 121 |
+
'vertebrae T1', 'vertebrae T10', 'vertebrae T11', 'vertebrae T12', 'vertebrae T2', 'vertebrae T3',
|
| 122 |
+
'vertebrae T4', 'vertebrae T5', 'vertebrae T6', 'vertebrae T7', 'vertebrae T8', 'vertebrae T9'],
|
| 123 |
+
'0012': ['liver', 'bladder', 'lungs', 'kidneys', 'bone', 'brain'],
|
| 124 |
+
'0013': ['liver', 'spleen', 'kidney left', 'kidney right', 'stomach', 'gallbladder', 'esophagus',
|
| 125 |
+
'pancreas',
|
| 126 |
+
'duodenum', 'colon', 'intestine', 'adrenal', 'rectum', 'bladder', 'head of femur left',
|
| 127 |
+
'head of femur right'],
|
| 128 |
+
'0014': ['cervical spine C1', 'cervical spine C2', 'cervical spine C3', 'cervical spine C4',
|
| 129 |
+
'cervical spine C5', 'cervical spine C6', 'cervical spine C7', 'thoracic spine T1',
|
| 130 |
+
|
| 131 |
+
'thoracic spine T2', 'thoracic spine T3', 'thoracic spine T4', 'thoracic spine T5',
|
| 132 |
+
'thoracic spine T6', 'thoracic spine T7', 'thoracic spine T8', 'thoracic spine T9',
|
| 133 |
+
|
| 134 |
+
'thoracic spine T10', 'thoracic spine T11', 'thoracic spine T12', 'lumbar spine L1',
|
| 135 |
+
'lumbar spine L1', 'lumbar spine L3', 'lumbar spine L4', 'lumbar spine L5', 'lumbar spine L6',
|
| 136 |
+
'sacrum', 'coccygis', 'additional 13th thoracic vertebra, T13', ],
|
| 137 |
+
'0015': ['cervical spine C1', 'cervical spine C2', 'cervical spine C3', 'cervical spine C4',
|
| 138 |
+
'cervical spine C5', 'cervical spine C6', 'cervical spine C7', 'thoracic spine T1',
|
| 139 |
+
'thoracic spine T2', 'thoracic spine T3', 'thoracic spine T4', 'thoracic spine T5',
|
| 140 |
+
'thoracic spine T6', 'thoracic spine T7', 'thoracic spine T8', 'thoracic spine T9',
|
| 141 |
+
'thoracic spine T10', 'thoracic spine T11', 'thoracic spine T12', 'lumbar spine L1',
|
| 142 |
+
'lumbar spine L1', 'lumbar spine L3', 'lumbar spine L4', 'lumbar spine L5', 'lumbar spine L6',
|
| 143 |
+
'sacrum', 'coccygis', 'additional 13th thoracic vertebra T13', ],
|
| 144 |
+
'0016': ['liver'],
|
| 145 |
+
'0017': ['kidney', 'pancreas', 'pancreatic lesion'],
|
| 146 |
+
'0018': ['colon cancer'],
|
| 147 |
+
'0019': ['hepatic vessels', 'tumour'],
|
| 148 |
+
'0020': ['liver', 'tumour'],
|
| 149 |
+
'0021': ['lung tumours'],
|
| 150 |
+
'0022': ['pancreas', 'tumour'],
|
| 151 |
+
'0023': ['spleen'],
|
| 152 |
+
'0024': ['left lung', 'right lung', 'trachea'],
|
| 153 |
+
}
|
term_dictionary.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|