complete the model package
Browse files- .gitattributes +1 -0
- README.md +171 -0
- configs/inference.json +136 -0
- configs/logging.conf +21 -0
- configs/metadata.json +210 -0
- docs/3DSlicer_use.png +0 -0
- docs/README.md +164 -0
- docs/demo.png +3 -0
- docs/license.txt +4 -0
- docs/unest.png +0 -0
- docs/wholebrain.png +0 -0
- models/model.pt +3 -0
- scripts/__init__.py +10 -0
- scripts/networks/__init__.py +10 -0
- scripts/networks/nest/__init__.py +16 -0
- scripts/networks/nest/utils.py +485 -0
- scripts/networks/nest_transformer_3D.py +489 -0
- scripts/networks/patchEmbed3D.py +190 -0
- scripts/networks/unest_base_patch_4.py +249 -0
- scripts/networks/unest_block.py +245 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
docs/demo.png filter=lfs diff=lfs merge=lfs -text
|
README.md
ADDED
|
@@ -0,0 +1,171 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
tags:
|
| 3 |
+
- monai
|
| 4 |
+
- medical
|
| 5 |
+
library_name: monai
|
| 6 |
+
license: unknown
|
| 7 |
+
---
|
| 8 |
+
# Description
|
| 9 |
+
Detailed whole brain segmentation is an essential quantitative technique in medical image analysis, which provides a non-invasive way of measuring brain regions from a clinical acquired structural magnetic resonance imaging (MRI).
|
| 10 |
+
We provide the pre-trained model for inferencing whole brain segmentation with 133 structures.
|
| 11 |
+
|
| 12 |
+
A tutorial and release of model for whole brain segmentation using the 3D transformer-based segmentation model UNEST.
|
| 13 |
+
|
| 14 |
+
Authors:
|
| 15 |
+
Xin Yu ([email protected]) (Primary)
|
| 16 |
+
|
| 17 |
+
Yinchi Zhou ([email protected]) | Yucheng Tang ([email protected])
|
| 18 |
+
|
| 19 |
+
<p align="center">
|
| 20 |
+
-------------------------------------------------------------------------------------
|
| 21 |
+
</p>
|
| 22 |
+
|
| 23 |
+
 <br>
|
| 24 |
+
<p align="center">
|
| 25 |
+
Fig.1 - The demonstration of T1w MRI images registered in MNI space and the whole brain segmentation labels with 133 classes</p>
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
|
| 29 |
+
# Model Overview
|
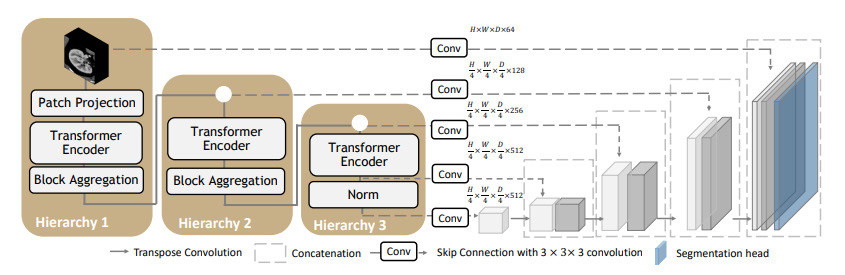
| 30 |
+
A pre-trained larger UNEST base model [1] for volumetric (3D) whole brain segmentation with T1w MR images.
|
| 31 |
+
To leverage information across embedded sequences, ”shifted window” transformers
|
| 32 |
+
are proposed for dense predictions and modeling multi-scale features. However, these
|
| 33 |
+
attempts that aim to complicate the self-attention range often yield high computation
|
| 34 |
+
complexity and data inefficiency. Inspired by the aggregation function in the nested
|
| 35 |
+
ViT, we propose a new design of a 3D U-shape medical segmentation model with
|
| 36 |
+
Nested Transformers (UNesT) hierarchically with the 3D block aggregation function,
|
| 37 |
+
that learn locality behaviors for small structures or small dataset. This design retains
|
| 38 |
+
the original global self-attention mechanism and achieves information communication
|
| 39 |
+
across patches by stacking transformer encoders hierarchically.
|
| 40 |
+
|
| 41 |
+
 <br>
|
| 42 |
+
<p align="center">
|
| 43 |
+
Fig.2 - The network architecture of UNEST Base model
|
| 44 |
+
</p>
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
## Data
|
| 48 |
+
The training data is from the Vanderbilt University and Vanderbilt University Medical Center with public released OASIS and CANDI datsets.
|
| 49 |
+
Training and testing data are MRI T1-weighted (T1w) 3D volumes coming from 3 different sites. There are a total of 133 classes in the whole brain segmentation task.
|
| 50 |
+
Among 50 T1w MRI scans from Open Access Series on Imaging Studies (OASIS) (Marcus et al., 2007) dataset, 45 scans are used for training and the other 5 for validation.
|
| 51 |
+
The testing cohort contains Colin27 T1w scan (Aubert-Broche et al., 2006) and 13 T1w MRI scans from the Child and Adolescent Neuro Development Initiative (CANDI)
|
| 52 |
+
(Kennedy et al., 2012). All data are registered to the MNI space using the MNI305 (Evans et al., 1993) template and preprocessed follow the method in (Huo et al., 2019). Input images are randomly cropped to the size of 96 × 96 × 96.
|
| 53 |
+
|
| 54 |
+
### Important
|
| 55 |
+
|
| 56 |
+
```diff
|
| 57 |
+
+ All the brain MRI images for training are registered to Affine registration from the target image to the MNI305 template using NiftyReg.
|
| 58 |
+
+ The data should be in the MNI305 space before inference.
|
| 59 |
+
|
| 60 |
+
|
| 61 |
+
Registration to MNI Space: Sample suggestion. E.g., use ANTS or other tools for registering T1 MRI image to MNI305 Space.
|
| 62 |
+
|
| 63 |
+
|
| 64 |
+
```
|
| 65 |
+
pip install antspyx
|
| 66 |
+
```
|
| 67 |
+
Sample ANTS registration
|
| 68 |
+
```
|
| 69 |
+
|
| 70 |
+
import ants
|
| 71 |
+
import sys
|
| 72 |
+
import os
|
| 73 |
+
|
| 74 |
+
fixed_image = ants.image_read('<fixed_image_path>')
|
| 75 |
+
moving_image = ants.image_read('<moving_image_path>')
|
| 76 |
+
transform = ants.registration(fixed_image,moving_image,'Affine')
|
| 77 |
+
|
| 78 |
+
reg3t = ants.apply_transforms(fixed_image,moving_image,transform['fwdtransforms'][0])
|
| 79 |
+
ants.image_write(reg3t,output_image_path)
|
| 80 |
+
|
| 81 |
+
```
|
| 82 |
+
## Training configuration
|
| 83 |
+
The training and inference was performed with at least one 24GB-memory GPU.
|
| 84 |
+
|
| 85 |
+
Actual Model Input: 96 x 96 x 96
|
| 86 |
+
|
| 87 |
+
## Input and output formats
|
| 88 |
+
Input: 1 channel T1w MRI image in MNI305 Space.
|
| 89 |
+
|
| 90 |
+
|
| 91 |
+
## commands example
|
| 92 |
+
Download trained checkpoint model to ./model/model.pt:
|
| 93 |
+
|
| 94 |
+
|
| 95 |
+
Add scripts component: To run the workflow with customized components, PYTHONPATH should be revised to include the path to the customized component:
|
| 96 |
+
|
| 97 |
+
```
|
| 98 |
+
export PYTHONPATH=$PYTHONPATH: '<path to the bundle root dir>/scripts'
|
| 99 |
+
|
| 100 |
+
```
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
Execute inference:
|
| 104 |
+
|
| 105 |
+
```
|
| 106 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 107 |
+
```
|
| 108 |
+
|
| 109 |
+
|
| 110 |
+
## More examples output
|
| 111 |
+
 <br>
|
| 112 |
+
<p align="center">
|
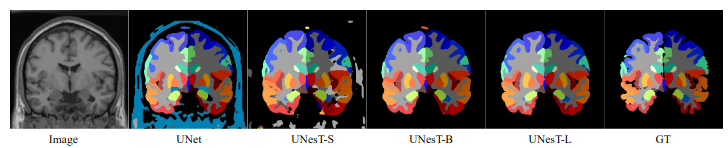
| 113 |
+
Fig.3 - The output prediction comparison with variant and ground truth
|
| 114 |
+
</p>
|
| 115 |
+
|
| 116 |
+
|
| 117 |
+
## Complete ROI of the whole brain segmentation
|
| 118 |
+
133 brain structures are segmented.
|
| 119 |
+
|
| 120 |
+
| #1 | #2 | #3 | #4 |
|
| 121 |
+
| :------------ | :---------- | :-------- | :-------- |
|
| 122 |
+
| 0: background | 1 : 3rd-Ventricle | 2 : 4th-Ventricle | 3 : Right-Accumbens-Area |
|
| 123 |
+
| 4 : Left-Accumbens-Area | 5 : Right-Amygdala | 6 : Left-Amygdala | 7 : Brain-Stem |
|
| 124 |
+
| 8 : Right-Caudate | 9 : Left-Caudate | 10 : Right-Cerebellum-Exterior | 11 : Left-Cerebellum-Exterior |
|
| 125 |
+
| 12 : Right-Cerebellum-White-Matter | 13 : Left-Cerebellum-White-Matter | 14 : Right-Cerebral-White-Matter | 15 : Left-Cerebral-White-Matter |
|
| 126 |
+
| 16 : Right-Hippocampus | 17 : Left-Hippocampus | 18 : Right-Inf-Lat-Vent | 19 : Left-Inf-Lat-Vent |
|
| 127 |
+
| 20 : Right-Lateral-Ventricle | 21 : Left-Lateral-Ventricle | 22 : Right-Pallidum | 23 : Left-Pallidum |
|
| 128 |
+
| 24 : Right-Putamen | 25 : Left-Putamen | 26 : Right-Thalamus-Proper | 27 : Left-Thalamus-Proper |
|
| 129 |
+
| 28 : Right-Ventral-DC | 29 : Left-Ventral-DC | 30 : Cerebellar-Vermal-Lobules-I-V | 31 : Cerebellar-Vermal-Lobules-VI-VII |
|
| 130 |
+
| 32 : Cerebellar-Vermal-Lobules-VIII-X | 33 : Left-Basal-Forebrain | 34 : Right-Basal-Forebrain | 35 : Right-ACgG--anterior-cingulate-gyrus |
|
| 131 |
+
| 36 : Left-ACgG--anterior-cingulate-gyrus | 37 : Right-AIns--anterior-insula | 38 : Left-AIns--anterior-insula | 39 : Right-AOrG--anterior-orbital-gyrus |
|
| 132 |
+
| 40 : Left-AOrG--anterior-orbital-gyrus | 41 : Right-AnG---angular-gyrus | 42 : Left-AnG---angular-gyrus | 43 : Right-Calc--calcarine-cortex |
|
| 133 |
+
| 44 : Left-Calc--calcarine-cortex | 45 : Right-CO----central-operculum | 46 : Left-CO----central-operculum | 47 : Right-Cun---cuneus |
|
| 134 |
+
| 48 : Left-Cun---cuneus | 49 : Right-Ent---entorhinal-area | 50 : Left-Ent---entorhinal-area | 51 : Right-FO----frontal-operculum |
|
| 135 |
+
| 52 : Left-FO----frontal-operculum | 53 : Right-FRP---frontal-pole | 54 : Left-FRP---frontal-pole | 55 : Right-FuG---fusiform-gyrus |
|
| 136 |
+
| 56 : Left-FuG---fusiform-gyrus | 57 : Right-GRe---gyrus-rectus | 58 : Left-GRe---gyrus-rectus | 59 : Right-IOG---inferior-occipital-gyrus ,
|
| 137 |
+
| 60 : Left-IOG---inferior-occipital-gyrus | 61 : Right-ITG---inferior-temporal-gyrus | 62 : Left-ITG---inferior-temporal-gyrus | 63 : Right-LiG---lingual-gyrus |
|
| 138 |
+
| 64 : Left-LiG---lingual-gyrus | 65 : Right-LOrG--lateral-orbital-gyrus | 66 : Left-LOrG--lateral-orbital-gyrus | 67 : Right-MCgG--middle-cingulate-gyrus |
|
| 139 |
+
| 68 : Left-MCgG--middle-cingulate-gyrus | 69 : Right-MFC---medial-frontal-cortex | 70 : Left-MFC---medial-frontal-cortex | 71 : Right-MFG---middle-frontal-gyrus |
|
| 140 |
+
| 72 : Left-MFG---middle-frontal-gyrus | 73 : Right-MOG---middle-occipital-gyrus | 74 : Left-MOG---middle-occipital-gyrus | 75 : Right-MOrG--medial-orbital-gyrus |
|
| 141 |
+
| 76 : Left-MOrG--medial-orbital-gyrus | 77 : Right-MPoG--postcentral-gyrus | 78 : Left-MPoG--postcentral-gyrus | 79 : Right-MPrG--precentral-gyrus |
|
| 142 |
+
| 80 : Left-MPrG--precentral-gyrus | 81 : Right-MSFG--superior-frontal-gyrus | 82 : Left-MSFG--superior-frontal-gyrus | 83 : Right-MTG---middle-temporal-gyrus |
|
| 143 |
+
| 84 : Left-MTG---middle-temporal-gyrus | 85 : Right-OCP---occipital-pole | 86 : Left-OCP---occipital-pole | 87 : Right-OFuG--occipital-fusiform-gyrus |
|
| 144 |
+
| 88 : Left-OFuG--occipital-fusiform-gyrus | 89 : Right-OpIFG-opercular-part-of-the-IFG | 90 : Left-OpIFG-opercular-part-of-the-IFG | 91 : Right-OrIFG-orbital-part-of-the-IFG |
|
| 145 |
+
| 92 : Left-OrIFG-orbital-part-of-the-IFG | 93 : Right-PCgG--posterior-cingulate-gyrus | 94 : Left-PCgG--posterior-cingulate-gyrus | 95 : Right-PCu---precuneus |
|
| 146 |
+
| 96 : Left-PCu---precuneus | 97 : Right-PHG---parahippocampal-gyrus | 98 : Left-PHG---parahippocampal-gyrus | 99 : Right-PIns--posterior-insula |
|
| 147 |
+
| 100 : Left-PIns--posterior-insula | 101 : Right-PO----parietal-operculum | 102 : Left-PO----parietal-operculum | 103 : Right-PoG---postcentral-gyrus |
|
| 148 |
+
| 104 : Left-PoG---postcentral-gyrus | 105 : Right-POrG--posterior-orbital-gyrus | 106 : Left-POrG--posterior-orbital-gyrus | 107 : Right-PP----planum-polare |
|
| 149 |
+
| 108 : Left-PP----planum-polare | 109 : Right-PrG---precentral-gyrus | 110 : Left-PrG---precentral-gyrus | 111 : Right-PT----planum-temporale |
|
| 150 |
+
| 112 : Left-PT----planum-temporale | 113 : Right-SCA---subcallosal-area | 114 : Left-SCA---subcallosal-area | 115 : Right-SFG---superior-frontal-gyrus |
|
| 151 |
+
| 116 : Left-SFG---superior-frontal-gyrus | 117 : Right-SMC---supplementary-motor-cortex | 118 : Left-SMC---supplementary-motor-cortex | 119 : Right-SMG---supramarginal-gyrus |
|
| 152 |
+
| 120 : Left-SMG---supramarginal-gyrus | 121 : Right-SOG---superior-occipital-gyrus | 122 : Left-SOG---superior-occipital-gyrus | 123 : Right-SPL---superior-parietal-lobule |
|
| 153 |
+
| 124 : Left-SPL---superior-parietal-lobule | 125 : Right-STG---superior-temporal-gyrus | 126 : Left-STG---superior-temporal-gyrus | 127 : Right-TMP---temporal-pole |
|
| 154 |
+
| 128 : Left-TMP---temporal-pole | 129 : Right-TrIFG-triangular-part-of-the-IFG | 130 : Left-TrIFG-triangular-part-of-the-IFG | 131 : Right-TTG---transverse-temporal-gyrus |
|
| 155 |
+
| 132 : Left-TTG---transverse-temporal-gyrus |
|
| 156 |
+
|
| 157 |
+
|
| 158 |
+
## Bundle Integration in MONAI Lable
|
| 159 |
+
The inference pipleine can be easily used by the MONAI Label server and 3D Slicer for fast labeling T1w MRI images in MNI space.
|
| 160 |
+
|
| 161 |
+
 <br>
|
| 162 |
+
|
| 163 |
+
# Disclaimer
|
| 164 |
+
This is an example, not to be used for diagnostic purposes.
|
| 165 |
+
|
| 166 |
+
# References
|
| 167 |
+
[1] Yu, Xin, Yinchi Zhou, Yucheng Tang et al. Characterizing Renal Structures with 3D Block Aggregate Transformers. arXiv preprint arXiv:2203.02430 (2022). https://arxiv.org/pdf/2203.02430.pdf
|
| 168 |
+
|
| 169 |
+
[2] Zizhao Zhang et al. Nested Hierarchical Transformer: Towards Accurate, Data-Efficient and Interpretable Visual Understanding. AAAI Conference on Artificial Intelligence (AAAI) 2022
|
| 170 |
+
|
| 171 |
+
[3] Huo, Yuankai, et al. 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage 194 (2019): 105-119.
|
configs/inference.json
ADDED
|
@@ -0,0 +1,136 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import glob",
|
| 4 |
+
"$import os"
|
| 5 |
+
],
|
| 6 |
+
"bundle_root": ".",
|
| 7 |
+
"output_dir": "$@bundle_root + '/eval'",
|
| 8 |
+
"dataset_dir": "$@bundle_root + '/dataset/images'",
|
| 9 |
+
"datalist": "$list(sorted(glob.glob(@dataset_dir + '/*.nii.gz')))",
|
| 10 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 11 |
+
"network_def": {
|
| 12 |
+
"_target_": "scripts.networks.unest_base_patch_4.UNesT",
|
| 13 |
+
"in_channels": 1,
|
| 14 |
+
"out_channels": 133,
|
| 15 |
+
"patch_size": 4,
|
| 16 |
+
"depths": [
|
| 17 |
+
2,
|
| 18 |
+
2,
|
| 19 |
+
8
|
| 20 |
+
],
|
| 21 |
+
"embed_dim": [
|
| 22 |
+
128,
|
| 23 |
+
256,
|
| 24 |
+
512
|
| 25 |
+
],
|
| 26 |
+
"num_heads": [
|
| 27 |
+
4,
|
| 28 |
+
8,
|
| 29 |
+
16
|
| 30 |
+
]
|
| 31 |
+
},
|
| 32 |
+
"network": "$@network_def.to(@device)",
|
| 33 |
+
"preprocessing": {
|
| 34 |
+
"_target_": "Compose",
|
| 35 |
+
"transforms": [
|
| 36 |
+
{
|
| 37 |
+
"_target_": "LoadImaged",
|
| 38 |
+
"keys": "image"
|
| 39 |
+
},
|
| 40 |
+
{
|
| 41 |
+
"_target_": "EnsureChannelFirstd",
|
| 42 |
+
"keys": "image"
|
| 43 |
+
},
|
| 44 |
+
{
|
| 45 |
+
"_target_": "NormalizeIntensityd",
|
| 46 |
+
"keys": "image",
|
| 47 |
+
"nonzero": "True",
|
| 48 |
+
"channel_wise": "True"
|
| 49 |
+
},
|
| 50 |
+
{
|
| 51 |
+
"_target_": "EnsureTyped",
|
| 52 |
+
"keys": "image"
|
| 53 |
+
}
|
| 54 |
+
]
|
| 55 |
+
},
|
| 56 |
+
"dataset": {
|
| 57 |
+
"_target_": "Dataset",
|
| 58 |
+
"data": "$[{'image': i} for i in @datalist]",
|
| 59 |
+
"transform": "@preprocessing"
|
| 60 |
+
},
|
| 61 |
+
"dataloader": {
|
| 62 |
+
"_target_": "DataLoader",
|
| 63 |
+
"dataset": "@dataset",
|
| 64 |
+
"batch_size": 1,
|
| 65 |
+
"shuffle": false,
|
| 66 |
+
"num_workers": 4
|
| 67 |
+
},
|
| 68 |
+
"inferer": {
|
| 69 |
+
"_target_": "SlidingWindowInferer",
|
| 70 |
+
"roi_size": [
|
| 71 |
+
96,
|
| 72 |
+
96,
|
| 73 |
+
96
|
| 74 |
+
],
|
| 75 |
+
"sw_batch_size": 4,
|
| 76 |
+
"overlap": 0.7
|
| 77 |
+
},
|
| 78 |
+
"postprocessing": {
|
| 79 |
+
"_target_": "Compose",
|
| 80 |
+
"transforms": [
|
| 81 |
+
{
|
| 82 |
+
"_target_": "Activationsd",
|
| 83 |
+
"keys": "pred",
|
| 84 |
+
"softmax": true
|
| 85 |
+
},
|
| 86 |
+
{
|
| 87 |
+
"_target_": "Invertd",
|
| 88 |
+
"keys": "pred",
|
| 89 |
+
"transform": "@preprocessing",
|
| 90 |
+
"orig_keys": "image",
|
| 91 |
+
"meta_key_postfix": "meta_dict",
|
| 92 |
+
"nearest_interp": false,
|
| 93 |
+
"to_tensor": true
|
| 94 |
+
},
|
| 95 |
+
{
|
| 96 |
+
"_target_": "AsDiscreted",
|
| 97 |
+
"keys": "pred",
|
| 98 |
+
"argmax": true
|
| 99 |
+
},
|
| 100 |
+
{
|
| 101 |
+
"_target_": "SaveImaged",
|
| 102 |
+
"keys": "pred",
|
| 103 |
+
"meta_keys": "pred_meta_dict",
|
| 104 |
+
"output_dir": "@output_dir"
|
| 105 |
+
}
|
| 106 |
+
]
|
| 107 |
+
},
|
| 108 |
+
"handlers": [
|
| 109 |
+
{
|
| 110 |
+
"_target_": "CheckpointLoader",
|
| 111 |
+
"load_path": "$@bundle_root + '/models/model.pt'",
|
| 112 |
+
"load_dict": {
|
| 113 |
+
"model": "@network"
|
| 114 |
+
},
|
| 115 |
+
"strict": "True"
|
| 116 |
+
},
|
| 117 |
+
{
|
| 118 |
+
"_target_": "StatsHandler",
|
| 119 |
+
"iteration_log": false
|
| 120 |
+
}
|
| 121 |
+
],
|
| 122 |
+
"evaluator": {
|
| 123 |
+
"_target_": "SupervisedEvaluator",
|
| 124 |
+
"device": "@device",
|
| 125 |
+
"val_data_loader": "@dataloader",
|
| 126 |
+
"network": "@network",
|
| 127 |
+
"inferer": "@inferer",
|
| 128 |
+
"postprocessing": "@postprocessing",
|
| 129 |
+
"val_handlers": "@handlers",
|
| 130 |
+
"amp": false
|
| 131 |
+
},
|
| 132 |
+
"evaluating": [
|
| 133 |
+
"$setattr(torch.backends.cudnn, 'benchmark', True)",
|
| 134 |
+
"[email protected]()"
|
| 135 |
+
]
|
| 136 |
+
}
|
configs/logging.conf
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[loggers]
|
| 2 |
+
keys=root
|
| 3 |
+
|
| 4 |
+
[handlers]
|
| 5 |
+
keys=consoleHandler
|
| 6 |
+
|
| 7 |
+
[formatters]
|
| 8 |
+
keys=fullFormatter
|
| 9 |
+
|
| 10 |
+
[logger_root]
|
| 11 |
+
level=INFO
|
| 12 |
+
handlers=consoleHandler
|
| 13 |
+
|
| 14 |
+
[handler_consoleHandler]
|
| 15 |
+
class=StreamHandler
|
| 16 |
+
level=INFO
|
| 17 |
+
formatter=fullFormatter
|
| 18 |
+
args=(sys.stdout,)
|
| 19 |
+
|
| 20 |
+
[formatter_fullFormatter]
|
| 21 |
+
format=%(asctime)s - %(name)s - %(levelname)s - %(message)s
|
configs/metadata.json
ADDED
|
@@ -0,0 +1,210 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.1.0",
|
| 4 |
+
"changelog": {
|
| 5 |
+
"0.1.0": "complete the model package"
|
| 6 |
+
},
|
| 7 |
+
"monai_version": "0.9.1",
|
| 8 |
+
"pytorch_version": "1.10.0",
|
| 9 |
+
"numpy_version": "1.21.2",
|
| 10 |
+
"optional_packages_version": {
|
| 11 |
+
"nibabel": "3.2.1",
|
| 12 |
+
"pytorch-ignite": "0.4.8",
|
| 13 |
+
"einops": "0.4.1",
|
| 14 |
+
"fire": "0.4.0",
|
| 15 |
+
"timm": "0.6.7"
|
| 16 |
+
},
|
| 17 |
+
"task": "Whole Brain Segmentation",
|
| 18 |
+
"description": "A 3D transformer-based model for whole brain segmentation from T1W MRI image",
|
| 19 |
+
"authors": "Vanderbilt University + MONAI team",
|
| 20 |
+
"copyright": "Copyright (c) MONAI Consortium",
|
| 21 |
+
"data_source": "",
|
| 22 |
+
"data_type": "nibabel",
|
| 23 |
+
"image_classes": "single channel data, intensity scaled to [0, 1]",
|
| 24 |
+
"label_classes": "133 Classes",
|
| 25 |
+
"pred_classes": "133 Classes",
|
| 26 |
+
"eval_metrics": {
|
| 27 |
+
"mean_dice": 0.71
|
| 28 |
+
},
|
| 29 |
+
"intended_use": "This is an example, not to be used for diagnostic purposes",
|
| 30 |
+
"references": [
|
| 31 |
+
"Xin, et al. Characterizing Renal Structures with 3D Block Aggregate Transformers. arXiv preprint arXiv:2203.02430 (2022). https://arxiv.org/pdf/2203.02430.pdf"
|
| 32 |
+
],
|
| 33 |
+
"network_data_format": {
|
| 34 |
+
"inputs": {
|
| 35 |
+
"image": {
|
| 36 |
+
"type": "image",
|
| 37 |
+
"format": "hounsfield",
|
| 38 |
+
"modality": "MRI",
|
| 39 |
+
"num_channels": 1,
|
| 40 |
+
"spatial_shape": [
|
| 41 |
+
96,
|
| 42 |
+
96,
|
| 43 |
+
96
|
| 44 |
+
],
|
| 45 |
+
"dtype": "float32",
|
| 46 |
+
"value_range": [
|
| 47 |
+
0,
|
| 48 |
+
1
|
| 49 |
+
],
|
| 50 |
+
"is_patch_data": true,
|
| 51 |
+
"channel_def": {
|
| 52 |
+
"0": "image"
|
| 53 |
+
}
|
| 54 |
+
}
|
| 55 |
+
},
|
| 56 |
+
"outputs": {
|
| 57 |
+
"pred": {
|
| 58 |
+
"type": "image",
|
| 59 |
+
"format": "segmentation",
|
| 60 |
+
"num_channels": 133,
|
| 61 |
+
"spatial_shape": [
|
| 62 |
+
96,
|
| 63 |
+
96,
|
| 64 |
+
96
|
| 65 |
+
],
|
| 66 |
+
"dtype": "float32",
|
| 67 |
+
"value_range": [
|
| 68 |
+
0,
|
| 69 |
+
1
|
| 70 |
+
],
|
| 71 |
+
"is_patch_data": true,
|
| 72 |
+
"channel_def": {
|
| 73 |
+
"0": "background",
|
| 74 |
+
"1": "3rd-Ventricle",
|
| 75 |
+
"2": "4th-Ventricle",
|
| 76 |
+
"3": "Right-Accumbens-Area",
|
| 77 |
+
"4": "Left-Accumbens-Area",
|
| 78 |
+
"5": "Right-Amygdala",
|
| 79 |
+
"6": "Left-Amygdala",
|
| 80 |
+
"7": "Brain-Stem",
|
| 81 |
+
"8": "Right-Caudate",
|
| 82 |
+
"9": "Left-Caudate",
|
| 83 |
+
"10": "Right-Cerebellum-Exterior",
|
| 84 |
+
"11": "Left-Cerebellum-Exterior",
|
| 85 |
+
"12": "Right-Cerebellum-White-Matter",
|
| 86 |
+
"13": "Left-Cerebellum-White-Matter",
|
| 87 |
+
"14": "Right-Cerebral-White-Matter",
|
| 88 |
+
"15": "Left-Cerebral-White-Matter",
|
| 89 |
+
"16": "Right-Hippocampus",
|
| 90 |
+
"17": "Left-Hippocampus",
|
| 91 |
+
"18": "Right-Inf-Lat-Vent",
|
| 92 |
+
"19": "Left-Inf-Lat-Vent",
|
| 93 |
+
"20": "Right-Lateral-Ventricle",
|
| 94 |
+
"21": "Left-Lateral-Ventricle",
|
| 95 |
+
"22": "Right-Pallidum",
|
| 96 |
+
"23": "Left-Pallidum",
|
| 97 |
+
"24": "Right-Putamen",
|
| 98 |
+
"25": "Left-Putamen",
|
| 99 |
+
"26": "Right-Thalamus-Proper",
|
| 100 |
+
"27": "Left-Thalamus-Proper",
|
| 101 |
+
"28": "Right-Ventral-DC",
|
| 102 |
+
"29": "Left-Ventral-DC",
|
| 103 |
+
"30": "Cerebellar-Vermal-Lobules-I-V",
|
| 104 |
+
"31": "Cerebellar-Vermal-Lobules-VI-VII",
|
| 105 |
+
"32": "Cerebellar-Vermal-Lobules-VIII-X",
|
| 106 |
+
"33": "Left-Basal-Forebrain",
|
| 107 |
+
"34": "Right-Basal-Forebrain",
|
| 108 |
+
"35": "Right-ACgG--anterior-cingulate-gyrus",
|
| 109 |
+
"36": "Left-ACgG--anterior-cingulate-gyrus",
|
| 110 |
+
"37": "Right-AIns--anterior-insula",
|
| 111 |
+
"38": "Left-AIns--anterior-insula",
|
| 112 |
+
"39": "Right-AOrG--anterior-orbital-gyrus",
|
| 113 |
+
"40": "Left-AOrG--anterior-orbital-gyrus",
|
| 114 |
+
"41": "Right-AnG---angular-gyrus",
|
| 115 |
+
"42": "Left-AnG---angular-gyrus",
|
| 116 |
+
"43": "Right-Calc--calcarine-cortex",
|
| 117 |
+
"44": "Left-Calc--calcarine-cortex",
|
| 118 |
+
"45": "Right-CO----central-operculum",
|
| 119 |
+
"46": "Left-CO----central-operculum",
|
| 120 |
+
"47": "Right-Cun---cuneus",
|
| 121 |
+
"48": "Left-Cun---cuneus",
|
| 122 |
+
"49": "Right-Ent---entorhinal-area",
|
| 123 |
+
"50": "Left-Ent---entorhinal-area",
|
| 124 |
+
"51": "Right-FO----frontal-operculum",
|
| 125 |
+
"52": "Left-FO----frontal-operculum",
|
| 126 |
+
"53": "Right-FRP---frontal-pole",
|
| 127 |
+
"54": "Left-FRP---frontal-pole",
|
| 128 |
+
"55": "Right-FuG---fusiform-gyrus ",
|
| 129 |
+
"56": "Left-FuG---fusiform-gyrus",
|
| 130 |
+
"57": "Right-GRe---gyrus-rectus",
|
| 131 |
+
"58": "Left-GRe---gyrus-rectus",
|
| 132 |
+
"59": "Right-IOG---inferior-occipital-gyrus",
|
| 133 |
+
"60": "Left-IOG---inferior-occipital-gyrus",
|
| 134 |
+
"61": "Right-ITG---inferior-temporal-gyrus",
|
| 135 |
+
"62": "Left-ITG---inferior-temporal-gyrus",
|
| 136 |
+
"63": "Right-LiG---lingual-gyrus",
|
| 137 |
+
"64": "Left-LiG---lingual-gyrus",
|
| 138 |
+
"65": "Right-LOrG--lateral-orbital-gyrus",
|
| 139 |
+
"66": "Left-LOrG--lateral-orbital-gyrus",
|
| 140 |
+
"67": "Right-MCgG--middle-cingulate-gyrus",
|
| 141 |
+
"68": "Left-MCgG--middle-cingulate-gyrus",
|
| 142 |
+
"69": "Right-MFC---medial-frontal-cortex",
|
| 143 |
+
"70": "Left-MFC---medial-frontal-cortex",
|
| 144 |
+
"71": "Right-MFG---middle-frontal-gyrus",
|
| 145 |
+
"72": "Left-MFG---middle-frontal-gyrus",
|
| 146 |
+
"73": "Right-MOG---middle-occipital-gyrus",
|
| 147 |
+
"74": "Left-MOG---middle-occipital-gyrus",
|
| 148 |
+
"75": "Right-MOrG--medial-orbital-gyrus",
|
| 149 |
+
"76": "Left-MOrG--medial-orbital-gyrus",
|
| 150 |
+
"77": "Right-MPoG--postcentral-gyrus",
|
| 151 |
+
"78": "Left-MPoG--postcentral-gyrus",
|
| 152 |
+
"79": "Right-MPrG--precentral-gyrus",
|
| 153 |
+
"80": "Left-MPrG--precentral-gyrus",
|
| 154 |
+
"81": "Right-MSFG--superior-frontal-gyrus",
|
| 155 |
+
"82": "Left-MSFG--superior-frontal-gyrus",
|
| 156 |
+
"83": "Right-MTG---middle-temporal-gyrus",
|
| 157 |
+
"84": "Left-MTG---middle-temporal-gyrus",
|
| 158 |
+
"85": "Right-OCP---occipital-pole",
|
| 159 |
+
"86": "Left-OCP---occipital-pole",
|
| 160 |
+
"87": "Right-OFuG--occipital-fusiform-gyrus",
|
| 161 |
+
"88": "Left-OFuG--occipital-fusiform-gyrus",
|
| 162 |
+
"89": "Right-OpIFG-opercular-part-of-the-IFG",
|
| 163 |
+
"90": "Left-OpIFG-opercular-part-of-the-IFG",
|
| 164 |
+
"91": "Right-OrIFG-orbital-part-of-the-IFG",
|
| 165 |
+
"92": "Left-OrIFG-orbital-part-of-the-IFG",
|
| 166 |
+
"93": "Right-PCgG--posterior-cingulate-gyrus",
|
| 167 |
+
"94": "Left-PCgG--posterior-cingulate-gyrus",
|
| 168 |
+
"95": "Right-PCu---precuneus",
|
| 169 |
+
"96": "Left-PCu---precuneus",
|
| 170 |
+
"97": "Right-PHG---parahippocampal-gyrus",
|
| 171 |
+
"98": "Left-PHG---parahippocampal-gyrus",
|
| 172 |
+
"99": "Right-PIns--posterior-insula",
|
| 173 |
+
"100": "Left-PIns--posterior-insula",
|
| 174 |
+
"101": "Right-PO----parietal-operculum",
|
| 175 |
+
"102": "Left-PO----parietal-operculum",
|
| 176 |
+
"103": "Right-PoG---postcentral-gyrus",
|
| 177 |
+
"104": "Left-PoG---postcentral-gyrus",
|
| 178 |
+
"105": "Right-POrG--posterior-orbital-gyrus",
|
| 179 |
+
"106": "Left-POrG--posterior-orbital-gyrus",
|
| 180 |
+
"107": "Right-PP----planum-polare",
|
| 181 |
+
"108": "Left-PP----planum-polare",
|
| 182 |
+
"109": "Right-PrG---precentral-gyrus",
|
| 183 |
+
"110": "Left-PrG---precentral-gyrus",
|
| 184 |
+
"111": "Right-PT----planum-temporale",
|
| 185 |
+
"112": "Left-PT----planum-temporale",
|
| 186 |
+
"113": "Right-SCA---subcallosal-area",
|
| 187 |
+
"114": "Left-SCA---subcallosal-area",
|
| 188 |
+
"115": "Right-SFG---superior-frontal-gyrus",
|
| 189 |
+
"116": "Left-SFG---superior-frontal-gyrus",
|
| 190 |
+
"117": "Right-SMC---supplementary-motor-cortex",
|
| 191 |
+
"118": "Left-SMC---supplementary-motor-cortex",
|
| 192 |
+
"119": "Right-SMG---supramarginal-gyrus",
|
| 193 |
+
"120": "Left-SMG---supramarginal-gyrus",
|
| 194 |
+
"121": "Right-SOG---superior-occipital-gyrus",
|
| 195 |
+
"122": "Left-SOG---superior-occipital-gyrus",
|
| 196 |
+
"123": "Right-SPL---superior-parietal-lobule",
|
| 197 |
+
"124": "Left-SPL---superior-parietal-lobule",
|
| 198 |
+
"125": "Right-STG---superior-temporal-gyrus",
|
| 199 |
+
"126": "Left-STG---superior-temporal-gyrus",
|
| 200 |
+
"127": "Right-TMP---temporal-pole",
|
| 201 |
+
"128": "Left-TMP---temporal-pole",
|
| 202 |
+
"129": "Right-TrIFG-triangular-part-of-the-IFG",
|
| 203 |
+
"130": "Left-TrIFG-triangular-part-of-the-IFG",
|
| 204 |
+
"131": "Right-TTG---transverse-temporal-gyrus",
|
| 205 |
+
"132": "Left-TTG---transverse-temporal-gyrus"
|
| 206 |
+
}
|
| 207 |
+
}
|
| 208 |
+
}
|
| 209 |
+
}
|
| 210 |
+
}
|
docs/3DSlicer_use.png
ADDED

|
docs/README.md
ADDED
|
@@ -0,0 +1,164 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Description
|
| 2 |
+
Detailed whole brain segmentation is an essential quantitative technique in medical image analysis, which provides a non-invasive way of measuring brain regions from a clinical acquired structural magnetic resonance imaging (MRI).
|
| 3 |
+
We provide the pre-trained model for inferencing whole brain segmentation with 133 structures.
|
| 4 |
+
|
| 5 |
+
A tutorial and release of model for whole brain segmentation using the 3D transformer-based segmentation model UNEST.
|
| 6 |
+
|
| 7 |
+
Authors:
|
| 8 |
+
Xin Yu ([email protected]) (Primary)
|
| 9 |
+
|
| 10 |
+
Yinchi Zhou ([email protected]) | Yucheng Tang ([email protected])
|
| 11 |
+
|
| 12 |
+
<p align="center">
|
| 13 |
+
-------------------------------------------------------------------------------------
|
| 14 |
+
</p>
|
| 15 |
+
|
| 16 |
+
 <br>
|
| 17 |
+
<p align="center">
|
| 18 |
+
Fig.1 - The demonstration of T1w MRI images registered in MNI space and the whole brain segmentation labels with 133 classes</p>
|
| 19 |
+
|
| 20 |
+
|
| 21 |
+
|
| 22 |
+
# Model Overview
|
| 23 |
+
A pre-trained larger UNEST base model [1] for volumetric (3D) whole brain segmentation with T1w MR images.
|
| 24 |
+
To leverage information across embedded sequences, ”shifted window” transformers
|
| 25 |
+
are proposed for dense predictions and modeling multi-scale features. However, these
|
| 26 |
+
attempts that aim to complicate the self-attention range often yield high computation
|
| 27 |
+
complexity and data inefficiency. Inspired by the aggregation function in the nested
|
| 28 |
+
ViT, we propose a new design of a 3D U-shape medical segmentation model with
|
| 29 |
+
Nested Transformers (UNesT) hierarchically with the 3D block aggregation function,
|
| 30 |
+
that learn locality behaviors for small structures or small dataset. This design retains
|
| 31 |
+
the original global self-attention mechanism and achieves information communication
|
| 32 |
+
across patches by stacking transformer encoders hierarchically.
|
| 33 |
+
|
| 34 |
+
 <br>
|
| 35 |
+
<p align="center">
|
| 36 |
+
Fig.2 - The network architecture of UNEST Base model
|
| 37 |
+
</p>
|
| 38 |
+
|
| 39 |
+
|
| 40 |
+
## Data
|
| 41 |
+
The training data is from the Vanderbilt University and Vanderbilt University Medical Center with public released OASIS and CANDI datsets.
|
| 42 |
+
Training and testing data are MRI T1-weighted (T1w) 3D volumes coming from 3 different sites. There are a total of 133 classes in the whole brain segmentation task.
|
| 43 |
+
Among 50 T1w MRI scans from Open Access Series on Imaging Studies (OASIS) (Marcus et al., 2007) dataset, 45 scans are used for training and the other 5 for validation.
|
| 44 |
+
The testing cohort contains Colin27 T1w scan (Aubert-Broche et al., 2006) and 13 T1w MRI scans from the Child and Adolescent Neuro Development Initiative (CANDI)
|
| 45 |
+
(Kennedy et al., 2012). All data are registered to the MNI space using the MNI305 (Evans et al., 1993) template and preprocessed follow the method in (Huo et al., 2019). Input images are randomly cropped to the size of 96 × 96 × 96.
|
| 46 |
+
|
| 47 |
+
### Important
|
| 48 |
+
|
| 49 |
+
```diff
|
| 50 |
+
+ All the brain MRI images for training are registered to Affine registration from the target image to the MNI305 template using NiftyReg.
|
| 51 |
+
+ The data should be in the MNI305 space before inference.
|
| 52 |
+
|
| 53 |
+
|
| 54 |
+
Registration to MNI Space: Sample suggestion. E.g., use ANTS or other tools for registering T1 MRI image to MNI305 Space.
|
| 55 |
+
|
| 56 |
+
|
| 57 |
+
```
|
| 58 |
+
pip install antspyx
|
| 59 |
+
```
|
| 60 |
+
Sample ANTS registration
|
| 61 |
+
```
|
| 62 |
+
|
| 63 |
+
import ants
|
| 64 |
+
import sys
|
| 65 |
+
import os
|
| 66 |
+
|
| 67 |
+
fixed_image = ants.image_read('<fixed_image_path>')
|
| 68 |
+
moving_image = ants.image_read('<moving_image_path>')
|
| 69 |
+
transform = ants.registration(fixed_image,moving_image,'Affine')
|
| 70 |
+
|
| 71 |
+
reg3t = ants.apply_transforms(fixed_image,moving_image,transform['fwdtransforms'][0])
|
| 72 |
+
ants.image_write(reg3t,output_image_path)
|
| 73 |
+
|
| 74 |
+
```
|
| 75 |
+
## Training configuration
|
| 76 |
+
The training and inference was performed with at least one 24GB-memory GPU.
|
| 77 |
+
|
| 78 |
+
Actual Model Input: 96 x 96 x 96
|
| 79 |
+
|
| 80 |
+
## Input and output formats
|
| 81 |
+
Input: 1 channel T1w MRI image in MNI305 Space.
|
| 82 |
+
|
| 83 |
+
|
| 84 |
+
## commands example
|
| 85 |
+
Download trained checkpoint model to ./model/model.pt:
|
| 86 |
+
|
| 87 |
+
|
| 88 |
+
Add scripts component: To run the workflow with customized components, PYTHONPATH should be revised to include the path to the customized component:
|
| 89 |
+
|
| 90 |
+
```
|
| 91 |
+
export PYTHONPATH=$PYTHONPATH: '<path to the bundle root dir>/scripts'
|
| 92 |
+
|
| 93 |
+
```
|
| 94 |
+
|
| 95 |
+
|
| 96 |
+
Execute inference:
|
| 97 |
+
|
| 98 |
+
```
|
| 99 |
+
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
|
| 100 |
+
```
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
## More examples output
|
| 104 |
+
 <br>
|
| 105 |
+
<p align="center">
|
| 106 |
+
Fig.3 - The output prediction comparison with variant and ground truth
|
| 107 |
+
</p>
|
| 108 |
+
|
| 109 |
+
|
| 110 |
+
## Complete ROI of the whole brain segmentation
|
| 111 |
+
133 brain structures are segmented.
|
| 112 |
+
|
| 113 |
+
| #1 | #2 | #3 | #4 |
|
| 114 |
+
| :------------ | :---------- | :-------- | :-------- |
|
| 115 |
+
| 0: background | 1 : 3rd-Ventricle | 2 : 4th-Ventricle | 3 : Right-Accumbens-Area |
|
| 116 |
+
| 4 : Left-Accumbens-Area | 5 : Right-Amygdala | 6 : Left-Amygdala | 7 : Brain-Stem |
|
| 117 |
+
| 8 : Right-Caudate | 9 : Left-Caudate | 10 : Right-Cerebellum-Exterior | 11 : Left-Cerebellum-Exterior |
|
| 118 |
+
| 12 : Right-Cerebellum-White-Matter | 13 : Left-Cerebellum-White-Matter | 14 : Right-Cerebral-White-Matter | 15 : Left-Cerebral-White-Matter |
|
| 119 |
+
| 16 : Right-Hippocampus | 17 : Left-Hippocampus | 18 : Right-Inf-Lat-Vent | 19 : Left-Inf-Lat-Vent |
|
| 120 |
+
| 20 : Right-Lateral-Ventricle | 21 : Left-Lateral-Ventricle | 22 : Right-Pallidum | 23 : Left-Pallidum |
|
| 121 |
+
| 24 : Right-Putamen | 25 : Left-Putamen | 26 : Right-Thalamus-Proper | 27 : Left-Thalamus-Proper |
|
| 122 |
+
| 28 : Right-Ventral-DC | 29 : Left-Ventral-DC | 30 : Cerebellar-Vermal-Lobules-I-V | 31 : Cerebellar-Vermal-Lobules-VI-VII |
|
| 123 |
+
| 32 : Cerebellar-Vermal-Lobules-VIII-X | 33 : Left-Basal-Forebrain | 34 : Right-Basal-Forebrain | 35 : Right-ACgG--anterior-cingulate-gyrus |
|
| 124 |
+
| 36 : Left-ACgG--anterior-cingulate-gyrus | 37 : Right-AIns--anterior-insula | 38 : Left-AIns--anterior-insula | 39 : Right-AOrG--anterior-orbital-gyrus |
|
| 125 |
+
| 40 : Left-AOrG--anterior-orbital-gyrus | 41 : Right-AnG---angular-gyrus | 42 : Left-AnG---angular-gyrus | 43 : Right-Calc--calcarine-cortex |
|
| 126 |
+
| 44 : Left-Calc--calcarine-cortex | 45 : Right-CO----central-operculum | 46 : Left-CO----central-operculum | 47 : Right-Cun---cuneus |
|
| 127 |
+
| 48 : Left-Cun---cuneus | 49 : Right-Ent---entorhinal-area | 50 : Left-Ent---entorhinal-area | 51 : Right-FO----frontal-operculum |
|
| 128 |
+
| 52 : Left-FO----frontal-operculum | 53 : Right-FRP---frontal-pole | 54 : Left-FRP---frontal-pole | 55 : Right-FuG---fusiform-gyrus |
|
| 129 |
+
| 56 : Left-FuG---fusiform-gyrus | 57 : Right-GRe---gyrus-rectus | 58 : Left-GRe---gyrus-rectus | 59 : Right-IOG---inferior-occipital-gyrus ,
|
| 130 |
+
| 60 : Left-IOG---inferior-occipital-gyrus | 61 : Right-ITG---inferior-temporal-gyrus | 62 : Left-ITG---inferior-temporal-gyrus | 63 : Right-LiG---lingual-gyrus |
|
| 131 |
+
| 64 : Left-LiG---lingual-gyrus | 65 : Right-LOrG--lateral-orbital-gyrus | 66 : Left-LOrG--lateral-orbital-gyrus | 67 : Right-MCgG--middle-cingulate-gyrus |
|
| 132 |
+
| 68 : Left-MCgG--middle-cingulate-gyrus | 69 : Right-MFC---medial-frontal-cortex | 70 : Left-MFC---medial-frontal-cortex | 71 : Right-MFG---middle-frontal-gyrus |
|
| 133 |
+
| 72 : Left-MFG---middle-frontal-gyrus | 73 : Right-MOG---middle-occipital-gyrus | 74 : Left-MOG---middle-occipital-gyrus | 75 : Right-MOrG--medial-orbital-gyrus |
|
| 134 |
+
| 76 : Left-MOrG--medial-orbital-gyrus | 77 : Right-MPoG--postcentral-gyrus | 78 : Left-MPoG--postcentral-gyrus | 79 : Right-MPrG--precentral-gyrus |
|
| 135 |
+
| 80 : Left-MPrG--precentral-gyrus | 81 : Right-MSFG--superior-frontal-gyrus | 82 : Left-MSFG--superior-frontal-gyrus | 83 : Right-MTG---middle-temporal-gyrus |
|
| 136 |
+
| 84 : Left-MTG---middle-temporal-gyrus | 85 : Right-OCP---occipital-pole | 86 : Left-OCP---occipital-pole | 87 : Right-OFuG--occipital-fusiform-gyrus |
|
| 137 |
+
| 88 : Left-OFuG--occipital-fusiform-gyrus | 89 : Right-OpIFG-opercular-part-of-the-IFG | 90 : Left-OpIFG-opercular-part-of-the-IFG | 91 : Right-OrIFG-orbital-part-of-the-IFG |
|
| 138 |
+
| 92 : Left-OrIFG-orbital-part-of-the-IFG | 93 : Right-PCgG--posterior-cingulate-gyrus | 94 : Left-PCgG--posterior-cingulate-gyrus | 95 : Right-PCu---precuneus |
|
| 139 |
+
| 96 : Left-PCu---precuneus | 97 : Right-PHG---parahippocampal-gyrus | 98 : Left-PHG---parahippocampal-gyrus | 99 : Right-PIns--posterior-insula |
|
| 140 |
+
| 100 : Left-PIns--posterior-insula | 101 : Right-PO----parietal-operculum | 102 : Left-PO----parietal-operculum | 103 : Right-PoG---postcentral-gyrus |
|
| 141 |
+
| 104 : Left-PoG---postcentral-gyrus | 105 : Right-POrG--posterior-orbital-gyrus | 106 : Left-POrG--posterior-orbital-gyrus | 107 : Right-PP----planum-polare |
|
| 142 |
+
| 108 : Left-PP----planum-polare | 109 : Right-PrG---precentral-gyrus | 110 : Left-PrG---precentral-gyrus | 111 : Right-PT----planum-temporale |
|
| 143 |
+
| 112 : Left-PT----planum-temporale | 113 : Right-SCA---subcallosal-area | 114 : Left-SCA---subcallosal-area | 115 : Right-SFG---superior-frontal-gyrus |
|
| 144 |
+
| 116 : Left-SFG---superior-frontal-gyrus | 117 : Right-SMC---supplementary-motor-cortex | 118 : Left-SMC---supplementary-motor-cortex | 119 : Right-SMG---supramarginal-gyrus |
|
| 145 |
+
| 120 : Left-SMG---supramarginal-gyrus | 121 : Right-SOG---superior-occipital-gyrus | 122 : Left-SOG---superior-occipital-gyrus | 123 : Right-SPL---superior-parietal-lobule |
|
| 146 |
+
| 124 : Left-SPL---superior-parietal-lobule | 125 : Right-STG---superior-temporal-gyrus | 126 : Left-STG---superior-temporal-gyrus | 127 : Right-TMP---temporal-pole |
|
| 147 |
+
| 128 : Left-TMP---temporal-pole | 129 : Right-TrIFG-triangular-part-of-the-IFG | 130 : Left-TrIFG-triangular-part-of-the-IFG | 131 : Right-TTG---transverse-temporal-gyrus |
|
| 148 |
+
| 132 : Left-TTG---transverse-temporal-gyrus |
|
| 149 |
+
|
| 150 |
+
|
| 151 |
+
## Bundle Integration in MONAI Lable
|
| 152 |
+
The inference pipleine can be easily used by the MONAI Label server and 3D Slicer for fast labeling T1w MRI images in MNI space.
|
| 153 |
+
|
| 154 |
+
 <br>
|
| 155 |
+
|
| 156 |
+
# Disclaimer
|
| 157 |
+
This is an example, not to be used for diagnostic purposes.
|
| 158 |
+
|
| 159 |
+
# References
|
| 160 |
+
[1] Yu, Xin, Yinchi Zhou, Yucheng Tang et al. Characterizing Renal Structures with 3D Block Aggregate Transformers. arXiv preprint arXiv:2203.02430 (2022). https://arxiv.org/pdf/2203.02430.pdf
|
| 161 |
+
|
| 162 |
+
[2] Zizhao Zhang et al. Nested Hierarchical Transformer: Towards Accurate, Data-Efficient and Interpretable Visual Understanding. AAAI Conference on Artificial Intelligence (AAAI) 2022
|
| 163 |
+
|
| 164 |
+
[3] Huo, Yuankai, et al. 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage 194 (2019): 105-119.
|
docs/demo.png
ADDED

|
Git LFS Details
|
docs/license.txt
ADDED
|
@@ -0,0 +1,4 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Third Party Licenses
|
| 2 |
+
-----------------------------------------------------------------------
|
| 3 |
+
|
| 4 |
+
/*********************************************************************/
|
docs/unest.png
ADDED
|
docs/wholebrain.png
ADDED
|
models/model.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:79a52ccd77bc35d05410f39788a1b063af3eb3b809b42241335c18aed27ec422
|
| 3 |
+
size 348901503
|
scripts/__init__.py
ADDED
|
@@ -0,0 +1,10 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
scripts/networks/__init__.py
ADDED
|
@@ -0,0 +1,10 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
scripts/networks/nest/__init__.py
ADDED
|
@@ -0,0 +1,16 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
from .utils import (
|
| 3 |
+
Conv3dSame,
|
| 4 |
+
DropPath,
|
| 5 |
+
Linear,
|
| 6 |
+
Mlp,
|
| 7 |
+
_assert,
|
| 8 |
+
conv3d_same,
|
| 9 |
+
create_conv3d,
|
| 10 |
+
create_pool3d,
|
| 11 |
+
get_padding,
|
| 12 |
+
get_same_padding,
|
| 13 |
+
pad_same,
|
| 14 |
+
to_ntuple,
|
| 15 |
+
trunc_normal_,
|
| 16 |
+
)
|
scripts/networks/nest/utils.py
ADDED
|
@@ -0,0 +1,485 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
|
| 3 |
+
|
| 4 |
+
import collections.abc
|
| 5 |
+
import math
|
| 6 |
+
import warnings
|
| 7 |
+
from itertools import repeat
|
| 8 |
+
from typing import List, Optional, Tuple
|
| 9 |
+
|
| 10 |
+
import torch
|
| 11 |
+
import torch.nn as nn
|
| 12 |
+
import torch.nn.functional as F
|
| 13 |
+
|
| 14 |
+
try:
|
| 15 |
+
from torch import _assert
|
| 16 |
+
except ImportError:
|
| 17 |
+
|
| 18 |
+
def _assert(condition: bool, message: str):
|
| 19 |
+
assert condition, message
|
| 20 |
+
|
| 21 |
+
|
| 22 |
+
def drop_block_2d(
|
| 23 |
+
x,
|
| 24 |
+
drop_prob: float = 0.1,
|
| 25 |
+
block_size: int = 7,
|
| 26 |
+
gamma_scale: float = 1.0,
|
| 27 |
+
with_noise: bool = False,
|
| 28 |
+
inplace: bool = False,
|
| 29 |
+
batchwise: bool = False,
|
| 30 |
+
):
|
| 31 |
+
"""DropBlock. See https://arxiv.org/pdf/1810.12890.pdf
|
| 32 |
+
|
| 33 |
+
DropBlock with an experimental gaussian noise option. This layer has been tested on a few training
|
| 34 |
+
runs with success, but needs further validation and possibly optimization for lower runtime impact.
|
| 35 |
+
"""
|
| 36 |
+
b, c, h, w = x.shape
|
| 37 |
+
total_size = w * h
|
| 38 |
+
clipped_block_size = min(block_size, min(w, h))
|
| 39 |
+
# seed_drop_rate, the gamma parameter
|
| 40 |
+
gamma = (
|
| 41 |
+
gamma_scale * drop_prob * total_size / clipped_block_size**2 / ((w - block_size + 1) * (h - block_size + 1))
|
| 42 |
+
)
|
| 43 |
+
|
| 44 |
+
# Forces the block to be inside the feature map.
|
| 45 |
+
w_i, h_i = torch.meshgrid(torch.arange(w).to(x.device), torch.arange(h).to(x.device))
|
| 46 |
+
valid_block = ((w_i >= clipped_block_size // 2) & (w_i < w - (clipped_block_size - 1) // 2)) & (
|
| 47 |
+
(h_i >= clipped_block_size // 2) & (h_i < h - (clipped_block_size - 1) // 2)
|
| 48 |
+
)
|
| 49 |
+
valid_block = torch.reshape(valid_block, (1, 1, h, w)).to(dtype=x.dtype)
|
| 50 |
+
|
| 51 |
+
if batchwise:
|
| 52 |
+
# one mask for whole batch, quite a bit faster
|
| 53 |
+
uniform_noise = torch.rand((1, c, h, w), dtype=x.dtype, device=x.device)
|
| 54 |
+
else:
|
| 55 |
+
uniform_noise = torch.rand_like(x)
|
| 56 |
+
block_mask = ((2 - gamma - valid_block + uniform_noise) >= 1).to(dtype=x.dtype)
|
| 57 |
+
block_mask = -F.max_pool2d(
|
| 58 |
+
-block_mask, kernel_size=clipped_block_size, stride=1, padding=clipped_block_size // 2 # block_size,
|
| 59 |
+
)
|
| 60 |
+
|
| 61 |
+
if with_noise:
|
| 62 |
+
normal_noise = torch.randn((1, c, h, w), dtype=x.dtype, device=x.device) if batchwise else torch.randn_like(x)
|
| 63 |
+
if inplace:
|
| 64 |
+
x.mul_(block_mask).add_(normal_noise * (1 - block_mask))
|
| 65 |
+
else:
|
| 66 |
+
x = x * block_mask + normal_noise * (1 - block_mask)
|
| 67 |
+
else:
|
| 68 |
+
normalize_scale = (block_mask.numel() / block_mask.to(dtype=torch.float32).sum().add(1e-7)).to(x.dtype)
|
| 69 |
+
if inplace:
|
| 70 |
+
x.mul_(block_mask * normalize_scale)
|
| 71 |
+
else:
|
| 72 |
+
x = x * block_mask * normalize_scale
|
| 73 |
+
return x
|
| 74 |
+
|
| 75 |
+
|
| 76 |
+
def drop_block_fast_2d(
|
| 77 |
+
x: torch.Tensor,
|
| 78 |
+
drop_prob: float = 0.1,
|
| 79 |
+
block_size: int = 7,
|
| 80 |
+
gamma_scale: float = 1.0,
|
| 81 |
+
with_noise: bool = False,
|
| 82 |
+
inplace: bool = False,
|
| 83 |
+
):
|
| 84 |
+
"""DropBlock. See https://arxiv.org/pdf/1810.12890.pdf
|
| 85 |
+
|
| 86 |
+
DropBlock with an experimental gaussian noise option. Simplied from above without concern for valid
|
| 87 |
+
block mask at edges.
|
| 88 |
+
"""
|
| 89 |
+
b, c, h, w = x.shape
|
| 90 |
+
total_size = w * h
|
| 91 |
+
clipped_block_size = min(block_size, min(w, h))
|
| 92 |
+
gamma = (
|
| 93 |
+
gamma_scale * drop_prob * total_size / clipped_block_size**2 / ((w - block_size + 1) * (h - block_size + 1))
|
| 94 |
+
)
|
| 95 |
+
|
| 96 |
+
block_mask = torch.empty_like(x).bernoulli_(gamma)
|
| 97 |
+
block_mask = F.max_pool2d(
|
| 98 |
+
block_mask.to(x.dtype), kernel_size=clipped_block_size, stride=1, padding=clipped_block_size // 2
|
| 99 |
+
)
|
| 100 |
+
|
| 101 |
+
if with_noise:
|
| 102 |
+
normal_noise = torch.empty_like(x).normal_()
|
| 103 |
+
if inplace:
|
| 104 |
+
x.mul_(1.0 - block_mask).add_(normal_noise * block_mask)
|
| 105 |
+
else:
|
| 106 |
+
x = x * (1.0 - block_mask) + normal_noise * block_mask
|
| 107 |
+
else:
|
| 108 |
+
block_mask = 1 - block_mask
|
| 109 |
+
normalize_scale = (block_mask.numel() / block_mask.to(dtype=torch.float32).sum().add(1e-6)).to(dtype=x.dtype)
|
| 110 |
+
if inplace:
|
| 111 |
+
x.mul_(block_mask * normalize_scale)
|
| 112 |
+
else:
|
| 113 |
+
x = x * block_mask * normalize_scale
|
| 114 |
+
return x
|
| 115 |
+
|
| 116 |
+
|
| 117 |
+
class DropBlock2d(nn.Module):
|
| 118 |
+
"""DropBlock. See https://arxiv.org/pdf/1810.12890.pdf"""
|
| 119 |
+
|
| 120 |
+
def __init__(
|
| 121 |
+
self, drop_prob=0.1, block_size=7, gamma_scale=1.0, with_noise=False, inplace=False, batchwise=False, fast=True
|
| 122 |
+
):
|
| 123 |
+
super(DropBlock2d, self).__init__()
|
| 124 |
+
self.drop_prob = drop_prob
|
| 125 |
+
self.gamma_scale = gamma_scale
|
| 126 |
+
self.block_size = block_size
|
| 127 |
+
self.with_noise = with_noise
|
| 128 |
+
self.inplace = inplace
|
| 129 |
+
self.batchwise = batchwise
|
| 130 |
+
self.fast = fast # FIXME finish comparisons of fast vs not
|
| 131 |
+
|
| 132 |
+
def forward(self, x):
|
| 133 |
+
if not self.training or not self.drop_prob:
|
| 134 |
+
return x
|
| 135 |
+
if self.fast:
|
| 136 |
+
return drop_block_fast_2d(
|
| 137 |
+
x, self.drop_prob, self.block_size, self.gamma_scale, self.with_noise, self.inplace
|
| 138 |
+
)
|
| 139 |
+
else:
|
| 140 |
+
return drop_block_2d(
|
| 141 |
+
x, self.drop_prob, self.block_size, self.gamma_scale, self.with_noise, self.inplace, self.batchwise
|
| 142 |
+
)
|
| 143 |
+
|
| 144 |
+
|
| 145 |
+
def drop_path(x, drop_prob: float = 0.0, training: bool = False, scale_by_keep: bool = True):
|
| 146 |
+
"""Drop paths (Stochastic Depth) per sample (when applied in main path of residual blocks).
|
| 147 |
+
|
| 148 |
+
This is the same as the DropConnect impl I created for EfficientNet, etc networks, however,
|
| 149 |
+
the original name is misleading as 'Drop Connect' is a different form of dropout in a separate paper...
|
| 150 |
+
See discussion: https://github.com/tensorflow/tpu/issues/494#issuecomment-532968956 ... I've opted for
|
| 151 |
+
changing the layer and argument names to 'drop path' rather than mix DropConnect as a layer name and use
|
| 152 |
+
'survival rate' as the argument.
|
| 153 |
+
|
| 154 |
+
"""
|
| 155 |
+
if drop_prob == 0.0 or not training:
|
| 156 |
+
return x
|
| 157 |
+
keep_prob = 1 - drop_prob
|
| 158 |
+
shape = (x.shape[0],) + (1,) * (x.ndim - 1) # work with diff dim tensors, not just 2D ConvNets
|
| 159 |
+
random_tensor = x.new_empty(shape).bernoulli_(keep_prob)
|
| 160 |
+
if keep_prob > 0.0 and scale_by_keep:
|
| 161 |
+
random_tensor.div_(keep_prob)
|
| 162 |
+
return x * random_tensor
|
| 163 |
+
|
| 164 |
+
|
| 165 |
+
class DropPath(nn.Module):
|
| 166 |
+
"""Drop paths (Stochastic Depth) per sample (when applied in main path of residual blocks)."""
|
| 167 |
+
|
| 168 |
+
def __init__(self, drop_prob=None, scale_by_keep=True):
|
| 169 |
+
super(DropPath, self).__init__()
|
| 170 |
+
self.drop_prob = drop_prob
|
| 171 |
+
self.scale_by_keep = scale_by_keep
|
| 172 |
+
|
| 173 |
+
def forward(self, x):
|
| 174 |
+
return drop_path(x, self.drop_prob, self.training, self.scale_by_keep)
|
| 175 |
+
|
| 176 |
+
|
| 177 |
+
def create_conv3d(in_channels, out_channels, kernel_size, **kwargs):
|
| 178 |
+
"""Select a 2d convolution implementation based on arguments
|
| 179 |
+
Creates and returns one of torch.nn.Conv2d, Conv2dSame, MixedConv3d, or CondConv2d.
|
| 180 |
+
|
| 181 |
+
Used extensively by EfficientNet, MobileNetv3 and related networks.
|
| 182 |
+
"""
|
| 183 |
+
|
| 184 |
+
depthwise = kwargs.pop("depthwise", False)
|
| 185 |
+
# for DW out_channels must be multiple of in_channels as must have out_channels % groups == 0
|
| 186 |
+
groups = in_channels if depthwise else kwargs.pop("groups", 1)
|
| 187 |
+
|
| 188 |
+
m = create_conv3d_pad(in_channels, out_channels, kernel_size, groups=groups, **kwargs)
|
| 189 |
+
return m
|
| 190 |
+
|
| 191 |
+
|
| 192 |
+
def conv3d_same(
|
| 193 |
+
x,
|
| 194 |
+
weight: torch.Tensor,
|
| 195 |
+
bias: Optional[torch.Tensor] = None,
|
| 196 |
+
stride: Tuple[int, int] = (1, 1, 1),
|
| 197 |
+
padding: Tuple[int, int] = (0, 0, 0),
|
| 198 |
+
dilation: Tuple[int, int] = (1, 1, 1),
|
| 199 |
+
groups: int = 1,
|
| 200 |
+
):
|
| 201 |
+
x = pad_same(x, weight.shape[-3:], stride, dilation)
|
| 202 |
+
return F.conv3d(x, weight, bias, stride, (0, 0, 0), dilation, groups)
|
| 203 |
+
|
| 204 |
+
|
| 205 |
+
class Conv3dSame(nn.Conv2d):
|
| 206 |
+
"""Tensorflow like 'SAME' convolution wrapper for 2D convolutions"""
|
| 207 |
+
|
| 208 |
+
def __init__(self, in_channels, out_channels, kernel_size, stride=1, padding=0, dilation=1, groups=1, bias=True):
|
| 209 |
+
super(Conv3dSame, self).__init__(in_channels, out_channels, kernel_size, stride, 0, dilation, groups, bias)
|
| 210 |
+
|
| 211 |
+
def forward(self, x):
|
| 212 |
+
return conv3d_same(x, self.weight, self.bias, self.stride, self.padding, self.dilation, self.groups)
|
| 213 |
+
|
| 214 |
+
|
| 215 |
+
def create_conv3d_pad(in_chs, out_chs, kernel_size, **kwargs):
|
| 216 |
+
padding = kwargs.pop("padding", "")
|
| 217 |
+
kwargs.setdefault("bias", False)
|
| 218 |
+
padding, is_dynamic = get_padding_value(padding, kernel_size, **kwargs)
|
| 219 |
+
if is_dynamic:
|
| 220 |
+
return Conv3dSame(in_chs, out_chs, kernel_size, **kwargs)
|
| 221 |
+
else:
|
| 222 |
+
return nn.Conv3d(in_chs, out_chs, kernel_size, padding=padding, **kwargs)
|
| 223 |
+
|
| 224 |
+
|
| 225 |
+
# Calculate symmetric padding for a convolution
|
| 226 |
+
def get_padding(kernel_size: int, stride: int = 1, dilation: int = 1, **_) -> int:
|
| 227 |
+
padding = ((stride - 1) + dilation * (kernel_size - 1)) // 2
|
| 228 |
+
return padding
|
| 229 |
+
|
| 230 |
+
|
| 231 |
+
# Calculate asymmetric TensorFlow-like 'SAME' padding for a convolution
|
| 232 |
+
def get_same_padding(x: int, k: int, s: int, d: int):
|
| 233 |
+
return max((math.ceil(x / s) - 1) * s + (k - 1) * d + 1 - x, 0)
|
| 234 |
+
|
| 235 |
+
|
| 236 |
+
# Can SAME padding for given args be done statically?
|
| 237 |
+
def is_static_pad(kernel_size: int, stride: int = 1, dilation: int = 1, **_):
|
| 238 |
+
return stride == 1 and (dilation * (kernel_size - 1)) % 2 == 0
|
| 239 |
+
|
| 240 |
+
|
| 241 |
+
# Dynamically pad input x with 'SAME' padding for conv with specified args
|
| 242 |
+
def pad_same(x, k: List[int], s: List[int], d: List[int] = (1, 1, 1), value: float = 0):
|
| 243 |
+
id, ih, iw = x.size()[-3:]
|
| 244 |
+
pad_d, pad_h, pad_w = (
|
| 245 |
+
get_same_padding(id, k[0], s[0], d[0]),
|
| 246 |
+
get_same_padding(ih, k[1], s[1], d[1]),
|
| 247 |
+
get_same_padding(iw, k[2], s[2], d[2]),
|
| 248 |
+
)
|
| 249 |
+
if pad_d > 0 or pad_h > 0 or pad_w > 0:
|
| 250 |
+
x = F.pad(
|
| 251 |
+
x,
|
| 252 |
+
[pad_d // 2, pad_d - pad_d // 2, pad_w // 2, pad_w - pad_w // 2, pad_h // 2, pad_h - pad_h // 2],
|
| 253 |
+
value=value,
|
| 254 |
+
)
|
| 255 |
+
return x
|
| 256 |
+
|
| 257 |
+
|
| 258 |
+
def get_padding_value(padding, kernel_size, **kwargs) -> Tuple[Tuple, bool]:
|
| 259 |
+
dynamic = False
|
| 260 |
+
if isinstance(padding, str):
|
| 261 |
+
# for any string padding, the padding will be calculated for you, one of three ways
|
| 262 |
+
padding = padding.lower()
|
| 263 |
+
if padding == "same":
|
| 264 |
+
# TF compatible 'SAME' padding, has a performance and GPU memory allocation impact
|
| 265 |
+
if is_static_pad(kernel_size, **kwargs):
|
| 266 |
+
# static case, no extra overhead
|
| 267 |
+
padding = get_padding(kernel_size, **kwargs)
|
| 268 |
+
else:
|
| 269 |
+
# dynamic 'SAME' padding, has runtime/GPU memory overhead
|
| 270 |
+
padding = 0
|
| 271 |
+
dynamic = True
|
| 272 |
+
elif padding == "valid":
|
| 273 |
+
# 'VALID' padding, same as padding=0
|
| 274 |
+
padding = 0
|
| 275 |
+
else:
|
| 276 |
+
# Default to PyTorch style 'same'-ish symmetric padding
|
| 277 |
+
padding = get_padding(kernel_size, **kwargs)
|
| 278 |
+
return padding, dynamic
|
| 279 |
+
|
| 280 |
+
|
| 281 |
+
# From PyTorch internals
|
| 282 |
+
def _ntuple(n):
|
| 283 |
+
def parse(x):
|
| 284 |
+
if isinstance(x, collections.abc.Iterable):
|
| 285 |
+
return x
|
| 286 |
+
return tuple(repeat(x, n))
|
| 287 |
+
|
| 288 |
+
return parse
|
| 289 |
+
|
| 290 |
+
|
| 291 |
+
to_1tuple = _ntuple(1)
|
| 292 |
+
to_2tuple = _ntuple(2)
|
| 293 |
+
to_3tuple = _ntuple(3)
|
| 294 |
+
to_4tuple = _ntuple(4)
|
| 295 |
+
to_ntuple = _ntuple
|
| 296 |
+
|
| 297 |
+
|
| 298 |
+
def make_divisible(v, divisor=8, min_value=None, round_limit=0.9):
|
| 299 |
+
min_value = min_value or divisor
|
| 300 |
+
new_v = max(min_value, int(v + divisor / 2) // divisor * divisor)
|
| 301 |
+
# Make sure that round down does not go down by more than 10%.
|
| 302 |
+
if new_v < round_limit * v:
|
| 303 |
+
new_v += divisor
|
| 304 |
+
return new_v
|
| 305 |
+
|
| 306 |
+
|
| 307 |
+
class Linear(nn.Linear):
|
| 308 |
+
r"""Applies a linear transformation to the incoming data: :math:`y = xA^T + b`
|
| 309 |
+
|
| 310 |
+
Wraps torch.nn.Linear to support AMP + torchscript usage by manually casting
|
| 311 |
+
weight & bias to input.dtype to work around an issue w/ torch.addmm in this use case.
|
| 312 |
+
"""
|
| 313 |
+
|
| 314 |
+
def forward(self, input: torch.Tensor) -> torch.Tensor:
|
| 315 |
+
if torch.jit.is_scripting():
|
| 316 |
+
bias = self.bias.to(dtype=input.dtype) if self.bias is not None else None
|
| 317 |
+
return F.linear(input, self.weight.to(dtype=input.dtype), bias=bias)
|
| 318 |
+
else:
|
| 319 |
+
return F.linear(input, self.weight, self.bias)
|
| 320 |
+
|
| 321 |
+
|
| 322 |
+
class Mlp(nn.Module):
|
| 323 |
+
"""MLP as used in Vision Transformer, MLP-Mixer and related networks"""
|
| 324 |
+
|
| 325 |
+
def __init__(self, in_features, hidden_features=None, out_features=None, act_layer=nn.GELU, drop=0.0):
|
| 326 |
+
super().__init__()
|
| 327 |
+
out_features = out_features or in_features
|
| 328 |
+
hidden_features = hidden_features or in_features
|
| 329 |
+
drop_probs = to_2tuple(drop)
|
| 330 |
+
|
| 331 |
+
self.fc1 = nn.Linear(in_features, hidden_features)
|
| 332 |
+
self.act = act_layer()
|
| 333 |
+
self.drop1 = nn.Dropout(drop_probs[0])
|
| 334 |
+
self.fc2 = nn.Linear(hidden_features, out_features)
|
| 335 |
+
self.drop2 = nn.Dropout(drop_probs[1])
|
| 336 |
+
|
| 337 |
+
def forward(self, x):
|
| 338 |
+
x = self.fc1(x)
|
| 339 |
+
x = self.act(x)
|
| 340 |
+
x = self.drop1(x)
|
| 341 |
+
x = self.fc2(x)
|
| 342 |
+
x = self.drop2(x)
|
| 343 |
+
return x
|
| 344 |
+
|
| 345 |
+
|
| 346 |
+
def avg_pool3d_same(
|
| 347 |
+
x,
|
| 348 |
+
kernel_size: List[int],
|
| 349 |
+
stride: List[int],
|
| 350 |
+
padding: List[int] = (0, 0, 0),
|
| 351 |
+
ceil_mode: bool = False,
|
| 352 |
+
count_include_pad: bool = True,
|
| 353 |
+
):
|
| 354 |
+
# FIXME how to deal with count_include_pad vs not for external padding?
|
| 355 |
+
x = pad_same(x, kernel_size, stride)
|
| 356 |
+
return F.avg_pool3d(x, kernel_size, stride, (0, 0, 0), ceil_mode, count_include_pad)
|
| 357 |
+
|
| 358 |
+
|
| 359 |
+
class AvgPool3dSame(nn.AvgPool2d):
|
| 360 |
+
"""Tensorflow like 'SAME' wrapper for 2D average pooling"""
|
| 361 |
+
|
| 362 |
+
def __init__(self, kernel_size: int, stride=None, padding=0, ceil_mode=False, count_include_pad=True):
|
| 363 |
+
kernel_size = to_2tuple(kernel_size)
|
| 364 |
+
stride = to_2tuple(stride)
|
| 365 |
+
super(AvgPool3dSame, self).__init__(kernel_size, stride, (0, 0, 0), ceil_mode, count_include_pad)
|
| 366 |
+
|
| 367 |
+
def forward(self, x):
|
| 368 |
+
x = pad_same(x, self.kernel_size, self.stride)
|
| 369 |
+
return F.avg_pool3d(x, self.kernel_size, self.stride, self.padding, self.ceil_mode, self.count_include_pad)
|
| 370 |
+
|
| 371 |
+
|
| 372 |
+
def max_pool3d_same(
|
| 373 |
+
x,
|
| 374 |
+
kernel_size: List[int],
|
| 375 |
+
stride: List[int],
|
| 376 |
+
padding: List[int] = (0, 0, 0),
|
| 377 |
+
dilation: List[int] = (1, 1, 1),
|
| 378 |
+
ceil_mode: bool = False,
|
| 379 |
+
):
|
| 380 |
+
x = pad_same(x, kernel_size, stride, value=-float("inf"))
|
| 381 |
+
return F.max_pool3d(x, kernel_size, stride, (0, 0, 0), dilation, ceil_mode)
|
| 382 |
+
|
| 383 |
+
|
| 384 |
+
class MaxPool3dSame(nn.MaxPool2d):
|
| 385 |
+
"""Tensorflow like 'SAME' wrapper for 3D max pooling"""
|
| 386 |
+
|
| 387 |
+
def __init__(self, kernel_size: int, stride=None, padding=0, dilation=1, ceil_mode=False):
|
| 388 |
+
kernel_size = to_2tuple(kernel_size)
|
| 389 |
+
stride = to_2tuple(stride)
|
| 390 |
+
dilation = to_2tuple(dilation)
|
| 391 |
+
super(MaxPool3dSame, self).__init__(kernel_size, stride, (0, 0, 0), dilation, ceil_mode)
|
| 392 |
+
|
| 393 |
+
def forward(self, x):
|
| 394 |
+
x = pad_same(x, self.kernel_size, self.stride, value=-float("inf"))
|
| 395 |
+
return F.max_pool3d(x, self.kernel_size, self.stride, (0, 0, 0), self.dilation, self.ceil_mode)
|
| 396 |
+
|
| 397 |
+
|
| 398 |
+
def create_pool3d(pool_type, kernel_size, stride=None, **kwargs):
|
| 399 |
+
stride = stride or kernel_size
|
| 400 |
+
padding = kwargs.pop("padding", "")
|
| 401 |
+
padding, is_dynamic = get_padding_value(padding, kernel_size, stride=stride, **kwargs)
|
| 402 |
+
if is_dynamic:
|
| 403 |
+
if pool_type == "avg":
|
| 404 |
+
return AvgPool3dSame(kernel_size, stride=stride, **kwargs)
|
| 405 |
+
elif pool_type == "max":
|
| 406 |
+
return MaxPool3dSame(kernel_size, stride=stride, **kwargs)
|
| 407 |
+
else:
|
| 408 |
+
raise AssertionError()
|
| 409 |
+
|
| 410 |
+
# assert False, f"Unsupported pool type {pool_type}"
|
| 411 |
+
else:
|
| 412 |
+
if pool_type == "avg":
|
| 413 |
+
return nn.AvgPool3d(kernel_size, stride=stride, padding=padding, **kwargs)
|
| 414 |
+
elif pool_type == "max":
|
| 415 |
+
return nn.MaxPool3d(kernel_size, stride=stride, padding=padding, **kwargs)
|
| 416 |
+
else:
|
| 417 |
+
raise AssertionError()
|
| 418 |
+
|
| 419 |
+
# assert False, f"Unsupported pool type {pool_type}"
|
| 420 |
+
|
| 421 |
+
|
| 422 |
+
def _float_to_int(x: float) -> int:
|
| 423 |
+
"""
|
| 424 |
+
Symbolic tracing helper to substitute for inbuilt `int`.
|
| 425 |
+
Hint: Inbuilt `int` can't accept an argument of type `Proxy`
|
| 426 |
+
"""
|
| 427 |
+
return int(x)
|
| 428 |
+
|
| 429 |
+
|
| 430 |
+
def _no_grad_trunc_normal_(tensor, mean, std, a, b):
|
| 431 |
+
# Cut & paste from PyTorch official master until it's in a few official releases - RW
|
| 432 |
+
# Method based on https://people.sc.fsu.edu/~jburkardt/presentations/truncated_normal.pdf
|
| 433 |
+
def norm_cdf(x):
|
| 434 |
+
# Computes standard normal cumulative distribution function
|
| 435 |
+
return (1.0 + math.erf(x / math.sqrt(2.0))) / 2.0
|
| 436 |
+
|
| 437 |
+
if (mean < a - 2 * std) or (mean > b + 2 * std):
|
| 438 |
+
warnings.warn(
|
| 439 |
+
"mean is more than 2 std from [a, b] in nn.init.trunc_normal_. "
|
| 440 |
+
"The distribution of values may be incorrect.",
|
| 441 |
+
stacklevel=2,
|
| 442 |
+
)
|
| 443 |
+
|
| 444 |
+
with torch.no_grad():
|
| 445 |
+
# Values are generated by using a truncated uniform distribution and
|
| 446 |
+
# then using the inverse CDF for the normal distribution.
|
| 447 |
+
# Get upper and lower cdf values
|
| 448 |
+
l = norm_cdf((a - mean) / std)
|
| 449 |
+
u = norm_cdf((b - mean) / std)
|
| 450 |
+
|
| 451 |
+
# Uniformly fill tensor with values from [l, u], then translate to
|
| 452 |
+
# [2l-1, 2u-1].
|
| 453 |
+
tensor.uniform_(2 * l - 1, 2 * u - 1)
|
| 454 |
+
|
| 455 |
+
# Use inverse cdf transform for normal distribution to get truncated
|
| 456 |
+
# standard normal
|
| 457 |
+
tensor.erfinv_()
|
| 458 |
+
|
| 459 |
+
# Transform to proper mean, std
|
| 460 |
+
tensor.mul_(std * math.sqrt(2.0))
|
| 461 |
+
tensor.add_(mean)
|
| 462 |
+
|
| 463 |
+
# Clamp to ensure it's in the proper range
|
| 464 |
+
tensor.clamp_(min=a, max=b)
|
| 465 |
+
return tensor
|
| 466 |
+
|
| 467 |
+
|
| 468 |
+
def trunc_normal_(tensor, mean=0.0, std=1.0, a=-2.0, b=2.0):
|
| 469 |
+
r"""Fills the input Tensor with values drawn from a truncated
|
| 470 |
+
normal distribution. The values are effectively drawn from the
|
| 471 |
+
normal distribution :math:`\mathcal{N}(\text{mean}, \text{std}^2)`
|
| 472 |
+
with values outside :math:`[a, b]` redrawn until they are within
|
| 473 |
+
the bounds. The method used for generating the random values works
|
| 474 |
+
best when :math:`a \leq \text{mean} \leq b`.
|
| 475 |
+
Args:
|
| 476 |
+
tensor: an n-dimensional `torch.Tensor`
|
| 477 |
+
mean: the mean of the normal distribution
|
| 478 |
+
std: the standard deviation of the normal distribution
|
| 479 |
+
a: the minimum cutoff value
|
| 480 |
+
b: the maximum cutoff value
|
| 481 |
+
Examples:
|
| 482 |
+
>>> w = torch.empty(3, 5)
|
| 483 |
+
>>> nn.init.trunc_normal_(w)
|
| 484 |
+
"""
|
| 485 |
+
return _no_grad_trunc_normal_(tensor, mean, std, a, b)
|
scripts/networks/nest_transformer_3D.py
ADDED
|
@@ -0,0 +1,489 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
|
| 3 |
+
# =========================================================================
|
| 4 |
+
# Adapted from https://github.com/google-research/nested-transformer.
|
| 5 |
+
# which has the following license...
|
| 6 |
+
# https://github.com/pytorch/vision/blob/main/LICENSE
|
| 7 |
+
#
|
| 8 |
+
# BSD 3-Clause License
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
# Redistribution and use in source and binary forms, with or without
|
| 12 |
+
# modification, are permitted provided that the following conditions are met:
|
| 13 |
+
|
| 14 |
+
# * Redistributions of source code must retain the above copyright notice, this
|
| 15 |
+
# list of conditions and the following disclaimer.
|
| 16 |
+
|
| 17 |
+
# * Redistributions in binary form must reproduce the above copyright notice,
|
| 18 |
+
# this list of conditions and the following disclaimer in the documentation
|
| 19 |
+
# and/or other materials provided with the distribution.
|
| 20 |
+
|
| 21 |
+
# * Neither the name of the copyright holder nor the names of its
|
| 22 |
+
# contributors may be used to endorse or promote products derived from
|
| 23 |
+
# this software without specific prior written permission.
|
| 24 |
+
|
| 25 |
+
# THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
|
| 26 |
+
# AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
|
| 27 |
+
# IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
|
| 28 |
+
# DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
|
| 29 |
+
# FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
|
| 30 |
+
# DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
|
| 31 |
+
# SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
|
| 32 |
+
# CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
|
| 33 |
+
# OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
|
| 34 |
+
# OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
|
| 35 |
+
|
| 36 |
+
""" Nested Transformer (NesT) in PyTorch
|
| 37 |
+
A PyTorch implement of Aggregating Nested Transformers as described in:
|
| 38 |
+
'Aggregating Nested Transformers'
|
| 39 |
+
- https://arxiv.org/abs/2105.12723
|
| 40 |
+
The official Jax code is released and available at https://github.com/google-research/nested-transformer.
|
| 41 |
+
The weights have been converted with convert/convert_nest_flax.py
|
| 42 |
+
Acknowledgments:
|
| 43 |
+
* The paper authors for sharing their research, code, and model weights
|
| 44 |
+
* Ross Wightman's existing code off which I based this
|
| 45 |
+
Copyright 2021 Alexander Soare
|
| 46 |
+
|
| 47 |
+
"""
|
| 48 |
+
|
| 49 |
+
import collections.abc
|
| 50 |
+
import logging
|
| 51 |
+
import math
|
| 52 |
+
from functools import partial
|
| 53 |
+
from typing import Callable, Sequence
|
| 54 |
+
|
| 55 |
+
import torch
|
| 56 |
+
import torch.nn.functional as F
|
| 57 |
+
from torch import nn
|
| 58 |
+
|
| 59 |
+
from .nest import DropPath, Mlp, _assert, create_conv3d, create_pool3d, to_ntuple, trunc_normal_
|
| 60 |
+
from .patchEmbed3D import PatchEmbed3D
|
| 61 |
+
|
| 62 |
+
_logger = logging.getLogger(__name__)
|
| 63 |
+
|
| 64 |
+
|
| 65 |
+
class Attention(nn.Module):
|
| 66 |
+
"""
|
| 67 |
+
This is much like `.vision_transformer.Attention` but uses *localised* self attention by accepting an input with
|
| 68 |
+
an extra "image block" dim
|
| 69 |
+
"""
|
| 70 |
+
|
| 71 |
+
def __init__(self, dim, num_heads=8, qkv_bias=False, attn_drop=0.0, proj_drop=0.0):
|
| 72 |
+
super().__init__()
|
| 73 |
+
self.num_heads = num_heads
|
| 74 |
+
head_dim = dim // num_heads
|
| 75 |
+
self.scale = head_dim**-0.5
|
| 76 |
+
|
| 77 |
+
self.qkv = nn.Linear(dim, 3 * dim, bias=qkv_bias)
|
| 78 |
+
self.attn_drop = nn.Dropout(attn_drop)
|
| 79 |
+
self.proj = nn.Linear(dim, dim)
|
| 80 |
+
self.proj_drop = nn.Dropout(proj_drop)
|
| 81 |
+
|
| 82 |
+
def forward(self, x):
|
| 83 |
+
"""
|
| 84 |
+
x is shape: B (batch_size), T (image blocks), N (seq length per image block), C (embed dim)
|
| 85 |
+
"""
|
| 86 |
+
b, t, n, c = x.shape
|
| 87 |
+
# result of next line is (qkv, B, num (H)eads, T, N, (C')hannels per head)
|
| 88 |
+
qkv = self.qkv(x).reshape(b, t, n, 3, self.num_heads, c // self.num_heads).permute(3, 0, 4, 1, 2, 5)
|
| 89 |
+
q, k, v = qkv.unbind(0) # make torchscript happy (cannot use tensor as tuple)
|
| 90 |
+
|
| 91 |
+
attn = (q @ k.transpose(-2, -1)) * self.scale # (B, H, T, N, N)
|
| 92 |
+
attn = attn.softmax(dim=-1)
|
| 93 |
+
attn = self.attn_drop(attn)
|
| 94 |
+
|
| 95 |
+
x = (attn @ v).permute(0, 2, 3, 4, 1).reshape(b, t, n, c)
|
| 96 |
+
x = self.proj(x)
|
| 97 |
+
x = self.proj_drop(x)
|
| 98 |
+
return x # (B, T, N, C)
|
| 99 |
+
|
| 100 |
+
|
| 101 |
+
class TransformerLayer(nn.Module):
|
| 102 |
+
"""
|
| 103 |
+
This is much like `.vision_transformer.Block` but:
|
| 104 |
+
- Called TransformerLayer here to allow for "block" as defined in the paper ("non-overlapping image blocks")
|
| 105 |
+
- Uses modified Attention layer that handles the "block" dimension
|
| 106 |
+
"""
|
| 107 |
+
|
| 108 |
+
def __init__(
|
| 109 |
+
self,
|
| 110 |
+
dim,
|
| 111 |
+
num_heads,
|
| 112 |
+
mlp_ratio=4.0,
|
| 113 |
+
qkv_bias=False,
|
| 114 |
+
drop=0.0,
|
| 115 |
+
attn_drop=0.0,
|
| 116 |
+
drop_path=0.0,
|
| 117 |
+
act_layer=nn.GELU,
|
| 118 |
+
norm_layer=nn.LayerNorm,
|
| 119 |
+
):
|
| 120 |
+
super().__init__()
|
| 121 |
+
self.norm1 = norm_layer(dim)
|
| 122 |
+
self.attn = Attention(dim, num_heads=num_heads, qkv_bias=qkv_bias, attn_drop=attn_drop, proj_drop=drop)
|
| 123 |
+
self.drop_path = DropPath(drop_path) if drop_path > 0.0 else nn.Identity()
|
| 124 |
+
self.norm2 = norm_layer(dim)
|
| 125 |
+
mlp_hidden_dim = int(dim * mlp_ratio)
|
| 126 |
+
self.mlp = Mlp(in_features=dim, hidden_features=mlp_hidden_dim, act_layer=act_layer, drop=drop)
|
| 127 |
+
|
| 128 |
+
def forward(self, x):
|
| 129 |
+
y = self.norm1(x)
|
| 130 |
+
x = x + self.drop_path(self.attn(y))
|
| 131 |
+
x = x + self.drop_path(self.mlp(self.norm2(x)))
|
| 132 |
+
return x
|
| 133 |
+
|
| 134 |
+
|
| 135 |
+
class ConvPool(nn.Module):
|
| 136 |
+
def __init__(self, in_channels, out_channels, norm_layer, pad_type=""):
|
| 137 |
+
super().__init__()
|
| 138 |
+
self.conv = create_conv3d(in_channels, out_channels, kernel_size=3, padding=pad_type, bias=True)
|
| 139 |
+
self.norm = norm_layer(out_channels)
|
| 140 |
+
self.pool = create_pool3d("max", kernel_size=3, stride=2, padding=pad_type)
|
| 141 |
+
|
| 142 |
+
def forward(self, x):
|
| 143 |
+
"""
|
| 144 |
+
x is expected to have shape (B, C, D, H, W)
|
| 145 |
+
"""
|
| 146 |
+
_assert(x.shape[-3] % 2 == 0, "BlockAggregation requires even input spatial dims")
|
| 147 |
+
_assert(x.shape[-2] % 2 == 0, "BlockAggregation requires even input spatial dims")
|
| 148 |
+
_assert(x.shape[-1] % 2 == 0, "BlockAggregation requires even input spatial dims")
|
| 149 |
+
|
| 150 |
+
# print('In ConvPool x : {}'.format(x.shape))
|
| 151 |
+
x = self.conv(x)
|
| 152 |
+
# Layer norm done over channel dim only
|
| 153 |
+
x = self.norm(x.permute(0, 2, 3, 4, 1)).permute(0, 4, 1, 2, 3)
|
| 154 |
+
x = self.pool(x)
|
| 155 |
+
return x # (B, C, D//2, H//2, W//2)
|
| 156 |
+
|
| 157 |
+
|
| 158 |
+
def blockify(x, block_size: int):
|
| 159 |
+
"""image to blocks
|
| 160 |
+
Args:
|
| 161 |
+
x (Tensor): with shape (B, D, H, W, C)
|
| 162 |
+
block_size (int): edge length of a single square block in units of D, H, W
|
| 163 |
+
"""
|
| 164 |
+
b, d, h, w, c = x.shape
|
| 165 |
+
_assert(d % block_size == 0, "`block_size` must divide input depth evenly")
|
| 166 |
+
_assert(h % block_size == 0, "`block_size` must divide input height evenly")
|
| 167 |
+
_assert(w % block_size == 0, "`block_size` must divide input width evenly")
|
| 168 |
+
grid_depth = d // block_size
|
| 169 |
+
grid_height = h // block_size
|
| 170 |
+
grid_width = w // block_size
|
| 171 |
+
x = x.reshape(b, grid_depth, block_size, grid_height, block_size, grid_width, block_size, c)
|
| 172 |
+
|
| 173 |
+
x = x.permute(0, 1, 3, 5, 2, 4, 6, 7).reshape(
|
| 174 |
+
b, grid_depth * grid_height * grid_width, -1, c
|
| 175 |
+
) # shape [2, 512, 27, 128]
|
| 176 |
+
|
| 177 |
+
return x # (B, T, N, C)
|
| 178 |
+
|
| 179 |
+
|
| 180 |
+
# @register_notrace_function # reason: int receives Proxy
|
| 181 |
+
def deblockify(x, block_size: int):
|
| 182 |
+
"""blocks to image
|
| 183 |
+
Args:
|
| 184 |
+
x (Tensor): with shape (B, T, N, C) where T is number of blocks and N is sequence size per block
|
| 185 |
+
block_size (int): edge length of a single square block in units of desired D, H, W
|
| 186 |
+
"""
|
| 187 |
+
b, t, _, c = x.shape
|
| 188 |
+
grid_size = round(math.pow(t, 1 / 3))
|
| 189 |
+
depth = height = width = grid_size * block_size
|
| 190 |
+
x = x.reshape(b, grid_size, grid_size, grid_size, block_size, block_size, block_size, c)
|
| 191 |
+
|
| 192 |
+
x = x.permute(0, 1, 4, 2, 5, 3, 6, 7).reshape(b, depth, height, width, c)
|
| 193 |
+
|
| 194 |
+
return x # (B, D, H, W, C)
|
| 195 |
+
|
| 196 |
+
|
| 197 |
+
class NestLevel(nn.Module):
|
| 198 |
+
"""Single hierarchical level of a Nested Transformer"""
|
| 199 |
+
|
| 200 |
+
def __init__(
|
| 201 |
+
self,
|
| 202 |
+
num_blocks,
|
| 203 |
+
block_size,
|
| 204 |
+
seq_length,
|
| 205 |
+
num_heads,
|
| 206 |
+
depth,
|
| 207 |
+
embed_dim,
|
| 208 |
+
prev_embed_dim=None,
|
| 209 |
+
mlp_ratio=4.0,
|
| 210 |
+
qkv_bias=True,
|
| 211 |
+
drop_rate=0.0,
|
| 212 |
+
attn_drop_rate=0.0,
|
| 213 |
+
drop_path_rates: Sequence[int] = (),
|
| 214 |
+
norm_layer=None,
|
| 215 |
+
act_layer=None,
|
| 216 |
+
pad_type="",
|
| 217 |
+
):
|
| 218 |
+
super().__init__()
|
| 219 |
+
self.block_size = block_size
|
| 220 |
+
self.pos_embed = nn.Parameter(torch.zeros(1, num_blocks, seq_length, embed_dim))
|
| 221 |
+
|
| 222 |
+
if prev_embed_dim is not None:
|
| 223 |
+
self.pool = ConvPool(prev_embed_dim, embed_dim, norm_layer=norm_layer, pad_type=pad_type)
|
| 224 |
+
else:
|
| 225 |
+
self.pool = nn.Identity()
|
| 226 |
+
|
| 227 |
+
# Transformer encoder
|
| 228 |
+
if len(drop_path_rates):
|
| 229 |
+
assert len(drop_path_rates) == depth, "Must provide as many drop path rates as there are transformer layers"
|
| 230 |
+
self.transformer_encoder = nn.Sequential(
|
| 231 |
+
*[
|
| 232 |
+
TransformerLayer(
|
| 233 |
+
dim=embed_dim,
|
| 234 |
+
num_heads=num_heads,
|
| 235 |
+
mlp_ratio=mlp_ratio,
|
| 236 |
+
qkv_bias=qkv_bias,
|
| 237 |
+
drop=drop_rate,
|
| 238 |
+
attn_drop=attn_drop_rate,
|
| 239 |
+
drop_path=drop_path_rates[i],
|
| 240 |
+
norm_layer=norm_layer,
|
| 241 |
+
act_layer=act_layer,
|
| 242 |
+
)
|
| 243 |
+
for i in range(depth)
|
| 244 |
+
]
|
| 245 |
+
)
|
| 246 |
+
|
| 247 |
+
def forward(self, x):
|
| 248 |
+
"""
|
| 249 |
+
expects x as (B, C, D, H, W)
|
| 250 |
+
"""
|
| 251 |
+
x = self.pool(x)
|
| 252 |
+
x = x.permute(0, 2, 3, 4, 1) # (B, H', W', C), switch to channels last for transformer
|
| 253 |
+
|
| 254 |
+
x = blockify(x, self.block_size) # (B, T, N, C')
|
| 255 |
+
x = x + self.pos_embed
|
| 256 |
+
|
| 257 |
+
x = self.transformer_encoder(x) # (B, ,T, N, C')
|
| 258 |
+
|
| 259 |
+
x = deblockify(x, self.block_size) # (B, D', H', W', C') [2, 24, 24, 24, 128]
|
| 260 |
+
# Channel-first for block aggregation, and generally to replicate convnet feature map at each stage
|
| 261 |
+
return x.permute(0, 4, 1, 2, 3) # (B, C, D', H', W')
|
| 262 |
+
|
| 263 |
+
|
| 264 |
+
class NestTransformer3D(nn.Module):
|
| 265 |
+
"""Nested Transformer (NesT)
|
| 266 |
+
A PyTorch impl of : `Aggregating Nested Transformers`
|
| 267 |
+
- https://arxiv.org/abs/2105.12723
|
| 268 |
+
"""
|
| 269 |
+
|
| 270 |
+
def __init__(
|
| 271 |
+
self,
|
| 272 |
+
img_size=96,
|
| 273 |
+
in_chans=1,
|
| 274 |
+
patch_size=2,
|
| 275 |
+
num_levels=3,
|
| 276 |
+
embed_dims=(128, 256, 512),
|
| 277 |
+
num_heads=(4, 8, 16),
|
| 278 |
+
depths=(2, 2, 20),
|
| 279 |
+
num_classes=1000,
|
| 280 |
+
mlp_ratio=4.0,
|
| 281 |
+
qkv_bias=True,
|
| 282 |
+
drop_rate=0.0,
|
| 283 |
+
attn_drop_rate=0.0,
|
| 284 |
+
drop_path_rate=0.5,
|
| 285 |
+
norm_layer=None,
|
| 286 |
+
act_layer=None,
|
| 287 |
+
pad_type="",
|
| 288 |
+
weight_init="",
|
| 289 |
+
global_pool="avg",
|
| 290 |
+
):
|
| 291 |
+
"""
|
| 292 |
+
Args:
|
| 293 |
+
img_size (int, tuple): input image size
|
| 294 |
+
in_chans (int): number of input channels
|
| 295 |
+
patch_size (int): patch size
|
| 296 |
+
num_levels (int): number of block hierarchies (T_d in the paper)
|
| 297 |
+
embed_dims (int, tuple): embedding dimensions of each level
|
| 298 |
+
num_heads (int, tuple): number of attention heads for each level
|
| 299 |
+
depths (int, tuple): number of transformer layers for each level
|
| 300 |
+
num_classes (int): number of classes for classification head
|
| 301 |
+
mlp_ratio (int): ratio of mlp hidden dim to embedding dim for MLP of transformer layers
|
| 302 |
+
qkv_bias (bool): enable bias for qkv if True
|
| 303 |
+
drop_rate (float): dropout rate for MLP of transformer layers, MSA final projection layer, and classifier
|
| 304 |
+
attn_drop_rate (float): attention dropout rate
|
| 305 |
+
drop_path_rate (float): stochastic depth rate
|
| 306 |
+
norm_layer: (nn.Module): normalization layer for transformer layers
|
| 307 |
+
act_layer: (nn.Module): activation layer in MLP of transformer layers
|
| 308 |
+
pad_type: str: Type of padding to use '' for PyTorch symmetric, 'same' for TF SAME
|
| 309 |
+
weight_init: (str): weight init scheme
|
| 310 |
+
global_pool: (str): type of pooling operation to apply to final feature map
|
| 311 |
+
Notes:
|
| 312 |
+
- Default values follow NesT-B from the original Jax code.
|
| 313 |
+
- `embed_dims`, `num_heads`, `depths` should be ints or tuples with length `num_levels`.
|
| 314 |
+
- For those following the paper, Table A1 may have errors!
|
| 315 |
+
- https://github.com/google-research/nested-transformer/issues/2
|
| 316 |
+
"""
|
| 317 |
+
super().__init__()
|
| 318 |
+
|
| 319 |
+
for param_name in ["embed_dims", "num_heads", "depths"]:
|
| 320 |
+
param_value = locals()[param_name]
|
| 321 |
+
if isinstance(param_value, collections.abc.Sequence):
|
| 322 |
+
assert len(param_value) == num_levels, f"Require `len({param_name}) == num_levels`"
|
| 323 |
+
|
| 324 |
+
embed_dims = to_ntuple(num_levels)(embed_dims)
|
| 325 |
+
num_heads = to_ntuple(num_levels)(num_heads)
|
| 326 |
+
depths = to_ntuple(num_levels)(depths)
|
| 327 |
+
self.num_classes = num_classes
|
| 328 |
+
self.num_features = embed_dims[-1]
|
| 329 |
+
self.feature_info = []
|
| 330 |
+
norm_layer = norm_layer or partial(nn.LayerNorm, eps=1e-6)
|
| 331 |
+
act_layer = act_layer or nn.GELU
|
| 332 |
+
self.drop_rate = drop_rate
|
| 333 |
+
self.num_levels = num_levels
|
| 334 |
+
if isinstance(img_size, collections.abc.Sequence):
|
| 335 |
+
assert img_size[0] == img_size[1], "Model only handles square inputs"
|
| 336 |
+
img_size = img_size[0]
|
| 337 |
+
assert img_size % patch_size == 0, "`patch_size` must divide `img_size` evenly"
|
| 338 |
+
self.patch_size = patch_size
|
| 339 |
+
|
| 340 |
+
# Number of blocks at each level
|
| 341 |
+
self.num_blocks = (8 ** torch.arange(num_levels)).flip(0).tolist()
|
| 342 |
+
assert (img_size // patch_size) % round(
|
| 343 |
+
math.pow(self.num_blocks[0], 1 / 3)
|
| 344 |
+
) == 0, "First level blocks don't fit evenly. Check `img_size`, `patch_size`, and `num_levels`"
|
| 345 |
+
|
| 346 |
+
# Block edge size in units of patches
|
| 347 |
+
# Hint: (img_size // patch_size) gives number of patches along edge of image. sqrt(self.num_blocks[0]) is the
|
| 348 |
+
# number of blocks along edge of image
|
| 349 |
+
self.block_size = int((img_size // patch_size) // round(math.pow(self.num_blocks[0], 1 / 3)))
|
| 350 |
+
|
| 351 |
+
# Patch embedding
|
| 352 |
+
self.patch_embed = PatchEmbed3D(
|
| 353 |
+
img_size=[img_size, img_size, img_size],
|
| 354 |
+
patch_size=[patch_size, patch_size, patch_size],
|
| 355 |
+
in_chans=in_chans,
|
| 356 |
+
embed_dim=embed_dims[0],
|
| 357 |
+
)
|
| 358 |
+
self.num_patches = self.patch_embed.num_patches
|
| 359 |
+
self.seq_length = self.num_patches // self.num_blocks[0]
|
| 360 |
+
# Build up each hierarchical level
|
| 361 |
+
levels = []
|
| 362 |
+
|
| 363 |
+
dp_rates = [x.tolist() for x in torch.linspace(0, drop_path_rate, sum(depths)).split(depths)]
|
| 364 |
+
prev_dim = None
|
| 365 |
+
curr_stride = 4
|
| 366 |
+
for i in range(len(self.num_blocks)):
|
| 367 |
+
dim = embed_dims[i]
|
| 368 |
+
levels.append(
|
| 369 |
+
NestLevel(
|
| 370 |
+
self.num_blocks[i],
|
| 371 |
+
self.block_size,
|
| 372 |
+
self.seq_length,
|
| 373 |
+
num_heads[i],
|
| 374 |
+
depths[i],
|
| 375 |
+
dim,
|
| 376 |
+
prev_dim,
|
| 377 |
+
mlp_ratio,
|
| 378 |
+
qkv_bias,
|
| 379 |
+
drop_rate,
|
| 380 |
+
attn_drop_rate,
|
| 381 |
+
dp_rates[i],
|
| 382 |
+
norm_layer,
|
| 383 |
+
act_layer,
|
| 384 |
+
pad_type=pad_type,
|
| 385 |
+
)
|
| 386 |
+
)
|
| 387 |
+
self.feature_info += [dict(num_chs=dim, reduction=curr_stride, module=f"levels.{i}")]
|
| 388 |
+
prev_dim = dim
|
| 389 |
+
curr_stride *= 2
|
| 390 |
+
|
| 391 |
+
self.levels = nn.ModuleList([levels[i] for i in range(num_levels)])
|
| 392 |
+
|
| 393 |
+
# Final normalization layer
|
| 394 |
+
self.norm = norm_layer(embed_dims[-1])
|
| 395 |
+
|
| 396 |
+
self.init_weights(weight_init)
|
| 397 |
+
|
| 398 |
+
def init_weights(self, mode=""):
|
| 399 |
+
assert mode in ("nlhb", "")
|
| 400 |
+
head_bias = -math.log(self.num_classes) if "nlhb" in mode else 0.0
|
| 401 |
+
for level in self.levels:
|
| 402 |
+
trunc_normal_(level.pos_embed, std=0.02, a=-2, b=2)
|
| 403 |
+
named_apply(partial(_init_nest_weights, head_bias=head_bias), self)
|
| 404 |
+
|
| 405 |
+
@torch.jit.ignore
|
| 406 |
+
def no_weight_decay(self):
|
| 407 |
+
return {f"level.{i}.pos_embed" for i in range(len(self.levels))}
|
| 408 |
+
|
| 409 |
+
def get_classifier(self):
|
| 410 |
+
return self.head
|
| 411 |
+
|
| 412 |
+
def forward_features(self, x):
|
| 413 |
+
"""x shape (B, C, D, H, W)"""
|
| 414 |
+
x = self.patch_embed(x)
|
| 415 |
+
|
| 416 |
+
hidden_states_out = [x]
|
| 417 |
+
|
| 418 |
+
for _, level in enumerate(self.levels):
|
| 419 |
+
x = level(x)
|
| 420 |
+
hidden_states_out.append(x)
|
| 421 |
+
# Layer norm done over channel dim only (to NDHWC and back)
|
| 422 |
+
x = self.norm(x.permute(0, 2, 3, 4, 1)).permute(0, 4, 1, 2, 3)
|
| 423 |
+
return x, hidden_states_out
|
| 424 |
+
|
| 425 |
+
def forward(self, x):
|
| 426 |
+
"""x shape (B, C, D, H, W)"""
|
| 427 |
+
x = self.forward_features(x)
|
| 428 |
+
|
| 429 |
+
if self.drop_rate > 0.0:
|
| 430 |
+
x = F.dropout(x, p=self.drop_rate, training=self.training)
|
| 431 |
+
return x
|
| 432 |
+
|
| 433 |
+
|
| 434 |
+
def named_apply(fn: Callable, module: nn.Module, name="", depth_first=True, include_root=False) -> nn.Module:
|
| 435 |
+
if not depth_first and include_root:
|
| 436 |
+
fn(module=module, name=name)
|
| 437 |
+
for child_name, child_module in module.named_children():
|
| 438 |
+
child_name = ".".join((name, child_name)) if name else child_name
|
| 439 |
+
named_apply(fn=fn, module=child_module, name=child_name, depth_first=depth_first, include_root=True)
|
| 440 |
+
if depth_first and include_root:
|
| 441 |
+
fn(module=module, name=name)
|
| 442 |
+
return module
|
| 443 |
+
|
| 444 |
+
|
| 445 |
+
def _init_nest_weights(module: nn.Module, name: str = "", head_bias: float = 0.0):
|
| 446 |
+
"""NesT weight initialization
|
| 447 |
+
Can replicate Jax implementation. Otherwise follows vision_transformer.py
|
| 448 |
+
"""
|
| 449 |
+
if isinstance(module, nn.Linear):
|
| 450 |
+
if name.startswith("head"):
|
| 451 |
+
trunc_normal_(module.weight, std=0.02, a=-2, b=2)
|
| 452 |
+
nn.init.constant_(module.bias, head_bias)
|
| 453 |
+
else:
|
| 454 |
+
trunc_normal_(module.weight, std=0.02, a=-2, b=2)
|
| 455 |
+
if module.bias is not None:
|
| 456 |
+
nn.init.zeros_(module.bias)
|
| 457 |
+
elif isinstance(module, nn.Conv2d):
|
| 458 |
+
trunc_normal_(module.weight, std=0.02, a=-2, b=2)
|
| 459 |
+
if module.bias is not None:
|
| 460 |
+
nn.init.zeros_(module.bias)
|
| 461 |
+
elif isinstance(module, (nn.LayerNorm, nn.GroupNorm, nn.BatchNorm2d)):
|
| 462 |
+
nn.init.zeros_(module.bias)
|
| 463 |
+
nn.init.ones_(module.weight)
|
| 464 |
+
|
| 465 |
+
|
| 466 |
+
def resize_pos_embed(posemb, posemb_new):
|
| 467 |
+
"""
|
| 468 |
+
Rescale the grid of position embeddings when loading from state_dict
|
| 469 |
+
Expected shape of position embeddings is (1, T, N, C), and considers only square images
|
| 470 |
+
"""
|
| 471 |
+
_logger.info("Resized position embedding: %s to %s", posemb.shape, posemb_new.shape)
|
| 472 |
+
seq_length_old = posemb.shape[2]
|
| 473 |
+
num_blocks_new, seq_length_new = posemb_new.shape[1:3]
|
| 474 |
+
size_new = int(math.sqrt(num_blocks_new * seq_length_new))
|
| 475 |
+
# First change to (1, C, H, W)
|
| 476 |
+
posemb = deblockify(posemb, int(math.sqrt(seq_length_old))).permute(0, 3, 1, 2)
|
| 477 |
+
posemb = F.interpolate(posemb, size=[size_new, size_new], mode="bicubic", align_corners=False)
|
| 478 |
+
# Now change to new (1, T, N, C)
|
| 479 |
+
posemb = blockify(posemb.permute(0, 2, 3, 1), int(math.sqrt(seq_length_new)))
|
| 480 |
+
return posemb
|
| 481 |
+
|
| 482 |
+
|
| 483 |
+
def checkpoint_filter_fn(state_dict, model):
|
| 484 |
+
"""resize positional embeddings of pretrained weights"""
|
| 485 |
+
pos_embed_keys = [k for k in state_dict.keys() if k.startswith("pos_embed_")]
|
| 486 |
+
for k in pos_embed_keys:
|
| 487 |
+
if state_dict[k].shape != getattr(model, k).shape:
|
| 488 |
+
state_dict[k] = resize_pos_embed(state_dict[k], getattr(model, k))
|
| 489 |
+
return state_dict
|
scripts/networks/patchEmbed3D.py
ADDED
|
@@ -0,0 +1,190 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
|
| 3 |
+
# Copyright 2020 - 2021 MONAI Consortium
|
| 4 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 5 |
+
# you may not use this file except in compliance with the License.
|
| 6 |
+
# You may obtain a copy of the License at
|
| 7 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 8 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 9 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 10 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 11 |
+
# See the License for the specific language governing permissions and
|
| 12 |
+
# limitations under the License.
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
import math
|
| 16 |
+
from typing import Sequence, Tuple, Union
|
| 17 |
+
|
| 18 |
+
import torch
|
| 19 |
+
import torch.nn as nn
|
| 20 |
+
import torch.nn.functional as F
|
| 21 |
+
from monai.utils import optional_import
|
| 22 |
+
|
| 23 |
+
Rearrange, _ = optional_import("einops.layers.torch", name="Rearrange")
|
| 24 |
+
|
| 25 |
+
|
| 26 |
+
class PatchEmbeddingBlock(nn.Module):
|
| 27 |
+
"""
|
| 28 |
+
A patch embedding block, based on: "Dosovitskiy et al.,
|
| 29 |
+
An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale <https://arxiv.org/abs/2010.11929>"
|
| 30 |
+
"""
|
| 31 |
+
|
| 32 |
+
def __init__(
|
| 33 |
+
self,
|
| 34 |
+
in_channels: int,
|
| 35 |
+
img_size: Tuple[int, int, int],
|
| 36 |
+
patch_size: Tuple[int, int, int],
|
| 37 |
+
hidden_size: int,
|
| 38 |
+
num_heads: int,
|
| 39 |
+
pos_embed: str,
|
| 40 |
+
dropout_rate: float = 0.0,
|
| 41 |
+
) -> None:
|
| 42 |
+
"""
|
| 43 |
+
Args:
|
| 44 |
+
in_channels: dimension of input channels.
|
| 45 |
+
img_size: dimension of input image.
|
| 46 |
+
patch_size: dimension of patch size.
|
| 47 |
+
hidden_size: dimension of hidden layer.
|
| 48 |
+
num_heads: number of attention heads.
|
| 49 |
+
pos_embed: position embedding layer type.
|
| 50 |
+
dropout_rate: faction of the input units to drop.
|
| 51 |
+
|
| 52 |
+
"""
|
| 53 |
+
|
| 54 |
+
super().__init__()
|
| 55 |
+
|
| 56 |
+
if not (0 <= dropout_rate <= 1):
|
| 57 |
+
raise AssertionError("dropout_rate should be between 0 and 1.")
|
| 58 |
+
|
| 59 |
+
if hidden_size % num_heads != 0:
|
| 60 |
+
raise AssertionError("hidden size should be divisible by num_heads.")
|
| 61 |
+
|
| 62 |
+
for m, p in zip(img_size, patch_size):
|
| 63 |
+
if m < p:
|
| 64 |
+
raise AssertionError("patch_size should be smaller than img_size.")
|
| 65 |
+
|
| 66 |
+
if pos_embed not in ["conv", "perceptron"]:
|
| 67 |
+
raise KeyError(f"Position embedding layer of type {pos_embed} is not supported.")
|
| 68 |
+
|
| 69 |
+
if pos_embed == "perceptron":
|
| 70 |
+
if img_size[0] % patch_size[0] != 0:
|
| 71 |
+
raise AssertionError("img_size should be divisible by patch_size for perceptron patch embedding.")
|
| 72 |
+
|
| 73 |
+
self.n_patches = (
|
| 74 |
+
(img_size[0] // patch_size[0]) * (img_size[1] // patch_size[1]) * (img_size[2] // patch_size[2])
|
| 75 |
+
)
|
| 76 |
+
self.patch_dim = in_channels * patch_size[0] * patch_size[1] * patch_size[2]
|
| 77 |
+
|
| 78 |
+
self.pos_embed = pos_embed
|
| 79 |
+
self.patch_embeddings: Union[nn.Conv3d, nn.Sequential]
|
| 80 |
+
if self.pos_embed == "conv":
|
| 81 |
+
self.patch_embeddings = nn.Conv3d(
|
| 82 |
+
in_channels=in_channels, out_channels=hidden_size, kernel_size=patch_size, stride=patch_size
|
| 83 |
+
)
|
| 84 |
+
elif self.pos_embed == "perceptron":
|
| 85 |
+
self.patch_embeddings = nn.Sequential(
|
| 86 |
+
Rearrange(
|
| 87 |
+
"b c (h p1) (w p2) (d p3)-> b (h w d) (p1 p2 p3 c)",
|
| 88 |
+
p1=patch_size[0],
|
| 89 |
+
p2=patch_size[1],
|
| 90 |
+
p3=patch_size[2],
|
| 91 |
+
),
|
| 92 |
+
nn.Linear(self.patch_dim, hidden_size),
|
| 93 |
+
)
|
| 94 |
+
self.position_embeddings = nn.Parameter(torch.zeros(1, self.n_patches, hidden_size))
|
| 95 |
+
self.cls_token = nn.Parameter(torch.zeros(1, 1, hidden_size))
|
| 96 |
+
self.dropout = nn.Dropout(dropout_rate)
|
| 97 |
+
self.trunc_normal_(self.position_embeddings, mean=0.0, std=0.02, a=-2.0, b=2.0)
|
| 98 |
+
self.apply(self._init_weights)
|
| 99 |
+
|
| 100 |
+
def _init_weights(self, m):
|
| 101 |
+
if isinstance(m, nn.Linear):
|
| 102 |
+
self.trunc_normal_(m.weight, mean=0.0, std=0.02, a=-2.0, b=2.0)
|
| 103 |
+
if isinstance(m, nn.Linear) and m.bias is not None:
|
| 104 |
+
nn.init.constant_(m.bias, 0)
|
| 105 |
+
elif isinstance(m, nn.LayerNorm):
|
| 106 |
+
nn.init.constant_(m.bias, 0)
|
| 107 |
+
nn.init.constant_(m.weight, 1.0)
|
| 108 |
+
|
| 109 |
+
def trunc_normal_(self, tensor, mean, std, a, b):
|
| 110 |
+
# From PyTorch official master until it's in a few official releases - RW
|
| 111 |
+
# Method based on https://people.sc.fsu.edu/~jburkardt/presentations/truncated_normal.pdf
|
| 112 |
+
def norm_cdf(x):
|
| 113 |
+
return (1.0 + math.erf(x / math.sqrt(2.0))) / 2.0
|
| 114 |
+
|
| 115 |
+
with torch.no_grad():
|
| 116 |
+
l = norm_cdf((a - mean) / std)
|
| 117 |
+
u = norm_cdf((b - mean) / std)
|
| 118 |
+
tensor.uniform_(2 * l - 1, 2 * u - 1)
|
| 119 |
+
tensor.erfinv_()
|
| 120 |
+
tensor.mul_(std * math.sqrt(2.0))
|
| 121 |
+
tensor.add_(mean)
|
| 122 |
+
tensor.clamp_(min=a, max=b)
|
| 123 |
+
return tensor
|
| 124 |
+
|
| 125 |
+
def forward(self, x):
|
| 126 |
+
if self.pos_embed == "conv":
|
| 127 |
+
x = self.patch_embeddings(x)
|
| 128 |
+
x = x.flatten(2)
|
| 129 |
+
x = x.transpose(-1, -2)
|
| 130 |
+
elif self.pos_embed == "perceptron":
|
| 131 |
+
x = self.patch_embeddings(x)
|
| 132 |
+
embeddings = x + self.position_embeddings
|
| 133 |
+
embeddings = self.dropout(embeddings)
|
| 134 |
+
return embeddings
|
| 135 |
+
|
| 136 |
+
|
| 137 |
+
class PatchEmbed3D(nn.Module):
|
| 138 |
+
"""Video to Patch Embedding.
|
| 139 |
+
|
| 140 |
+
Args:
|
| 141 |
+
patch_size (int): Patch token size. Default: (2,4,4).
|
| 142 |
+
in_chans (int): Number of input video channels. Default: 3.
|
| 143 |
+
embed_dim (int): Number of linear projection output channels. Default: 96.
|
| 144 |
+
norm_layer (nn.Module, optional): Normalization layer. Default: None
|
| 145 |
+
"""
|
| 146 |
+
|
| 147 |
+
def __init__(
|
| 148 |
+
self,
|
| 149 |
+
img_size: Sequence[int] = (96, 96, 96),
|
| 150 |
+
patch_size=(4, 4, 4),
|
| 151 |
+
in_chans: int = 1,
|
| 152 |
+
embed_dim: int = 96,
|
| 153 |
+
norm_layer=None,
|
| 154 |
+
):
|
| 155 |
+
super().__init__()
|
| 156 |
+
self.patch_size = patch_size
|
| 157 |
+
|
| 158 |
+
self.in_chans = in_chans
|
| 159 |
+
self.embed_dim = embed_dim
|
| 160 |
+
|
| 161 |
+
self.grid_size = (img_size[0] // patch_size[0], img_size[1] // patch_size[1], img_size[2] // patch_size[2])
|
| 162 |
+
self.num_patches = self.grid_size[0] * self.grid_size[1] * self.grid_size[2]
|
| 163 |
+
|
| 164 |
+
self.proj = nn.Conv3d(in_chans, embed_dim, kernel_size=patch_size, stride=patch_size)
|
| 165 |
+
|
| 166 |
+
if norm_layer is not None:
|
| 167 |
+
self.norm = norm_layer(embed_dim)
|
| 168 |
+
else:
|
| 169 |
+
self.norm = None
|
| 170 |
+
|
| 171 |
+
def forward(self, x):
|
| 172 |
+
"""Forward function."""
|
| 173 |
+
# padding
|
| 174 |
+
_, _, d, h, w = x.size()
|
| 175 |
+
if w % self.patch_size[2] != 0:
|
| 176 |
+
x = F.pad(x, (0, self.patch_size[2] - w % self.patch_size[2]))
|
| 177 |
+
if h % self.patch_size[1] != 0:
|
| 178 |
+
x = F.pad(x, (0, 0, 0, self.patch_size[1] - h % self.patch_size[1]))
|
| 179 |
+
if d % self.patch_size[0] != 0:
|
| 180 |
+
x = F.pad(x, (0, 0, 0, 0, 0, self.patch_size[0] - d % self.patch_size[0]))
|
| 181 |
+
|
| 182 |
+
x = self.proj(x) # B C D Wh Ww
|
| 183 |
+
if self.norm is not None:
|
| 184 |
+
d, wh, ww = x.size(2), x.size(3), x.size(4)
|
| 185 |
+
x = x.flatten(2).transpose(1, 2)
|
| 186 |
+
x = self.norm(x)
|
| 187 |
+
x = x.transpose(1, 2).view(-1, self.embed_dim, d, wh, ww)
|
| 188 |
+
# pdb.set_trace()
|
| 189 |
+
|
| 190 |
+
return x
|
scripts/networks/unest_base_patch_4.py
ADDED
|
@@ -0,0 +1,249 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
# limitations under the License.
|
| 3 |
+
"""
|
| 4 |
+
The 3D NEST transformer based segmentation model
|
| 5 |
+
|
| 6 |
+
MASI Lab, Vanderbilty University
|
| 7 |
+
|
| 8 |
+
|
| 9 |
+
Authors: Xin Yu, Yinchi Zhou, Yucheng Tang, Bennett Landman
|
| 10 |
+
|
| 11 |
+
|
| 12 |
+
The NEST code is partly from
|
| 13 |
+
|
| 14 |
+
Nested Hierarchical Transformer: Towards Accurate, Data-Efficient and
|
| 15 |
+
Interpretable Visual Understanding
|
| 16 |
+
https://arxiv.org/pdf/2105.12723.pdf
|
| 17 |
+
|
| 18 |
+
"""
|
| 19 |
+
from typing import Sequence, Tuple, Union
|
| 20 |
+
|
| 21 |
+
import torch
|
| 22 |
+
import torch.nn as nn
|
| 23 |
+
from monai.networks.blocks import Convolution
|
| 24 |
+
from monai.networks.blocks.dynunet_block import UnetOutBlock
|
| 25 |
+
from scripts.networks.nest_transformer_3D import NestTransformer3D
|
| 26 |
+
from scripts.networks.unest_block import UNesTBlock, UNesTConvBlock, UNestUpBlock
|
| 27 |
+
|
| 28 |
+
|
| 29 |
+
class UNesT(nn.Module):
|
| 30 |
+
"""
|
| 31 |
+
UNesT model implementation
|
| 32 |
+
"""
|
| 33 |
+
|
| 34 |
+
def __init__(
|
| 35 |
+
self,
|
| 36 |
+
in_channels: int,
|
| 37 |
+
out_channels: int,
|
| 38 |
+
img_size: Sequence[int] = (96, 96, 96),
|
| 39 |
+
feature_size: int = 16,
|
| 40 |
+
patch_size: int = 2,
|
| 41 |
+
depths: Sequence[int] = (2, 2, 2, 2),
|
| 42 |
+
num_heads: Sequence[int] = (3, 6, 12, 24),
|
| 43 |
+
embed_dim: Sequence[int] = (128, 256, 512),
|
| 44 |
+
window_size: Sequence[int] = (7, 7, 7),
|
| 45 |
+
norm_name: Union[Tuple, str] = "instance",
|
| 46 |
+
conv_block: bool = False,
|
| 47 |
+
res_block: bool = True,
|
| 48 |
+
dropout_rate: float = 0.0,
|
| 49 |
+
) -> None:
|
| 50 |
+
"""
|
| 51 |
+
Args:
|
| 52 |
+
in_channels: dimension of input channels.
|
| 53 |
+
out_channels: dimension of output channels.
|
| 54 |
+
img_size: dimension of input image.
|
| 55 |
+
feature_size: dimension of network feature size.
|
| 56 |
+
hidden_size: dimension of hidden layer.
|
| 57 |
+
mlp_dim: dimension of feedforward layer.
|
| 58 |
+
num_heads: number of attention heads.
|
| 59 |
+
pos_embed: position embedding layer type.
|
| 60 |
+
norm_name: feature normalization type and arguments.
|
| 61 |
+
conv_block: bool argument to determine if convolutional block is used.
|
| 62 |
+
res_block: bool argument to determine if residual block is used.
|
| 63 |
+
dropout_rate: faction of the input units to drop.
|
| 64 |
+
|
| 65 |
+
Examples:
|
| 66 |
+
|
| 67 |
+
# for single channel input 4-channel output with patch size of (96,96,96), feature size of 32 and batch norm
|
| 68 |
+
>>> net = UNETR(in_channels=1, out_channels=4, img_size=(96,96,96), feature_size=32, norm_name='batch')
|
| 69 |
+
|
| 70 |
+
# for 4-channel input 3-channel output with patch size of (128,128,128), conv position embedding and instance norm
|
| 71 |
+
>>> net = UNETR(in_channels=4, out_channels=3, img_size=(128,128,128), pos_embed='conv', norm_name='instance')
|
| 72 |
+
|
| 73 |
+
"""
|
| 74 |
+
|
| 75 |
+
super().__init__()
|
| 76 |
+
if not (0 <= dropout_rate <= 1):
|
| 77 |
+
raise AssertionError("dropout_rate should be between 0 and 1.")
|
| 78 |
+
self.embed_dim = embed_dim
|
| 79 |
+
self.nestViT = NestTransformer3D(
|
| 80 |
+
img_size=96,
|
| 81 |
+
in_chans=1,
|
| 82 |
+
patch_size=patch_size,
|
| 83 |
+
num_levels=3,
|
| 84 |
+
embed_dims=embed_dim,
|
| 85 |
+
num_heads=num_heads,
|
| 86 |
+
depths=depths,
|
| 87 |
+
num_classes=1000,
|
| 88 |
+
mlp_ratio=4.0,
|
| 89 |
+
qkv_bias=True,
|
| 90 |
+
drop_rate=0.0,
|
| 91 |
+
attn_drop_rate=0.0,
|
| 92 |
+
drop_path_rate=0.5,
|
| 93 |
+
norm_layer=None,
|
| 94 |
+
act_layer=None,
|
| 95 |
+
pad_type="",
|
| 96 |
+
weight_init="",
|
| 97 |
+
global_pool="avg",
|
| 98 |
+
)
|
| 99 |
+
self.encoder1 = UNesTConvBlock(
|
| 100 |
+
spatial_dims=3,
|
| 101 |
+
in_channels=1,
|
| 102 |
+
out_channels=feature_size * 2,
|
| 103 |
+
kernel_size=3,
|
| 104 |
+
stride=1,
|
| 105 |
+
norm_name=norm_name,
|
| 106 |
+
res_block=res_block,
|
| 107 |
+
)
|
| 108 |
+
self.encoder2 = UNestUpBlock(
|
| 109 |
+
spatial_dims=3,
|
| 110 |
+
in_channels=self.embed_dim[0],
|
| 111 |
+
out_channels=feature_size * 4,
|
| 112 |
+
num_layer=1,
|
| 113 |
+
kernel_size=3,
|
| 114 |
+
stride=1,
|
| 115 |
+
upsample_kernel_size=2,
|
| 116 |
+
norm_name=norm_name,
|
| 117 |
+
conv_block=False,
|
| 118 |
+
res_block=False,
|
| 119 |
+
)
|
| 120 |
+
|
| 121 |
+
self.encoder3 = UNesTConvBlock(
|
| 122 |
+
spatial_dims=3,
|
| 123 |
+
in_channels=self.embed_dim[0],
|
| 124 |
+
out_channels=8 * feature_size,
|
| 125 |
+
kernel_size=3,
|
| 126 |
+
stride=1,
|
| 127 |
+
norm_name=norm_name,
|
| 128 |
+
res_block=res_block,
|
| 129 |
+
)
|
| 130 |
+
self.encoder4 = UNesTConvBlock(
|
| 131 |
+
spatial_dims=3,
|
| 132 |
+
in_channels=self.embed_dim[1],
|
| 133 |
+
out_channels=16 * feature_size,
|
| 134 |
+
kernel_size=3,
|
| 135 |
+
stride=1,
|
| 136 |
+
norm_name=norm_name,
|
| 137 |
+
res_block=res_block,
|
| 138 |
+
)
|
| 139 |
+
self.decoder5 = UNesTBlock(
|
| 140 |
+
spatial_dims=3,
|
| 141 |
+
in_channels=2 * self.embed_dim[2],
|
| 142 |
+
out_channels=feature_size * 32,
|
| 143 |
+
stride=1,
|
| 144 |
+
kernel_size=3,
|
| 145 |
+
upsample_kernel_size=2,
|
| 146 |
+
norm_name=norm_name,
|
| 147 |
+
res_block=res_block,
|
| 148 |
+
)
|
| 149 |
+
self.decoder4 = UNesTBlock(
|
| 150 |
+
spatial_dims=3,
|
| 151 |
+
in_channels=self.embed_dim[2],
|
| 152 |
+
out_channels=feature_size * 16,
|
| 153 |
+
stride=1,
|
| 154 |
+
kernel_size=3,
|
| 155 |
+
upsample_kernel_size=2,
|
| 156 |
+
norm_name=norm_name,
|
| 157 |
+
res_block=res_block,
|
| 158 |
+
)
|
| 159 |
+
self.decoder3 = UNesTBlock(
|
| 160 |
+
spatial_dims=3,
|
| 161 |
+
in_channels=feature_size * 16,
|
| 162 |
+
out_channels=feature_size * 8,
|
| 163 |
+
stride=1,
|
| 164 |
+
kernel_size=3,
|
| 165 |
+
upsample_kernel_size=2,
|
| 166 |
+
norm_name=norm_name,
|
| 167 |
+
res_block=res_block,
|
| 168 |
+
)
|
| 169 |
+
self.decoder2 = UNesTBlock(
|
| 170 |
+
spatial_dims=3,
|
| 171 |
+
in_channels=feature_size * 8,
|
| 172 |
+
out_channels=feature_size * 4,
|
| 173 |
+
stride=1,
|
| 174 |
+
kernel_size=3,
|
| 175 |
+
upsample_kernel_size=2,
|
| 176 |
+
norm_name=norm_name,
|
| 177 |
+
res_block=res_block,
|
| 178 |
+
)
|
| 179 |
+
self.decoder1 = UNesTBlock(
|
| 180 |
+
spatial_dims=3,
|
| 181 |
+
in_channels=feature_size * 4,
|
| 182 |
+
out_channels=feature_size * 2,
|
| 183 |
+
stride=1,
|
| 184 |
+
kernel_size=3,
|
| 185 |
+
upsample_kernel_size=2,
|
| 186 |
+
norm_name=norm_name,
|
| 187 |
+
res_block=res_block,
|
| 188 |
+
)
|
| 189 |
+
self.encoder10 = Convolution(
|
| 190 |
+
dimensions=3,
|
| 191 |
+
in_channels=32 * feature_size,
|
| 192 |
+
out_channels=64 * feature_size,
|
| 193 |
+
strides=2,
|
| 194 |
+
adn_ordering="ADN",
|
| 195 |
+
dropout=0.0,
|
| 196 |
+
)
|
| 197 |
+
self.out = UnetOutBlock(spatial_dims=3, in_channels=feature_size * 2, out_channels=out_channels) # type: ignore
|
| 198 |
+
|
| 199 |
+
def proj_feat(self, x, hidden_size, feat_size):
|
| 200 |
+
x = x.view(x.size(0), feat_size[0], feat_size[1], feat_size[2], hidden_size)
|
| 201 |
+
x = x.permute(0, 4, 1, 2, 3).contiguous()
|
| 202 |
+
return x
|
| 203 |
+
|
| 204 |
+
def load_from(self, weights):
|
| 205 |
+
with torch.no_grad():
|
| 206 |
+
# copy weights from patch embedding
|
| 207 |
+
for i in weights["state_dict"]:
|
| 208 |
+
print(i)
|
| 209 |
+
self.vit.patch_embedding.position_embeddings.copy_(
|
| 210 |
+
weights["state_dict"]["module.transformer.patch_embedding.position_embeddings_3d"]
|
| 211 |
+
)
|
| 212 |
+
self.vit.patch_embedding.cls_token.copy_(
|
| 213 |
+
weights["state_dict"]["module.transformer.patch_embedding.cls_token"]
|
| 214 |
+
)
|
| 215 |
+
self.vit.patch_embedding.patch_embeddings[1].weight.copy_(
|
| 216 |
+
weights["state_dict"]["module.transformer.patch_embedding.patch_embeddings_3d.1.weight"]
|
| 217 |
+
)
|
| 218 |
+
self.vit.patch_embedding.patch_embeddings[1].bias.copy_(
|
| 219 |
+
weights["state_dict"]["module.transformer.patch_embedding.patch_embeddings_3d.1.bias"]
|
| 220 |
+
)
|
| 221 |
+
|
| 222 |
+
# copy weights from encoding blocks (default: num of blocks: 12)
|
| 223 |
+
for bname, block in self.vit.blocks.named_children():
|
| 224 |
+
print(block)
|
| 225 |
+
block.loadFrom(weights, n_block=bname)
|
| 226 |
+
# last norm layer of transformer
|
| 227 |
+
self.vit.norm.weight.copy_(weights["state_dict"]["module.transformer.norm.weight"])
|
| 228 |
+
self.vit.norm.bias.copy_(weights["state_dict"]["module.transformer.norm.bias"])
|
| 229 |
+
|
| 230 |
+
def forward(self, x_in):
|
| 231 |
+
x, hidden_states_out = self.nestViT(x_in)
|
| 232 |
+
enc0 = self.encoder1(x_in) # 2, 32, 96, 96, 96
|
| 233 |
+
x1 = hidden_states_out[0] # 2, 128, 24, 24, 24 2, 128, 12, 12, 12
|
| 234 |
+
enc1 = self.encoder2(x1) # 2, 64, 48, 48, 48 torch.Size([2, 64, 24, 24, 24])
|
| 235 |
+
x2 = hidden_states_out[1] # 2, 128, 24, 24, 24
|
| 236 |
+
enc2 = self.encoder3(x2) # 2, 128, 24, 24, 24 torch.Size([2, 128, 12, 12, 12])
|
| 237 |
+
x3 = hidden_states_out[2] # 2, 256, 12, 12, 12 torch.Size([2, 256, 6, 6, 6])
|
| 238 |
+
enc3 = self.encoder4(x3) # 2, 256, 12, 12, 12 torch.Size([2, 256, 6, 6, 6])
|
| 239 |
+
x4 = hidden_states_out[3]
|
| 240 |
+
enc4 = x4 # 2, 512, 6, 6, 6 torch.Size([2, 512, 3, 3, 3])
|
| 241 |
+
dec4 = x # 2, 512, 6, 6, 6 torch.Size([2, 512, 3, 3, 3])
|
| 242 |
+
dec4 = self.encoder10(dec4) # 2, 1024, 3, 3, 3 torch.Size([2, 1024, 2, 2, 2])
|
| 243 |
+
dec3 = self.decoder5(dec4, enc4) # 2, 512, 6, 6, 6
|
| 244 |
+
dec2 = self.decoder4(dec3, enc3) # 2, 256, 12, 12, 12
|
| 245 |
+
dec1 = self.decoder3(dec2, enc2) # 2, 128, 24, 24, 24
|
| 246 |
+
dec0 = self.decoder2(dec1, enc1) # 2, 64, 48, 48, 48
|
| 247 |
+
out = self.decoder1(dec0, enc0) # 2, 32, 96, 96, 96
|
| 248 |
+
logits = self.out(out)
|
| 249 |
+
return logits
|
scripts/networks/unest_block.py
ADDED
|
@@ -0,0 +1,245 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
|
| 3 |
+
from typing import Sequence, Tuple, Union
|
| 4 |
+
|
| 5 |
+
import torch
|
| 6 |
+
import torch.nn as nn
|
| 7 |
+
from monai.networks.blocks.dynunet_block import UnetBasicBlock, UnetResBlock, get_conv_layer
|
| 8 |
+
|
| 9 |
+
|
| 10 |
+
class UNesTBlock(nn.Module):
|
| 11 |
+
""" """
|
| 12 |
+
|
| 13 |
+
def __init__(
|
| 14 |
+
self,
|
| 15 |
+
spatial_dims: int,
|
| 16 |
+
in_channels: int,
|
| 17 |
+
out_channels: int, # type: ignore
|
| 18 |
+
kernel_size: Union[Sequence[int], int],
|
| 19 |
+
stride: Union[Sequence[int], int],
|
| 20 |
+
upsample_kernel_size: Union[Sequence[int], int],
|
| 21 |
+
norm_name: Union[Tuple, str],
|
| 22 |
+
res_block: bool = False,
|
| 23 |
+
) -> None:
|
| 24 |
+
"""
|
| 25 |
+
Args:
|
| 26 |
+
spatial_dims: number of spatial dimensions.
|
| 27 |
+
in_channels: number of input channels.
|
| 28 |
+
out_channels: number of output channels.
|
| 29 |
+
kernel_size: convolution kernel size.
|
| 30 |
+
stride: convolution stride.
|
| 31 |
+
upsample_kernel_size: convolution kernel size for transposed convolution layers.
|
| 32 |
+
norm_name: feature normalization type and arguments.
|
| 33 |
+
res_block: bool argument to determine if residual block is used.
|
| 34 |
+
|
| 35 |
+
"""
|
| 36 |
+
|
| 37 |
+
super(UNesTBlock, self).__init__()
|
| 38 |
+
upsample_stride = upsample_kernel_size
|
| 39 |
+
self.transp_conv = get_conv_layer(
|
| 40 |
+
spatial_dims,
|
| 41 |
+
in_channels,
|
| 42 |
+
out_channels,
|
| 43 |
+
kernel_size=upsample_kernel_size,
|
| 44 |
+
stride=upsample_stride,
|
| 45 |
+
conv_only=True,
|
| 46 |
+
is_transposed=True,
|
| 47 |
+
)
|
| 48 |
+
|
| 49 |
+
if res_block:
|
| 50 |
+
self.conv_block = UnetResBlock(
|
| 51 |
+
spatial_dims,
|
| 52 |
+
out_channels + out_channels,
|
| 53 |
+
out_channels,
|
| 54 |
+
kernel_size=kernel_size,
|
| 55 |
+
stride=1,
|
| 56 |
+
norm_name=norm_name,
|
| 57 |
+
)
|
| 58 |
+
else:
|
| 59 |
+
self.conv_block = UnetBasicBlock( # type: ignore
|
| 60 |
+
spatial_dims,
|
| 61 |
+
out_channels + out_channels,
|
| 62 |
+
out_channels,
|
| 63 |
+
kernel_size=kernel_size,
|
| 64 |
+
stride=1,
|
| 65 |
+
norm_name=norm_name,
|
| 66 |
+
)
|
| 67 |
+
|
| 68 |
+
def forward(self, inp, skip):
|
| 69 |
+
# number of channels for skip should equals to out_channels
|
| 70 |
+
out = self.transp_conv(inp)
|
| 71 |
+
# print(out.shape)
|
| 72 |
+
# print(skip.shape)
|
| 73 |
+
out = torch.cat((out, skip), dim=1)
|
| 74 |
+
out = self.conv_block(out)
|
| 75 |
+
return out
|
| 76 |
+
|
| 77 |
+
|
| 78 |
+
class UNestUpBlock(nn.Module):
|
| 79 |
+
""" """
|
| 80 |
+
|
| 81 |
+
def __init__(
|
| 82 |
+
self,
|
| 83 |
+
spatial_dims: int,
|
| 84 |
+
in_channels: int,
|
| 85 |
+
out_channels: int,
|
| 86 |
+
num_layer: int,
|
| 87 |
+
kernel_size: Union[Sequence[int], int],
|
| 88 |
+
stride: Union[Sequence[int], int],
|
| 89 |
+
upsample_kernel_size: Union[Sequence[int], int],
|
| 90 |
+
norm_name: Union[Tuple, str],
|
| 91 |
+
conv_block: bool = False,
|
| 92 |
+
res_block: bool = False,
|
| 93 |
+
) -> None:
|
| 94 |
+
"""
|
| 95 |
+
Args:
|
| 96 |
+
spatial_dims: number of spatial dimensions.
|
| 97 |
+
in_channels: number of input channels.
|
| 98 |
+
out_channels: number of output channels.
|
| 99 |
+
num_layer: number of upsampling blocks.
|
| 100 |
+
kernel_size: convolution kernel size.
|
| 101 |
+
stride: convolution stride.
|
| 102 |
+
upsample_kernel_size: convolution kernel size for transposed convolution layers.
|
| 103 |
+
norm_name: feature normalization type and arguments.
|
| 104 |
+
conv_block: bool argument to determine if convolutional block is used.
|
| 105 |
+
res_block: bool argument to determine if residual block is used.
|
| 106 |
+
|
| 107 |
+
"""
|
| 108 |
+
|
| 109 |
+
super().__init__()
|
| 110 |
+
|
| 111 |
+
upsample_stride = upsample_kernel_size
|
| 112 |
+
self.transp_conv_init = get_conv_layer(
|
| 113 |
+
spatial_dims,
|
| 114 |
+
in_channels,
|
| 115 |
+
out_channels,
|
| 116 |
+
kernel_size=upsample_kernel_size,
|
| 117 |
+
stride=upsample_stride,
|
| 118 |
+
conv_only=True,
|
| 119 |
+
is_transposed=True,
|
| 120 |
+
)
|
| 121 |
+
if conv_block:
|
| 122 |
+
if res_block:
|
| 123 |
+
self.blocks = nn.ModuleList(
|
| 124 |
+
[
|
| 125 |
+
nn.Sequential(
|
| 126 |
+
get_conv_layer(
|
| 127 |
+
spatial_dims,
|
| 128 |
+
out_channels,
|
| 129 |
+
out_channels,
|
| 130 |
+
kernel_size=upsample_kernel_size,
|
| 131 |
+
stride=upsample_stride,
|
| 132 |
+
conv_only=True,
|
| 133 |
+
is_transposed=True,
|
| 134 |
+
),
|
| 135 |
+
UnetResBlock(
|
| 136 |
+
spatial_dims=3,
|
| 137 |
+
in_channels=out_channels,
|
| 138 |
+
out_channels=out_channels,
|
| 139 |
+
kernel_size=kernel_size,
|
| 140 |
+
stride=stride,
|
| 141 |
+
norm_name=norm_name,
|
| 142 |
+
),
|
| 143 |
+
)
|
| 144 |
+
for i in range(num_layer)
|
| 145 |
+
]
|
| 146 |
+
)
|
| 147 |
+
else:
|
| 148 |
+
self.blocks = nn.ModuleList(
|
| 149 |
+
[
|
| 150 |
+
nn.Sequential(
|
| 151 |
+
get_conv_layer(
|
| 152 |
+
spatial_dims,
|
| 153 |
+
out_channels,
|
| 154 |
+
out_channels,
|
| 155 |
+
kernel_size=upsample_kernel_size,
|
| 156 |
+
stride=upsample_stride,
|
| 157 |
+
conv_only=True,
|
| 158 |
+
is_transposed=True,
|
| 159 |
+
),
|
| 160 |
+
UnetBasicBlock(
|
| 161 |
+
spatial_dims=3,
|
| 162 |
+
in_channels=out_channels,
|
| 163 |
+
out_channels=out_channels,
|
| 164 |
+
kernel_size=kernel_size,
|
| 165 |
+
stride=stride,
|
| 166 |
+
norm_name=norm_name,
|
| 167 |
+
),
|
| 168 |
+
)
|
| 169 |
+
for i in range(num_layer)
|
| 170 |
+
]
|
| 171 |
+
)
|
| 172 |
+
else:
|
| 173 |
+
self.blocks = nn.ModuleList(
|
| 174 |
+
[
|
| 175 |
+
get_conv_layer(
|
| 176 |
+
spatial_dims,
|
| 177 |
+
out_channels,
|
| 178 |
+
out_channels,
|
| 179 |
+
kernel_size=1,
|
| 180 |
+
stride=1,
|
| 181 |
+
conv_only=True,
|
| 182 |
+
is_transposed=True,
|
| 183 |
+
)
|
| 184 |
+
for i in range(num_layer)
|
| 185 |
+
]
|
| 186 |
+
)
|
| 187 |
+
|
| 188 |
+
def forward(self, x):
|
| 189 |
+
x = self.transp_conv_init(x)
|
| 190 |
+
for blk in self.blocks:
|
| 191 |
+
x = blk(x)
|
| 192 |
+
return x
|
| 193 |
+
|
| 194 |
+
|
| 195 |
+
class UNesTConvBlock(nn.Module):
|
| 196 |
+
"""
|
| 197 |
+
UNesT block with skip connections
|
| 198 |
+
"""
|
| 199 |
+
|
| 200 |
+
def __init__(
|
| 201 |
+
self,
|
| 202 |
+
spatial_dims: int,
|
| 203 |
+
in_channels: int,
|
| 204 |
+
out_channels: int,
|
| 205 |
+
kernel_size: Union[Sequence[int], int],
|
| 206 |
+
stride: Union[Sequence[int], int],
|
| 207 |
+
norm_name: Union[Tuple, str],
|
| 208 |
+
res_block: bool = False,
|
| 209 |
+
) -> None:
|
| 210 |
+
"""
|
| 211 |
+
Args:
|
| 212 |
+
spatial_dims: number of spatial dimensions.
|
| 213 |
+
in_channels: number of input channels.
|
| 214 |
+
out_channels: number of output channels.
|
| 215 |
+
kernel_size: convolution kernel size.
|
| 216 |
+
stride: convolution stride.
|
| 217 |
+
norm_name: feature normalization type and arguments.
|
| 218 |
+
res_block: bool argument to determine if residual block is used.
|
| 219 |
+
|
| 220 |
+
"""
|
| 221 |
+
|
| 222 |
+
super().__init__()
|
| 223 |
+
|
| 224 |
+
if res_block:
|
| 225 |
+
self.layer = UnetResBlock(
|
| 226 |
+
spatial_dims=spatial_dims,
|
| 227 |
+
in_channels=in_channels,
|
| 228 |
+
out_channels=out_channels,
|
| 229 |
+
kernel_size=kernel_size,
|
| 230 |
+
stride=stride,
|
| 231 |
+
norm_name=norm_name,
|
| 232 |
+
)
|
| 233 |
+
else:
|
| 234 |
+
self.layer = UnetBasicBlock( # type: ignore
|
| 235 |
+
spatial_dims=spatial_dims,
|
| 236 |
+
in_channels=in_channels,
|
| 237 |
+
out_channels=out_channels,
|
| 238 |
+
kernel_size=kernel_size,
|
| 239 |
+
stride=stride,
|
| 240 |
+
norm_name=norm_name,
|
| 241 |
+
)
|
| 242 |
+
|
| 243 |
+
def forward(self, inp):
|
| 244 |
+
out = self.layer(inp)
|
| 245 |
+
return out
|