Spaces:
Running
on
T4
Running
on
T4
Simon Duerr
commited on
Commit
•
00aa807
1
Parent(s):
0291496
add proteinmpnn
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- ProteinMPNN +0 -1
- ProteinMPNN/LICENSE +21 -0
- ProteinMPNN/README.md +111 -0
- ProteinMPNN/ca_model_weights/v_48_002.pt +3 -0
- ProteinMPNN/ca_model_weights/v_48_010.pt +3 -0
- ProteinMPNN/ca_model_weights/v_48_020.pt +3 -0
- ProteinMPNN/colab_notebooks/README.md +1 -0
- ProteinMPNN/colab_notebooks/ca_only_quickdemo.ipynb +0 -0
- ProteinMPNN/colab_notebooks/quickdemo.ipynb +0 -0
- ProteinMPNN/colab_notebooks/quickdemo_wAF2.ipynb +612 -0
- ProteinMPNN/examples/submit_example_1.sh +28 -0
- ProteinMPNN/examples/submit_example_2.sh +34 -0
- ProteinMPNN/examples/submit_example_3.sh +27 -0
- ProteinMPNN/examples/submit_example_3_score_only.sh +28 -0
- ProteinMPNN/examples/submit_example_3_score_only_from_fasta.sh +30 -0
- ProteinMPNN/examples/submit_example_4.sh +40 -0
- ProteinMPNN/examples/submit_example_4_non_fixed.sh +40 -0
- ProteinMPNN/examples/submit_example_5.sh +44 -0
- ProteinMPNN/examples/submit_example_6.sh +34 -0
- ProteinMPNN/examples/submit_example_7.sh +29 -0
- ProteinMPNN/examples/submit_example_8.sh +34 -0
- ProteinMPNN/examples/submit_example_pssm.sh +49 -0
- ProteinMPNN/helper_scripts/assign_fixed_chains.py +39 -0
- ProteinMPNN/helper_scripts/make_bias_AA.py +27 -0
- ProteinMPNN/helper_scripts/make_bias_per_res_dict.py +53 -0
- ProteinMPNN/helper_scripts/make_fixed_positions_dict.py +59 -0
- ProteinMPNN/helper_scripts/make_pos_neg_tied_positions_dict.py +73 -0
- ProteinMPNN/helper_scripts/make_pssm_input_dict.py +36 -0
- ProteinMPNN/helper_scripts/make_tied_positions_dict.py +61 -0
- ProteinMPNN/helper_scripts/other_tools/make_omit_AA.py +39 -0
- ProteinMPNN/helper_scripts/other_tools/make_pssm_dict.py +64 -0
- ProteinMPNN/helper_scripts/parse_multiple_chains.out +1 -0
- ProteinMPNN/helper_scripts/parse_multiple_chains.py +163 -0
- ProteinMPNN/helper_scripts/parse_multiple_chains.sh +7 -0
- ProteinMPNN/inputs/PDB_complexes/pdbs/3HTN.pdb +0 -0
- ProteinMPNN/inputs/PDB_complexes/pdbs/4YOW.pdb +0 -0
- ProteinMPNN/inputs/PDB_homooligomers/pdbs/4GYT.pdb +0 -0
- ProteinMPNN/inputs/PDB_homooligomers/pdbs/6EHB.pdb +0 -0
- ProteinMPNN/inputs/PDB_monomers/pdbs/5L33.pdb +0 -0
- ProteinMPNN/inputs/PDB_monomers/pdbs/6MRR.pdb +0 -0
- ProteinMPNN/inputs/PSSM_inputs/3HTN.npz +0 -0
- ProteinMPNN/inputs/PSSM_inputs/4YOW.npz +0 -0
- ProteinMPNN/outputs/example_1_outputs/parsed_pdbs.jsonl +2 -0
- ProteinMPNN/outputs/example_1_outputs/seqs/5L33.fa +6 -0
- ProteinMPNN/outputs/example_1_outputs/seqs/6MRR.fa +6 -0
- ProteinMPNN/outputs/example_2_outputs/assigned_pdbs.jsonl +1 -0
- ProteinMPNN/outputs/example_2_outputs/parsed_pdbs.jsonl +0 -0
- ProteinMPNN/outputs/example_2_outputs/seqs/3HTN.fa +6 -0
- ProteinMPNN/outputs/example_2_outputs/seqs/4YOW.fa +6 -0
- ProteinMPNN/outputs/example_3_outputs/seqs/3HTN.fa +6 -0
ProteinMPNN
DELETED
|
@@ -1 +0,0 @@
|
|
| 1 |
-
Subproject commit 8907e6671bfbfc92303b5f79c4b5e6ce47cdef57
|
|
|
|
|
|
ProteinMPNN/LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2022 Justas Dauparas
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
ProteinMPNN/README.md
ADDED
|
@@ -0,0 +1,111 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# ProteinMPNN
|
| 2 |
+
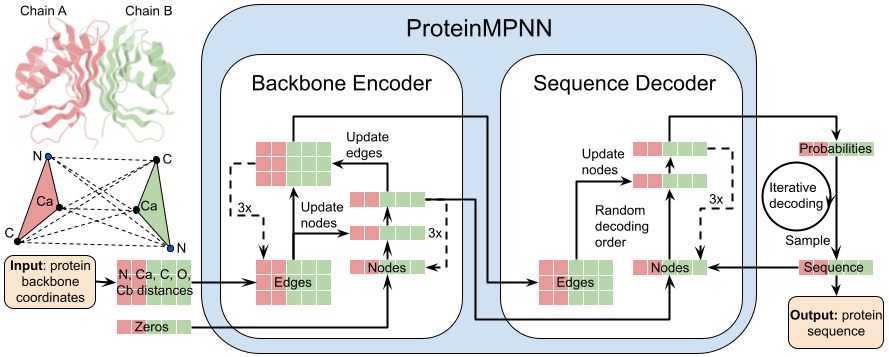
|
| 3 |
+
Read [ProteinMPNN paper](https://www.biorxiv.org/content/10.1101/2022.06.03.494563v1).
|
| 4 |
+
|
| 5 |
+
To run ProteinMPNN clone this github repo and install Python>=3.0, PyTorch, Numpy.
|
| 6 |
+
|
| 7 |
+
Full protein backbone models: `vanilla_model_weights/v_48_002.pt, v_48_010.pt, v_48_020.pt, v_48_030.pt`, `soluble_model_weights/v_48_010.pt, v_48_020.pt`.
|
| 8 |
+
|
| 9 |
+
CA only models: `ca_model_weights/v_48_002.pt, v_48_010.pt, v_48_020.pt`. Enable flag `--ca_only` to use these models.
|
| 10 |
+
|
| 11 |
+
Helper scripts: `helper_scripts` - helper functions to parse PDBs, assign which chains to design, which residues to fix, adding AA bias, tying residues etc.
|
| 12 |
+
|
| 13 |
+
Code organization:
|
| 14 |
+
* `protein_mpnn_run.py` - the main script to initialialize and run the model.
|
| 15 |
+
* `protein_mpnn_utils.py` - utility functions for the main script.
|
| 16 |
+
* `examples/` - simple code examples.
|
| 17 |
+
* `inputs/` - input PDB files for examples
|
| 18 |
+
* `outputs/` - outputs from examples
|
| 19 |
+
* `colab_notebooks/` - Google Colab examples
|
| 20 |
+
* `training/` - code and data to retrain the model
|
| 21 |
+
-----------------------------------------------------------------------------------------------------
|
| 22 |
+
Input flags for `protein_mpnn_run.py`:
|
| 23 |
+
```
|
| 24 |
+
argparser.add_argument("--suppress_print", type=int, default=0, help="0 for False, 1 for True")
|
| 25 |
+
argparser.add_argument("--ca_only", action="store_true", default=False, help="Parse CA-only structures and use CA-only models (default: false)")
|
| 26 |
+
argparser.add_argument("--path_to_model_weights", type=str, default="", help="Path to model weights folder;")
|
| 27 |
+
argparser.add_argument("--model_name", type=str, default="v_48_020", help="ProteinMPNN model name: v_48_002, v_48_010, v_48_020, v_48_030; v_48_010=version with 48 edges 0.10A noise")
|
| 28 |
+
argparser.add_argument("--use_soluble_model", action="store_true", default=False, help="Flag to load ProteinMPNN weights trained on soluble proteins only.")
|
| 29 |
+
argparser.add_argument("--seed", type=int, default=0, help="If set to 0 then a random seed will be picked;")
|
| 30 |
+
argparser.add_argument("--save_score", type=int, default=0, help="0 for False, 1 for True; save score=-log_prob to npy files")
|
| 31 |
+
argparser.add_argument("--path_to_fasta", type=str, default="", help="score provided input sequence in a fasta format; e.g. GGGGGG/PPPPS/WWW for chains A, B, C sorted alphabetically and separated by /")
|
| 32 |
+
argparser.add_argument("--save_probs", type=int, default=0, help="0 for False, 1 for True; save MPNN predicted probabilites per position")
|
| 33 |
+
argparser.add_argument("--score_only", type=int, default=0, help="0 for False, 1 for True; score input backbone-sequence pairs")
|
| 34 |
+
argparser.add_argument("--conditional_probs_only", type=int, default=0, help="0 for False, 1 for True; output conditional probabilities p(s_i given the rest of the sequence and backbone)")
|
| 35 |
+
argparser.add_argument("--conditional_probs_only_backbone", type=int, default=0, help="0 for False, 1 for True; if true output conditional probabilities p(s_i given backbone)")
|
| 36 |
+
argparser.add_argument("--unconditional_probs_only", type=int, default=0, help="0 for False, 1 for True; output unconditional probabilities p(s_i given backbone) in one forward pass")
|
| 37 |
+
argparser.add_argument("--backbone_noise", type=float, default=0.00, help="Standard deviation of Gaussian noise to add to backbone atoms")
|
| 38 |
+
argparser.add_argument("--num_seq_per_target", type=int, default=1, help="Number of sequences to generate per target")
|
| 39 |
+
argparser.add_argument("--batch_size", type=int, default=1, help="Batch size; can set higher for titan, quadro GPUs, reduce this if running out of GPU memory")
|
| 40 |
+
argparser.add_argument("--max_length", type=int, default=200000, help="Max sequence length")
|
| 41 |
+
argparser.add_argument("--sampling_temp", type=str, default="0.1", help="A string of temperatures, 0.2 0.25 0.5. Sampling temperature for amino acids. Suggested values 0.1, 0.15, 0.2, 0.25, 0.3. Higher values will lead to more diversity.")
|
| 42 |
+
argparser.add_argument("--out_folder", type=str, help="Path to a folder to output sequences, e.g. /home/out/")
|
| 43 |
+
argparser.add_argument("--pdb_path", type=str, default='', help="Path to a single PDB to be designed")
|
| 44 |
+
argparser.add_argument("--pdb_path_chains", type=str, default='', help="Define which chains need to be designed for a single PDB ")
|
| 45 |
+
argparser.add_argument("--jsonl_path", type=str, help="Path to a folder with parsed pdb into jsonl")
|
| 46 |
+
argparser.add_argument("--chain_id_jsonl",type=str, default='', help="Path to a dictionary specifying which chains need to be designed and which ones are fixed, if not specied all chains will be designed.")
|
| 47 |
+
argparser.add_argument("--fixed_positions_jsonl", type=str, default='', help="Path to a dictionary with fixed positions")
|
| 48 |
+
argparser.add_argument("--omit_AAs", type=list, default='X', help="Specify which amino acids should be omitted in the generated sequence, e.g. 'AC' would omit alanine and cystine.")
|
| 49 |
+
argparser.add_argument("--bias_AA_jsonl", type=str, default='', help="Path to a dictionary which specifies AA composion bias if neededi, e.g. {A: -1.1, F: 0.7} would make A less likely and F more likely.")
|
| 50 |
+
argparser.add_argument("--bias_by_res_jsonl", default='', help="Path to dictionary with per position bias.")
|
| 51 |
+
argparser.add_argument("--omit_AA_jsonl", type=str, default='', help="Path to a dictionary which specifies which amino acids need to be omited from design at specific chain indices")
|
| 52 |
+
argparser.add_argument("--pssm_jsonl", type=str, default='', help="Path to a dictionary with pssm")
|
| 53 |
+
argparser.add_argument("--pssm_multi", type=float, default=0.0, help="A value between [0.0, 1.0], 0.0 means do not use pssm, 1.0 ignore MPNN predictions")
|
| 54 |
+
argparser.add_argument("--pssm_threshold", type=float, default=0.0, help="A value between -inf + inf to restric per position AAs")
|
| 55 |
+
argparser.add_argument("--pssm_log_odds_flag", type=int, default=0, help="0 for False, 1 for True")
|
| 56 |
+
argparser.add_argument("--pssm_bias_flag", type=int, default=0, help="0 for False, 1 for True")
|
| 57 |
+
argparser.add_argument("--tied_positions_jsonl", type=str, default='', help="Path to a dictionary with tied positions")
|
| 58 |
+
|
| 59 |
+
```
|
| 60 |
+
-----------------------------------------------------------------------------------------------------
|
| 61 |
+
For example to make a conda environment to run ProteinMPNN:
|
| 62 |
+
* `conda create --name mlfold` - this creates conda environment called `mlfold`
|
| 63 |
+
* `source activate mlfold` - this activate environment
|
| 64 |
+
* `conda install pytorch torchvision torchaudio cudatoolkit=11.3 -c pytorch` - install pytorch following steps from https://pytorch.org/
|
| 65 |
+
-----------------------------------------------------------------------------------------------------
|
| 66 |
+
These are provided `examples/`:
|
| 67 |
+
* `submit_example_1.sh` - simple monomer example
|
| 68 |
+
* `submit_example_2.sh` - simple multi-chain example
|
| 69 |
+
* `submit_example_3.sh` - directly from the .pdb path
|
| 70 |
+
* `submit_example_3_score_only.sh` - return score only (model's uncertainty)
|
| 71 |
+
* `submit_example_3_score_only_from_fasta.sh` - return score only (model's uncertainty) loading sequence from fasta files
|
| 72 |
+
* `submit_example_4.sh` - fix some residue positions
|
| 73 |
+
* `submit_example_4_non_fixed.sh` - specify which positions to design
|
| 74 |
+
* `submit_example_5.sh` - tie some positions together (symmetry)
|
| 75 |
+
* `submit_example_6.sh` - homooligomer example
|
| 76 |
+
* `submit_example_7.sh` - return sequence unconditional probabilities (PSSM like)
|
| 77 |
+
* `submit_example_8.sh` - add amino acid bias
|
| 78 |
+
* `submit_example_pssm.sh` - use PSSM bias when designing sequences
|
| 79 |
+
-----------------------------------------------------------------------------------------------------
|
| 80 |
+
Output example:
|
| 81 |
+
```
|
| 82 |
+
>3HTN, score=1.1705, global_score=1.2045, fixed_chains=['B'], designed_chains=['A', 'C'], model_name=v_48_020, git_hash=015ff820b9b5741ead6ba6795258f35a9c15e94b, seed=37
|
| 83 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 84 |
+
>T=0.1, sample=1, score=0.7291, global_score=0.9330, seq_recovery=0.5736
|
| 85 |
+
NMYSYKKIGNKYIVSINNHTEIVKALKKFCEEKNIKSGSVNGIGSIGSVTLKFYNLETKEEELKTFNANFEISNLTGFISMHDNKVFLDLHITIGDENFSALAGHLVSAVVNGTCELIVEDFNELVSTKYNEELGLWLLDFEK/NMYSYKKIGNKYIVSINNHTDIVTAIKKFCEDKKIKSGTINGIGQVKEVTLEFRNFETGEKEEKTFKKQFTISNLTGFISTKDGKVFLDLHITFGDENFSALAGHLISAIVDGKCELIIEDYNEEINVKYNEELGLYLLDFNK
|
| 86 |
+
>T=0.1, sample=2, score=0.7414, global_score=0.9355, seq_recovery=0.6075
|
| 87 |
+
NMYKYKKIGNKYIVSINNHTEIVKAIKEFCKEKNIKSGTINGIGQVGKVTLRFYNPETKEYTEKTFNDNFEISNLTGFISTYKNEVFLHLHITFGKSDFSALAGHLLSAIVNGICELIVEDFKENLSMKYDEKTGLYLLDFEK/NMYKYKKIGNKYVVSINNHTEIVEALKAFCEDKKIKSGTVNGIGQVSKVTLKFFNIETKESKEKTFNKNFEISNLTGFISEINGEVFLHLHITIGDENFSALAGHLLSAVVNGEAILIVEDYKEKVNRKYNEELGLNLLDFNL
|
| 88 |
+
```
|
| 89 |
+
* `score` - average over residues that were designed negative log probability of sampled amino acids
|
| 90 |
+
* `global score` - average over all residues in all chains negative log probability of sampled/fixed amino acids
|
| 91 |
+
* `fixed_chains` - chains that were not designed (fixed)
|
| 92 |
+
* `designed_chains` - chains that were redesigned
|
| 93 |
+
* `model_name/CA_model_name` - model name that was used to generate results, e.g. `v_48_020`
|
| 94 |
+
* `git_hash` - github version that was used to generate outputs
|
| 95 |
+
* `seed` - random seed
|
| 96 |
+
* `T=0.1` - temperature equal to 0.1 was used to sample sequences
|
| 97 |
+
* `sample` - sequence sample number 1, 2, 3...etc
|
| 98 |
+
-----------------------------------------------------------------------------------------------------
|
| 99 |
+
```
|
| 100 |
+
@article{dauparas2022robust,
|
| 101 |
+
title={Robust deep learning--based protein sequence design using ProteinMPNN},
|
| 102 |
+
author={Dauparas, Justas and Anishchenko, Ivan and Bennett, Nathaniel and Bai, Hua and Ragotte, Robert J and Milles, Lukas F and Wicky, Basile IM and Courbet, Alexis and de Haas, Rob J and Bethel, Neville and others},
|
| 103 |
+
journal={Science},
|
| 104 |
+
volume={378},
|
| 105 |
+
number={6615},
|
| 106 |
+
pages={49--56},
|
| 107 |
+
year={2022},
|
| 108 |
+
publisher={American Association for the Advancement of Science}
|
| 109 |
+
}
|
| 110 |
+
```
|
| 111 |
+
-----------------------------------------------------------------------------------------------------
|
ProteinMPNN/ca_model_weights/v_48_002.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:ec038b44a987d7c8351b6ed887c82a2370d54e45e55a6bdaf508a729cef0340e
|
| 3 |
+
size 6624011
|
ProteinMPNN/ca_model_weights/v_48_010.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:cdb50498d45578d20b271fa7817b8cd8bfde3875ad69dbd3f5e4b5dd3e588301
|
| 3 |
+
size 6624011
|
ProteinMPNN/ca_model_weights/v_48_020.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f28f40170e21858c5ff31ef50b6e63414ff76dc331b19f85aa8586a12031744a
|
| 3 |
+
size 6624011
|
ProteinMPNN/colab_notebooks/README.md
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
<a href="https://colab.research.google.com/github/dauparas/ProteinMPNN/blob/main/colab_notebooks/quickdemo.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
|
ProteinMPNN/colab_notebooks/ca_only_quickdemo.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/colab_notebooks/quickdemo.ipynb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/colab_notebooks/quickdemo_wAF2.ipynb
ADDED
|
@@ -0,0 +1,612 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "markdown",
|
| 5 |
+
"metadata": {
|
| 6 |
+
"id": "view-in-github",
|
| 7 |
+
"colab_type": "text"
|
| 8 |
+
},
|
| 9 |
+
"source": [
|
| 10 |
+
"<a href=\"https://colab.research.google.com/github/dauparas/ProteinMPNN/blob/main/colab_notebooks/quickdemo_wAF2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>"
|
| 11 |
+
]
|
| 12 |
+
},
|
| 13 |
+
{
|
| 14 |
+
"cell_type": "markdown",
|
| 15 |
+
"metadata": {
|
| 16 |
+
"id": "AYZebfKn8gef"
|
| 17 |
+
},
|
| 18 |
+
"source": [
|
| 19 |
+
"#ProteinMPNN w/AF2\n",
|
| 20 |
+
"This notebook is intended as a quick demo, more features to come!\n",
|
| 21 |
+
"\n",
|
| 22 |
+
"Examples: \n",
|
| 23 |
+
"1. pdb: `6MRR`, homomer: `False`, designed_chain: `A`\n",
|
| 24 |
+
"2. pdb: `1X2I`, homomer: `True`, designed_chain: `A,B` \n",
|
| 25 |
+
" (for correct symmetric tying lenghts of homomer chains should be the same)"
|
| 26 |
+
]
|
| 27 |
+
},
|
| 28 |
+
{
|
| 29 |
+
"cell_type": "code",
|
| 30 |
+
"source": [
|
| 31 |
+
"#@title Setup ProteinMPNN\n",
|
| 32 |
+
"import warnings\n",
|
| 33 |
+
"warnings.simplefilter(action='ignore', category=FutureWarning)\n",
|
| 34 |
+
"\n",
|
| 35 |
+
"import json, time, os, sys, glob, re\n",
|
| 36 |
+
"from google.colab import files\n",
|
| 37 |
+
"import numpy as np\n",
|
| 38 |
+
"\n",
|
| 39 |
+
"if not os.path.isdir(\"ProteinMPNN\"):\n",
|
| 40 |
+
" os.system(\"git clone -q https://github.com/dauparas/ProteinMPNN.git\")\n",
|
| 41 |
+
"\n",
|
| 42 |
+
"if \"ProteinMPNN\" not in sys.path:\n",
|
| 43 |
+
" sys.path.append('/content/ProteinMPNN')\n",
|
| 44 |
+
"\n",
|
| 45 |
+
"import matplotlib.pyplot as plt\n",
|
| 46 |
+
"import shutil\n",
|
| 47 |
+
"import warnings\n",
|
| 48 |
+
"import torch\n",
|
| 49 |
+
"from torch import optim\n",
|
| 50 |
+
"from torch.utils.data import DataLoader\n",
|
| 51 |
+
"from torch.utils.data.dataset import random_split, Subset\n",
|
| 52 |
+
"import copy\n",
|
| 53 |
+
"import torch.nn as nn\n",
|
| 54 |
+
"import torch.nn.functional as F\n",
|
| 55 |
+
"import random\n",
|
| 56 |
+
"import os.path\n",
|
| 57 |
+
"from protein_mpnn_utils import loss_nll, loss_smoothed, gather_edges, gather_nodes, gather_nodes_t, cat_neighbors_nodes, _scores, _S_to_seq, tied_featurize, parse_PDB\n",
|
| 58 |
+
"from protein_mpnn_utils import StructureDataset, StructureDatasetPDB, ProteinMPNN\n",
|
| 59 |
+
"\n",
|
| 60 |
+
"device = torch.device(\"cpu\")\n",
|
| 61 |
+
"#v_48_010=version with 48 edges 0.10A noise\n",
|
| 62 |
+
"model_name = \"v_48_020\" #@param [\"v_48_002\", \"v_48_010\", \"v_48_020\", \"v_48_030\"]\n",
|
| 63 |
+
"\n",
|
| 64 |
+
"\n",
|
| 65 |
+
"backbone_noise=0.00 # Standard deviation of Gaussian noise to add to backbone atoms\n",
|
| 66 |
+
"\n",
|
| 67 |
+
"path_to_model_weights='/content/ProteinMPNN/vanilla_model_weights' \n",
|
| 68 |
+
"hidden_dim = 128\n",
|
| 69 |
+
"num_layers = 3 \n",
|
| 70 |
+
"model_folder_path = path_to_model_weights\n",
|
| 71 |
+
"if model_folder_path[-1] != '/':\n",
|
| 72 |
+
" model_folder_path = model_folder_path + '/'\n",
|
| 73 |
+
"checkpoint_path = model_folder_path + f'{model_name}.pt'\n",
|
| 74 |
+
"\n",
|
| 75 |
+
"checkpoint = torch.load(checkpoint_path, map_location=device) \n",
|
| 76 |
+
"print('Number of edges:', checkpoint['num_edges'])\n",
|
| 77 |
+
"noise_level_print = checkpoint['noise_level']\n",
|
| 78 |
+
"print(f'Training noise level: {noise_level_print}A')\n",
|
| 79 |
+
"model = ProteinMPNN(num_letters=21, node_features=hidden_dim, edge_features=hidden_dim, hidden_dim=hidden_dim, num_encoder_layers=num_layers, num_decoder_layers=num_layers, augment_eps=backbone_noise, k_neighbors=checkpoint['num_edges'])\n",
|
| 80 |
+
"model.to(device)\n",
|
| 81 |
+
"model.load_state_dict(checkpoint['model_state_dict'])\n",
|
| 82 |
+
"model.eval()\n",
|
| 83 |
+
"print(\"Model loaded\")\n",
|
| 84 |
+
"\n",
|
| 85 |
+
"def make_tied_positions_for_homomers(pdb_dict_list):\n",
|
| 86 |
+
" my_dict = {}\n",
|
| 87 |
+
" for result in pdb_dict_list:\n",
|
| 88 |
+
" all_chain_list = sorted([item[-1:] for item in list(result) if item[:9]=='seq_chain']) #A, B, C, ...\n",
|
| 89 |
+
" tied_positions_list = []\n",
|
| 90 |
+
" chain_length = len(result[f\"seq_chain_{all_chain_list[0]}\"])\n",
|
| 91 |
+
" for i in range(1,chain_length+1):\n",
|
| 92 |
+
" temp_dict = {}\n",
|
| 93 |
+
" for j, chain in enumerate(all_chain_list):\n",
|
| 94 |
+
" temp_dict[chain] = [i] #needs to be a list\n",
|
| 95 |
+
" tied_positions_list.append(temp_dict)\n",
|
| 96 |
+
" my_dict[result['name']] = tied_positions_list\n",
|
| 97 |
+
" return my_dict\n",
|
| 98 |
+
"\n",
|
| 99 |
+
"#########################\n",
|
| 100 |
+
"def get_pdb(pdb_code=\"\"):\n",
|
| 101 |
+
" if pdb_code is None or pdb_code == \"\":\n",
|
| 102 |
+
" upload_dict = files.upload()\n",
|
| 103 |
+
" pdb_string = upload_dict[list(upload_dict.keys())[0]]\n",
|
| 104 |
+
" with open(\"tmp.pdb\",\"wb\") as out: out.write(pdb_string)\n",
|
| 105 |
+
" return \"tmp.pdb\"\n",
|
| 106 |
+
" else:\n",
|
| 107 |
+
" os.system(f\"wget -qnc https://files.rcsb.org/view/{pdb_code}.pdb\")\n",
|
| 108 |
+
" return f\"{pdb_code}.pdb\""
|
| 109 |
+
],
|
| 110 |
+
"metadata": {
|
| 111 |
+
"id": "2nKSlaMlSpcf",
|
| 112 |
+
"cellView": "form"
|
| 113 |
+
},
|
| 114 |
+
"execution_count": null,
|
| 115 |
+
"outputs": []
|
| 116 |
+
},
|
| 117 |
+
{
|
| 118 |
+
"cell_type": "code",
|
| 119 |
+
"execution_count": null,
|
| 120 |
+
"metadata": {
|
| 121 |
+
"cellView": "form",
|
| 122 |
+
"id": "xMVlYh8Fv2of"
|
| 123 |
+
},
|
| 124 |
+
"outputs": [],
|
| 125 |
+
"source": [
|
| 126 |
+
"#@title #Run ProteinMPNN\n",
|
| 127 |
+
"\n",
|
| 128 |
+
"#@markdown #### Input Options\n",
|
| 129 |
+
"pdb='6MRR' #@param {type:\"string\"}\n",
|
| 130 |
+
"pdb = pdb.replace(\" \",\"\")\n",
|
| 131 |
+
"pdb_path = get_pdb(pdb)\n",
|
| 132 |
+
"#@markdown - pdb code (leave blank to get an upload prompt)\n",
|
| 133 |
+
"\n",
|
| 134 |
+
"homomer = False #@param {type:\"boolean\"}\n",
|
| 135 |
+
"designed_chain = \"A\" #@param {type:\"string\"}\n",
|
| 136 |
+
"fixed_chain = \"\" #@param {type:\"string\"}\n",
|
| 137 |
+
"\n",
|
| 138 |
+
"if designed_chain == \"\":\n",
|
| 139 |
+
" designed_chain_list = []\n",
|
| 140 |
+
"else:\n",
|
| 141 |
+
" designed_chain_list = re.sub(\"[^A-Za-z]+\",\",\", designed_chain).split(\",\")\n",
|
| 142 |
+
"\n",
|
| 143 |
+
"if fixed_chain == \"\":\n",
|
| 144 |
+
" fixed_chain_list = []\n",
|
| 145 |
+
"else:\n",
|
| 146 |
+
" fixed_chain_list = re.sub(\"[^A-Za-z]+\",\",\", fixed_chain).split(\",\")\n",
|
| 147 |
+
"\n",
|
| 148 |
+
"chain_list = list(set(designed_chain_list + fixed_chain_list))\n",
|
| 149 |
+
"\n",
|
| 150 |
+
"#@markdown - specified which chain(s) to design and which chain(s) to keep fixed. \n",
|
| 151 |
+
"#@markdown Use comma:`A,B` to specifiy more than one chain\n",
|
| 152 |
+
"\n",
|
| 153 |
+
"#chain = \"A\" #@param {type:\"string\"}\n",
|
| 154 |
+
"#pdb_path_chains = chain\n",
|
| 155 |
+
"##@markdown - Define which chain to redesign\n",
|
| 156 |
+
"\n",
|
| 157 |
+
"#@markdown #### Design Options\n",
|
| 158 |
+
"num_seqs = 8 #@param [\"1\", \"2\", \"4\", \"8\", \"16\", \"32\", \"64\"] {type:\"raw\"}\n",
|
| 159 |
+
"num_seq_per_target = num_seqs\n",
|
| 160 |
+
"\n",
|
| 161 |
+
"#@markdown - Sampling temperature for amino acids, T=0.0 means taking argmax, T>>1.0 means sample randomly.\n",
|
| 162 |
+
"sampling_temp = \"0.1\" #@param [\"0.0001\", \"0.1\", \"0.15\", \"0.2\", \"0.25\", \"0.3\", \"0.5\"]\n",
|
| 163 |
+
"\n",
|
| 164 |
+
"\n",
|
| 165 |
+
"\n",
|
| 166 |
+
"save_score=0 # 0 for False, 1 for True; save score=-log_prob to npy files\n",
|
| 167 |
+
"save_probs=0 # 0 for False, 1 for True; save MPNN predicted probabilites per position\n",
|
| 168 |
+
"score_only=0 # 0 for False, 1 for True; score input backbone-sequence pairs\n",
|
| 169 |
+
"conditional_probs_only=0 # 0 for False, 1 for True; output conditional probabilities p(s_i given the rest of the sequence and backbone)\n",
|
| 170 |
+
"conditional_probs_only_backbone=0 # 0 for False, 1 for True; if true output conditional probabilities p(s_i given backbone)\n",
|
| 171 |
+
" \n",
|
| 172 |
+
"batch_size=1 # Batch size; can set higher for titan, quadro GPUs, reduce this if running out of GPU memory\n",
|
| 173 |
+
"max_length=20000 # Max sequence length\n",
|
| 174 |
+
" \n",
|
| 175 |
+
"out_folder='.' # Path to a folder to output sequences, e.g. /home/out/\n",
|
| 176 |
+
"jsonl_path='' # Path to a folder with parsed pdb into jsonl\n",
|
| 177 |
+
"omit_AAs='X' # Specify which amino acids should be omitted in the generated sequence, e.g. 'AC' would omit alanine and cystine.\n",
|
| 178 |
+
" \n",
|
| 179 |
+
"pssm_multi=0.0 # A value between [0.0, 1.0], 0.0 means do not use pssm, 1.0 ignore MPNN predictions\n",
|
| 180 |
+
"pssm_threshold=0.0 # A value between -inf + inf to restric per position AAs\n",
|
| 181 |
+
"pssm_log_odds_flag=0 # 0 for False, 1 for True\n",
|
| 182 |
+
"pssm_bias_flag=0 # 0 for False, 1 for True\n",
|
| 183 |
+
"\n",
|
| 184 |
+
"\n",
|
| 185 |
+
"##############################################################\n",
|
| 186 |
+
"\n",
|
| 187 |
+
"folder_for_outputs = out_folder\n",
|
| 188 |
+
"\n",
|
| 189 |
+
"NUM_BATCHES = num_seq_per_target//batch_size\n",
|
| 190 |
+
"BATCH_COPIES = batch_size\n",
|
| 191 |
+
"temperatures = [float(item) for item in sampling_temp.split()]\n",
|
| 192 |
+
"omit_AAs_list = omit_AAs\n",
|
| 193 |
+
"alphabet = 'ACDEFGHIKLMNPQRSTVWYX'\n",
|
| 194 |
+
"\n",
|
| 195 |
+
"omit_AAs_np = np.array([AA in omit_AAs_list for AA in alphabet]).astype(np.float32)\n",
|
| 196 |
+
"\n",
|
| 197 |
+
"chain_id_dict = None\n",
|
| 198 |
+
"fixed_positions_dict = None\n",
|
| 199 |
+
"pssm_dict = None\n",
|
| 200 |
+
"omit_AA_dict = None\n",
|
| 201 |
+
"bias_AA_dict = None\n",
|
| 202 |
+
"tied_positions_dict = None\n",
|
| 203 |
+
"bias_by_res_dict = None\n",
|
| 204 |
+
"bias_AAs_np = np.zeros(len(alphabet))\n",
|
| 205 |
+
"\n",
|
| 206 |
+
"\n",
|
| 207 |
+
"###############################################################\n",
|
| 208 |
+
"pdb_dict_list = parse_PDB(pdb_path, input_chain_list=chain_list)\n",
|
| 209 |
+
"dataset_valid = StructureDatasetPDB(pdb_dict_list, truncate=None, max_length=max_length)\n",
|
| 210 |
+
"\n",
|
| 211 |
+
"chain_id_dict = {}\n",
|
| 212 |
+
"chain_id_dict[pdb_dict_list[0]['name']]= (designed_chain_list, fixed_chain_list)\n",
|
| 213 |
+
"\n",
|
| 214 |
+
"print(chain_id_dict)\n",
|
| 215 |
+
"for chain in chain_list:\n",
|
| 216 |
+
" l = len(pdb_dict_list[0][f\"seq_chain_{chain}\"])\n",
|
| 217 |
+
" print(f\"Length of chain {chain} is {l}\")\n",
|
| 218 |
+
"\n",
|
| 219 |
+
"if homomer:\n",
|
| 220 |
+
" tied_positions_dict = make_tied_positions_for_homomers(pdb_dict_list)\n",
|
| 221 |
+
"else:\n",
|
| 222 |
+
" tied_positions_dict = None\n",
|
| 223 |
+
"\n",
|
| 224 |
+
"#################################################################\n",
|
| 225 |
+
"sequences = []\n",
|
| 226 |
+
"with torch.no_grad():\n",
|
| 227 |
+
" print('Generating sequences...')\n",
|
| 228 |
+
" for ix, protein in enumerate(dataset_valid):\n",
|
| 229 |
+
" score_list = []\n",
|
| 230 |
+
" all_probs_list = []\n",
|
| 231 |
+
" all_log_probs_list = []\n",
|
| 232 |
+
" S_sample_list = []\n",
|
| 233 |
+
" batch_clones = [copy.deepcopy(protein) for i in range(BATCH_COPIES)]\n",
|
| 234 |
+
" X, S, mask, lengths, chain_M, chain_encoding_all, chain_list_list, visible_list_list, masked_list_list, masked_chain_length_list_list, chain_M_pos, omit_AA_mask, residue_idx, dihedral_mask, tied_pos_list_of_lists_list, pssm_coef, pssm_bias, pssm_log_odds_all, bias_by_res_all, tied_beta = tied_featurize(batch_clones, device, chain_id_dict, fixed_positions_dict, omit_AA_dict, tied_positions_dict, pssm_dict, bias_by_res_dict)\n",
|
| 235 |
+
" pssm_log_odds_mask = (pssm_log_odds_all > pssm_threshold).float() #1.0 for true, 0.0 for false\n",
|
| 236 |
+
" name_ = batch_clones[0]['name']\n",
|
| 237 |
+
"\n",
|
| 238 |
+
" randn_1 = torch.randn(chain_M.shape, device=X.device)\n",
|
| 239 |
+
" log_probs = model(X, S, mask, chain_M*chain_M_pos, residue_idx, chain_encoding_all, randn_1)\n",
|
| 240 |
+
" mask_for_loss = mask*chain_M*chain_M_pos\n",
|
| 241 |
+
" scores = _scores(S, log_probs, mask_for_loss)\n",
|
| 242 |
+
" native_score = scores.cpu().data.numpy()\n",
|
| 243 |
+
"\n",
|
| 244 |
+
" for temp in temperatures:\n",
|
| 245 |
+
" for j in range(NUM_BATCHES):\n",
|
| 246 |
+
" randn_2 = torch.randn(chain_M.shape, device=X.device)\n",
|
| 247 |
+
" if tied_positions_dict == None:\n",
|
| 248 |
+
" sample_dict = model.sample(X, randn_2, S, chain_M, chain_encoding_all, residue_idx, mask=mask, temperature=temp, omit_AAs_np=omit_AAs_np, bias_AAs_np=bias_AAs_np, chain_M_pos=chain_M_pos, omit_AA_mask=omit_AA_mask, pssm_coef=pssm_coef, pssm_bias=pssm_bias, pssm_multi=pssm_multi, pssm_log_odds_flag=bool(pssm_log_odds_flag), pssm_log_odds_mask=pssm_log_odds_mask, pssm_bias_flag=bool(pssm_bias_flag), bias_by_res=bias_by_res_all)\n",
|
| 249 |
+
" S_sample = sample_dict[\"S\"] \n",
|
| 250 |
+
" else:\n",
|
| 251 |
+
" sample_dict = model.tied_sample(X, randn_2, S, chain_M, chain_encoding_all, residue_idx, mask=mask, temperature=temp, omit_AAs_np=omit_AAs_np, bias_AAs_np=bias_AAs_np, chain_M_pos=chain_M_pos, omit_AA_mask=omit_AA_mask, pssm_coef=pssm_coef, pssm_bias=pssm_bias, pssm_multi=pssm_multi, pssm_log_odds_flag=bool(pssm_log_odds_flag), pssm_log_odds_mask=pssm_log_odds_mask, pssm_bias_flag=bool(pssm_bias_flag), tied_pos=tied_pos_list_of_lists_list[0], tied_beta=tied_beta, bias_by_res=bias_by_res_all)\n",
|
| 252 |
+
" # Compute scores\n",
|
| 253 |
+
" S_sample = sample_dict[\"S\"]\n",
|
| 254 |
+
" log_probs = model(X, S_sample, mask, chain_M*chain_M_pos, residue_idx, chain_encoding_all, randn_2, use_input_decoding_order=True, decoding_order=sample_dict[\"decoding_order\"])\n",
|
| 255 |
+
" mask_for_loss = mask*chain_M*chain_M_pos\n",
|
| 256 |
+
" scores = _scores(S_sample, log_probs, mask_for_loss)\n",
|
| 257 |
+
" scores = scores.cpu().data.numpy()\n",
|
| 258 |
+
" all_probs_list.append(sample_dict[\"probs\"].cpu().data.numpy())\n",
|
| 259 |
+
" all_log_probs_list.append(log_probs.cpu().data.numpy())\n",
|
| 260 |
+
" S_sample_list.append(S_sample.cpu().data.numpy())\n",
|
| 261 |
+
" for b_ix in range(BATCH_COPIES):\n",
|
| 262 |
+
" masked_chain_length_list = masked_chain_length_list_list[b_ix]\n",
|
| 263 |
+
" masked_list = masked_list_list[b_ix]\n",
|
| 264 |
+
" seq_recovery_rate = torch.sum(torch.sum(torch.nn.functional.one_hot(S[b_ix], 21)*torch.nn.functional.one_hot(S_sample[b_ix], 21),axis=-1)*mask_for_loss[b_ix])/torch.sum(mask_for_loss[b_ix])\n",
|
| 265 |
+
" seq = _S_to_seq(S_sample[b_ix], chain_M[b_ix])\n",
|
| 266 |
+
" score = scores[b_ix]\n",
|
| 267 |
+
" score_list.append(score)\n",
|
| 268 |
+
" native_seq = _S_to_seq(S[b_ix], chain_M[b_ix])\n",
|
| 269 |
+
" if b_ix == 0 and j==0 and temp==temperatures[0]:\n",
|
| 270 |
+
" start = 0\n",
|
| 271 |
+
" end = 0\n",
|
| 272 |
+
" list_of_AAs = []\n",
|
| 273 |
+
" for mask_l in masked_chain_length_list:\n",
|
| 274 |
+
" end += mask_l\n",
|
| 275 |
+
" list_of_AAs.append(native_seq[start:end])\n",
|
| 276 |
+
" start = end\n",
|
| 277 |
+
" native_seq = \"\".join(list(np.array(list_of_AAs)[np.argsort(masked_list)]))\n",
|
| 278 |
+
" l0 = 0\n",
|
| 279 |
+
" for mc_length in list(np.array(masked_chain_length_list)[np.argsort(masked_list)])[:-1]:\n",
|
| 280 |
+
" l0 += mc_length\n",
|
| 281 |
+
" native_seq = native_seq[:l0] + '/' + native_seq[l0:]\n",
|
| 282 |
+
" l0 += 1\n",
|
| 283 |
+
" sorted_masked_chain_letters = np.argsort(masked_list_list[0])\n",
|
| 284 |
+
" print_masked_chains = [masked_list_list[0][i] for i in sorted_masked_chain_letters]\n",
|
| 285 |
+
" sorted_visible_chain_letters = np.argsort(visible_list_list[0])\n",
|
| 286 |
+
" print_visible_chains = [visible_list_list[0][i] for i in sorted_visible_chain_letters]\n",
|
| 287 |
+
" native_score_print = np.format_float_positional(np.float32(native_score.mean()), unique=False, precision=4)\n",
|
| 288 |
+
" line = '>{}, score={}, fixed_chains={}, designed_chains={}, model_name={}\\n{}\\n'.format(name_, native_score_print, print_visible_chains, print_masked_chains, model_name, native_seq)\n",
|
| 289 |
+
" print(line.rstrip())\n",
|
| 290 |
+
" start = 0\n",
|
| 291 |
+
" end = 0\n",
|
| 292 |
+
" list_of_AAs = []\n",
|
| 293 |
+
" for mask_l in masked_chain_length_list:\n",
|
| 294 |
+
" end += mask_l\n",
|
| 295 |
+
" list_of_AAs.append(seq[start:end])\n",
|
| 296 |
+
" start = end\n",
|
| 297 |
+
"\n",
|
| 298 |
+
" seq = \"\".join(list(np.array(list_of_AAs)[np.argsort(masked_list)]))\n",
|
| 299 |
+
" l0 = 0\n",
|
| 300 |
+
" for mc_length in list(np.array(masked_chain_length_list)[np.argsort(masked_list)])[:-1]:\n",
|
| 301 |
+
" l0 += mc_length\n",
|
| 302 |
+
" seq = seq[:l0] + '/' + seq[l0:]\n",
|
| 303 |
+
" l0 += 1\n",
|
| 304 |
+
" score_print = np.format_float_positional(np.float32(score), unique=False, precision=4)\n",
|
| 305 |
+
" seq_rec_print = np.format_float_positional(np.float32(seq_recovery_rate.detach().cpu().numpy()), unique=False, precision=4)\n",
|
| 306 |
+
" line = '>T={}, sample={}, score={}, seq_recovery={}\\n{}\\n'.format(temp,b_ix,score_print,seq_rec_print,seq)\n",
|
| 307 |
+
" sequences.append(seq)\n",
|
| 308 |
+
" print(line.rstrip())\n",
|
| 309 |
+
"\n",
|
| 310 |
+
"\n",
|
| 311 |
+
"all_probs_concat = np.concatenate(all_probs_list)\n",
|
| 312 |
+
"all_log_probs_concat = np.concatenate(all_log_probs_list)\n",
|
| 313 |
+
"S_sample_concat = np.concatenate(S_sample_list)"
|
| 314 |
+
]
|
| 315 |
+
},
|
| 316 |
+
{
|
| 317 |
+
"cell_type": "markdown",
|
| 318 |
+
"source": [
|
| 319 |
+
"# Predict with AlphaFold2 (with single-sequence input)"
|
| 320 |
+
],
|
| 321 |
+
"metadata": {
|
| 322 |
+
"id": "5mQ4VLG1dPsd"
|
| 323 |
+
}
|
| 324 |
+
},
|
| 325 |
+
{
|
| 326 |
+
"cell_type": "code",
|
| 327 |
+
"source": [
|
| 328 |
+
"#@title Setup AlphaFold\n",
|
| 329 |
+
"\n",
|
| 330 |
+
"# import libraries\n",
|
| 331 |
+
"from IPython.utils import io\n",
|
| 332 |
+
"import os,sys,re\n",
|
| 333 |
+
"\n",
|
| 334 |
+
"if \"af_backprop\" not in sys.path:\n",
|
| 335 |
+
" import tensorflow as tf\n",
|
| 336 |
+
" import jax\n",
|
| 337 |
+
" import jax.numpy as jnp\n",
|
| 338 |
+
" import numpy as np\n",
|
| 339 |
+
" import matplotlib\n",
|
| 340 |
+
" from matplotlib import animation\n",
|
| 341 |
+
" import matplotlib.pyplot as plt\n",
|
| 342 |
+
" from IPython.display import HTML\n",
|
| 343 |
+
" import tqdm.notebook\n",
|
| 344 |
+
" TQDM_BAR_FORMAT = '{l_bar}{bar}| {n_fmt}/{total_fmt} [elapsed: {elapsed} remaining: {remaining}]'\n",
|
| 345 |
+
"\n",
|
| 346 |
+
" with io.capture_output() as captured:\n",
|
| 347 |
+
" # install ALPHAFOLD\n",
|
| 348 |
+
" if not os.path.isdir(\"af_backprop\"):\n",
|
| 349 |
+
" %shell git clone https://github.com/sokrypton/af_backprop.git\n",
|
| 350 |
+
" %shell pip -q install biopython dm-haiku ml-collections py3Dmol\n",
|
| 351 |
+
" %shell wget -qnc https://raw.githubusercontent.com/sokrypton/ColabFold/main/beta/colabfold.py\n",
|
| 352 |
+
" if not os.path.isdir(\"params\"):\n",
|
| 353 |
+
" %shell mkdir params\n",
|
| 354 |
+
" %shell curl -fsSL https://storage.googleapis.com/alphafold/alphafold_params_2021-07-14.tar | tar x -C params\n",
|
| 355 |
+
"\n",
|
| 356 |
+
" if not os.path.exists(\"MMalign\"):\n",
|
| 357 |
+
" # install MMalign\n",
|
| 358 |
+
" os.system(\"wget -qnc https://zhanggroup.org/MM-align/bin/module/MMalign.cpp\")\n",
|
| 359 |
+
" os.system(\"g++ -static -O3 -ffast-math -o MMalign MMalign.cpp\")\n",
|
| 360 |
+
"\n",
|
| 361 |
+
" def mmalign(pdb_a,pdb_b):\n",
|
| 362 |
+
" # pass to MMalign\n",
|
| 363 |
+
" output = os.popen(f'./MMalign {pdb_a} {pdb_b}')\n",
|
| 364 |
+
" # parse outputs\n",
|
| 365 |
+
" parse_float = lambda x: float(x.split(\"=\")[1].split()[0])\n",
|
| 366 |
+
" tms = []\n",
|
| 367 |
+
" for line in output:\n",
|
| 368 |
+
" line = line.rstrip()\n",
|
| 369 |
+
" if line.startswith(\"TM-score\"): tms.append(parse_float(line))\n",
|
| 370 |
+
" return tms\n",
|
| 371 |
+
"\n",
|
| 372 |
+
" # configure which device to use\n",
|
| 373 |
+
" try:\n",
|
| 374 |
+
" # check if TPU is available\n",
|
| 375 |
+
" import jax.tools.colab_tpu\n",
|
| 376 |
+
" jax.tools.colab_tpu.setup_tpu()\n",
|
| 377 |
+
" print('Running on TPU')\n",
|
| 378 |
+
" DEVICE = \"tpu\"\n",
|
| 379 |
+
" except:\n",
|
| 380 |
+
" if jax.local_devices()[0].platform == 'cpu':\n",
|
| 381 |
+
" print(\"WARNING: no GPU detected, will be using CPU\")\n",
|
| 382 |
+
" DEVICE = \"cpu\"\n",
|
| 383 |
+
" else:\n",
|
| 384 |
+
" print('Running on GPU')\n",
|
| 385 |
+
" DEVICE = \"gpu\"\n",
|
| 386 |
+
" # disable GPU on tensorflow\n",
|
| 387 |
+
" tf.config.set_visible_devices([], 'GPU')\n",
|
| 388 |
+
"\n",
|
| 389 |
+
" # import libraries\n",
|
| 390 |
+
" sys.path.append('af_backprop')\n",
|
| 391 |
+
" from utils import update_seq, update_aatype, get_plddt, get_pae\n",
|
| 392 |
+
" import colabfold as cf\n",
|
| 393 |
+
" from alphafold.common import protein as alphafold_protein\n",
|
| 394 |
+
" from alphafold.data import pipeline\n",
|
| 395 |
+
" from alphafold.model import data, config\n",
|
| 396 |
+
" from alphafold.common import residue_constants\n",
|
| 397 |
+
" from alphafold.model import model as alphafold_model\n",
|
| 398 |
+
"\n",
|
| 399 |
+
"# custom functions\n",
|
| 400 |
+
"def clear_mem():\n",
|
| 401 |
+
" backend = jax.lib.xla_bridge.get_backend()\n",
|
| 402 |
+
" for buf in backend.live_buffers(): buf.delete()\n",
|
| 403 |
+
"\n",
|
| 404 |
+
"def setup_model(max_len):\n",
|
| 405 |
+
" clear_mem()\n",
|
| 406 |
+
"\n",
|
| 407 |
+
" # setup model\n",
|
| 408 |
+
" cfg = config.model_config(\"model_3_ptm\")\n",
|
| 409 |
+
" cfg.model.num_recycle = 0\n",
|
| 410 |
+
" cfg.data.common.num_recycle = 0\n",
|
| 411 |
+
" cfg.data.eval.max_msa_clusters = 1\n",
|
| 412 |
+
" cfg.data.common.max_extra_msa = 1\n",
|
| 413 |
+
" cfg.data.eval.masked_msa_replace_fraction = 0\n",
|
| 414 |
+
" cfg.model.global_config.subbatch_size = None\n",
|
| 415 |
+
"\n",
|
| 416 |
+
" # get params\n",
|
| 417 |
+
" model_param = data.get_model_haiku_params(model_name=\"model_3_ptm\", data_dir=\".\")\n",
|
| 418 |
+
" model_runner = alphafold_model.RunModel(cfg, model_param, is_training=False, recycle_mode=\"none\")\n",
|
| 419 |
+
"\n",
|
| 420 |
+
" model_params = []\n",
|
| 421 |
+
" for k in [1,2,3,4,5]:\n",
|
| 422 |
+
" if k == 3:\n",
|
| 423 |
+
" model_params.append(model_param)\n",
|
| 424 |
+
" else:\n",
|
| 425 |
+
" params = data.get_model_haiku_params(model_name=f\"model_{k}_ptm\", data_dir=\".\")\n",
|
| 426 |
+
" model_params.append({k: params[k] for k in model_runner.params.keys()})\n",
|
| 427 |
+
"\n",
|
| 428 |
+
" seq = \"A\" * max_len\n",
|
| 429 |
+
" length = len(seq)\n",
|
| 430 |
+
" feature_dict = {\n",
|
| 431 |
+
" **pipeline.make_sequence_features(sequence=seq, description=\"none\", num_res=length),\n",
|
| 432 |
+
" **pipeline.make_msa_features(msas=[[seq]], deletion_matrices=[[[0]*length]])\n",
|
| 433 |
+
" }\n",
|
| 434 |
+
" inputs = model_runner.process_features(feature_dict,random_seed=0)\n",
|
| 435 |
+
"\n",
|
| 436 |
+
" def runner(I, params):\n",
|
| 437 |
+
" # update sequence\n",
|
| 438 |
+
" inputs = I[\"inputs\"]\n",
|
| 439 |
+
" inputs.update(I[\"prev\"])\n",
|
| 440 |
+
"\n",
|
| 441 |
+
" seq = jax.nn.one_hot(I[\"seq\"],20)\n",
|
| 442 |
+
" update_seq(seq, inputs)\n",
|
| 443 |
+
" update_aatype(inputs[\"target_feat\"][...,1:], inputs)\n",
|
| 444 |
+
"\n",
|
| 445 |
+
" # mask prediction\n",
|
| 446 |
+
" mask = jnp.arange(inputs[\"residue_index\"].shape[0]) < I[\"length\"]\n",
|
| 447 |
+
" inputs[\"seq_mask\"] = inputs[\"seq_mask\"].at[:].set(mask)\n",
|
| 448 |
+
" inputs[\"msa_mask\"] = inputs[\"msa_mask\"].at[:].set(mask)\n",
|
| 449 |
+
" inputs[\"residue_index\"] = jnp.where(mask, inputs[\"residue_index\"], 0)\n",
|
| 450 |
+
"\n",
|
| 451 |
+
" # get prediction\n",
|
| 452 |
+
" key = jax.random.PRNGKey(0)\n",
|
| 453 |
+
" outputs = model_runner.apply(params, key, inputs)\n",
|
| 454 |
+
"\n",
|
| 455 |
+
" prev = {\"init_msa_first_row\":outputs['representations']['msa_first_row'][None],\n",
|
| 456 |
+
" \"init_pair\":outputs['representations']['pair'][None],\n",
|
| 457 |
+
" \"init_pos\":outputs['structure_module']['final_atom_positions'][None]}\n",
|
| 458 |
+
" \n",
|
| 459 |
+
" aux = {\"final_atom_positions\":outputs[\"structure_module\"][\"final_atom_positions\"],\n",
|
| 460 |
+
" \"final_atom_mask\":outputs[\"structure_module\"][\"final_atom_mask\"],\n",
|
| 461 |
+
" \"plddt\":get_plddt(outputs),\"pae\":get_pae(outputs),\n",
|
| 462 |
+
" \"length\":I[\"length\"], \"seq\":I[\"seq\"], \"prev\":prev,\n",
|
| 463 |
+
" \"residue_idx\":inputs[\"residue_index\"][0]}\n",
|
| 464 |
+
" return aux\n",
|
| 465 |
+
"\n",
|
| 466 |
+
" return jax.jit(runner), model_params, {\"inputs\":inputs, \"length\":max_length}\n",
|
| 467 |
+
"\n",
|
| 468 |
+
"def save_pdb(outs, filename, Ls=None):\n",
|
| 469 |
+
" '''save pdb coordinates'''\n",
|
| 470 |
+
" p = {\"residue_index\":outs[\"residue_idx\"] + 1,\n",
|
| 471 |
+
" \"aatype\":outs[\"seq\"],\n",
|
| 472 |
+
" \"atom_positions\":outs[\"final_atom_positions\"],\n",
|
| 473 |
+
" \"atom_mask\":outs[\"final_atom_mask\"],\n",
|
| 474 |
+
" \"plddt\":outs[\"plddt\"]}\n",
|
| 475 |
+
" p = jax.tree_map(lambda x:x[:outs[\"length\"]], p)\n",
|
| 476 |
+
" b_factors = 100 * p.pop(\"plddt\")[:,None] * p[\"atom_mask\"]\n",
|
| 477 |
+
" p = alphafold_protein.Protein(**p,b_factors=b_factors)\n",
|
| 478 |
+
" pdb_lines = alphafold_protein.to_pdb(p)\n",
|
| 479 |
+
" with open(filename, 'w') as f:\n",
|
| 480 |
+
" f.write(pdb_lines)\n",
|
| 481 |
+
" if Ls is not None:\n",
|
| 482 |
+
" pdb_lines = cf.read_pdb_renum(filename, Ls)\n",
|
| 483 |
+
" with open(filename, 'w') as f:\n",
|
| 484 |
+
" f.write(pdb_lines)"
|
| 485 |
+
],
|
| 486 |
+
"metadata": {
|
| 487 |
+
"cellView": "form",
|
| 488 |
+
"id": "4ZBUThXU7yY8"
|
| 489 |
+
},
|
| 490 |
+
"execution_count": null,
|
| 491 |
+
"outputs": []
|
| 492 |
+
},
|
| 493 |
+
{
|
| 494 |
+
"cell_type": "code",
|
| 495 |
+
"source": [
|
| 496 |
+
"#@title Run AlphaFold\n",
|
| 497 |
+
"num_models = 1 #@param [\"1\",\"2\",\"3\",\"4\",\"5\"] {type:\"raw\"}\n",
|
| 498 |
+
"num_recycles = 1 #@param [\"0\",\"1\",\"2\",\"3\"] {type:\"raw\"}\n",
|
| 499 |
+
"num_sequences = len(sequences)\n",
|
| 500 |
+
"outs = []\n",
|
| 501 |
+
"positions = []\n",
|
| 502 |
+
"plddts = []\n",
|
| 503 |
+
"paes = []\n",
|
| 504 |
+
"LS = []\n",
|
| 505 |
+
"\n",
|
| 506 |
+
"with tqdm.notebook.tqdm(total=(num_recycles + 1) * num_models * num_sequences, bar_format=TQDM_BAR_FORMAT) as pbar:\n",
|
| 507 |
+
" print(f\"seq_num model_num avg_pLDDT avg_pAE TMscore\")\n",
|
| 508 |
+
" for s,ori_sequence in enumerate(sequences):\n",
|
| 509 |
+
" Ls = [len(s) for s in ori_sequence.replace(\":\",\"/\").split(\"/\")]\n",
|
| 510 |
+
" LS.append(Ls)\n",
|
| 511 |
+
" sequence = re.sub(\"[^A-Z]\",\"\",ori_sequence)\n",
|
| 512 |
+
" length = len(sequence)\n",
|
| 513 |
+
"\n",
|
| 514 |
+
" # avoid recompiling if length within 25\n",
|
| 515 |
+
" if \"max_len\" not in dir() or length > max_len or (max_len - length) > 25:\n",
|
| 516 |
+
" max_len = length + 25\n",
|
| 517 |
+
" runner, params, I = setup_model(max_len)\n",
|
| 518 |
+
"\n",
|
| 519 |
+
" outs.append([])\n",
|
| 520 |
+
" positions.append([])\n",
|
| 521 |
+
" plddts.append([])\n",
|
| 522 |
+
" paes.append([])\n",
|
| 523 |
+
"\n",
|
| 524 |
+
" r = -1\n",
|
| 525 |
+
" # pad sequence to max length\n",
|
| 526 |
+
" seq = np.array([residue_constants.restype_order.get(aa,0) for aa in sequence])\n",
|
| 527 |
+
" seq = np.pad(seq,[0,max_len-length],constant_values=-1)\n",
|
| 528 |
+
" I[\"inputs\"]['residue_index'][:] = cf.chain_break(np.arange(max_len), Ls, length=32)\n",
|
| 529 |
+
" I.update({\"seq\":seq, \"length\":length})\n",
|
| 530 |
+
" \n",
|
| 531 |
+
" # for each model\n",
|
| 532 |
+
" for n in range(num_models):\n",
|
| 533 |
+
" # restart recycle\n",
|
| 534 |
+
" I[\"prev\"] = {'init_msa_first_row': np.zeros([1, max_len, 256]),\n",
|
| 535 |
+
" 'init_pair': np.zeros([1, max_len, max_len, 128]),\n",
|
| 536 |
+
" 'init_pos': np.zeros([1, max_len, 37, 3])}\n",
|
| 537 |
+
" for r in range(num_recycles + 1):\n",
|
| 538 |
+
" O = runner(I, params[n])\n",
|
| 539 |
+
" O = jax.tree_map(lambda x:np.asarray(x), O)\n",
|
| 540 |
+
" I[\"prev\"] = O[\"prev\"]\n",
|
| 541 |
+
" pbar.update(1)\n",
|
| 542 |
+
" \n",
|
| 543 |
+
" positions[-1].append(O[\"final_atom_positions\"][:length])\n",
|
| 544 |
+
" plddts[-1].append(O[\"plddt\"][:length])\n",
|
| 545 |
+
" paes[-1].append(O[\"pae\"][:length,:length])\n",
|
| 546 |
+
" outs[-1].append(O)\n",
|
| 547 |
+
" save_pdb(outs[-1][-1], f\"out_seq_{s}_model_{n}.pdb\", Ls=LS[-1])\n",
|
| 548 |
+
" tmscores = mmalign(pdb_path, f\"out_seq_{s}_model_{n}.pdb\")\n",
|
| 549 |
+
" print(f\"{s} {n}\\t{plddts[-1][-1].mean():.3}\\t{paes[-1][-1].mean():.3}\\t{tmscores[-1]:.3}\")"
|
| 550 |
+
],
|
| 551 |
+
"metadata": {
|
| 552 |
+
"cellView": "form",
|
| 553 |
+
"id": "p2uNokqudTSH"
|
| 554 |
+
},
|
| 555 |
+
"execution_count": null,
|
| 556 |
+
"outputs": []
|
| 557 |
+
},
|
| 558 |
+
{
|
| 559 |
+
"cell_type": "code",
|
| 560 |
+
"source": [
|
| 561 |
+
"#@title Display 3D structure {run: \"auto\"}\n",
|
| 562 |
+
"#@markdown #### select which sequence to show (if more than one designed example)\n",
|
| 563 |
+
"seq_num = 0 #@param [\"0\",\"1\",\"2\",\"3\",\"4\",\"5\",\"6\",\"7\"] {type:\"raw\"}\n",
|
| 564 |
+
"assert seq_num < len(outs), f\"ERROR: seq_num ({seq_num}) exceeds number of designed sequences ({num_sequences})\"\n",
|
| 565 |
+
"model_num = 0 #@param [\"0\",\"1\",\"2\",\"3\",\"4\"] {type:\"raw\"}\n",
|
| 566 |
+
"assert model_num < len(outs[0]), f\"ERROR: model_num ({num_models}) exceeds number of model params used ({num_models})\"\n",
|
| 567 |
+
"#@markdown #### options\n",
|
| 568 |
+
"\n",
|
| 569 |
+
"color = \"confidence\" #@param [\"chain\", \"confidence\", \"rainbow\"]\n",
|
| 570 |
+
"if color == \"confidence\": color = \"lDDT\"\n",
|
| 571 |
+
"show_sidechains = False #@param {type:\"boolean\"}\n",
|
| 572 |
+
"show_mainchains = False #@param {type:\"boolean\"}\n",
|
| 573 |
+
"\n",
|
| 574 |
+
"v = cf.show_pdb(f\"out_seq_{seq_num}_model_{model_num}.pdb\", show_sidechains, show_mainchains, color,\n",
|
| 575 |
+
" color_HP=True, size=(800,480), Ls=LS[seq_num]) \n",
|
| 576 |
+
"v.setHoverable({}, True,\n",
|
| 577 |
+
" '''function(atom,viewer,event,container){if(!atom.label){atom.label=viewer.addLabel(\" \"+atom.resn+\":\"+atom.resi,{position:atom,backgroundColor:'mintcream',fontColor:'black'});}}''',\n",
|
| 578 |
+
" '''function(atom,viewer){if(atom.label){viewer.removeLabel(atom.label);delete atom.label;}}''')\n",
|
| 579 |
+
"v.show() \n",
|
| 580 |
+
"if color == \"lDDT\":\n",
|
| 581 |
+
" cf.plot_plddt_legend().show()\n",
|
| 582 |
+
"\n",
|
| 583 |
+
"# add confidence plots\n",
|
| 584 |
+
"cf.plot_confidence(plddts[seq_num][model_num]*100, paes[seq_num][model_num], Ls=LS[seq_num]).show()"
|
| 585 |
+
],
|
| 586 |
+
"metadata": {
|
| 587 |
+
"cellView": "form",
|
| 588 |
+
"id": "0TNhcwok8d_w"
|
| 589 |
+
},
|
| 590 |
+
"execution_count": null,
|
| 591 |
+
"outputs": []
|
| 592 |
+
}
|
| 593 |
+
],
|
| 594 |
+
"metadata": {
|
| 595 |
+
"colab": {
|
| 596 |
+
"name": "quickdemo_wAF2.ipynb",
|
| 597 |
+
"provenance": [],
|
| 598 |
+
"include_colab_link": true
|
| 599 |
+
},
|
| 600 |
+
"kernelspec": {
|
| 601 |
+
"display_name": "Python 3",
|
| 602 |
+
"name": "python3"
|
| 603 |
+
},
|
| 604 |
+
"language_info": {
|
| 605 |
+
"name": "python"
|
| 606 |
+
},
|
| 607 |
+
"accelerator": "GPU",
|
| 608 |
+
"gpuClass": "standard"
|
| 609 |
+
},
|
| 610 |
+
"nbformat": 4,
|
| 611 |
+
"nbformat_minor": 0
|
| 612 |
+
}
|
ProteinMPNN/examples/submit_example_1.sh
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 2
|
| 6 |
+
#SBATCH --output=example_1.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../inputs/PDB_monomers/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_1_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 19 |
+
|
| 20 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 21 |
+
|
| 22 |
+
python ../protein_mpnn_run.py \
|
| 23 |
+
--jsonl_path $path_for_parsed_chains \
|
| 24 |
+
--out_folder $output_dir \
|
| 25 |
+
--num_seq_per_target 2 \
|
| 26 |
+
--sampling_temp "0.1" \
|
| 27 |
+
--seed 37 \
|
| 28 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_2.sh
ADDED
|
@@ -0,0 +1,34 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 2
|
| 6 |
+
#SBATCH --output=example_2.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
folder_with_pdbs="../inputs/PDB_complexes/pdbs/"
|
| 12 |
+
|
| 13 |
+
output_dir="../outputs/example_2_outputs"
|
| 14 |
+
if [ ! -d $output_dir ]
|
| 15 |
+
then
|
| 16 |
+
mkdir -p $output_dir
|
| 17 |
+
fi
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 21 |
+
chains_to_design="A B"
|
| 22 |
+
|
| 23 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 24 |
+
|
| 25 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 26 |
+
|
| 27 |
+
python ../protein_mpnn_run.py \
|
| 28 |
+
--jsonl_path $path_for_parsed_chains \
|
| 29 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 30 |
+
--out_folder $output_dir \
|
| 31 |
+
--num_seq_per_target 2 \
|
| 32 |
+
--sampling_temp "0.1" \
|
| 33 |
+
--seed 37 \
|
| 34 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_3.sh
ADDED
|
@@ -0,0 +1,27 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_3.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
path_to_PDB="../inputs/PDB_complexes/pdbs/3HTN.pdb"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_3_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
chains_to_design="A B"
|
| 19 |
+
|
| 20 |
+
python ../protein_mpnn_run.py \
|
| 21 |
+
--pdb_path $path_to_PDB \
|
| 22 |
+
--pdb_path_chains "$chains_to_design" \
|
| 23 |
+
--out_folder $output_dir \
|
| 24 |
+
--num_seq_per_target 2 \
|
| 25 |
+
--sampling_temp "0.1" \
|
| 26 |
+
--seed 37 \
|
| 27 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_3_score_only.sh
ADDED
|
@@ -0,0 +1,28 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_3.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
path_to_PDB="../inputs/PDB_complexes/pdbs/3HTN.pdb"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_3_score_only_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
chains_to_design="A B"
|
| 19 |
+
|
| 20 |
+
python ../protein_mpnn_run.py \
|
| 21 |
+
--pdb_path $path_to_PDB \
|
| 22 |
+
--pdb_path_chains "$chains_to_design" \
|
| 23 |
+
--out_folder $output_dir \
|
| 24 |
+
--num_seq_per_target 10 \
|
| 25 |
+
--sampling_temp "0.1" \
|
| 26 |
+
--score_only 1 \
|
| 27 |
+
--seed 37 \
|
| 28 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_3_score_only_from_fasta.sh
ADDED
|
@@ -0,0 +1,30 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_3_from_fasta.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
path_to_PDB="../inputs/PDB_complexes/pdbs/3HTN.pdb"
|
| 11 |
+
path_to_fasta="/home/justas/projects/github/ProteinMPNN/outputs/example_3_outputs/seqs/3HTN.fa"
|
| 12 |
+
|
| 13 |
+
output_dir="../outputs/example_3_score_only_from_fasta_outputs"
|
| 14 |
+
if [ ! -d $output_dir ]
|
| 15 |
+
then
|
| 16 |
+
mkdir -p $output_dir
|
| 17 |
+
fi
|
| 18 |
+
|
| 19 |
+
chains_to_design="A B"
|
| 20 |
+
|
| 21 |
+
python ../protein_mpnn_run.py \
|
| 22 |
+
--path_to_fasta $path_to_fasta \
|
| 23 |
+
--pdb_path $path_to_PDB \
|
| 24 |
+
--pdb_path_chains "$chains_to_design" \
|
| 25 |
+
--out_folder $output_dir \
|
| 26 |
+
--num_seq_per_target 5 \
|
| 27 |
+
--sampling_temp "0.1" \
|
| 28 |
+
--score_only 1 \
|
| 29 |
+
--seed 13 \
|
| 30 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_4.sh
ADDED
|
@@ -0,0 +1,40 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_4.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../inputs/PDB_complexes/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_4_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 21 |
+
path_for_fixed_positions=$output_dir"/fixed_pdbs.jsonl"
|
| 22 |
+
chains_to_design="A C"
|
| 23 |
+
#The first amino acid in the chain corresponds to 1 and not PDB residues index for now.
|
| 24 |
+
fixed_positions="1 2 3 4 5 6 7 8 23 25, 10 11 12 13 14 15 16 17 18 19 20 40" #fixing/not designing residues 1 2 3...25 in chain A and residues 10 11 12...40 in chain C
|
| 25 |
+
|
| 26 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 27 |
+
|
| 28 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 29 |
+
|
| 30 |
+
python ../helper_scripts/make_fixed_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_fixed_positions --chain_list "$chains_to_design" --position_list "$fixed_positions"
|
| 31 |
+
|
| 32 |
+
python ../protein_mpnn_run.py \
|
| 33 |
+
--jsonl_path $path_for_parsed_chains \
|
| 34 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 35 |
+
--fixed_positions_jsonl $path_for_fixed_positions \
|
| 36 |
+
--out_folder $output_dir \
|
| 37 |
+
--num_seq_per_target 2 \
|
| 38 |
+
--sampling_temp "0.1" \
|
| 39 |
+
--seed 37 \
|
| 40 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_4_non_fixed.sh
ADDED
|
@@ -0,0 +1,40 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_4_non_fixed.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../inputs/PDB_complexes/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_4_non_fixed_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 21 |
+
path_for_fixed_positions=$output_dir"/fixed_pdbs.jsonl"
|
| 22 |
+
chains_to_design="A C"
|
| 23 |
+
#The first amino acid in the chain corresponds to 1 and not PDB residues index for now.
|
| 24 |
+
design_only_positions="1 2 3 4 5 6 7 8 9 10, 3 4 5 6 7 8" #design only these residues; use flag --specify_non_fixed
|
| 25 |
+
|
| 26 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 27 |
+
|
| 28 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 29 |
+
|
| 30 |
+
python ../helper_scripts/make_fixed_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_fixed_positions --chain_list "$chains_to_design" --position_list "$design_only_positions" --specify_non_fixed
|
| 31 |
+
|
| 32 |
+
python ../protein_mpnn_run.py \
|
| 33 |
+
--jsonl_path $path_for_parsed_chains \
|
| 34 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 35 |
+
--fixed_positions_jsonl $path_for_fixed_positions \
|
| 36 |
+
--out_folder $output_dir \
|
| 37 |
+
--num_seq_per_target 2 \
|
| 38 |
+
--sampling_temp "0.1" \
|
| 39 |
+
--seed 37 \
|
| 40 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_5.sh
ADDED
|
@@ -0,0 +1,44 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_5.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../inputs/PDB_complexes/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_5_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 21 |
+
path_for_fixed_positions=$output_dir"/fixed_pdbs.jsonl"
|
| 22 |
+
path_for_tied_positions=$output_dir"/tied_pdbs.jsonl"
|
| 23 |
+
chains_to_design="A C"
|
| 24 |
+
fixed_positions="9 10 11 12 13 14 15 16 17 18 19 20 21 22 23, 10 11 18 19 20 22"
|
| 25 |
+
tied_positions="1 2 3 4 5 6 7 8, 1 2 3 4 5 6 7 8" #two list must match in length; residue 1 in chain A and C will be sampled togther;
|
| 26 |
+
|
| 27 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 28 |
+
|
| 29 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 30 |
+
|
| 31 |
+
python ../helper_scripts/make_fixed_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_fixed_positions --chain_list "$chains_to_design" --position_list "$fixed_positions"
|
| 32 |
+
|
| 33 |
+
python ../helper_scripts/make_tied_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_tied_positions --chain_list "$chains_to_design" --position_list "$tied_positions"
|
| 34 |
+
|
| 35 |
+
python ../protein_mpnn_run.py \
|
| 36 |
+
--jsonl_path $path_for_parsed_chains \
|
| 37 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 38 |
+
--fixed_positions_jsonl $path_for_fixed_positions \
|
| 39 |
+
--tied_positions_jsonl $path_for_tied_positions \
|
| 40 |
+
--out_folder $output_dir \
|
| 41 |
+
--num_seq_per_target 2 \
|
| 42 |
+
--sampling_temp "0.1" \
|
| 43 |
+
--seed 37 \
|
| 44 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_6.sh
ADDED
|
@@ -0,0 +1,34 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 3
|
| 6 |
+
#SBATCH --output=example_6.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../inputs/PDB_homooligomers/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_6_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 20 |
+
path_for_tied_positions=$output_dir"/tied_pdbs.jsonl"
|
| 21 |
+
path_for_designed_sequences=$output_dir"/temp_0.1"
|
| 22 |
+
|
| 23 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 24 |
+
|
| 25 |
+
python ../helper_scripts/make_tied_positions_dict.py --input_path=$path_for_parsed_chains --output_path=$path_for_tied_positions --homooligomer 1
|
| 26 |
+
|
| 27 |
+
python ../protein_mpnn_run.py \
|
| 28 |
+
--jsonl_path $path_for_parsed_chains \
|
| 29 |
+
--tied_positions_jsonl $path_for_tied_positions \
|
| 30 |
+
--out_folder $output_dir \
|
| 31 |
+
--num_seq_per_target 2 \
|
| 32 |
+
--sampling_temp "0.2" \
|
| 33 |
+
--seed 37 \
|
| 34 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_7.sh
ADDED
|
@@ -0,0 +1,29 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 2
|
| 6 |
+
#SBATCH --output=example_7.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../inputs/PDB_monomers/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_7_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 19 |
+
|
| 20 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 21 |
+
|
| 22 |
+
python ../protein_mpnn_run.py \
|
| 23 |
+
--jsonl_path $path_for_parsed_chains \
|
| 24 |
+
--out_folder $output_dir \
|
| 25 |
+
--num_seq_per_target 1 \
|
| 26 |
+
--sampling_temp "0.1" \
|
| 27 |
+
--unconditional_probs_only 1 \
|
| 28 |
+
--seed 37 \
|
| 29 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_8.sh
ADDED
|
@@ -0,0 +1,34 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 2
|
| 6 |
+
#SBATCH --output=example_8.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
folder_with_pdbs="../inputs/PDB_monomers/pdbs/"
|
| 11 |
+
|
| 12 |
+
output_dir="../outputs/example_8_outputs"
|
| 13 |
+
if [ ! -d $output_dir ]
|
| 14 |
+
then
|
| 15 |
+
mkdir -p $output_dir
|
| 16 |
+
fi
|
| 17 |
+
|
| 18 |
+
path_for_bias=$output_dir"/bias_pdbs.jsonl"
|
| 19 |
+
#Adding global polar amino acid bias (Doug Tischer)
|
| 20 |
+
AA_list="D E H K N Q R S T W Y"
|
| 21 |
+
bias_list="1.39 1.39 1.39 1.39 1.39 1.39 1.39 1.39 1.39 1.39 1.39"
|
| 22 |
+
python ../helper_scripts/make_bias_AA.py --output_path=$path_for_bias --AA_list="$AA_list" --bias_list="$bias_list"
|
| 23 |
+
|
| 24 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 25 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 26 |
+
|
| 27 |
+
python ../protein_mpnn_run.py \
|
| 28 |
+
--jsonl_path $path_for_parsed_chains \
|
| 29 |
+
--out_folder $output_dir \
|
| 30 |
+
--bias_AA_jsonl $path_for_bias \
|
| 31 |
+
--num_seq_per_target 2 \
|
| 32 |
+
--sampling_temp "0.1" \
|
| 33 |
+
--seed 37 \
|
| 34 |
+
--batch_size 1
|
ProteinMPNN/examples/submit_example_pssm.sh
ADDED
|
@@ -0,0 +1,49 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH -p gpu
|
| 3 |
+
#SBATCH --mem=32g
|
| 4 |
+
#SBATCH --gres=gpu:rtx2080:1
|
| 5 |
+
#SBATCH -c 2
|
| 6 |
+
#SBATCH --output=example_2.out
|
| 7 |
+
|
| 8 |
+
source activate mlfold
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
#new_probabilities_using_PSSM = (1-pssm_multi*pssm_coef_gathered[:,None])*probs + pssm_multi*pssm_coef_gathered[:,None]*pssm_bias_gathered
|
| 12 |
+
#probs - predictions from MPNN
|
| 13 |
+
#pssm_bias_gathered - input PSSM bias (needs to be a probability distribution)
|
| 14 |
+
#pssm_multi - a number between 0.0 (no bias) and 1.0 (no MPNN) inputed via flag --pssm_multi; this is a global number equally applied to all the residues
|
| 15 |
+
#pssm_coef_gathered - a number between 0.0 (no bias) and 1.0 (no MPNN) inputed via ../helper_scripts/make_pssm_input_dict.py can be adjusted per residue level; i.e only apply PSSM bias to specific residues; or chains
|
| 16 |
+
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
pssm_input_path="../inputs/PSSM_inputs"
|
| 20 |
+
folder_with_pdbs="../inputs/PDB_complexes/pdbs/"
|
| 21 |
+
|
| 22 |
+
output_dir="../outputs/example_pssm_outputs"
|
| 23 |
+
if [ ! -d $output_dir ]
|
| 24 |
+
then
|
| 25 |
+
mkdir -p $output_dir
|
| 26 |
+
fi
|
| 27 |
+
|
| 28 |
+
path_for_parsed_chains=$output_dir"/parsed_pdbs.jsonl"
|
| 29 |
+
path_for_assigned_chains=$output_dir"/assigned_pdbs.jsonl"
|
| 30 |
+
pssm=$output_dir"/pssm.jsonl"
|
| 31 |
+
chains_to_design="A B"
|
| 32 |
+
|
| 33 |
+
python ../helper_scripts/parse_multiple_chains.py --input_path=$folder_with_pdbs --output_path=$path_for_parsed_chains
|
| 34 |
+
|
| 35 |
+
python ../helper_scripts/assign_fixed_chains.py --input_path=$path_for_parsed_chains --output_path=$path_for_assigned_chains --chain_list "$chains_to_design"
|
| 36 |
+
|
| 37 |
+
python ../helper_scripts/make_pssm_input_dict.py --jsonl_input_path=$path_for_parsed_chains --PSSM_input_path=$pssm_input_path --output_path=$pssm
|
| 38 |
+
|
| 39 |
+
python ../protein_mpnn_run.py \
|
| 40 |
+
--jsonl_path $path_for_parsed_chains \
|
| 41 |
+
--chain_id_jsonl $path_for_assigned_chains \
|
| 42 |
+
--out_folder $output_dir \
|
| 43 |
+
--num_seq_per_target 2 \
|
| 44 |
+
--sampling_temp "0.1" \
|
| 45 |
+
--seed 37 \
|
| 46 |
+
--batch_size 1 \
|
| 47 |
+
--pssm_jsonl $pssm \
|
| 48 |
+
--pssm_multi 0.3 \
|
| 49 |
+
--pssm_bias_flag 1
|
ProteinMPNN/helper_scripts/assign_fixed_chains.py
ADDED
|
@@ -0,0 +1,39 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
import json
|
| 5 |
+
|
| 6 |
+
with open(args.input_path, 'r') as json_file:
|
| 7 |
+
json_list = list(json_file)
|
| 8 |
+
|
| 9 |
+
global_designed_chain_list = []
|
| 10 |
+
if args.chain_list != '':
|
| 11 |
+
global_designed_chain_list = [str(item) for item in args.chain_list.split()]
|
| 12 |
+
my_dict = {}
|
| 13 |
+
for json_str in json_list:
|
| 14 |
+
result = json.loads(json_str)
|
| 15 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:9]=='seq_chain'] #['A','B', 'C',...]
|
| 16 |
+
if len(global_designed_chain_list) > 0:
|
| 17 |
+
designed_chain_list = global_designed_chain_list
|
| 18 |
+
else:
|
| 19 |
+
#manually specify, e.g.
|
| 20 |
+
designed_chain_list = ["A"]
|
| 21 |
+
fixed_chain_list = [letter for letter in all_chain_list if letter not in designed_chain_list] #fix/do not redesign these chains
|
| 22 |
+
my_dict[result['name']]= (designed_chain_list, fixed_chain_list)
|
| 23 |
+
|
| 24 |
+
with open(args.output_path, 'w') as f:
|
| 25 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
if __name__ == "__main__":
|
| 29 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 30 |
+
argparser.add_argument("--input_path", type=str, help="Path to the parsed PDBs")
|
| 31 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 32 |
+
argparser.add_argument("--chain_list", type=str, default='', help="List of the chains that need to be designed")
|
| 33 |
+
|
| 34 |
+
args = argparser.parse_args()
|
| 35 |
+
main(args)
|
| 36 |
+
|
| 37 |
+
# Output looks like this:
|
| 38 |
+
# {"5TTA": [["A"], ["B"]], "3LIS": [["A"], ["B"]]}
|
| 39 |
+
|
ProteinMPNN/helper_scripts/make_bias_AA.py
ADDED
|
@@ -0,0 +1,27 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
|
| 5 |
+
import numpy as np
|
| 6 |
+
import json
|
| 7 |
+
|
| 8 |
+
bias_list = [float(item) for item in args.bias_list.split()]
|
| 9 |
+
AA_list = [str(item) for item in args.AA_list.split()]
|
| 10 |
+
|
| 11 |
+
my_dict = dict(zip(AA_list, bias_list))
|
| 12 |
+
|
| 13 |
+
with open(args.output_path, 'w') as f:
|
| 14 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
if __name__ == "__main__":
|
| 18 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 19 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 20 |
+
argparser.add_argument("--AA_list", type=str, default='', help="List of AAs to be biased")
|
| 21 |
+
argparser.add_argument("--bias_list", type=str, default='', help="AA bias strengths")
|
| 22 |
+
|
| 23 |
+
args = argparser.parse_args()
|
| 24 |
+
main(args)
|
| 25 |
+
|
| 26 |
+
#e.g. output
|
| 27 |
+
#{"A": -0.01, "G": 0.02}
|
ProteinMPNN/helper_scripts/make_bias_per_res_dict.py
ADDED
|
@@ -0,0 +1,53 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
import glob
|
| 5 |
+
import random
|
| 6 |
+
import numpy as np
|
| 7 |
+
import json
|
| 8 |
+
|
| 9 |
+
mpnn_alphabet = 'ACDEFGHIKLMNPQRSTVWYX'
|
| 10 |
+
|
| 11 |
+
mpnn_alphabet_dict = {'A': 0,'C': 1,'D': 2,'E': 3,'F': 4,'G': 5,'H': 6,'I': 7,'K': 8,'L': 9,'M': 10,'N': 11,'P': 12,'Q': 13,'R': 14,'S': 15,'T': 16,'V': 17,'W': 18,'Y': 19,'X': 20}
|
| 12 |
+
|
| 13 |
+
with open(args.input_path, 'r') as json_file:
|
| 14 |
+
json_list = list(json_file)
|
| 15 |
+
|
| 16 |
+
my_dict = {}
|
| 17 |
+
for json_str in json_list:
|
| 18 |
+
result = json.loads(json_str)
|
| 19 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:10]=='seq_chain_']
|
| 20 |
+
bias_by_res_dict = {}
|
| 21 |
+
for chain in all_chain_list:
|
| 22 |
+
chain_length = len(result[f'seq_chain_{chain}'])
|
| 23 |
+
bias_per_residue = np.zeros([chain_length, 21])
|
| 24 |
+
|
| 25 |
+
|
| 26 |
+
if chain == 'A':
|
| 27 |
+
residues = [0, 1, 2, 3, 4, 5, 11, 12, 13, 14, 15]
|
| 28 |
+
amino_acids = [5, 9] #[G, L]
|
| 29 |
+
for res in residues:
|
| 30 |
+
for aa in amino_acids:
|
| 31 |
+
bias_per_residue[res, aa] = 100.5
|
| 32 |
+
|
| 33 |
+
if chain == 'C':
|
| 34 |
+
residues = [0, 1, 2, 3, 4, 5, 11, 12, 13, 14, 15]
|
| 35 |
+
amino_acids = range(21)[1:] #[G, L]
|
| 36 |
+
for res in residues:
|
| 37 |
+
for aa in amino_acids:
|
| 38 |
+
bias_per_residue[res, aa] = -100.5
|
| 39 |
+
|
| 40 |
+
bias_by_res_dict[chain] = bias_per_residue.tolist()
|
| 41 |
+
my_dict[result['name']] = bias_by_res_dict
|
| 42 |
+
|
| 43 |
+
with open(args.output_path, 'w') as f:
|
| 44 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
if __name__ == "__main__":
|
| 48 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 49 |
+
argparser.add_argument("--input_path", type=str, help="Path to the parsed PDBs")
|
| 50 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 51 |
+
|
| 52 |
+
args = argparser.parse_args()
|
| 53 |
+
main(args)
|
ProteinMPNN/helper_scripts/make_fixed_positions_dict.py
ADDED
|
@@ -0,0 +1,59 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
import glob
|
| 5 |
+
import random
|
| 6 |
+
import numpy as np
|
| 7 |
+
import json
|
| 8 |
+
import itertools
|
| 9 |
+
|
| 10 |
+
with open(args.input_path, 'r') as json_file:
|
| 11 |
+
json_list = list(json_file)
|
| 12 |
+
|
| 13 |
+
fixed_list = [[int(item) for item in one.split()] for one in args.position_list.split(",")]
|
| 14 |
+
global_designed_chain_list = [str(item) for item in args.chain_list.split()]
|
| 15 |
+
my_dict = {}
|
| 16 |
+
|
| 17 |
+
if not args.specify_non_fixed:
|
| 18 |
+
for json_str in json_list:
|
| 19 |
+
result = json.loads(json_str)
|
| 20 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:9]=='seq_chain']
|
| 21 |
+
fixed_position_dict = {}
|
| 22 |
+
for i, chain in enumerate(global_designed_chain_list):
|
| 23 |
+
fixed_position_dict[chain] = fixed_list[i]
|
| 24 |
+
for chain in all_chain_list:
|
| 25 |
+
if chain not in global_designed_chain_list:
|
| 26 |
+
fixed_position_dict[chain] = []
|
| 27 |
+
my_dict[result['name']] = fixed_position_dict
|
| 28 |
+
else:
|
| 29 |
+
for json_str in json_list:
|
| 30 |
+
result = json.loads(json_str)
|
| 31 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:9]=='seq_chain']
|
| 32 |
+
fixed_position_dict = {}
|
| 33 |
+
for chain in all_chain_list:
|
| 34 |
+
seq_length = len(result[f'seq_chain_{chain}'])
|
| 35 |
+
all_residue_list = (np.arange(seq_length)+1).tolist()
|
| 36 |
+
if chain not in global_designed_chain_list:
|
| 37 |
+
fixed_position_dict[chain] = all_residue_list
|
| 38 |
+
else:
|
| 39 |
+
idx = np.argwhere(np.array(global_designed_chain_list) == chain)[0][0]
|
| 40 |
+
fixed_position_dict[chain] = list(set(all_residue_list)-set(fixed_list[idx]))
|
| 41 |
+
my_dict[result['name']] = fixed_position_dict
|
| 42 |
+
|
| 43 |
+
with open(args.output_path, 'w') as f:
|
| 44 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 45 |
+
|
| 46 |
+
#e.g. output
|
| 47 |
+
#{"5TTA": {"A": [1, 2, 3, 7, 8, 9, 22, 25, 33], "B": []}, "3LIS": {"A": [], "B": []}}
|
| 48 |
+
|
| 49 |
+
if __name__ == "__main__":
|
| 50 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 51 |
+
argparser.add_argument("--input_path", type=str, help="Path to the parsed PDBs")
|
| 52 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 53 |
+
argparser.add_argument("--chain_list", type=str, default='', help="List of the chains that need to be fixed")
|
| 54 |
+
argparser.add_argument("--position_list", type=str, default='', help="Position lists, e.g. 11 12 14 18, 1 2 3 4 for first chain and the second chain")
|
| 55 |
+
argparser.add_argument("--specify_non_fixed", action="store_true", default=False, help="Allows specifying just residues that need to be designed (default: false)")
|
| 56 |
+
|
| 57 |
+
args = argparser.parse_args()
|
| 58 |
+
main(args)
|
| 59 |
+
|
ProteinMPNN/helper_scripts/make_pos_neg_tied_positions_dict.py
ADDED
|
@@ -0,0 +1,73 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
|
| 5 |
+
import glob
|
| 6 |
+
import random
|
| 7 |
+
import numpy as np
|
| 8 |
+
import json
|
| 9 |
+
import itertools
|
| 10 |
+
|
| 11 |
+
with open(args.input_path, 'r') as json_file:
|
| 12 |
+
json_list = list(json_file)
|
| 13 |
+
|
| 14 |
+
homooligomeric_state = args.homooligomer
|
| 15 |
+
|
| 16 |
+
if homooligomeric_state == 0:
|
| 17 |
+
tied_list = [[int(item) for item in one.split()] for one in args.position_list.split(",")]
|
| 18 |
+
global_designed_chain_list = [str(item) for item in args.chain_list.split()]
|
| 19 |
+
my_dict = {}
|
| 20 |
+
for json_str in json_list:
|
| 21 |
+
result = json.loads(json_str)
|
| 22 |
+
all_chain_list = sorted([item[-1:] for item in list(result) if item[:9]=='seq_chain']) #A, B, C, ...
|
| 23 |
+
tied_positions_list = []
|
| 24 |
+
for i, pos in enumerate(tied_list[0]):
|
| 25 |
+
temp_dict = {}
|
| 26 |
+
for j, chain in enumerate(global_designed_chain_list):
|
| 27 |
+
temp_dict[chain] = [tied_list[j][i]] #needs to be a list
|
| 28 |
+
tied_positions_list.append(temp_dict)
|
| 29 |
+
my_dict[result['name']] = tied_positions_list
|
| 30 |
+
else:
|
| 31 |
+
if args.pos_neg_chain_list:
|
| 32 |
+
chain_list_input = [[str(item) for item in one.split()] for one in args.pos_neg_chain_list.split(",")]
|
| 33 |
+
chain_betas_input = [[float(item) for item in one.split()] for one in args.pos_neg_chain_betas.split(",")]
|
| 34 |
+
chain_list_flat = [item for sublist in chain_list_input for item in sublist]
|
| 35 |
+
chain_betas_flat = [item for sublist in chain_betas_input for item in sublist]
|
| 36 |
+
chain_betas_dict = dict(zip(chain_list_flat, chain_betas_flat))
|
| 37 |
+
my_dict = {}
|
| 38 |
+
for json_str in json_list:
|
| 39 |
+
result = json.loads(json_str)
|
| 40 |
+
all_chain_list = sorted([item[-1:] for item in list(result) if item[:9]=='seq_chain']) #A, B, C, ...
|
| 41 |
+
tied_positions_list = []
|
| 42 |
+
chain_length = len(result[f"seq_chain_{all_chain_list[0]}"])
|
| 43 |
+
for chains in chain_list_input:
|
| 44 |
+
for i in range(1,chain_length+1):
|
| 45 |
+
temp_dict = {}
|
| 46 |
+
for j, chain in enumerate(chains):
|
| 47 |
+
if args.pos_neg_chain_list and chain in chain_list_flat:
|
| 48 |
+
temp_dict[chain] = [[i], [chain_betas_dict[chain]]]
|
| 49 |
+
else:
|
| 50 |
+
temp_dict[chain] = [[i], [1.0]] #first list is for residue numbers, second list is for weights for the energy, +ive and -ive design
|
| 51 |
+
tied_positions_list.append(temp_dict)
|
| 52 |
+
my_dict[result['name']] = tied_positions_list
|
| 53 |
+
|
| 54 |
+
with open(args.output_path, 'w') as f:
|
| 55 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 56 |
+
|
| 57 |
+
if __name__ == "__main__":
|
| 58 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 59 |
+
argparser.add_argument("--input_path", type=str, help="Path to the parsed PDBs")
|
| 60 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 61 |
+
argparser.add_argument("--chain_list", type=str, default='', help="List of the chains that need to be fixed")
|
| 62 |
+
argparser.add_argument("--position_list", type=str, default='', help="Position lists, e.g. 11 12 14 18, 1 2 3 4 for first chain and the second chain")
|
| 63 |
+
argparser.add_argument("--homooligomer", type=int, default=0, help="If 0 do not use, if 1 then design homooligomer")
|
| 64 |
+
argparser.add_argument("--pos_neg_chain_list", type=str, default='', help="Chain lists to be tied together")
|
| 65 |
+
argparser.add_argument("--pos_neg_chain_betas", type=str, default='', help="Chain beta list for the chain lists provided; 1.0 for the positive design, -0.1 or -0.5 for negative, 0.0 means do not use that chain info")
|
| 66 |
+
|
| 67 |
+
args = argparser.parse_args()
|
| 68 |
+
main(args)
|
| 69 |
+
|
| 70 |
+
|
| 71 |
+
#e.g. output
|
| 72 |
+
#{"5TTA": [], "3LIS": [{"A": [1], "B": [1]}, {"A": [2], "B": [2]}, {"A": [3], "B": [3]}, {"A": [4], "B": [4]}, {"A": [5], "B": [5]}, {"A": [6], "B": [6]}, {"A": [7], "B": [7]}, {"A": [8], "B": [8]}, {"A": [9], "B": [9]}, {"A": [10], "B": [10]}, {"A": [11], "B": [11]}, {"A": [12], "B": [12]}, {"A": [13], "B": [13]}, {"A": [14], "B": [14]}, {"A": [15], "B": [15]}, {"A": [16], "B": [16]}, {"A": [17], "B": [17]}, {"A": [18], "B": [18]}, {"A": [19], "B": [19]}, {"A": [20], "B": [20]}, {"A": [21], "B": [21]}, {"A": [22], "B": [22]}, {"A": [23], "B": [23]}, {"A": [24], "B": [24]}, {"A": [25], "B": [25]}, {"A": [26], "B": [26]}, {"A": [27], "B": [27]}, {"A": [28], "B": [28]}, {"A": [29], "B": [29]}, {"A": [30], "B": [30]}, {"A": [31], "B": [31]}, {"A": [32], "B": [32]}, {"A": [33], "B": [33]}, {"A": [34], "B": [34]}, {"A": [35], "B": [35]}, {"A": [36], "B": [36]}, {"A": [37], "B": [37]}, {"A": [38], "B": [38]}, {"A": [39], "B": [39]}, {"A": [40], "B": [40]}, {"A": [41], "B": [41]}, {"A": [42], "B": [42]}, {"A": [43], "B": [43]}, {"A": [44], "B": [44]}, {"A": [45], "B": [45]}, {"A": [46], "B": [46]}, {"A": [47], "B": [47]}, {"A": [48], "B": [48]}, {"A": [49], "B": [49]}, {"A": [50], "B": [50]}, {"A": [51], "B": [51]}, {"A": [52], "B": [52]}, {"A": [53], "B": [53]}, {"A": [54], "B": [54]}, {"A": [55], "B": [55]}, {"A": [56], "B": [56]}, {"A": [57], "B": [57]}, {"A": [58], "B": [58]}, {"A": [59], "B": [59]}, {"A": [60], "B": [60]}, {"A": [61], "B": [61]}, {"A": [62], "B": [62]}, {"A": [63], "B": [63]}, {"A": [64], "B": [64]}, {"A": [65], "B": [65]}, {"A": [66], "B": [66]}, {"A": [67], "B": [67]}, {"A": [68], "B": [68]}, {"A": [69], "B": [69]}, {"A": [70], "B": [70]}, {"A": [71], "B": [71]}, {"A": [72], "B": [72]}, {"A": [73], "B": [73]}, {"A": [74], "B": [74]}, {"A": [75], "B": [75]}, {"A": [76], "B": [76]}, {"A": [77], "B": [77]}, {"A": [78], "B": [78]}, {"A": [79], "B": [79]}, {"A": [80], "B": [80]}, {"A": [81], "B": [81]}, {"A": [82], "B": [82]}, {"A": [83], "B": [83]}, {"A": [84], "B": [84]}, {"A": [85], "B": [85]}, {"A": [86], "B": [86]}, {"A": [87], "B": [87]}, {"A": [88], "B": [88]}, {"A": [89], "B": [89]}, {"A": [90], "B": [90]}, {"A": [91], "B": [91]}, {"A": [92], "B": [92]}, {"A": [93], "B": [93]}, {"A": [94], "B": [94]}, {"A": [95], "B": [95]}, {"A": [96], "B": [96]}]}
|
| 73 |
+
|
ProteinMPNN/helper_scripts/make_pssm_input_dict.py
ADDED
|
@@ -0,0 +1,36 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
import json
|
| 5 |
+
import numpy as np
|
| 6 |
+
with open(args.jsonl_input_path, 'r') as json_file:
|
| 7 |
+
json_list = list(json_file)
|
| 8 |
+
|
| 9 |
+
my_dict = {}
|
| 10 |
+
for json_str in json_list:
|
| 11 |
+
result = json.loads(json_str)
|
| 12 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:9]=='seq_chain']
|
| 13 |
+
path_to_PSSM = args.PSSM_input_path+"/"+result['name'] + ".npz"
|
| 14 |
+
print(path_to_PSSM)
|
| 15 |
+
pssm_input = np.load(path_to_PSSM)
|
| 16 |
+
pssm_dict = {}
|
| 17 |
+
for chain in all_chain_list:
|
| 18 |
+
pssm_dict[chain] = {}
|
| 19 |
+
pssm_dict[chain]['pssm_coef'] = pssm_input[chain+'_coef'].tolist() #[L] per position coefficient to trust PSSM; 0.0 - do not use it; 1.0 - just use PSSM only
|
| 20 |
+
pssm_dict[chain]['pssm_bias'] = pssm_input[chain+'_bias'].tolist() #[L,21] probability (sums up to 1.0 over alphabet of size 21) from PSSM
|
| 21 |
+
pssm_dict[chain]['pssm_log_odds'] = pssm_input[chain+'_odds'].tolist() #[L,21] log_odds ratios coming from PSSM; optional/not needed
|
| 22 |
+
my_dict[result['name']] = pssm_dict
|
| 23 |
+
|
| 24 |
+
#Write output to:
|
| 25 |
+
with open(args.output_path, 'w') as f:
|
| 26 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 27 |
+
|
| 28 |
+
if __name__ == "__main__":
|
| 29 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 30 |
+
|
| 31 |
+
argparser.add_argument("--PSSM_input_path", type=str, help="Path to PSSMs saved as npz files.")
|
| 32 |
+
argparser.add_argument("--jsonl_input_path", type=str, help="Path where to load .jsonl dictionary of parsed pdbs.")
|
| 33 |
+
argparser.add_argument("--output_path", type=str, help="Path where to save .jsonl dictionary with PSSM bias.")
|
| 34 |
+
|
| 35 |
+
args = argparser.parse_args()
|
| 36 |
+
main(args)
|
ProteinMPNN/helper_scripts/make_tied_positions_dict.py
ADDED
|
@@ -0,0 +1,61 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
|
| 5 |
+
import glob
|
| 6 |
+
import random
|
| 7 |
+
import numpy as np
|
| 8 |
+
import json
|
| 9 |
+
import itertools
|
| 10 |
+
|
| 11 |
+
with open(args.input_path, 'r') as json_file:
|
| 12 |
+
json_list = list(json_file)
|
| 13 |
+
|
| 14 |
+
homooligomeric_state = args.homooligomer
|
| 15 |
+
|
| 16 |
+
if homooligomeric_state == 0:
|
| 17 |
+
tied_list = [[int(item) for item in one.split()] for one in args.position_list.split(",")]
|
| 18 |
+
global_designed_chain_list = [str(item) for item in args.chain_list.split()]
|
| 19 |
+
my_dict = {}
|
| 20 |
+
for json_str in json_list:
|
| 21 |
+
result = json.loads(json_str)
|
| 22 |
+
all_chain_list = sorted([item[-1:] for item in list(result) if item[:9]=='seq_chain']) #A, B, C, ...
|
| 23 |
+
tied_positions_list = []
|
| 24 |
+
for i, pos in enumerate(tied_list[0]):
|
| 25 |
+
temp_dict = {}
|
| 26 |
+
for j, chain in enumerate(global_designed_chain_list):
|
| 27 |
+
temp_dict[chain] = [tied_list[j][i]] #needs to be a list
|
| 28 |
+
tied_positions_list.append(temp_dict)
|
| 29 |
+
my_dict[result['name']] = tied_positions_list
|
| 30 |
+
else:
|
| 31 |
+
my_dict = {}
|
| 32 |
+
for json_str in json_list:
|
| 33 |
+
result = json.loads(json_str)
|
| 34 |
+
all_chain_list = sorted([item[-1:] for item in list(result) if item[:9]=='seq_chain']) #A, B, C, ...
|
| 35 |
+
tied_positions_list = []
|
| 36 |
+
chain_length = len(result[f"seq_chain_{all_chain_list[0]}"])
|
| 37 |
+
for i in range(1,chain_length+1):
|
| 38 |
+
temp_dict = {}
|
| 39 |
+
for j, chain in enumerate(all_chain_list):
|
| 40 |
+
temp_dict[chain] = [i] #needs to be a list
|
| 41 |
+
tied_positions_list.append(temp_dict)
|
| 42 |
+
my_dict[result['name']] = tied_positions_list
|
| 43 |
+
|
| 44 |
+
with open(args.output_path, 'w') as f:
|
| 45 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 46 |
+
|
| 47 |
+
if __name__ == "__main__":
|
| 48 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 49 |
+
argparser.add_argument("--input_path", type=str, help="Path to the parsed PDBs")
|
| 50 |
+
argparser.add_argument("--output_path", type=str, help="Path to the output dictionary")
|
| 51 |
+
argparser.add_argument("--chain_list", type=str, default='', help="List of the chains that need to be fixed")
|
| 52 |
+
argparser.add_argument("--position_list", type=str, default='', help="Position lists, e.g. 11 12 14 18, 1 2 3 4 for first chain and the second chain")
|
| 53 |
+
argparser.add_argument("--homooligomer", type=int, default=0, help="If 0 do not use, if 1 then design homooligomer")
|
| 54 |
+
|
| 55 |
+
args = argparser.parse_args()
|
| 56 |
+
main(args)
|
| 57 |
+
|
| 58 |
+
|
| 59 |
+
#e.g. output
|
| 60 |
+
#{"5TTA": [], "3LIS": [{"A": [1], "B": [1]}, {"A": [2], "B": [2]}, {"A": [3], "B": [3]}, {"A": [4], "B": [4]}, {"A": [5], "B": [5]}, {"A": [6], "B": [6]}, {"A": [7], "B": [7]}, {"A": [8], "B": [8]}, {"A": [9], "B": [9]}, {"A": [10], "B": [10]}, {"A": [11], "B": [11]}, {"A": [12], "B": [12]}, {"A": [13], "B": [13]}, {"A": [14], "B": [14]}, {"A": [15], "B": [15]}, {"A": [16], "B": [16]}, {"A": [17], "B": [17]}, {"A": [18], "B": [18]}, {"A": [19], "B": [19]}, {"A": [20], "B": [20]}, {"A": [21], "B": [21]}, {"A": [22], "B": [22]}, {"A": [23], "B": [23]}, {"A": [24], "B": [24]}, {"A": [25], "B": [25]}, {"A": [26], "B": [26]}, {"A": [27], "B": [27]}, {"A": [28], "B": [28]}, {"A": [29], "B": [29]}, {"A": [30], "B": [30]}, {"A": [31], "B": [31]}, {"A": [32], "B": [32]}, {"A": [33], "B": [33]}, {"A": [34], "B": [34]}, {"A": [35], "B": [35]}, {"A": [36], "B": [36]}, {"A": [37], "B": [37]}, {"A": [38], "B": [38]}, {"A": [39], "B": [39]}, {"A": [40], "B": [40]}, {"A": [41], "B": [41]}, {"A": [42], "B": [42]}, {"A": [43], "B": [43]}, {"A": [44], "B": [44]}, {"A": [45], "B": [45]}, {"A": [46], "B": [46]}, {"A": [47], "B": [47]}, {"A": [48], "B": [48]}, {"A": [49], "B": [49]}, {"A": [50], "B": [50]}, {"A": [51], "B": [51]}, {"A": [52], "B": [52]}, {"A": [53], "B": [53]}, {"A": [54], "B": [54]}, {"A": [55], "B": [55]}, {"A": [56], "B": [56]}, {"A": [57], "B": [57]}, {"A": [58], "B": [58]}, {"A": [59], "B": [59]}, {"A": [60], "B": [60]}, {"A": [61], "B": [61]}, {"A": [62], "B": [62]}, {"A": [63], "B": [63]}, {"A": [64], "B": [64]}, {"A": [65], "B": [65]}, {"A": [66], "B": [66]}, {"A": [67], "B": [67]}, {"A": [68], "B": [68]}, {"A": [69], "B": [69]}, {"A": [70], "B": [70]}, {"A": [71], "B": [71]}, {"A": [72], "B": [72]}, {"A": [73], "B": [73]}, {"A": [74], "B": [74]}, {"A": [75], "B": [75]}, {"A": [76], "B": [76]}, {"A": [77], "B": [77]}, {"A": [78], "B": [78]}, {"A": [79], "B": [79]}, {"A": [80], "B": [80]}, {"A": [81], "B": [81]}, {"A": [82], "B": [82]}, {"A": [83], "B": [83]}, {"A": [84], "B": [84]}, {"A": [85], "B": [85]}, {"A": [86], "B": [86]}, {"A": [87], "B": [87]}, {"A": [88], "B": [88]}, {"A": [89], "B": [89]}, {"A": [90], "B": [90]}, {"A": [91], "B": [91]}, {"A": [92], "B": [92]}, {"A": [93], "B": [93]}, {"A": [94], "B": [94]}, {"A": [95], "B": [95]}, {"A": [96], "B": [96]}]}
|
| 61 |
+
|
ProteinMPNN/helper_scripts/other_tools/make_omit_AA.py
ADDED
|
@@ -0,0 +1,39 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import glob
|
| 2 |
+
import random
|
| 3 |
+
import numpy as np
|
| 4 |
+
import json
|
| 5 |
+
import itertools
|
| 6 |
+
|
| 7 |
+
#MODIFY this path
|
| 8 |
+
with open('/home/justas/projects/lab_github/mpnn/data/pdbs.jsonl', 'r') as json_file:
|
| 9 |
+
json_list = list(json_file)
|
| 10 |
+
|
| 11 |
+
my_dict = {}
|
| 12 |
+
for json_str in json_list:
|
| 13 |
+
result = json.loads(json_str)
|
| 14 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:9]=='seq_chain']
|
| 15 |
+
fixed_position_dict = {}
|
| 16 |
+
print(result['name'])
|
| 17 |
+
if result['name'] == '5TTA':
|
| 18 |
+
for chain in all_chain_list:
|
| 19 |
+
if chain == 'A':
|
| 20 |
+
fixed_position_dict[chain] = [
|
| 21 |
+
[[int(item) for item in list(itertools.chain(list(np.arange(1,4)), list(np.arange(7,10)), [22, 25, 33]))], 'GPL'],
|
| 22 |
+
[[int(item) for item in list(itertools.chain([40, 41, 42, 43]))], 'WC'],
|
| 23 |
+
[[int(item) for item in list(itertools.chain(list(np.arange(50,150))))], 'ACEFGHIKLMNRSTVWYX'],
|
| 24 |
+
[[int(item) for item in list(itertools.chain(list(np.arange(160,200))))], 'FGHIKLPQDMNRSTVWYX']]
|
| 25 |
+
else:
|
| 26 |
+
fixed_position_dict[chain] = []
|
| 27 |
+
else:
|
| 28 |
+
for chain in all_chain_list:
|
| 29 |
+
fixed_position_dict[chain] = []
|
| 30 |
+
my_dict[result['name']] = fixed_position_dict
|
| 31 |
+
|
| 32 |
+
#MODIFY this path
|
| 33 |
+
with open('/home/justas/projects/lab_github/mpnn/data/omit_AA.jsonl', 'w') as f:
|
| 34 |
+
f.write(json.dumps(my_dict) + '\n')
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
print('Finished')
|
| 38 |
+
#e.g. output
|
| 39 |
+
#{"5TTA": {"A": [[[1, 2, 3, 7, 8, 9, 22, 25, 33], "GPL"], [[40, 41, 42, 43], "WC"], [[50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149], "ACEFGHIKLMNRSTVWYX"], [[160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199], "FGHIKLPQDMNRSTVWYX"]], "B": []}, "3LIS": {"A": [], "B": []}}
|
ProteinMPNN/helper_scripts/other_tools/make_pssm_dict.py
ADDED
|
@@ -0,0 +1,64 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pandas as pd
|
| 2 |
+
import numpy as np
|
| 3 |
+
|
| 4 |
+
import glob
|
| 5 |
+
import random
|
| 6 |
+
import numpy as np
|
| 7 |
+
import json
|
| 8 |
+
|
| 9 |
+
|
| 10 |
+
def softmax(x, T):
|
| 11 |
+
return np.exp(x/T)/np.sum(np.exp(x/T), -1, keepdims=True)
|
| 12 |
+
|
| 13 |
+
def parse_pssm(path):
|
| 14 |
+
data = pd.read_csv(path, skiprows=2)
|
| 15 |
+
floats_list_list = []
|
| 16 |
+
for i in range(data.values.shape[0]):
|
| 17 |
+
str1 = data.values[i][0][4:]
|
| 18 |
+
floats_list = []
|
| 19 |
+
for item in str1.split():
|
| 20 |
+
floats_list.append(float(item))
|
| 21 |
+
floats_list_list.append(floats_list)
|
| 22 |
+
np_lines = np.array(floats_list_list)
|
| 23 |
+
return np_lines
|
| 24 |
+
|
| 25 |
+
np_lines = parse_pssm('/home/swang523/RLcage/capsid/monomersfordesign/8-16-21/pssm_rainity_final_8-16-21_int/build_0.2089_0.98_0.4653_19_2.00_0.005745.pssm')
|
| 26 |
+
|
| 27 |
+
mpnn_alphabet = 'ACDEFGHIKLMNPQRSTVWYX'
|
| 28 |
+
input_alphabet = 'ARNDCQEGHILKMFPSTWYV'
|
| 29 |
+
|
| 30 |
+
permutation_matrix = np.zeros([20,21])
|
| 31 |
+
for i in range(20):
|
| 32 |
+
letter1 = input_alphabet[i]
|
| 33 |
+
for j in range(21):
|
| 34 |
+
letter2 = mpnn_alphabet[j]
|
| 35 |
+
if letter1 == letter2:
|
| 36 |
+
permutation_matrix[i,j]=1.
|
| 37 |
+
|
| 38 |
+
pssm_log_odds = np_lines[:,:20] @ permutation_matrix
|
| 39 |
+
pssm_probs = np_lines[:,20:40] @ permutation_matrix
|
| 40 |
+
|
| 41 |
+
X_mask = np.concatenate([np.zeros([1,20]), np.ones([1,1])], -1)
|
| 42 |
+
|
| 43 |
+
def softmax(x, T):
|
| 44 |
+
return np.exp(x/T)/np.sum(np.exp(x/T), -1, keepdims=True)
|
| 45 |
+
|
| 46 |
+
#Load parsed PDBs:
|
| 47 |
+
with open('/home/justas/projects/cages/parsed/test.jsonl', 'r') as json_file:
|
| 48 |
+
json_list = list(json_file)
|
| 49 |
+
|
| 50 |
+
my_dict = {}
|
| 51 |
+
for json_str in json_list:
|
| 52 |
+
result = json.loads(json_str)
|
| 53 |
+
all_chain_list = [item[-1:] for item in list(result) if item[:9]=='seq_chain']
|
| 54 |
+
pssm_dict = {}
|
| 55 |
+
for chain in all_chain_list:
|
| 56 |
+
pssm_dict[chain] = {}
|
| 57 |
+
pssm_dict[chain]['pssm_coef'] = (np.ones(len(result['seq_chain_A']))).tolist() #a number between 0.0 and 1.0 specifying how much attention put to PSSM, can be adjusted later as a flag
|
| 58 |
+
pssm_dict[chain]['pssm_bias'] = (softmax(pssm_log_odds-X_mask*1e8, 1.0)).tolist() #PSSM like, [length, 21] such that sum over the last dimension adds up to 1.0
|
| 59 |
+
pssm_dict[chain]['pssm_log_odds'] = (pssm_log_odds).tolist()
|
| 60 |
+
my_dict[result['name']] = pssm_dict
|
| 61 |
+
|
| 62 |
+
#Write output to:
|
| 63 |
+
with open('/home/justas/projects/lab_github/mpnn/data/pssm_dict.jsonl', 'w') as f:
|
| 64 |
+
f.write(json.dumps(my_dict) + '\n')
|
ProteinMPNN/helper_scripts/parse_multiple_chains.out
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
Successfully finished: 2 pdbs
|
ProteinMPNN/helper_scripts/parse_multiple_chains.py
ADDED
|
@@ -0,0 +1,163 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
|
| 3 |
+
def main(args):
|
| 4 |
+
|
| 5 |
+
import numpy as np
|
| 6 |
+
import os, time, gzip, json
|
| 7 |
+
import glob
|
| 8 |
+
|
| 9 |
+
folder_with_pdbs_path = args.input_path
|
| 10 |
+
save_path = args.output_path
|
| 11 |
+
ca_only = args.ca_only
|
| 12 |
+
|
| 13 |
+
alpha_1 = list("ARNDCQEGHILKMFPSTWYV-")
|
| 14 |
+
states = len(alpha_1)
|
| 15 |
+
alpha_3 = ['ALA','ARG','ASN','ASP','CYS','GLN','GLU','GLY','HIS','ILE',
|
| 16 |
+
'LEU','LYS','MET','PHE','PRO','SER','THR','TRP','TYR','VAL','GAP']
|
| 17 |
+
|
| 18 |
+
aa_1_N = {a:n for n,a in enumerate(alpha_1)}
|
| 19 |
+
aa_3_N = {a:n for n,a in enumerate(alpha_3)}
|
| 20 |
+
aa_N_1 = {n:a for n,a in enumerate(alpha_1)}
|
| 21 |
+
aa_1_3 = {a:b for a,b in zip(alpha_1,alpha_3)}
|
| 22 |
+
aa_3_1 = {b:a for a,b in zip(alpha_1,alpha_3)}
|
| 23 |
+
|
| 24 |
+
def AA_to_N(x):
|
| 25 |
+
# ["ARND"] -> [[0,1,2,3]]
|
| 26 |
+
x = np.array(x);
|
| 27 |
+
if x.ndim == 0: x = x[None]
|
| 28 |
+
return [[aa_1_N.get(a, states-1) for a in y] for y in x]
|
| 29 |
+
|
| 30 |
+
def N_to_AA(x):
|
| 31 |
+
# [[0,1,2,3]] -> ["ARND"]
|
| 32 |
+
x = np.array(x);
|
| 33 |
+
if x.ndim == 1: x = x[None]
|
| 34 |
+
return ["".join([aa_N_1.get(a,"-") for a in y]) for y in x]
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
def parse_PDB_biounits(x, atoms=['N','CA','C'], chain=None):
|
| 38 |
+
'''
|
| 39 |
+
input: x = PDB filename
|
| 40 |
+
atoms = atoms to extract (optional)
|
| 41 |
+
output: (length, atoms, coords=(x,y,z)), sequence
|
| 42 |
+
'''
|
| 43 |
+
xyz,seq,min_resn,max_resn = {},{},1e6,-1e6
|
| 44 |
+
for line in open(x,"rb"):
|
| 45 |
+
line = line.decode("utf-8","ignore").rstrip()
|
| 46 |
+
|
| 47 |
+
if line[:6] == "HETATM" and line[17:17+3] == "MSE":
|
| 48 |
+
line = line.replace("HETATM","ATOM ")
|
| 49 |
+
line = line.replace("MSE","MET")
|
| 50 |
+
|
| 51 |
+
if line[:4] == "ATOM":
|
| 52 |
+
ch = line[21:22]
|
| 53 |
+
if ch == chain or chain is None:
|
| 54 |
+
atom = line[12:12+4].strip()
|
| 55 |
+
resi = line[17:17+3]
|
| 56 |
+
resn = line[22:22+5].strip()
|
| 57 |
+
x,y,z = [float(line[i:(i+8)]) for i in [30,38,46]]
|
| 58 |
+
|
| 59 |
+
if resn[-1].isalpha():
|
| 60 |
+
resa,resn = resn[-1],int(resn[:-1])-1
|
| 61 |
+
else:
|
| 62 |
+
resa,resn = "",int(resn)-1
|
| 63 |
+
# resn = int(resn)
|
| 64 |
+
if resn < min_resn:
|
| 65 |
+
min_resn = resn
|
| 66 |
+
if resn > max_resn:
|
| 67 |
+
max_resn = resn
|
| 68 |
+
if resn not in xyz:
|
| 69 |
+
xyz[resn] = {}
|
| 70 |
+
if resa not in xyz[resn]:
|
| 71 |
+
xyz[resn][resa] = {}
|
| 72 |
+
if resn not in seq:
|
| 73 |
+
seq[resn] = {}
|
| 74 |
+
if resa not in seq[resn]:
|
| 75 |
+
seq[resn][resa] = resi
|
| 76 |
+
|
| 77 |
+
if atom not in xyz[resn][resa]:
|
| 78 |
+
xyz[resn][resa][atom] = np.array([x,y,z])
|
| 79 |
+
|
| 80 |
+
# convert to numpy arrays, fill in missing values
|
| 81 |
+
seq_,xyz_ = [],[]
|
| 82 |
+
try:
|
| 83 |
+
for resn in range(min_resn,max_resn+1):
|
| 84 |
+
if resn in seq:
|
| 85 |
+
for k in sorted(seq[resn]): seq_.append(aa_3_N.get(seq[resn][k],20))
|
| 86 |
+
else: seq_.append(20)
|
| 87 |
+
if resn in xyz:
|
| 88 |
+
for k in sorted(xyz[resn]):
|
| 89 |
+
for atom in atoms:
|
| 90 |
+
if atom in xyz[resn][k]: xyz_.append(xyz[resn][k][atom])
|
| 91 |
+
else: xyz_.append(np.full(3,np.nan))
|
| 92 |
+
else:
|
| 93 |
+
for atom in atoms: xyz_.append(np.full(3,np.nan))
|
| 94 |
+
return np.array(xyz_).reshape(-1,len(atoms),3), N_to_AA(np.array(seq_))
|
| 95 |
+
except TypeError:
|
| 96 |
+
return 'no_chain', 'no_chain'
|
| 97 |
+
|
| 98 |
+
|
| 99 |
+
|
| 100 |
+
pdb_dict_list = []
|
| 101 |
+
c = 0
|
| 102 |
+
|
| 103 |
+
if folder_with_pdbs_path[-1]!='/':
|
| 104 |
+
folder_with_pdbs_path = folder_with_pdbs_path+'/'
|
| 105 |
+
|
| 106 |
+
|
| 107 |
+
init_alphabet = ['A', 'B', 'C', 'D', 'E', 'F', 'G','H', 'I', 'J','K', 'L', 'M', 'N', 'O', 'P', 'Q', 'R', 'S', 'T','U', 'V','W','X', 'Y', 'Z', 'a', 'b', 'c', 'd', 'e', 'f', 'g','h', 'i', 'j','k', 'l', 'm', 'n', 'o', 'p', 'q', 'r', 's', 't','u', 'v','w','x', 'y', 'z']
|
| 108 |
+
extra_alphabet = [str(item) for item in list(np.arange(300))]
|
| 109 |
+
chain_alphabet = init_alphabet + extra_alphabet
|
| 110 |
+
|
| 111 |
+
biounit_names = glob.glob(folder_with_pdbs_path+'*.pdb')
|
| 112 |
+
for biounit in biounit_names:
|
| 113 |
+
my_dict = {}
|
| 114 |
+
s = 0
|
| 115 |
+
concat_seq = ''
|
| 116 |
+
concat_N = []
|
| 117 |
+
concat_CA = []
|
| 118 |
+
concat_C = []
|
| 119 |
+
concat_O = []
|
| 120 |
+
concat_mask = []
|
| 121 |
+
coords_dict = {}
|
| 122 |
+
for letter in chain_alphabet:
|
| 123 |
+
if ca_only:
|
| 124 |
+
sidechain_atoms = ['CA']
|
| 125 |
+
else:
|
| 126 |
+
sidechain_atoms = ['N', 'CA', 'C', 'O']
|
| 127 |
+
xyz, seq = parse_PDB_biounits(biounit, atoms=sidechain_atoms, chain=letter)
|
| 128 |
+
if type(xyz) != str:
|
| 129 |
+
concat_seq += seq[0]
|
| 130 |
+
my_dict['seq_chain_'+letter]=seq[0]
|
| 131 |
+
coords_dict_chain = {}
|
| 132 |
+
if ca_only:
|
| 133 |
+
coords_dict_chain['CA_chain_'+letter]=xyz.tolist()
|
| 134 |
+
else:
|
| 135 |
+
coords_dict_chain['N_chain_' + letter] = xyz[:, 0, :].tolist()
|
| 136 |
+
coords_dict_chain['CA_chain_' + letter] = xyz[:, 1, :].tolist()
|
| 137 |
+
coords_dict_chain['C_chain_' + letter] = xyz[:, 2, :].tolist()
|
| 138 |
+
coords_dict_chain['O_chain_' + letter] = xyz[:, 3, :].tolist()
|
| 139 |
+
my_dict['coords_chain_'+letter]=coords_dict_chain
|
| 140 |
+
s += 1
|
| 141 |
+
fi = biounit.rfind("/")
|
| 142 |
+
my_dict['name']=biounit[(fi+1):-4]
|
| 143 |
+
my_dict['num_of_chains'] = s
|
| 144 |
+
my_dict['seq'] = concat_seq
|
| 145 |
+
if s < len(chain_alphabet):
|
| 146 |
+
pdb_dict_list.append(my_dict)
|
| 147 |
+
c+=1
|
| 148 |
+
|
| 149 |
+
|
| 150 |
+
with open(save_path, 'w') as f:
|
| 151 |
+
for entry in pdb_dict_list:
|
| 152 |
+
f.write(json.dumps(entry) + '\n')
|
| 153 |
+
|
| 154 |
+
|
| 155 |
+
if __name__ == "__main__":
|
| 156 |
+
argparser = argparse.ArgumentParser(formatter_class=argparse.ArgumentDefaultsHelpFormatter)
|
| 157 |
+
|
| 158 |
+
argparser.add_argument("--input_path", type=str, help="Path to a folder with pdb files, e.g. /home/my_pdbs/")
|
| 159 |
+
argparser.add_argument("--output_path", type=str, help="Path where to save .jsonl dictionary of parsed pdbs")
|
| 160 |
+
argparser.add_argument("--ca_only", action="store_true", default=False, help="parse a backbone-only structure (default: false)")
|
| 161 |
+
|
| 162 |
+
args = argparser.parse_args()
|
| 163 |
+
main(args)
|
ProteinMPNN/helper_scripts/parse_multiple_chains.sh
ADDED
|
@@ -0,0 +1,7 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/bin/bash
|
| 2 |
+
#SBATCH --mem=32g
|
| 3 |
+
#SBATCH -c 2
|
| 4 |
+
#SBATCH --output=parse_multiple_chains.out
|
| 5 |
+
|
| 6 |
+
source activate mlfold
|
| 7 |
+
python parse_multiple_chains.py --input_path='../PDB_complexes/pdbs/' --output_path='../PDB_complexes/parsed_pdbs.jsonl'
|
ProteinMPNN/inputs/PDB_complexes/pdbs/3HTN.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/inputs/PDB_complexes/pdbs/4YOW.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/inputs/PDB_homooligomers/pdbs/4GYT.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/inputs/PDB_homooligomers/pdbs/6EHB.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/inputs/PDB_monomers/pdbs/5L33.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/inputs/PDB_monomers/pdbs/6MRR.pdb
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/inputs/PSSM_inputs/3HTN.npz
ADDED
|
Binary file (148 kB). View file
|
|
|
ProteinMPNN/inputs/PSSM_inputs/4YOW.npz
ADDED
|
Binary file (240 kB). View file
|
|
|
ProteinMPNN/outputs/example_1_outputs/parsed_pdbs.jsonl
ADDED
|
@@ -0,0 +1,2 @@
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{"seq_chain_A": "GWSTELEKHREELKEFLKKEGITNVEIRIDNGRLEVRVEGGTERLKRFLEELRQKLEKKGYTVDIKIE", "coords_chain_A": {"N_chain_A": [[-15.113, 4.641, 12.533], [-13.275, 3.42, 10.93], [-10.741, 1.675, 9.445], [-7.432, 1.448, 9.871], [-5.644, -0.548, 8.854], [-7.205, -1.96, 6.899], [-7.793, 0.183, 5.237], [-5.26, 0.685, 4.162], [-4.958, -1.588, 2.516], [-7.177, -1.19, 0.885], [-6.291, 1.085, -0.528], [-4.142, -0.079, -1.945], [-5.518, -1.938, -3.517], [-7.137, -0.17, -5.019], [-5.151, 1.232, -6.404], [-4.087, -0.835, -8.02], [-6.444, -1.411, -9.542], [-6.711, 1.026, -10.874], [-4.295, 1.105, -12.41], [-4.894, -1.039, -14.08], [-7.091, -0.121, -15.482], [-9.668, -0.781, -14.596], [-12.955, -0.348, -13.741], [-15.301, -1.272, -12.622], [-15.546, -2.334, -10.044], [-16.758, -2.673, -6.77], [-16.165, -1.622, -3.469], [-16.378, -2.183, -0.036], [-15.967, -1.433, 3.409], [-16.551, -2.631, 6.58], [-16.843, -2.78, 9.914], [-14.464, -4.361, 10.3], [-14.612, -6.107, 8.198], [-13.921, -6.735, 4.835], [-15.439, -6.161, 1.804], [-15.834, -7.017, -1.536], [-17.223, -6.523, -4.647], [-17.77, -7.513, -7.955], [-18.767, -7.543, -11.345], [-17.695, -9.931, -13.085], [-14.398, -10.398, -14.406], [-10.967, -10.184, -15.446], [-8.395, -12.722, -15.59], [-6.205, -11.462, -14.409], [-7.298, -9.643, -12.523], [-8.704, -11.528, -10.949], [-6.649, -13.146, -9.873], [-5.274, -11.233, -8.368], [-7.233, -10.585, -6.439], [-7.641, -13.078, -5.203], [-5.146, -13.427, -3.951], [-5.083, -11.169, -2.282], [-7.277, -11.766, -0.57], [-6.278, -14.12, 0.645], [-4.045, -13.155, 2.06], [-5.234, -11.366, 3.89], [-7.155, -12.951, 5.25], [-5.415, -14.589, 6.647], [-4.611, -12.942, 8.681], [-6.891, -12.779, 10.155], [-9.004, -11.198, 9.278], [-12.305, -10.775, 7.928], [-13.164, -11.51, 4.65], [-15.046, -10.898, 1.956], [-15.534, -11.745, -1.147], [-17.085, -11.4, -4.207], [-17.565, -12.423, -7.43], [-19.448, -11.812, -10.016]], "CA_chain_A": [[-15.455, 3.353, 11.854], [-12.239, 3.522, 9.924], [-9.735, 0.662, 9.74], [-6.128, 1.8, 9.322], [-5.074, -1.624, 8.054], [-7.991, -2.219, 5.697], [-7.623, 1.317, 4.337], [-4.025, 0.475, 3.411], [-5.233, -2.549, 1.457], [-8.065, -0.527, -0.059], [-5.465, 1.902, -1.408], [-3.396, -0.941, -2.853], [-6.467, -2.459, -4.49], [-7.527, 0.902, -5.927], [-4.022, 1.506, -7.283], [-4.098, -1.901, -9.02], [-7.565, -1.214, -10.455], [-6.381, 2.179, -11.705], [-3.302, 0.671, -13.388], [-5.533, -1.961, -15.007], [-8.251, 0.462, -16.125], [-10.865, -1.492, -14.176], [-13.808, 0.635, -13.093], [-16.571, -1.877, -12.233], [-15.4, -3.01, -8.758], [-17.257, -1.959, -5.603], [-15.423, -1.976, -2.265], [-16.955, -1.707, 1.214], [-15.316, -1.837, 4.653], [-17.596, -2.568, 7.598], [-16.558, -3.484, 11.165], [-13.399, -5.329, 10.144], [-14.87, -6.942, 7.044], [-13.72, -6.182, 3.503], [-16.342, -6.715, 0.811], [-15.531, -6.62, -2.905], [-18.17, -7.079, -5.604], [-17.534, -7.115, -9.33], [-19.369, -8.381, -12.371], [-16.73, -10.481, -13.989], [-13.092, -9.792, -14.467], [-9.913, -10.939, -16.082], [-7.519, -13.504, -14.741], [-5.339, -10.574, -13.637], [-8.066, -9.217, -11.357], [-9.037, -12.676, -10.118], [-5.482, -13.557, -9.098], [-5.007, -10.2, -7.373], [-8.209, -10.695, -5.362], [-7.356, -14.301, -4.461], [-3.977, -13.177, -3.117], [-5.501, -10.237, -1.239], [-8.087, -12.495, 0.396], [-5.447, -15.036, 1.423], [-3.208, -12.371, 2.957], [-6.035, -10.78, 4.959], [-7.726, -14.004, 6.078], [-4.371, -15.076, 7.539], [-4.587, -12.019, 9.803], [-8.15, -12.851, 10.866], [-10.051, -10.372, 8.703], [-13.312, -11.38, 7.069], [-13.029, -10.997, 3.289], [-16.162, -11.398, 1.161], [-15.314, -11.41, -2.547], [-18.077, -11.972, -5.106], [-17.355, -12.096, -8.833], [-20.724, -12.228, -10.578]], "C_chain_A": [[-14.525, 3.068, 10.696], [-11.128, 2.581, 10.337], [-8.423, 1.057, 9.074], [-5.594, 0.705, 8.401], [-5.884, -1.859, 6.782], [-7.943, -1.043, 4.732], [-6.325, 1.21, 3.548], [-4.256, -0.489, 2.257], [-6.223, -2.003, 0.447], [-7.273, 0.337, -1.032], [-4.696, 1.044, -2.401], [-4.311, -1.534, -3.911], [-6.855, -1.387, -5.493], [-6.396, 1.25, -6.885], [-3.907, 0.448, -8.373], [-5.226, -1.69, -10.026], [-7.297, -0.052, -11.4], [-5.369, 1.804, -12.788], [-3.949, -0.183, -14.469], [-6.661, -1.332, -15.813], [-9.553, -0.226, -15.794], [-11.707, -0.553, -13.334], [-15.118, 0.051, -12.584], [-16.509, -2.642, -10.91], [-15.935, -2.111, -7.648], [-16.587, -2.519, -4.359], [-16.155, -1.378, -1.072], [-16.124, -2.248, 2.369], [-16.312, -1.604, 5.773], [-17.173, -3.42, 8.787], [-15.579, -4.644, 10.974], [-13.57, -6.281, 8.988], [-14.622, -6.128, 5.784], [-14.64, -6.92, 2.547], [-15.981, -6.143, -0.547], [-16.543, -7.305, -3.811], [-17.806, -6.597, -6.999], [-18.266, -8.076, -10.237], [-18.258, -8.767, -13.326], [-15.43, -9.727, -13.946], [-12.137, -10.73, -15.157], [-9.201, -11.812, -15.06], [-6.628, -12.616, -13.888], [-6.046, -10.077, -12.38], [-8.321, -10.379, -10.406], [-7.846, -13.1, -9.277], [-5.133, -12.518, -8.043], [-5.947, -10.345, -6.178], [-8.016, -11.971, -4.552], [-6.204, -14.094, -3.491], [-4.309, -12.212, -1.986], [-6.31, -10.947, -0.159], [-7.225, -13.41, 1.257], [-4.622, -14.283, 2.46], [-4.01, -11.836, 4.136], [-6.693, -11.844, 5.834], [-6.69, -14.56, 7.045], [-4.246, -14.212, 8.781], [-5.907, -11.982, 10.559], [-9.234, -11.922, 10.372], [-11.067, -11.248, 7.98], [-13.236, -10.706, 5.708], [-14.076, -11.68, 2.422], [-15.976, -10.869, -0.254], [-16.369, -12.137, -3.367], [-17.771, -11.49, -6.511], [-18.538, -12.681, -9.594], [-20.658, -12.365, -12.09]], "O_chain_A": [[-14.897, 2.519, 9.662], [-10.68, 2.634, 11.485], [-8.304, 0.991, 7.855], [-5.143, 0.977, 7.279], [-5.323, -1.971, 5.685], [-8.0, -1.245, 3.513], [-6.273, 1.603, 2.377], [-3.814, -0.247, 1.129], [-6.118, -2.31, -0.74], [-7.536, 0.331, -2.241], [-4.583, 1.398, -3.577], [-3.94, -1.609, -5.083], [-6.892, -1.646, -6.703], [-6.638, 1.55, -8.059], [-3.651, 0.78, -9.537], [-5.01, -1.776, -11.239], [-7.634, -0.111, -12.591], [-5.549, 2.134, -13.966], [-3.596, -0.085, -15.651], [-7.156, -1.972, -16.745], [-10.461, -0.263, -16.626], [-11.246, -0.047, -12.305], [-15.961, 0.814, -12.103], [-17.363, -3.495, -10.656], [-15.586, -0.928, -7.571], [-16.44, -3.737, -4.225], [-16.472, -0.181, -1.077], [-15.638, -3.382, 2.317], [-16.864, -0.507, 5.887], [-17.148, -4.645, 8.685], [-15.802, -5.764, 11.436], [-12.747, -7.189, 8.815], [-15.045, -4.971, 5.688], [-14.627, -8.155, 2.491], [-15.862, -4.925, -0.694], [-16.72, -8.526, -3.736], [-17.604, -5.4, -7.224], [-18.383, -9.265, -9.953], [-17.93, -8.033, -14.267], [-15.345, -8.577, -13.499], [-12.441, -11.902, -15.409], [-9.352, -11.668, -13.836], [-6.32, -12.97, -12.748], [-5.482, -10.097, -11.272], [-8.202, -10.239, -9.186], [-7.993, -13.366, -8.078], [-4.776, -12.872, -6.917], [-5.52, -10.263, -5.018], [-8.189, -11.956, -3.329], [-6.257, -14.545, -2.341], [-3.883, -12.411, -0.84], [-6.071, -10.762, 1.042], [-7.393, -13.461, 2.478], [-4.512, -14.709, 3.621], [-3.538, -11.859, 5.278], [-6.787, -11.672, 7.055], [-7.035, -14.941, 8.167], [-3.788, -14.689, 9.822], [-6.022, -11.231, 11.533], [-10.311, -11.879, 10.981], [-10.746, -12.329, 7.484], [-13.222, -9.474, 5.623], [-14.005, -12.897, 2.196], [-16.231, -9.692, -0.528], [-16.5, -13.357, -3.266], [-17.731, -10.283, -6.76], [-18.639, -13.904, -9.761], [-21.628, -12.801, -12.712]]}, "name": "6MRR", "num_of_chains": 1, "seq": "GWSTELEKHREELKEFLKKEGITNVEIRIDNGRLEVRVEGGTERLKRFLEELRQKLEKKGYTVDIKIE"}
|
| 2 |
+
{"seq_chain_A": "HMPEEEKAARLFIEALEKGDPELMRKVISPDTRMEDNGREFTGDEVVEYVKEIQKRGEQWHLRRYTKEGNSWRFEVQVDNNGQTEQWEVQIEVRNGRIKRVTITHV", "coords_chain_A": {"N_chain_A": [[37.0, 18.222, 51.819], [35.18, 19.045, 54.805], [33.142, 21.39, 56.357], [32.697, 22.256, 59.882], [30.075, 22.366, 60.868], [28.465, 21.048, 58.967], [29.669, 18.568, 59.079], [29.059, 17.634, 61.702], [26.271, 17.24, 61.58], [26.225, 15.306, 59.622], [27.541, 13.181, 60.918], [25.603, 12.501, 62.842], [23.621, 11.465, 61.194], [25.073, 9.367, 60.115], [25.367, 7.722, 62.376], [22.785, 6.789, 62.655], [22.499, 5.42, 60.214], [24.449, 3.414, 60.569], [23.344, 2.25, 62.7], [24.374, 2.554, 65.225], [24.763, 2.964, 68.494], [26.944, 3.77, 70.035], [28.442, 5.552, 68.362], [26.446, 7.553, 68.106], [26.246, 8.499, 70.748], [28.563, 9.948, 71.018], [28.108, 12.096, 69.352], [25.861, 13.648, 70.164], [24.504, 16.068, 72.578], [23.777, 16.707, 76.082], [21.518, 18.351, 75.963], [20.455, 18.057, 73.397], [17.593, 17.855, 71.366], [15.114, 15.757, 69.978], [12.531, 15.031, 67.686], [10.309, 12.633, 66.571], [7.796, 11.95, 64.523], [5.869, 12.982, 66.497], [6.728, 11.686, 68.914], [8.927, 12.6, 71.49], [12.386, 13.091, 72.157], [15.152, 13.567, 74.34], [18.607, 14.262, 74.992], [20.249, 11.415, 76.286], [18.136, 9.735, 75.635], [18.012, 9.917, 72.876], [20.144, 8.337, 72.204], [19.126, 5.939, 73.133], [17.028, 5.681, 71.32], [18.585, 4.982, 69.05], [19.592, 2.533, 69.856], [17.261, 1.04, 69.728], [16.741, 1.045, 67.012], [18.819, -0.538, 66.081], [18.18, -2.888, 67.459], [15.617, -3.575, 66.242], [16.392, -4.089, 63.726], [16.097, -2.152, 61.8], [16.701, -0.407, 58.831], [18.613, 2.321, 57.804], [19.364, 4.606, 55.165], [21.224, 7.164, 53.799], [20.201, 9.35, 51.263], [20.133, 11.886, 49.855], [20.945, 15.377, 50.442], [20.462, 18.834, 51.494], [21.245, 22.207, 52.499], [18.911, 23.991, 54.2], [17.94, 27.07, 54.88], [18.965, 27.602, 58.12], [19.227, 24.935, 58.758], [19.98, 21.458, 58.206], [18.725, 19.353, 55.737], [17.945, 16.136, 55.375], [17.279, 13.513, 53.17], [16.307, 10.372, 53.735], [16.304, 6.85, 53.277], [14.36, 4.368, 54.732], [14.218, 1.096, 55.78], [12.396, -1.822, 56.732], [11.509, -5.193, 57.372], [10.625, -5.836, 54.804], [9.326, -3.567, 53.956], [9.713, -0.36, 52.528], [10.495, 2.99, 53.244], [10.35, 6.575, 52.736], [11.862, 9.008, 54.514], [12.232, 12.164, 56.203], [14.836, 14.014, 57.563], [15.572, 16.566, 59.658], [18.051, 18.728, 60.656], [19.208, 20.958, 63.088], [22.156, 22.563, 63.663], [23.645, 24.166, 66.175], [26.134, 26.306, 67.255], [28.264, 24.429, 66.79], [27.157, 22.089, 67.871], [24.652, 19.433, 68.054], [21.475, 20.587, 67.962], [18.615, 20.547, 67.532], [16.573, 18.007, 66.237], [14.476, 16.857, 63.707], [12.331, 14.421, 62.706], [10.453, 13.297, 60.044], [7.846, 11.327, 58.803], [5.177, 10.579, 57.057]], "CA_chain_A": [[36.936, 18.773, 53.168], [33.829, 19.307, 55.268], [33.003, 22.335, 57.475], [32.383, 21.616, 61.147], [28.63, 22.278, 61.041], [27.969, 19.998, 58.095], [30.255, 17.336, 59.605], [28.319, 17.193, 62.883], [24.978, 16.74, 61.124], [26.544, 14.088, 58.891], [27.832, 12.133, 61.893], [24.312, 12.112, 63.413], [23.007, 10.631, 60.175], [25.91, 8.164, 60.045], [25.045, 6.895, 63.536], [21.519, 6.158, 62.308], [22.821, 4.501, 59.135], [25.19, 2.299, 61.114], [22.592, 1.729, 63.824], [25.424, 2.426, 66.209], [24.548, 3.825, 69.667], [28.216, 4.325, 70.466], [28.703, 6.763, 67.587], [25.452, 8.572, 68.459], [26.576, 9.042, 72.062], [29.65, 10.906, 70.821], [27.638, 13.226, 68.553], [24.834, 14.469, 70.801], [24.761, 16.77, 73.826], [22.683, 16.47, 77.021], [20.351, 19.16, 75.595], [19.926, 17.722, 72.07], [16.203, 17.421, 71.347], [14.694, 15.122, 68.734], [11.136, 14.692, 67.52], [10.007, 11.689, 65.501], [6.346, 11.777, 64.428], [5.194, 13.191, 67.767], [7.53, 11.223, 70.04], [10.037, 13.505, 71.773], [13.571, 12.523, 72.816], [16.186, 14.512, 74.768], [19.828, 13.667, 75.507], [20.521, 9.981, 76.157], [17.012, 9.105, 74.937], [18.376, 9.832, 71.466], [20.98, 7.138, 72.128], [18.19, 4.839, 73.298], [16.414, 5.594, 69.984], [19.465, 4.246, 68.135], [19.602, 1.154, 70.325], [16.045, 0.407, 69.247], [16.922, 0.85, 65.574], [19.706, -1.684, 66.003], [17.363, -4.009, 67.906], [14.558, -3.966, 65.32], [17.02, -4.306, 62.438], [15.647, -1.091, 60.918], [17.816, 0.015, 57.986], [18.627, 3.687, 57.284], [20.338, 4.921, 54.124], [21.169, 8.544, 53.342], [20.076, 9.473, 49.814], [20.579, 13.205, 49.457], [20.616, 16.466, 51.352], [20.718, 20.236, 51.207], [21.091, 23.102, 53.628], [17.708, 24.807, 54.042], [18.051, 28.065, 55.926], [20.011, 27.214, 59.057], [19.161, 23.525, 59.1], [20.28, 20.496, 57.163], [17.581, 18.51, 55.443], [18.39, 14.859, 54.825], [16.355, 12.518, 52.643], [16.831, 9.069, 54.073], [15.447, 5.708, 53.01], [14.308, 3.533, 55.936], [13.632, -0.203, 55.453], [12.292, -2.933, 57.665], [10.532, -6.258, 57.193], [10.11, -5.81, 53.444], [8.72, -2.286, 53.633], [10.651, 0.732, 52.332], [9.888, 4.302, 53.432], [11.125, 7.801, 52.577], [11.761, 9.852, 55.689], [12.975, 13.407, 56.146], [15.508, 14.281, 58.831], [15.997, 17.954, 59.618], [18.953, 18.92, 61.785], [19.776, 22.273, 63.355], [23.418, 22.556, 64.378], [23.916, 25.488, 66.721], [27.402, 26.364, 67.981], [28.974, 23.169, 66.639], [26.507, 20.955, 68.531], [23.321, 19.08, 67.562], [20.524, 21.296, 68.816], [17.262, 20.046, 67.347], [16.415, 17.077, 65.126], [13.085, 16.599, 63.409], [12.153, 13.313, 61.776], [9.088, 13.272, 59.537], [7.438, 10.301, 57.858], [3.735, 10.392, 57.071]], "C_chain_A": [[35.516, 19.181, 53.529], [33.835, 20.246, 56.472], [32.424, 21.668, 58.723], [30.875, 21.424, 61.352], [28.045, 21.144, 60.221], [28.428, 18.625, 58.602], [29.459, 16.766, 60.777], [26.977, 16.58, 62.491], [25.152, 15.408, 60.402], [26.83, 12.921, 59.83], [26.562, 11.609, 62.585], [23.641, 11.12, 62.474], [23.742, 9.311, 60.031], [25.597, 7.187, 61.182], [23.732, 6.145, 63.321], [21.733, 5.074, 61.25], [23.514, 3.243, 59.644], [24.422, 1.619, 62.24], [23.493, 1.584, 65.033], [25.271, 3.432, 67.346], [25.829, 4.473, 70.157], [28.568, 5.591, 69.685], [27.729, 7.873, 67.973], [25.765, 9.271, 69.778], [27.63, 10.133, 71.946], [29.225, 12.164, 70.07], [26.725, 14.189, 69.313], [25.32, 15.147, 72.075], [23.586, 16.509, 74.765], [21.4, 17.227, 76.67], [19.883, 19.001, 74.14], [18.479, 17.252, 72.148], [15.814, 16.879, 69.984], [13.215, 14.791, 68.796], [11.019, 13.732, 66.357], [8.511, 11.383, 65.495], [5.617, 11.906, 65.766], [5.97, 12.779, 69.006], [8.693, 12.188, 70.248], [11.14, 12.773, 72.507], [14.571, 13.629, 73.143], [17.412, 13.767, 75.307], [19.981, 12.179, 75.234], [19.346, 9.184, 75.593], [17.272, 8.974, 73.437], [19.264, 8.61, 71.249], [20.105, 5.892, 72.234], [17.42, 4.6, 71.996], [17.271, 4.767, 69.027], [19.569, 2.782, 68.557], [18.408, 0.378, 69.783], [16.076, 0.161, 67.749], [17.732, -0.411, 65.332], [18.974, -2.968, 66.397], [16.379, -4.492, 66.836], [15.096, -4.299, 63.928], [16.594, -3.31, 61.369], [16.837, -0.53, 60.144], [17.671, 1.442, 57.459], [19.7, 3.886, 56.222], [20.172, 6.366, 53.668], [21.132, 8.588, 51.81], [20.646, 10.787, 49.314], [20.267, 14.245, 50.522], [21.046, 17.824, 50.859], [20.686, 21.006, 52.51], [19.918, 24.019, 53.329], [17.639, 25.811, 55.176], [19.214, 27.689, 56.819], [19.807, 25.792, 59.584], [19.526, 22.664, 57.913], [19.11, 19.552, 56.986], [18.028, 17.274, 54.701], [17.2, 14.029, 54.397], [16.961, 11.15, 52.89], [15.812, 8.012, 53.681], [15.513, 4.779, 54.22], [13.552, 2.243, 55.625], [13.475, -1.054, 56.689], [11.196, -3.906, 57.263], [9.861, -6.201, 55.824], [9.414, -4.523, 53.038], [9.757, -1.167, 53.588], [9.951, 2.083, 52.441], [10.884, 5.43, 53.152], [10.835, 8.756, 53.723], [12.571, 11.114, 55.473], [13.554, 13.659, 57.513], [16.084, 15.679, 58.809], [16.836, 18.239, 60.853], [19.393, 20.364, 61.913], [21.031, 22.116, 64.2], [23.671, 23.966, 64.867], [25.19, 25.419, 67.548], [28.163, 25.044, 67.963], [28.334, 21.936, 67.267], [25.122, 20.677, 67.946], [22.274, 19.664, 68.485], [19.119, 20.714, 68.743], [17.234, 19.151, 66.116], [14.943, 16.776, 64.944], [12.96, 15.527, 62.351], [10.703, 13.266, 61.349], [8.891, 12.117, 58.572], [5.949, 10.026, 57.982], [3.267, 9.765, 55.77]], "O_chain_A": [[34.75, 19.627, 52.679], [34.466, 19.951, 57.486], [31.745, 20.64, 58.632], [30.444, 20.43, 61.936], [27.223, 20.37, 60.71], [27.666, 17.644, 58.56], [29.228, 15.556, 60.851], [26.587, 15.54, 63.01], [24.344, 14.49, 60.563], [26.43, 11.793, 59.561], [26.466, 10.42, 62.876], [23.133, 10.086, 62.899], [23.11, 8.253, 59.861], [25.565, 5.978, 60.976], [23.587, 5.004, 63.732], [21.208, 3.961, 61.365], [23.214, 2.143, 59.19], [24.797, 0.534, 62.681], [23.414, 0.602, 65.768], [25.628, 4.597, 67.202], [25.794, 5.601, 70.653], [28.946, 6.599, 70.283], [28.147, 9.011, 68.154], [25.567, 10.478, 69.921], [27.585, 11.13, 72.657], [29.904, 13.182, 70.145], [26.795, 15.401, 69.13], [26.4, 14.846, 72.582], [22.509, 16.12, 74.303], [20.32, 16.76, 77.023], [19.016, 19.744, 73.693], [18.171, 16.339, 72.914], [16.109, 17.485, 68.964], [12.702, 14.379, 69.847], [11.564, 13.984, 65.281], [8.025, 10.649, 66.357], [4.861, 11.02, 66.147], [5.876, 13.441, 70.042], [9.374, 12.559, 69.286], [10.86, 11.923, 73.362], [14.807, 14.512, 72.329], [17.271, 12.751, 75.992], [19.846, 11.725, 74.084], [19.535, 8.068, 75.133], [16.802, 8.045, 72.793], [19.141, 7.904, 70.238], [20.301, 4.917, 71.505], [17.186, 3.453, 71.614], [16.744, 3.928, 68.26], [19.568, 1.88, 67.714], [18.527, -0.796, 69.411], [15.514, -0.823, 67.273], [17.375, -1.244, 64.501], [19.117, -3.994, 65.744], [16.312, -5.687, 66.557], [14.345, -4.733, 63.062], [16.717, -3.592, 60.189], [17.874, -0.25, 60.724], [16.729, 1.724, 56.722], [20.833, 3.459, 56.399], [19.101, 6.761, 53.213], [21.922, 7.924, 51.14], [21.54, 10.807, 48.463], [19.441, 14.03, 51.403], [21.88, 17.95, 49.948], [20.154, 20.521, 53.506], [19.91, 24.707, 52.301], [17.308, 25.452, 56.305], [20.323, 27.467, 56.337], [20.162, 25.477, 60.718], [19.425, 23.084, 56.754], [18.544, 19.031, 57.959], [18.427, 17.355, 53.535], [16.231, 13.89, 55.157], [18.022, 10.811, 52.366], [14.608, 8.257, 53.699], [16.601, 4.461, 54.699], [12.391, 2.3, 55.229], [14.338, -1.043, 57.557], [10.101, -3.503, 56.86], [8.668, -6.468, 55.697], [8.959, -4.394, 51.905], [10.569, -1.042, 54.499], [8.934, 2.298, 51.817], [12.103, 5.252, 53.305], [9.722, 9.241, 53.893], [13.499, 11.135, 54.676], [12.845, 13.511, 58.513], [16.983, 15.964, 58.026], [16.401, 17.994, 61.976], [19.882, 20.939, 60.945], [20.986, 21.581, 65.294], [23.86, 24.873, 64.065], [25.32, 24.562, 68.418], [28.641, 24.59, 68.998], [28.902, 20.843, 67.213], [24.492, 21.583, 67.413], [22.176, 19.279, 69.665], [18.494, 20.423, 69.772], [17.819, 19.495, 65.079], [14.236, 16.503, 65.911], [13.432, 15.7, 61.239], [9.814, 13.225, 62.192], [9.69, 11.926, 57.664], [5.503, 9.336, 58.905], [3.96, 9.844, 54.754]]}, "name": "5L33", "num_of_chains": 1, "seq": "HMPEEEKAARLFIEALEKGDPELMRKVISPDTRMEDNGREFTGDEVVEYVKEIQKRGEQWHLRRYTKEGNSWRFEVQVDNNGQTEQWEVQIEVRNGRIKRVTITHV"}
|
ProteinMPNN/outputs/example_1_outputs/seqs/5L33.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>5L33, score=1.5874, global_score=1.5874, fixed_chains=[], designed_chains=['A'], model_name=v_48_020, git_hash=015ff820b9b5741ead6ba6795258f35a9c15e94b, seed=37
|
| 2 |
+
HMPEEEKAARLFIEALEKGDPELMRKVISPDTRMEDNGREFTGDEVVEYVKEIQKRGEQWHLRRYTKEGNSWRFEVQVDNNGQTEQWEVQIEVRNGRIKRVTITHV
|
| 3 |
+
>T=0.1, sample=1, score=0.8221, global_score=0.8221, seq_recovery=0.5094
|
| 4 |
+
MINEEEKKALDFIEALEKADPELMKKVIEPDTKMEVNGKKYEGEEIVEFVKKLKEEGVKYKLLSYKKEGNKYVFEVEKSKNGVTKKITIEIEVENGKVKKIVITEK
|
| 5 |
+
>T=0.1, sample=2, score=0.8356, global_score=0.8356, seq_recovery=0.4434
|
| 6 |
+
SINEEEQKALDYIKALEKADPELMKKVITPDTKMTVNGKEYEGEEIVEYVKELKERGIKYKLLSYKKEGDKYVFTVERSENGKTYTITIEVKVKDGKVEEIVIKEE
|
ProteinMPNN/outputs/example_1_outputs/seqs/6MRR.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>6MRR, score=1.4683, global_score=1.4683, fixed_chains=[], designed_chains=['A'], model_name=v_48_020, git_hash=015ff820b9b5741ead6ba6795258f35a9c15e94b, seed=37
|
| 2 |
+
GWSTELEKHREELKEFLKKEGITNVEIRIDNGRLEVRVEGGTERLKRFLEELRQKLEKKGYTVDIKIE
|
| 3 |
+
>T=0.1, sample=1, score=0.9617, global_score=0.9617, seq_recovery=0.5000
|
| 4 |
+
GMDEELEKYVKELKAFLKEKGINNVEIKIENGTLTIKMNGASKETREFLEKLKKELEEKGYKVNIEIS
|
| 5 |
+
>T=0.1, sample=2, score=0.9513, global_score=0.9513, seq_recovery=0.4853
|
| 6 |
+
GKDEELEKYVKELKKFLKEKGINNVKIEVKDGTLTIEMKGCSKETKDFLKKLKKELEKKGYKVNIKIY
|
ProteinMPNN/outputs/example_2_outputs/assigned_pdbs.jsonl
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{"3HTN": [["A", "B"], ["C"]], "4YOW": [["A", "B"], ["C", "D", "E", "F"]]}
|
ProteinMPNN/outputs/example_2_outputs/parsed_pdbs.jsonl
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
ProteinMPNN/outputs/example_2_outputs/seqs/3HTN.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>3HTN, score=1.4405, global_score=1.4946, fixed_chains=['C'], designed_chains=['A', 'B'], CA_model_name=v_48_020, git_hash=015ff820b9b5741ead6ba6795258f35a9c15e94b, seed=37
|
| 2 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKXXXXDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 3 |
+
>T=0.1, sample=1, score=0.8450, global_score=1.0949, seq_recovery=0.5071
|
| 4 |
+
KLYSYKEIGNKYIVSINVGTDLVEALKKFCEEKNIKSGTINGIGEVSKLTLKFYDFETKETELKTFEGNFTISNLTGLIYTYNGKIFLHLHVTFGDEDFSALAGHLVSATVLQEALLKVENYNENITAKFDEKLGLYLLDFNS/MSYKYKKIGNKYLVSINIGKDLVESLKEFVKEKNIKSGTINGIGGVSEVTLRFFDPEXXXXKERTFKGLFDISNLTGFISTKDGEPFLHLHATFGDEDFSALAGHLVSAKVSTGAELLVENYNVELTRKYDEKLGVYLLDFNA
|
| 5 |
+
>T=0.1, sample=2, score=0.8471, global_score=1.0996, seq_recovery=0.5000
|
| 6 |
+
MLYDYKKIGNKYFVKVNVDQDLVEALKEFCEELGIKSGTINGIGEVSEVTLRFFDFETKESVDKTFKEPFTISNLTGLISTYNGKIHLHLHITFSDKEFSALAGHLVSAKVLQEALLIVEDYGENITRKYDKETGLLLLDFNS/MLYKYKKIGNKYLIEINIGKDLVEALKEFVEEKNIKAGTINGIGMVEEVTLEYYDPKXXXXEKKTFEGLFEISNLTGFIYTKDGKPVLHLHVTFGDEDFSALAGHLVSAKVLGEAELLVEDYNVELTVKYDEERGEDLLDFNS
|
ProteinMPNN/outputs/example_2_outputs/seqs/4YOW.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>4YOW, score=1.3574, global_score=1.3913, fixed_chains=['C', 'D', 'E', 'F'], designed_chains=['A', 'B'], CA_model_name=v_48_020, git_hash=015ff820b9b5741ead6ba6795258f35a9c15e94b, seed=37
|
| 2 |
+
MRIVAADTGGAVLDESFQPVGLIATVAVLVEKPYKTSKRFLVKYADPYNYDLSGRQAIRDEIELAIELAREVSPDVIHLNSTLGGIEVRKLDESTIDALQISDRGKEIWKELSKDLQPLAKKFWEETGIEIIAIGKSSVPVRIAEIYAGIFSVKWALDNVKEKGGLLVGLPRYMEVEIKKDKIIGKSLDPREGGLYGEVKTEVPQGIKWELYPNPLVRRFMVFEITS/XXXX
|
| 3 |
+
>T=0.1, sample=1, score=0.8241, global_score=1.2059, seq_recovery=0.5154
|
| 4 |
+
MKIVASDAGGYLLDEELKPIGRIAVVAVLVEKPFTSAKEYKVEYLDPEKYNLEGNDDLIKEFELAVELAKKYKPDVILLDLNLGGVELSELNPEVIEKLQISEETKEFLIKLSEILSPKAKEFKKETGIPILLAGGNSTAVKIAELLASAAAVKWALENVKEKGKLLIGLERAVEIEIEEDKIRARDLDPRYGGLYAEIDIKIPEGLKYEQYPNPFKPGEMVFEIEK/XXXX
|
| 5 |
+
>T=0.1, sample=2, score=0.8195, global_score=1.2174, seq_recovery=0.5419
|
| 6 |
+
MKIVAADAGGYLVDEDLKPIGRIAVVAVLVEKPFTSSKVYKVKYIDPEKADLNGNEDLRLELELAIELAKEYKPDIILLDLNLGGVELSELNEETIKKLQISEEAKKKLIELSKELSPLAKKFKEETGIPILLAGDNSVPVHIAEILASAEAVKWALENVKEKGEVKVLLHESVSIEIEEDKIKARSLDPRLGGLEAEIEIKIPEGIEYEQEPNPFRPHHMVFTAKV/XXXX
|
ProteinMPNN/outputs/example_3_outputs/seqs/3HTN.fa
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
>3HTN, score=1.1550, global_score=1.1955, fixed_chains=['C'], designed_chains=['A', 'B'], model_name=v_48_020, git_hash=015ff820b9b5741ead6ba6795258f35a9c15e94b, seed=37
|
| 2 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKXXXXDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 3 |
+
>T=0.1, sample=1, score=0.7339, global_score=0.9189, seq_recovery=0.5390
|
| 4 |
+
KLYDYEKIGNKYIVSIYNNTDIVKALKKFCEEKNIKSGTVNGIGQVKEVTLKFYNFETKESEEKTFKKNFTISNLTGFISEHDGKIFLDLHITFGDENFSALAGHLVSAIVNGECKLVIEDYKEKVSTKYDEELGLWLLDFNK/ETYKYKKIGNKYLVSINNGKDLVDSIKKFCKDKKIKSGTVNGIGSISKLTLEFFDPDXXXXKTKTLEKNLEISNLTGFISTKDGEVFLDLHITIGDENFSALAGHLISAIVNGIAELKIEDYNKEINVKYDEKLGLYLLDFNK
|
| 5 |
+
>T=0.1, sample=2, score=0.7064, global_score=0.9034, seq_recovery=0.5993
|
| 6 |
+
HMYEYKKIGNKYIVSVKNNTELVEALKAFCEEKKIKSGTVNGIGQVKSVTLRFYDFKTKTSKDTTFNQNLEISNLTGFISEYNNKVFLDLHITFGDSNFSALAGHLLSAVVGGEAIFVVEDYKEKISRKYDEKLGLYLLDFNK/NMYKYKKIGNKYIVSINNGKNLVKALKKFCEDKNIKSGTINGIGMISKVTLYFFDPEXXXXTTKTFNELLEISNLTGFISEKNGKVFLHLHITIGDSNFSALAGHLIDAVVNGIAEVIVEDFNEKINVKYNEETGLWLLDFNK
|