Update ProteinMPNN-main/README.md
Browse files- ProteinMPNN-main/README.md +92 -0
ProteinMPNN-main/README.md
CHANGED
|
@@ -0,0 +1,92 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# ProteinMPNN
|
| 2 |
+
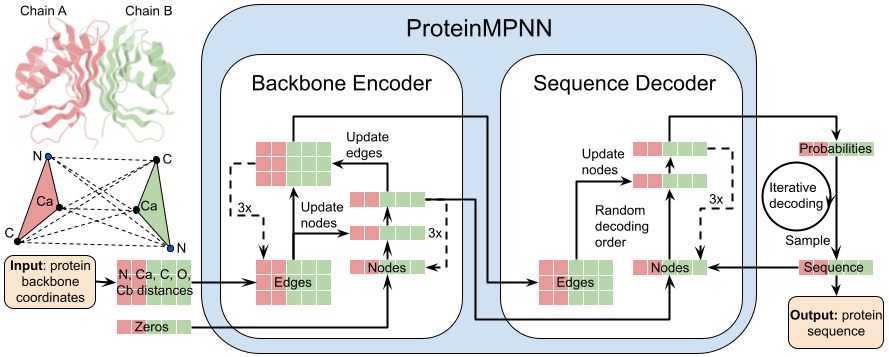
|
| 3 |
+
Read [ProteinMPNN paper](https://www.biorxiv.org/content/10.1101/2022.06.03.494563v1).
|
| 4 |
+
|
| 5 |
+
To run ProteinMPNN clone this github repo and install Python>=3.0, PyTorch, Numpy.
|
| 6 |
+
|
| 7 |
+
Full protein backbone models: `vanilla_model_weights/v_48_002.pt, v_48_010.pt, v_48_020.pt, v_48_030.pt`.
|
| 8 |
+
|
| 9 |
+
CA only models: `ca_model_weights/v_48_002.pt, v_48_010.pt, v_48_020.pt`. Enable flag `--ca_only` to use these models.
|
| 10 |
+
|
| 11 |
+
Helper scripts: `helper_scripts` - helper functions to parse PDBs, assign which chains to design, which residues to fix, adding AA bias, tying residues etc.
|
| 12 |
+
|
| 13 |
+
Code organization:
|
| 14 |
+
* `protein_mpnn_run.py` - the main script to initialialize and run the model.
|
| 15 |
+
* `protein_mpnn_utils.py` - utility functions for the main script.
|
| 16 |
+
* `examples/` - simple code examples.
|
| 17 |
+
* `inputs/` - input PDB files for examples
|
| 18 |
+
* `outputs/` - outputs from examples
|
| 19 |
+
* `colab_notebooks/` - Google Colab examples
|
| 20 |
+
-----------------------------------------------------------------------------------------------------
|
| 21 |
+
Input flags for `protein_mpnn_run.py`:
|
| 22 |
+
```
|
| 23 |
+
argparser.add_argument("--ca_only", action="store_true", default=False, help="Parse CA-only structures and use CA-only models (default: false)")
|
| 24 |
+
argparser.add_argument("--path_to_model_weights", type=str, default="", help="Path to model weights folder;")
|
| 25 |
+
argparser.add_argument("--model_name", type=str, default="v_48_020", help="ProteinMPNN model name: v_48_002, v_48_010, v_48_020, v_48_030; v_48_010=version with 48 edges 0.10A noise")
|
| 26 |
+
argparser.add_argument("--seed", type=int, default=0, help="If set to 0 then a random seed will be picked;")
|
| 27 |
+
argparser.add_argument("--save_score", type=int, default=0, help="0 for False, 1 for True; save score=-log_prob to npy files")
|
| 28 |
+
argparser.add_argument("--save_probs", type=int, default=0, help="0 for False, 1 for True; save MPNN predicted probabilites per position")
|
| 29 |
+
argparser.add_argument("--score_only", type=int, default=0, help="0 for False, 1 for True; score input backbone-sequence pairs")
|
| 30 |
+
argparser.add_argument("--conditional_probs_only", type=int, default=0, help="0 for False, 1 for True; output conditional probabilities p(s_i given the rest of the sequence and backbone)")
|
| 31 |
+
argparser.add_argument("--conditional_probs_only_backbone", type=int, default=0, help="0 for False, 1 for True; if true output conditional probabilities p(s_i given backbone)")
|
| 32 |
+
argparser.add_argument("--unconditional_probs_only", type=int, default=0, help="0 for False, 1 for True; output unconditional probabilities p(s_i given backbone) in one forward pass")
|
| 33 |
+
argparser.add_argument("--backbone_noise", type=float, default=0.00, help="Standard deviation of Gaussian noise to add to backbone atoms")
|
| 34 |
+
argparser.add_argument("--num_seq_per_target", type=int, default=1, help="Number of sequences to generate per target")
|
| 35 |
+
argparser.add_argument("--batch_size", type=int, default=1, help="Batch size; can set higher for titan, quadro GPUs, reduce this if running out of GPU memory")
|
| 36 |
+
argparser.add_argument("--max_length", type=int, default=200000, help="Max sequence length")
|
| 37 |
+
argparser.add_argument("--sampling_temp", type=str, default="0.1", help="A string of temperatures, 0.2 0.25 0.5. Sampling temperature for amino acids. Suggested values 0.1, 0.15, 0.2, 0.25, 0.3. Higher values will lead to more diversity.")
|
| 38 |
+
argparser.add_argument("--out_folder", type=str, help="Path to a folder to output sequences, e.g. /home/out/")
|
| 39 |
+
argparser.add_argument("--pdb_path", type=str, default='', help="Path to a single PDB to be designed")
|
| 40 |
+
argparser.add_argument("--pdb_path_chains", type=str, default='', help="Define which chains need to be designed for a single PDB ")
|
| 41 |
+
argparser.add_argument("--jsonl_path", type=str, help="Path to a folder with parsed pdb into jsonl")
|
| 42 |
+
argparser.add_argument("--chain_id_jsonl",type=str, default='', help="Path to a dictionary specifying which chains need to be designed and which ones are fixed, if not specied all chains will be designed.")
|
| 43 |
+
argparser.add_argument("--fixed_positions_jsonl", type=str, default='', help="Path to a dictionary with fixed positions")
|
| 44 |
+
argparser.add_argument("--omit_AAs", type=list, default='X', help="Specify which amino acids should be omitted in the generated sequence, e.g. 'AC' would omit alanine and cystine.")
|
| 45 |
+
argparser.add_argument("--bias_AA_jsonl", type=str, default='', help="Path to a dictionary which specifies AA composion bias if neededi, e.g. {A: -1.1, F: 0.7} would make A less likely and F more likely.")
|
| 46 |
+
argparser.add_argument("--bias_by_res_jsonl", default='', help="Path to dictionary with per position bias.")
|
| 47 |
+
argparser.add_argument("--omit_AA_jsonl", type=str, default='', help="Path to a dictionary which specifies which amino acids need to be omited from design at specific chain indices")
|
| 48 |
+
argparser.add_argument("--pssm_jsonl", type=str, default='', help="Path to a dictionary with pssm")
|
| 49 |
+
argparser.add_argument("--pssm_multi", type=float, default=0.0, help="A value between [0.0, 1.0], 0.0 means do not use pssm, 1.0 ignore MPNN predictions")
|
| 50 |
+
argparser.add_argument("--pssm_threshold", type=float, default=0.0, help="A value between -inf + inf to restric per position AAs")
|
| 51 |
+
argparser.add_argument("--pssm_log_odds_flag", type=int, default=0, help="0 for False, 1 for True")
|
| 52 |
+
argparser.add_argument("--pssm_bias_flag", type=int, default=0, help="0 for False, 1 for True")
|
| 53 |
+
argparser.add_argument("--tied_positions_jsonl", type=str, default='', help="Path to a dictionary with tied positions")
|
| 54 |
+
|
| 55 |
+
```
|
| 56 |
+
-----------------------------------------------------------------------------------------------------
|
| 57 |
+
For example to make a conda environment to run ProteinMPNN:
|
| 58 |
+
* `conda create --name mlfold` - this creates conda environment called `mlfold`
|
| 59 |
+
* `source activate mlfold` - this activate environment
|
| 60 |
+
* `conda install pytorch torchvision torchaudio cudatoolkit=11.3 -c pytorch` - install pytorch following steps from https://pytorch.org/
|
| 61 |
+
-----------------------------------------------------------------------------------------------------
|
| 62 |
+
These are provided `examples/`:
|
| 63 |
+
* `submit_example_1.sh` - simple monomer example
|
| 64 |
+
* `submit_example_2.sh` - simple multi-chain example
|
| 65 |
+
* `submit_example_3.sh` - directly from the .pdb path
|
| 66 |
+
* `submit_example_3_score_only.sh` - return score only (model's uncertainty)
|
| 67 |
+
* `submit_example_4.sh` - fix some residue positions
|
| 68 |
+
* `submit_example_4_non_fixed.sh` - specify which positions to design
|
| 69 |
+
* `submit_example_5.sh` - tie some positions together (symmetry)
|
| 70 |
+
* `submit_example_6.sh` - homooligomer example
|
| 71 |
+
* `submit_example_7.sh` - return sequence unconditional probabilities (PSSM like)
|
| 72 |
+
* `submit_example_8.sh` - add amino acid bias
|
| 73 |
+
-----------------------------------------------------------------------------------------------------
|
| 74 |
+
Output example:
|
| 75 |
+
```
|
| 76 |
+
>3HTN, score=1.1705, global_score=1.2045, fixed_chains=['B'], designed_chains=['A', 'C'], model_name=v_48_020, git_hash=015ff820b9b5741ead6ba6795258f35a9c15e94b, seed=37
|
| 77 |
+
NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER/NMYSYKKIGNKYIVSINNHTEIVKALNAFCKEKGILSGSINGIGAIGELTLRFFNPKTKAYDDKTFREQMEISNLTGNISSMNEQVYLHLHITVGRSDYSALAGHLLSAIQNGAGEFVVEDYSERISRTYNPDLGLNIYDFER
|
| 78 |
+
>T=0.1, sample=1, score=0.7291, global_score=0.9330, seq_recovery=0.5736
|
| 79 |
+
NMYSYKKIGNKYIVSINNHTEIVKALKKFCEEKNIKSGSVNGIGSIGSVTLKFYNLETKEEELKTFNANFEISNLTGFISMHDNKVFLDLHITIGDENFSALAGHLVSAVVNGTCELIVEDFNELVSTKYNEELGLWLLDFEK/NMYSYKKIGNKYIVSINNHTDIVTAIKKFCEDKKIKSGTINGIGQVKEVTLEFRNFETGEKEEKTFKKQFTISNLTGFISTKDGKVFLDLHITFGDENFSALAGHLISAIVDGKCELIIEDYNEEINVKYNEELGLYLLDFNK
|
| 80 |
+
>T=0.1, sample=2, score=0.7414, global_score=0.9355, seq_recovery=0.6075
|
| 81 |
+
NMYKYKKIGNKYIVSINNHTEIVKAIKEFCKEKNIKSGTINGIGQVGKVTLRFYNPETKEYTEKTFNDNFEISNLTGFISTYKNEVFLHLHITFGKSDFSALAGHLLSAIVNGICELIVEDFKENLSMKYDEKTGLYLLDFEK/NMYKYKKIGNKYVVSINNHTEIVEALKAFCEDKKIKSGTVNGIGQVSKVTLKFFNIETKESKEKTFNKNFEISNLTGFISEINGEVFLHLHITIGDENFSALAGHLLSAVVNGEAILIVEDYKEKVNRKYNEELGLNLLDFNL
|
| 82 |
+
```
|
| 83 |
+
* `score` - average over residues that were designed negative log probability of sampled amino acids
|
| 84 |
+
* `global score` - average over all residues in all chains negative log probability of sampled/fixed amino acids
|
| 85 |
+
* `fixed_chains` - chains that were not designed (fixed)
|
| 86 |
+
* `designed_chains` - chains that were redesigned
|
| 87 |
+
* `model_name/CA_model_name` - model name that was used to generate results, e.g. `v_48_020`
|
| 88 |
+
* `git_hash` - github version that was used to generate outputs
|
| 89 |
+
* `seed` - random seed
|
| 90 |
+
* `T=0.1` - temperature equal to 0.1 was used to sample sequences
|
| 91 |
+
* `sample` - sequence sample number 1, 2, 3...etc
|
| 92 |
+
-----------------------------------------------------------------------------------------------------
|