Spaces:
Sleeping
Sleeping
initial commit
Browse files- .gitattributes +1 -0
- README.md +28 -13
- app.py +98 -0
- assests/screenshot.png +0 -0
- dataloader.py +18 -0
- requirements.txt +5 -0
- segmentation.py +90 -0
- utils.py +79 -0
- vectors.kv +3 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
vectors.kv filter=lfs diff=lfs merge=lfs -text
|
README.md
CHANGED
|
@@ -1,13 +1,28 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
| 4 |
-
|
| 5 |
-
|
| 6 |
-
|
| 7 |
-
|
| 8 |
-
|
| 9 |
-
|
| 10 |
-
|
| 11 |
-
|
| 12 |
-
|
| 13 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# SNOMED-Entity-Linking
|
| 2 |
+
A [Gradio](https://www.gradio.app/) app for Entity linking on the [SNOMED CT](https://www.snomed.org/five-step-briefing), a knowledge graph of clinical healthcare terminology.
|
| 3 |
+
|
| 4 |
+

|
| 5 |
+
|
| 6 |
+
## Motivation
|
| 7 |
+
Much of the world's healthcare data is stored in free-text documents, usually clinical notes taken by doctors. This unstructured data can be challenging to analyze and extract meaningful insights from.
|
| 8 |
+
However, by applying a standardized terminology like SNOMED CT, we can the interpretability of these notes for patients and individuals outside the organization of origin.
|
| 9 |
+
Moreover, healthcare organizations can convert this free-text data into a structured format that can be readily analyzed by computers, in turn stimulating the development of new medicines, treatment pathways, and better patient outcomes.
|
| 10 |
+
|
| 11 |
+
Here, we use entity linking to analyze clinical notes identifing and labeling the portions of each note that correspond to specific medical concepts.
|
| 12 |
+
|
| 13 |
+
# Methodology
|
| 14 |
+
The pipline involves two models, one for segmentation and the other for disambiguation (classification of the segmentations).
|
| 15 |
+
The segmentation model is a [CANINE-s](https://huggingface.co/google/canine-s) character-level transformer model finetuned to optimise the BCE, Dice, and Focal loss each weighted 1, 1, .1 respectively. The objective function is then optimised using Adam with a learning rate of 1e-5.
|
| 16 |
+
The classification model uses the [BioBERT](https://huggingface.co/dmis-lab/biosyn-biobert-bc5cdr-disease) model. Here, the model is trained similarly using Adan and a learning rate of 2e-5. We train using the [MultipleNegativesRankingLoss](https://arxiv.org/pdf/1705.00652) using the [SentenceTransformers](https://sbert.net/) library.
|
| 17 |
+
|
| 18 |
+
## Dataset
|
| 19 |
+
The dataset used to train the models is the dataset used for the [SNOMED CT Entity Linking Challenge](https://physionet.org/content/snomed-ct-entity-challenge/1.0.0/), which is a subset of [MIMIC-IV-Note](https://physionet.org/content/mimic-iv-note/2.2/) of 75,000 entity annotations across about 300 discharge notes.
|
| 20 |
+
For the sake of simplicity we only include entities with more than 10 mentions.
|
| 21 |
+
|
| 22 |
+
|
| 23 |
+
## References
|
| 24 |
+
- Hardman, W., Banks, M., Davidson, R., Truran, D., Ayuningtyas, N. W., Ngo, H., Johnson, A., & Pollard, T. (2023). SNOMED CT Entity Linking Challenge (version 1.0.0). PhysioNet. https://doi.org/10.13026/s48e-sp45.
|
| 25 |
+
- Goldberger, A., Amaral, L., Glass, L., Hausdorff, J., Ivanov, P. C., Mark, R., ... & Stanley, H. E. (2000). PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation [Online]. 101 (23), pp. e215–e220.
|
| 26 |
+
- Jinhyuk Lee, Wonjin Yoon, Sungdong Kim, Donghyeon Kim, Sunkyu Kim, Chan Ho So, Jaewoo Kang, BioBERT: a pre-trained biomedical language representation model for biomedical text mining, Bioinformatics, Volume 36, Issue 4, February 2020, Pages 1234–1240, https://doi.org/10.1093/bioinformatics/btz682
|
| 27 |
+
- Henderson, M., Al-Rfou, R., Strope, B., Sung, Y., Lukács, L., Guo, R., Kumar, S., Miklos, B., & Kurzweil, R. (2017). Efficient Natural Language Response Suggestion for Smart Reply. ArXiv, abs/1705.00652.
|
| 28 |
+
- Reimers, N., & Gurevych, I. (2019). Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks. Conference on Empirical Methods in Natural Language Processing.
|
app.py
ADDED
|
@@ -0,0 +1,98 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import pandas as pd
|
| 3 |
+
import configparser
|
| 4 |
+
import gradio as gr
|
| 5 |
+
from gensim.models import KeyedVectors
|
| 6 |
+
from sentence_transformers import SentenceTransformer
|
| 7 |
+
from transformers import AutoModelForTokenClassification, AutoTokenizer
|
| 8 |
+
|
| 9 |
+
from segmentation import segment
|
| 10 |
+
from utils import clean_entity
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
class Linker:
|
| 14 |
+
def __init__(self, config: dict[str, object],
|
| 15 |
+
context_window_width: int = -1):
|
| 16 |
+
self._vectors = None
|
| 17 |
+
self._emb_model = None
|
| 18 |
+
if context_window_width <= 0:
|
| 19 |
+
context_window_width = config['context_window_width']
|
| 20 |
+
self.context_window_width = context_window_width
|
| 21 |
+
self.config = config
|
| 22 |
+
|
| 23 |
+
def add_context(self, row: pd.Series) -> str:
|
| 24 |
+
window_start = max(0, row.start - self.context_window_width)
|
| 25 |
+
window_end = min(row.end + self.context_window_width, len(row.text))
|
| 26 |
+
return clean_entity(row.text[window_start:window_end])
|
| 27 |
+
|
| 28 |
+
def _load_embeddings(self):
|
| 29 |
+
self._vectors = KeyedVectors.load(self.config['keyed_vectors_file'])
|
| 30 |
+
|
| 31 |
+
def _load_model(self):
|
| 32 |
+
self._emb_model = SentenceTransformer(config['embedding_model'])
|
| 33 |
+
|
| 34 |
+
@property
|
| 35 |
+
def embeddings(self):
|
| 36 |
+
if self._vectors is None:
|
| 37 |
+
self._load_embeddings()
|
| 38 |
+
return self._vectors
|
| 39 |
+
|
| 40 |
+
@property
|
| 41 |
+
def embedding_model(self):
|
| 42 |
+
if self._emb_model is None:
|
| 43 |
+
self._load_model()
|
| 44 |
+
return self._emb_model
|
| 45 |
+
|
| 46 |
+
def link(self, df: pd.DataFrame) -> list[dict]:
|
| 47 |
+
mention_emb = self.embedding_model.encode(df.mention.str.lower().values)
|
| 48 |
+
|
| 49 |
+
concepts = [self.embeddings.most_similar(m, topn=1)[0][0]
|
| 50 |
+
for m in mention_emb]
|
| 51 |
+
return concepts
|
| 52 |
+
|
| 53 |
+
|
| 54 |
+
def highlight_text(spans: pd.DataFrame, text: str) -> list[tuple[str, object]]:
|
| 55 |
+
token_concepts = [None for _ in text]
|
| 56 |
+
|
| 57 |
+
for row in spans.itertuples():
|
| 58 |
+
for k in range(row.start, row.end):
|
| 59 |
+
token_concepts[k] = row.concept
|
| 60 |
+
|
| 61 |
+
return list(zip(list(text), token_concepts))
|
| 62 |
+
|
| 63 |
+
|
| 64 |
+
def entity_link(query: str) -> list[tuple[str, object]]:
|
| 65 |
+
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
|
| 66 |
+
seg_model = AutoModelForTokenClassification.from_pretrained(
|
| 67 |
+
config['segmentation_model']
|
| 68 |
+
)
|
| 69 |
+
seg_tokenizer = AutoTokenizer.from_pretrained(
|
| 70 |
+
config['segmentation_tokenizer']
|
| 71 |
+
)
|
| 72 |
+
thresh = float(config['thresh'])
|
| 73 |
+
query_df = pd.DataFrame({'note_id': [0], 'text': [query]})
|
| 74 |
+
|
| 75 |
+
seg = segment(query_df, seg_model, seg_tokenizer, device, thresh)
|
| 76 |
+
linked_concepts = []
|
| 77 |
+
if len(seg) > 0:
|
| 78 |
+
seg = seg.sort_values('start')
|
| 79 |
+
linked_concepts = linker.link(seg)
|
| 80 |
+
seg['concept'] = linked_concepts
|
| 81 |
+
|
| 82 |
+
return highlight_text(seg, query)
|
| 83 |
+
|
| 84 |
+
|
| 85 |
+
config_parser = configparser.ConfigParser()
|
| 86 |
+
config_parser.read('config.ini')
|
| 87 |
+
config = config_parser['DEFAULT']
|
| 88 |
+
linker = Linker(config)
|
| 89 |
+
|
| 90 |
+
demo = gr.Interface(
|
| 91 |
+
fn=entity_link,
|
| 92 |
+
inputs=["text"],
|
| 93 |
+
outputs=gr.HighlightedText(
|
| 94 |
+
label="linking",
|
| 95 |
+
combine_adjacent=True,
|
| 96 |
+
),
|
| 97 |
+
theme=gr.themes.Base()
|
| 98 |
+
)
|
assests/screenshot.png
ADDED
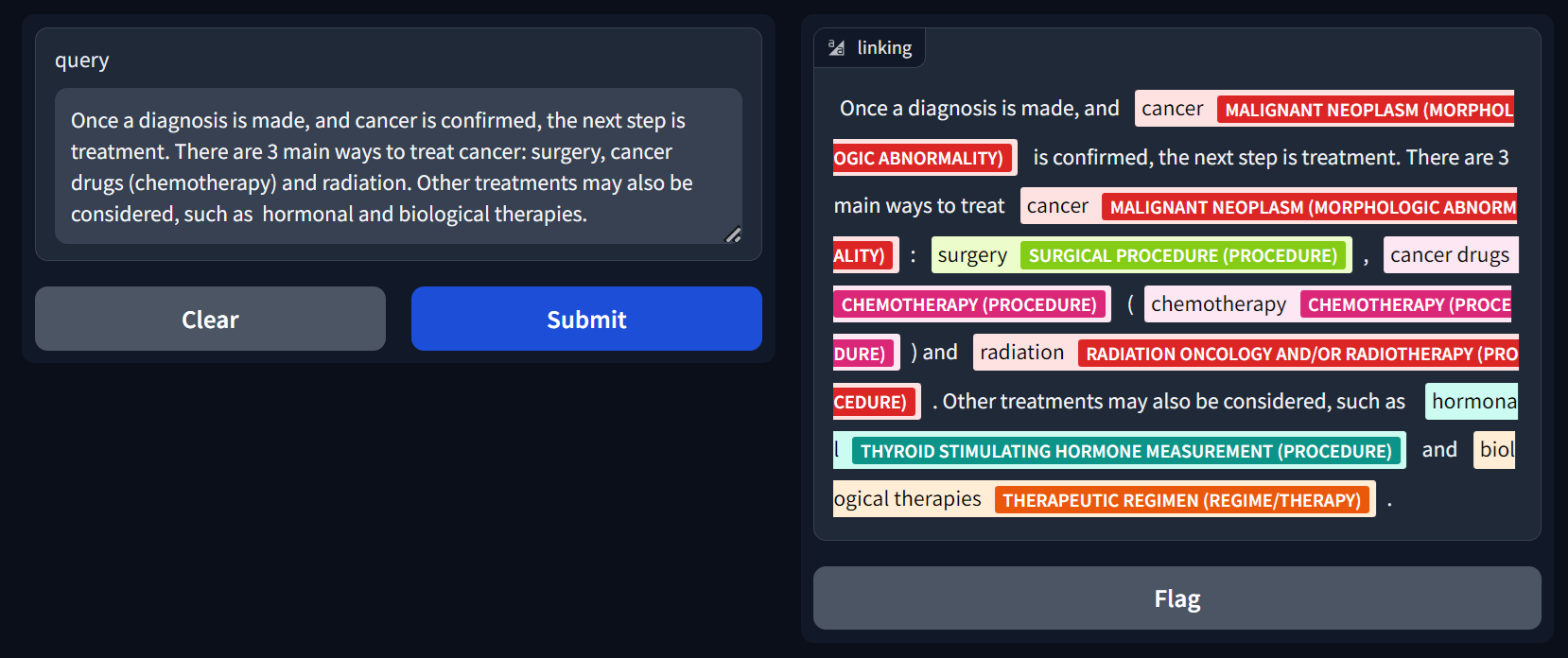
|
dataloader.py
ADDED
|
@@ -0,0 +1,18 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
from torch.utils.data import DataLoader
|
| 3 |
+
|
| 4 |
+
|
| 5 |
+
class TestDataset(torch.utils.data.Dataset):
|
| 6 |
+
def __init__(self, encodings: list[dict[str, list]]):
|
| 7 |
+
self.encodings = encodings
|
| 8 |
+
|
| 9 |
+
def __getitem__(self, idx):
|
| 10 |
+
item = {key: torch.tensor(val) for key, val in self.encodings[idx].items()}
|
| 11 |
+
return item
|
| 12 |
+
|
| 13 |
+
def __len__(self):
|
| 14 |
+
return len(self.encodings)
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
def create_dataloader(dat: list[dict[str, list]], batch_size: int) -> DataLoader:
|
| 18 |
+
return DataLoader(TestDataset(dat), batch_size=batch_size, shuffle=False)
|
requirements.txt
ADDED
|
@@ -0,0 +1,5 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
torch==2.2.1
|
| 2 |
+
pandas==2.2.0
|
| 3 |
+
sentence_transformers==2.6.1
|
| 4 |
+
transformers==4.39.1
|
| 5 |
+
numpy==1.26.4
|
segmentation.py
ADDED
|
@@ -0,0 +1,90 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch.nn.functional as F
|
| 2 |
+
import pandas as pd
|
| 3 |
+
|
| 4 |
+
from dataloader import create_dataloader
|
| 5 |
+
from utils import *
|
| 6 |
+
|
| 7 |
+
|
| 8 |
+
def predict_segmentation(inp, model, device, batch_size=8):
|
| 9 |
+
test_loader = create_dataloader(inp, batch_size)
|
| 10 |
+
|
| 11 |
+
predictions = []
|
| 12 |
+
for batch in test_loader:
|
| 13 |
+
batch = {k: v.to(device) for k, v in batch.items()}
|
| 14 |
+
p = F.sigmoid(model(**batch).logits).detach().cpu().numpy()
|
| 15 |
+
predictions.append(p)
|
| 16 |
+
|
| 17 |
+
return np.concatenate(predictions, axis=0)
|
| 18 |
+
|
| 19 |
+
|
| 20 |
+
def create_data(text, tokenizer, seq_len=512):
|
| 21 |
+
tokens = tokenizer(text, add_special_tokens=False)
|
| 22 |
+
_token_batches = {k: [pad_seq(x, seq_len) for x in batch_list(v, seq_len)]
|
| 23 |
+
for (k, v) in tokens.items()}
|
| 24 |
+
n_batches = len(_token_batches['input_ids'])
|
| 25 |
+
return [{k: v[i] for k, v in _token_batches.items()}
|
| 26 |
+
for i in range(n_batches)]
|
| 27 |
+
|
| 28 |
+
|
| 29 |
+
def segment_tokens(notes, model, tokenizer, device, batch_size=8):
|
| 30 |
+
predictions = {}
|
| 31 |
+
for note in notes.itertuples():
|
| 32 |
+
note_id = note.note_id
|
| 33 |
+
raw_text = note.text.lower()
|
| 34 |
+
|
| 35 |
+
inp = create_data(raw_text, tokenizer)
|
| 36 |
+
pred_probs = predict_segmentation(inp, model, device, batch_size=batch_size)
|
| 37 |
+
pred_probs = np.squeeze(pred_probs, -1)
|
| 38 |
+
pred_probs = np.concatenate(pred_probs)
|
| 39 |
+
|
| 40 |
+
predictions[note_id] = pred_probs
|
| 41 |
+
|
| 42 |
+
return predictions
|
| 43 |
+
|
| 44 |
+
|
| 45 |
+
def segment(notes, model, tokenizer, device, thresh, batch_size=8):
|
| 46 |
+
predictions = []
|
| 47 |
+
|
| 48 |
+
predictions_prob_map = segment_tokens(notes, model, tokenizer, device, batch_size)
|
| 49 |
+
|
| 50 |
+
for note in notes.itertuples():
|
| 51 |
+
|
| 52 |
+
note_id = note.note_id
|
| 53 |
+
raw_text = note.text
|
| 54 |
+
|
| 55 |
+
decoded_text = tokenizer.decode(tokenizer.encode(raw_text, add_special_tokens=False))
|
| 56 |
+
|
| 57 |
+
pred_probs = predictions_prob_map[note_id]
|
| 58 |
+
|
| 59 |
+
_, pred_probs = align_decoded(raw_text, decoded_text, pred_probs)
|
| 60 |
+
pred_probs = np.array(pred_probs, 'float32')
|
| 61 |
+
pred = (pred_probs > thresh).astype('uint8')
|
| 62 |
+
|
| 63 |
+
spans = get_sequential_spans(pred)
|
| 64 |
+
|
| 65 |
+
note_predictions = {'note_id': [], 'start': [], 'end': [], 'mention': [], 'score': []}
|
| 66 |
+
for (start, end) in spans:
|
| 67 |
+
note_predictions['note_id'].append(note_id)
|
| 68 |
+
note_predictions['score'].append(pred_probs[start:end].mean())
|
| 69 |
+
note_predictions['start'].append(start)
|
| 70 |
+
note_predictions['end'].append(end)
|
| 71 |
+
note_predictions['mention'].append(raw_text[start:end])
|
| 72 |
+
|
| 73 |
+
note_predictions = pd.DataFrame(note_predictions)
|
| 74 |
+
note_predictions = note_predictions.sort_values('score', ascending=False)
|
| 75 |
+
|
| 76 |
+
# remove overlapping spans
|
| 77 |
+
seen_spans = set()
|
| 78 |
+
unseen = []
|
| 79 |
+
for span in note_predictions[['start', 'end']].values:
|
| 80 |
+
span = tuple(span)
|
| 81 |
+
s = False
|
| 82 |
+
if not is_overlap(seen_spans, span):
|
| 83 |
+
seen_spans.add(span)
|
| 84 |
+
s = True
|
| 85 |
+
unseen.append(s)
|
| 86 |
+
note_predictions = note_predictions[unseen]
|
| 87 |
+
|
| 88 |
+
predictions.append(note_predictions)
|
| 89 |
+
predictions = pd.concat(predictions).reset_index(drop=True)
|
| 90 |
+
return predictions
|
utils.py
ADDED
|
@@ -0,0 +1,79 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
|
| 3 |
+
|
| 4 |
+
def is_overlap(existing_spans, new_span):
|
| 5 |
+
for span in existing_spans:
|
| 6 |
+
# Check if either end of the new span is within an existing span
|
| 7 |
+
if (span[0] <= new_span[0] <= span[1]) or \
|
| 8 |
+
(span[0] <= new_span[1] <= span[1]):
|
| 9 |
+
return True
|
| 10 |
+
# Check if the new span entirely covers an existing span
|
| 11 |
+
if new_span[0] <= span[0] and new_span[1] >= span[1]:
|
| 12 |
+
return True
|
| 13 |
+
return False
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
def get_sequential_spans(a):
|
| 17 |
+
spans = []
|
| 18 |
+
|
| 19 |
+
prev = False
|
| 20 |
+
start = 0
|
| 21 |
+
|
| 22 |
+
for i, x in enumerate(a):
|
| 23 |
+
if not prev and x:
|
| 24 |
+
start = i
|
| 25 |
+
elif prev and not x:
|
| 26 |
+
spans.append((start, i))
|
| 27 |
+
|
| 28 |
+
prev = x
|
| 29 |
+
|
| 30 |
+
if x:
|
| 31 |
+
spans.append((start, i + 1))
|
| 32 |
+
|
| 33 |
+
return spans
|
| 34 |
+
|
| 35 |
+
|
| 36 |
+
def batch_list(iterable, n=1):
|
| 37 |
+
l = len(iterable)
|
| 38 |
+
for ndx in range(0, l, n):
|
| 39 |
+
yield iterable[ndx:min(ndx + n, l)]
|
| 40 |
+
|
| 41 |
+
|
| 42 |
+
def pad_seq(seq, max_len):
|
| 43 |
+
n = len(seq)
|
| 44 |
+
if n >= max_len:
|
| 45 |
+
return seq
|
| 46 |
+
else:
|
| 47 |
+
return np.pad(seq, (0, max_len - n))
|
| 48 |
+
|
| 49 |
+
|
| 50 |
+
def align_decoded(x, d, y):
|
| 51 |
+
clean_text = ""
|
| 52 |
+
clean_label = []
|
| 53 |
+
j = 0
|
| 54 |
+
for i in range(len(d)):
|
| 55 |
+
found = False
|
| 56 |
+
for delim in [',', '.', '?', "'"]:
|
| 57 |
+
if (x[j:j + 2] == f" {delim}") and (d[i] == f"{delim}"):
|
| 58 |
+
found = True
|
| 59 |
+
clean_text += f' {delim}'
|
| 60 |
+
clean_label += [y[j], y[j]]
|
| 61 |
+
j += 1
|
| 62 |
+
|
| 63 |
+
if not found:
|
| 64 |
+
clean_text += x[j]
|
| 65 |
+
clean_label += [y[j]]
|
| 66 |
+
j += 1
|
| 67 |
+
|
| 68 |
+
if (clean_text != x) and (x[-1:] == "\n"):
|
| 69 |
+
clean_text += "\n"
|
| 70 |
+
clean_label += [0, 0]
|
| 71 |
+
|
| 72 |
+
return clean_text, clean_label
|
| 73 |
+
|
| 74 |
+
|
| 75 |
+
def clean_entity(t):
|
| 76 |
+
t = t.lower()
|
| 77 |
+
t = t.replace(' \n', " ")
|
| 78 |
+
t = t.replace('\n', " ")
|
| 79 |
+
return t
|
vectors.kv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:55c8f6f379646d6ddb06d4f33d615e09f3354ce229271113e2ce57ae6164c673
|
| 3 |
+
size 4914710
|