Model Details
We introduce a suite of neural language model tools for pre-training, fine-tuning SMILES-based molecular language models. Furthermore, we also provide recipes for semi-supervised recipes for fine-tuning these languages in low-data settings using Semi-supervised learning.
Enumeration-aware Molecular Transformers
Introduces contrastive learning alongside multi-task regression, and masked language modelling as pre-training objectives to inject enumeration knowledge into pre-trained language models.
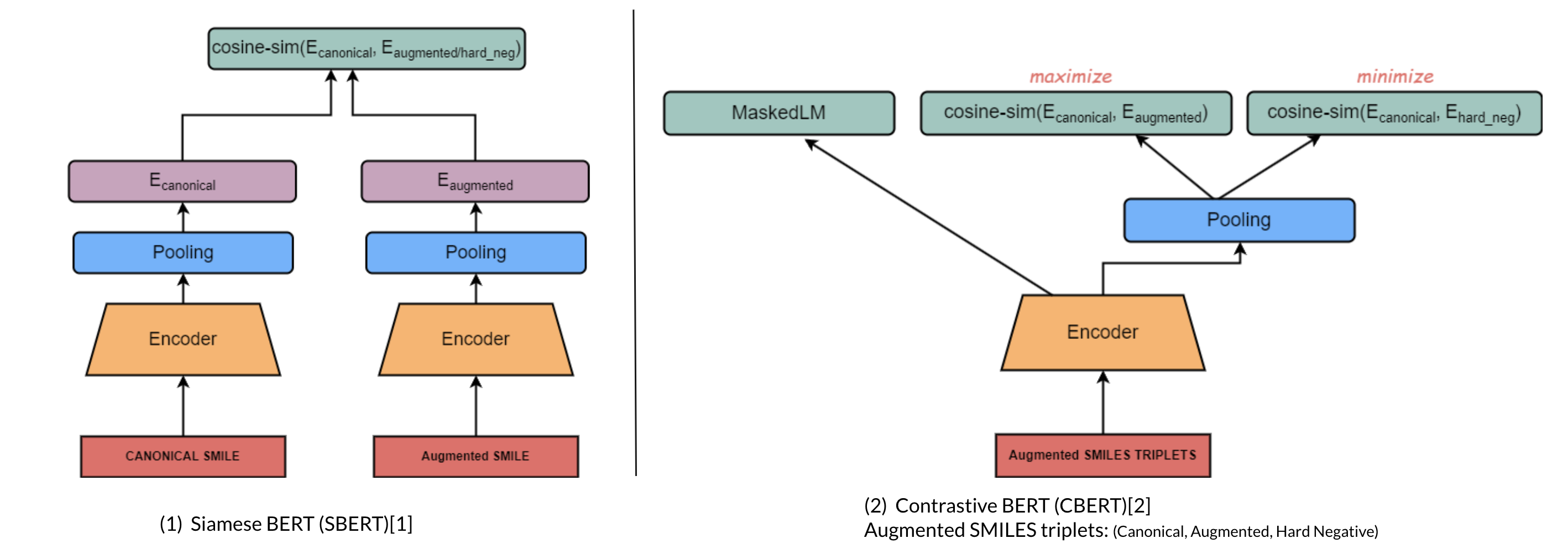
a. Molecular Domain Adaptation (Contrastive Encoder-based)
i. Architecture
ii. Contrastive Learning
b. Canonicalization Encoder-decoder (Denoising Encoder-decoder)
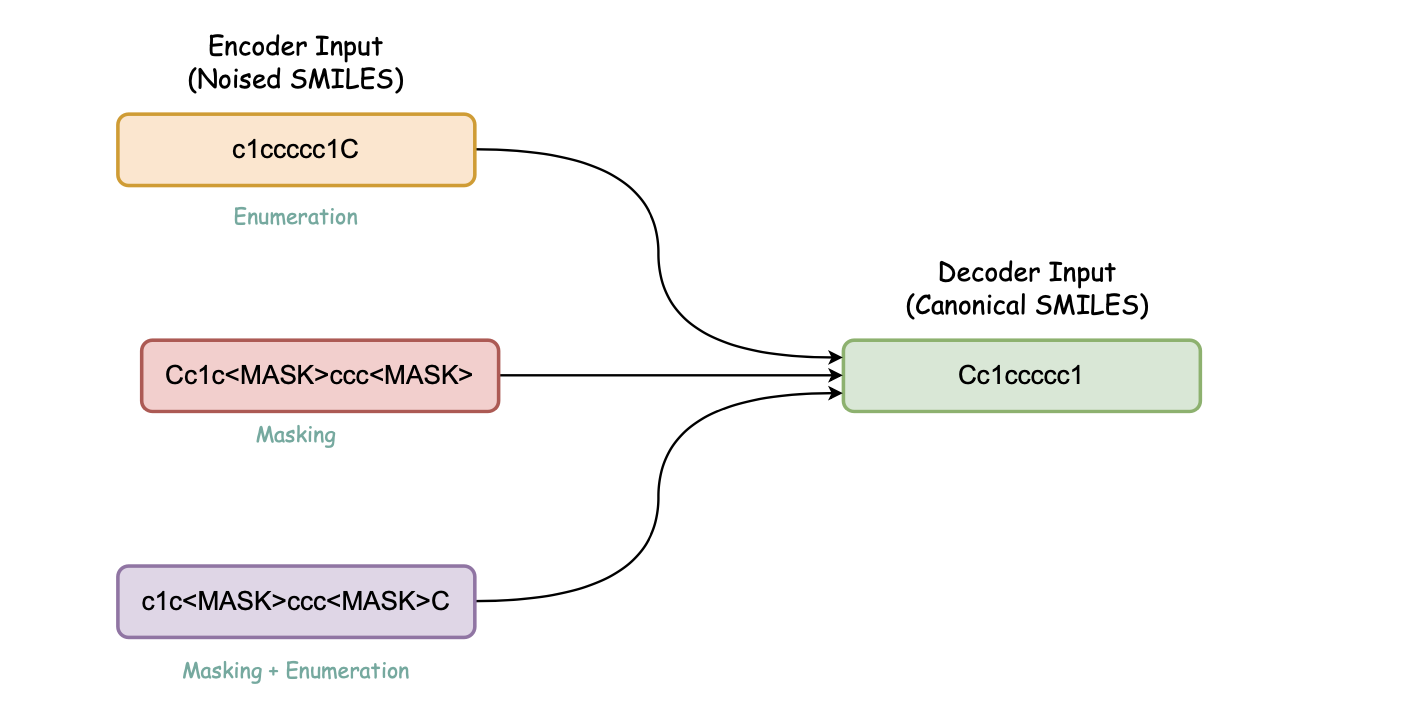
Pretraining steps for this model:
- Pretrain BART model with Denoising objective on noised Guacamol dataset
Fore more details please see our github repository.
Virtual Screening Benchmark (Github Repository)
original version presented in S. Riniker, G. Landrum, J. Cheminf., 5, 26 (2013), DOI: 10.1186/1758-2946-5-26, URL: http://www.jcheminf.com/content/5/1/26
extended version presented in S. Riniker, N. Fechner, G. Landrum, J. Chem. Inf. Model., 53, 2829, (2013), DOI: 10.1021/ci400466r, URL: http://pubs.acs.org/doi/abs/10.1021/ci400466r
Model List
Our released models are listed as following. You can import these models by using the smiles-featurizers package or using HuggingFace's Transformers.
| Model | Type | AUROC | BEDROC |
|---|---|---|---|
| UdS-LSV/smole-bert | Bert |
0.615 | 0.225 |
| UdS-LSV/smole-bert-mtr | Bert |
0.621 | 0.262 |
| UdS-LSV/smole-bart | Bart |
0.660 | 0.263 |
| UdS-LSV/muv2x-simcse-smole-bart | Simcse |
0.697 | 0.270 |
| UdS-LSV/siamese-smole-bert-muv-1x | SentenceTransformer |
0.673 | 0.274 |
- Downloads last month
- 2