license: apache-2.0
tags:
- multi-modal
- 3D medical segmentation
size_categories:
- 1K<n<10K
Dataset Description
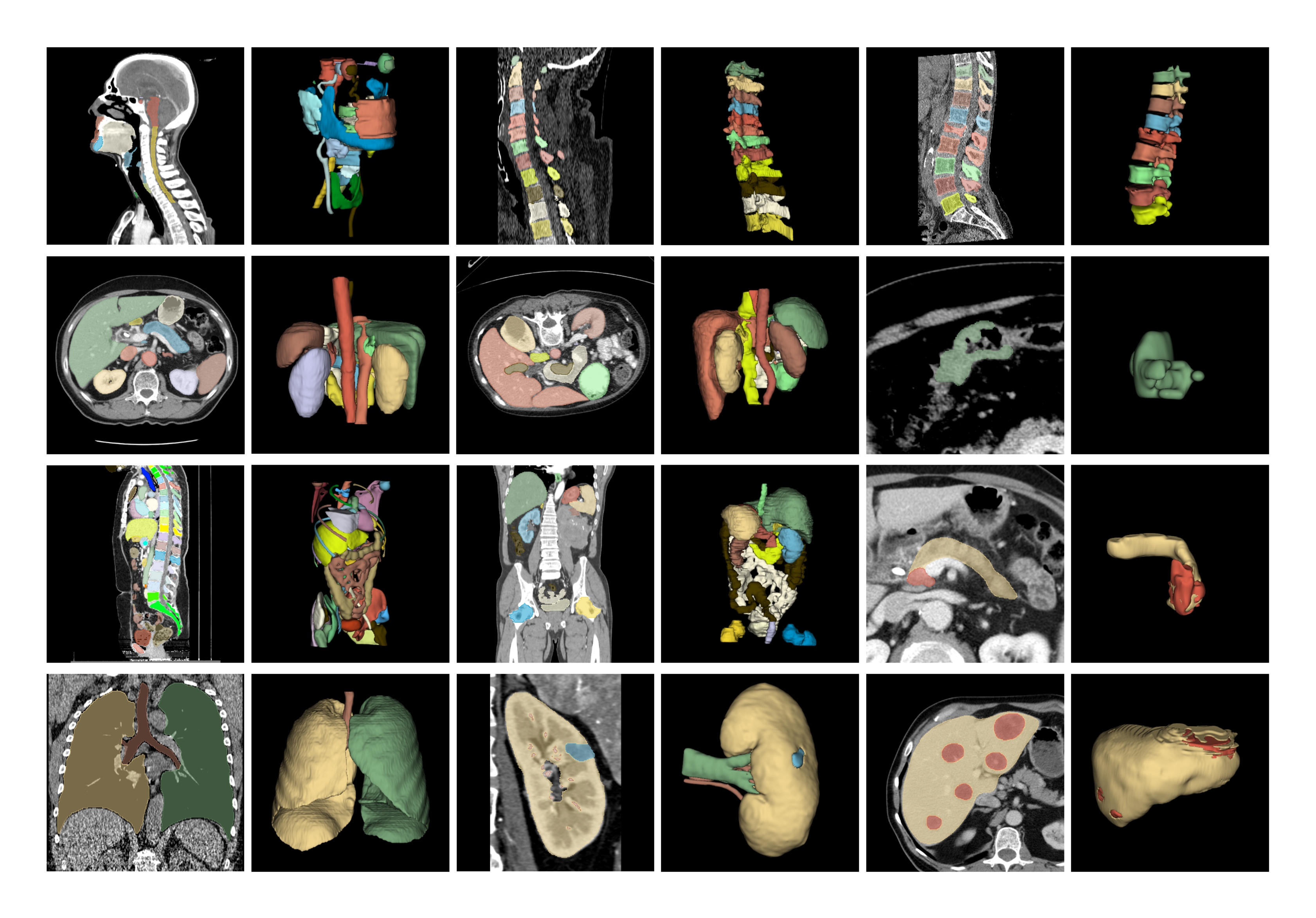
Large-scale General 3D Medical Image Segmentation Dataset (M3D-Seg)
Dataset Introduction
3D medical image segmentation poses a significant challenge in medical image analysis. Currently, due to privacy and cost constraints, publicly available large-scale 3D medical images and their annotated data are scarce. To address this, we have collected 25 publicly available 3D CT segmentation datasets, including CHAOS, HaN-Seg, AMOS22, AbdomenCT-1k, KiTS23, KiPA22, KiTS19, BTCV, Pancreas-CT, 3D-IRCADB, FLARE22, TotalSegmentator, CT-ORG, WORD, VerSe19, VerSe20, SLIVER07, QUBIQ, MSD-Colon, MSD-HepaticVessel, MSD-Liver, MSD-lung, MSD-pancreas, MSD-spleen, LUNA16. These datasets are uniformly encoded from 0000 to 0024, totaling 5,772 3D images and 149,196 3D mask annotations. The semantic labels corresponding to each mask can be represented in text. Within each sub-dataset folder, there are multiple data folders (containing image and mask files), and each sub-dataset independently utilizes its JSON file to split.
- data_load_demo.py: Provides an example code on reading images and masks from the dataset.
- data_preocess.py: Describes how to convert raw
nii.gzor other format data into a more efficientnpyformat and preprocess them, saving them in a unified format. This dataset has already been preprocessed, so there is no need to use data_preocess.py again. If adding new datasets, please follow a unified processing approach. - dataset_info.json & dataset_info.txt: Contain the names of each dataset and their label texts.
- term_dictionary.json: Provides multiple definitions or descriptions for each semantic label in the dataset,
generated by
ChatGPTfor each term. Researchers can convert category IDs in the dataset to label texts using the information in dataset_info.txt and further convert them into text descriptions using term_dictionary.json as text inputs for segmentation models, enabling tasks such as segmentation based on text prompts and referring segmentation.
This dataset supports not only traditional semantic segmentation tasks but also text-based segmentation tasks. For detailed methods, please refer to SegVol and M3D. As a general segmentation dataset, we provide a convenient, unified, and structured dataset organization that allows for the uniform integration of more public and private datasets in the same format as this dataset, thereby constructing a larger-scale general 3D medical image segmentation dataset.
Supported Tasks
This dataset not only supports traditional image-mask semantic segmentation tasks but also represents data in the form of image-mask-text, where masks can be converted into box coordinates through bounding boxes. Based on this, the dataset can effectively support a series of image segmentation and positioning tasks, as follows:
- 3D Segmentation: Semantic segmentation, text-based segmentation, referring segmentation, reasoning segmentation, etc.
- 3D Positioning: Visual grounding/referring expression comprehension, referring expression generation.
Dataset Format and Structure
Data Format
M3D_Seg/
0000/
1/
image.npy
mask_(1, 512, 512, 96).npz
2/
......
0000.json
0001/
......
Dataset Download
The total dataset size is approximately 224G.
Clone with HTTP
git clone https://huggingface.co/datasets/GoodBaiBai88/M3D-Seg
SDK Download
from datasets import load_dataset
dataset = load_dataset("GoodBaiBai88/M3D-Seg")
Manual Download
Manually download all files from the dataset files. It is recommended to use batch download tools for efficient downloading. Please note the following:
Downloading in Parts and Merging: Since dataset 0024 has a large volume, the original compressed file has been split into two parts:
0024_1and0024_2. Make sure to download these two files separately and unzip them in the same directory to ensure data integrity.Masks with Sparse Matrices: To save storage space effectively, foreground information in masks is stored in sparse matrix format and saved with the extension
.npz. The name of each mask file typically includes its shape information for identification and loading purposes.Data Load Demo: There is a script named data_load_demo.py, which serves as a reference for correctly reading the sparse matrix format of masks and other related data. Please refer to this script for specific loading procedures and required dependencies.
Dataset Loading Method
1. Direct Usage of Preprocessed Data
If you have already downloaded the preprocessed dataset, no additional data processing steps are required.
You can directly jump to step 2 to build and load the dataset.
Please note that the contents provided by this dataset have been transformed and numbered through data_process.py,
differing from the original nii.gz files. To understand the specific preprocessing process,
refer to the data_process.py file for detailed information. If adding new datasets or modifying existing ones,
please refer to data_process.py for data preprocessing and uniform formatting.
2. Build Dataset
To facilitate model training and evaluation using this dataset, we provide an example code for the Dataset class. Wrap the dataset in your project according to the following example:
Data Splitting
Each sub-dataset folder is splitted into train and test parts through a JSON file,
facilitating model training and testing.
Dataset Sources
Dataset Copyright Information
All datasets in this dataset are publicly available. For detailed copyright information, please refer to the corresponding dataset links.
Citation
If you use this dataset, please cite the following works:
@misc{bai2024m3d,
title={M3D: Advancing 3D Medical Image Analysis with Multi-Modal Large Language Models},
author={Fan Bai and Yuxin Du and Tiejun Huang and Max Q. -H. Meng and Bo Zhao},
year={2024},
eprint={2404.00578},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
@misc{du2024segvol,
title={SegVol: Universal and Interactive Volumetric Medical Image Segmentation},
author={Yuxin Du and Fan Bai and Tiejun Huang and Bo Zhao},
year={2024},
eprint={2311.13385},
archivePrefix={arXiv},
primaryClass={cs.CV}
}