hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
sequence | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
sequence | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
sequence | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
sequence | cell_types
sequence | cell_type_groups
sequence |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
e7f5645db450dd14e705dfc28aba1bc97621c8a6 | 726 | ipynb | Jupyter Notebook | tensorflow_basics.ipynb | sanattaori/Tensorflow-basics | 4abc428cab3319fd95dbaa268de2da0186510c68 | [
"MIT"
] | null | null | null | tensorflow_basics.ipynb | sanattaori/Tensorflow-basics | 4abc428cab3319fd95dbaa268de2da0186510c68 | [
"MIT"
] | null | null | null | tensorflow_basics.ipynb | sanattaori/Tensorflow-basics | 4abc428cab3319fd95dbaa268de2da0186510c68 | [
"MIT"
] | null | null | null | 17.707317 | 40 | 0.413223 | [
[
[
"# Tensorflow Basics",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
e7f573b703771e29c4dc2600c06d3b18d9dceef6 | 31,266 | ipynb | Jupyter Notebook | src/demoHydraulicsSizing_1.ipynb | kevindorma/Hydraulics2.jl | 331009cc9417b560eb9442fdf8e1e85d1c5d416f | [
"MIT"
] | null | null | null | src/demoHydraulicsSizing_1.ipynb | kevindorma/Hydraulics2.jl | 331009cc9417b560eb9442fdf8e1e85d1c5d416f | [
"MIT"
] | null | null | null | src/demoHydraulicsSizing_1.ipynb | kevindorma/Hydraulics2.jl | 331009cc9417b560eb9442fdf8e1e85d1c5d416f | [
"MIT"
] | null | null | null | 43.066116 | 2,040 | 0.491908 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e7f57dce2cae6cf605c2614d4545ffd6f3a7b29a | 2,977 | ipynb | Jupyter Notebook | 02-Advanced_Examples_of_Spark_Programs/Movie-Similarities.ipynb | egbertli/Apache-Spark-Projects-with-Python | fe805a13dfe29d70fd233580c96334de59a10128 | [
"MIT"
] | 1 | 2017-12-29T17:24:34.000Z | 2017-12-29T17:24:34.000Z | 02-Advanced_Examples_of_Spark_Programs/Movie-Similarities.ipynb | egbertli/Apache-Spark-Projects-with-Python | fe805a13dfe29d70fd233580c96334de59a10128 | [
"MIT"
] | null | null | null | 02-Advanced_Examples_of_Spark_Programs/Movie-Similarities.ipynb | egbertli/Apache-Spark-Projects-with-Python | fe805a13dfe29d70fd233580c96334de59a10128 | [
"MIT"
] | null | null | null | 25.886957 | 120 | 0.57474 | [
[
[
"# ITEM-BASED COLLABORATIVE FILTERING\n```\nFinding similar movies using Spark and the MovieLens data set\n\nIntroducing caching RDD's\n```",
"_____no_output_____"
],
[
"- Find every pair of movies that were watched by the same person\n- Measure the similarity of their ratings across all users who watched both\n- Sort by movie, then by similarity strength",
"_____no_output_____"
],
[
"## Turn into Spark Problem\n- Map input ratings to (userID, (movieID, rating))\n- Find every movie pair rated by the same user\n- - This can be done with a 'self-join' operation\n- - At this point we have (userID, (movieID1, rating1), (movieID2, rating2))\n- Filter out duplicated pairs\n- Make the movie pairs the key\n- - map to (movieID1, movieID2), (rating1, rating2))\n- groupByKey() to get every rating pair found for each movie pair\n- Compute similarity between ratings for each movie in the pair\n- Sor, Save and display results",
"_____no_output_____"
],
[
"- This is some heavy lifting! Let's use every core of computer\n```python\nconf = SparkConf().setMaster('local[*]').setAppName('MovieSimilarities')\nsc = SparkContext(conf=conf)\n```",
"_____no_output_____"
],
[
"#### CACHING RDD'S\n- In this example, we'll query the final RDD of movie similarities a couple of times\n- Any time you will perform more than one action on an RDD, you must cache it!\n- - Otherwise, Spark might re-evaluate the entire RDD all over again!\n- Use .cache() or .persist() to do this.\n- - What's the difference?\n- - Persisit() optionally lets you cache it to dist instead of just memory, just in case a node fails or something",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e7f585f6683c1ba93b7af81a646c536c1b3dad67 | 8,106 | ipynb | Jupyter Notebook | Project_2_VQE_Molecules/S4_Measurement.ipynb | HermanniH/CohortProject_2020_Week2 | 8b18676a4460fe93757a295a7692d1a4d7fcb28a | [
"MIT"
] | 1 | 2020-07-16T03:27:28.000Z | 2020-07-16T03:27:28.000Z | Project_2_VQE_Molecules/S4_Measurement.ipynb | HermanniH/CohortProject_2020_Week2 | 8b18676a4460fe93757a295a7692d1a4d7fcb28a | [
"MIT"
] | null | null | null | Project_2_VQE_Molecules/S4_Measurement.ipynb | HermanniH/CohortProject_2020_Week2 | 8b18676a4460fe93757a295a7692d1a4d7fcb28a | [
"MIT"
] | null | null | null | 31.176923 | 229 | 0.546139 | [
[
[
"# Measurement Grouping\n\nSince current quantum hardware is limited to single-qubit projective measurement, only terms commuting within individual qubit's subspace can be measured together. These terms are said to be qubit-wise commuting (QWC). \n\nThus, one can not measure the entire electronic Hamiltonian $\\hat H$ at once, and instead needs to separate it into fragments. \n$$\\hat H = \\sum_n \\hat H_n$$\nwhere each $\\hat H_n$ is a QWC fragment. ",
"_____no_output_____"
]
],
[
[
"from utility import * ",
"_____no_output_____"
]
],
[
[
"Here we use $H_2$ as an example for finding QWC fragments. Notice below that each fragment has the same terms on all qubits. \n\nTo show differences between QWC and more advanced grouping, we didn't use the qubit-tappering techinique shown in step 2.",
"_____no_output_____"
]
],
[
[
"h2 = get_qubit_hamiltonian(mol='h2', geometry=1, basis='sto3g', qubit_transf='jw')\n\nqwc_list = get_qwc_group(h2)\nprint('Fragments 1: \\n{}\\n'.format(qwc_list[4]))\nprint('Fragments 2:\\n{}\\n'.format(qwc_list[1]))\nprint('Number of fragments: {}'.format(len(qwc_list)))",
"Fragments 1: \n0.13716572937099508 [Z0] +\n0.15660062488237947 [Z0 Z1] +\n0.10622904490856075 [Z0 Z2] +\n0.15542669077992832 [Z0 Z3] +\n0.13716572937099503 [Z1] +\n0.15542669077992832 [Z1 Z2] +\n0.10622904490856075 [Z1 Z3] +\n-0.13036292057109117 [Z2] +\n0.16326768673564346 [Z2 Z3] +\n-0.13036292057109117 [Z3]\n\nFragments 2:\n-0.04919764587136755 [X0 X1 Y2 Y3]\n\nNumber of fragments: 5\n"
]
],
[
[
"By applying extra unitaries, one may rotate more terms of $\\hat H$ into a QWC fragment. \n\nRecall that in digital quantum computing, the expectation value of $\\hat H_n$ given a trial wavefunction $|\\psi\\rangle$ is \n$$ E_n =\\ \\langle\\psi| \\hat H_n | \\psi\\rangle$$\nInserting unitary transformation $\\hat U_n$ does not change the expectation value.\n$$ E_n =\\ \\langle\\psi| \\hat U_n^\\dagger \\hat U_n \\hat H_n \\hat U_n^\\dagger \\hat U_n |\\psi\\rangle$$ \nThis nonetheless changes the trial wavefunction and the terms to be measured. \n$$ |\\psi\\rangle \\rightarrow \\hat U_n |\\psi\\rangle = |\\phi\\rangle$$\n$$ \\hat H_n \\rightarrow \\hat U_n \\hat H_n \\hat U_n^\\dagger = \\hat A_n$$\nThe transformation of $|\\psi \\rangle$ can be done on the quantum computer, and the transformation of $\\hat H_n$ is possible on the classical computer. \n\nNow, although $\\hat A_n$ needs to be a QWC fragment to be measurable on a quantum computer, $\\hat H_n$ does not. \nInstead, if we restrict $\\hat U_n$ to be a clifford operation, the terms in $\\hat H$ need only mutually commute. \n\nHere, we obtain measurable parts of $H_2$ by partitioning its terms into mutually commuting fragments. ",
"_____no_output_____"
]
],
[
[
"comm_groups = get_commuting_group(h2)\nprint('Number of mutually commuting fragments: {}'.format(len(comm_groups)))\nprint('The first commuting group')\nprint(comm_groups[1])",
"Number of mutually commuting fragments: 2\nThe first commuting group\n-0.32760818967480887 [] +\n-0.04919764587136755 [X0 X1 Y2 Y3] +\n0.04919764587136755 [X0 Y1 Y2 X3] +\n0.04919764587136755 [Y0 X1 X2 Y3] +\n-0.04919764587136755 [Y0 Y1 X2 X3] +\n0.15660062488237947 [Z0 Z1] +\n0.10622904490856075 [Z0 Z2] +\n0.15542669077992832 [Z0 Z3] +\n0.15542669077992832 [Z1 Z2] +\n0.10622904490856075 [Z1 Z3] +\n0.16326768673564346 [Z2 Z3]\n"
]
],
[
[
"To see this fragment is indeed measurable, one can construct the corresponding unitary operator $\\hat U_n$.",
"_____no_output_____"
]
],
[
[
"uqwc = get_qwc_unitary(comm_groups[1])\nprint('This is unitary, U * U^+ = I ')\nprint(uqwc * uqwc)",
"This is unitary, U * U^+ = I \n(0.9999999999999996+0j) []\n"
]
],
[
[
"Applying this unitary gives the qubit-wise commuting form of the first mutually commuting group",
"_____no_output_____"
]
],
[
[
"qwc = remove_complex(uqwc * comm_groups[1] * uqwc)\nprint(qwc)",
"-0.32760818967480876 [] +\n0.1554266907799282 [X0] +\n0.1566006248823793 [X0 X1] +\n0.04919764587136754 [X0 X1 Z3] +\n0.1062290449085607 [X0 X2] +\n-0.04919764587136754 [X0 Z3] +\n0.1062290449085607 [X1] +\n0.1554266907799282 [X1 X2] +\n-0.04919764587136754 [X1 X2 Z3] +\n0.16326768673564332 [X2] +\n0.04919764587136754 [X2 Z3]\n"
]
],
[
[
"In addition, current quantum computer can measure only the $z$ operators. Thus, QWC fragments with $x$ or $y$ operators require extra single-qubit unitaries that rotate them into $z$. ",
"_____no_output_____"
]
],
[
[
"uz = get_zform_unitary(qwc)\nprint(\"Checking whether U * U^+ is identity: {}\".format(uz * uz))\n\nallz = remove_complex(uz * qwc * uz)\nprint(\"\\nThe all-z form of qwc fragment:\\n{}\".format(allz))",
"Checking whether U * U^+ is identity: 0.9999999999999998 []\n\nThe all-z form of qwc fragment:\n-0.3276081896748086 [] +\n0.15542669077992813 [Z0] +\n0.15660062488237922 [Z0 Z1] +\n0.049197645871367504 [Z0 Z1 Z3] +\n0.10622904490856065 [Z0 Z2] +\n-0.049197645871367504 [Z0 Z3] +\n0.10622904490856065 [Z1] +\n0.15542669077992813 [Z1 Z2] +\n-0.049197645871367504 [Z1 Z2 Z3] +\n0.16326768673564326 [Z2] +\n0.049197645871367504 [Z2 Z3]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f587fa7685855bf2491df1f3a3d9ad0e18e103 | 495,529 | ipynb | Jupyter Notebook | Evaluating Model Profitability.ipynb | gschwaeb/NHL_Game_Prediction | c7a4dd46fcb2c96a25f6fbc8c2fdbf7b33528aef | [
"CC0-1.0"
] | 3 | 2021-09-21T22:48:13.000Z | 2022-02-19T21:32:13.000Z | Evaluating Model Profitability.ipynb | gschwaeb/NHL_Game_Prediction | c7a4dd46fcb2c96a25f6fbc8c2fdbf7b33528aef | [
"CC0-1.0"
] | null | null | null | Evaluating Model Profitability.ipynb | gschwaeb/NHL_Game_Prediction | c7a4dd46fcb2c96a25f6fbc8c2fdbf7b33528aef | [
"CC0-1.0"
] | 3 | 2022-02-04T21:49:30.000Z | 2022-03-23T03:18:08.000Z | 136.284103 | 124,972 | 0.794993 | [
[
[
"# Overview\n\nIn this notebook, I will compare predictions on the 2021 season from my final model against historical odds. Data for the historical odds was gathered from [Sportsbook Reviews Online](https://www.sportsbookreviewsonline.com/scoresoddsarchives/nhl/nhloddsarchives.htm). Per their website: Data is sourced from various online sportsbooks including 5dimes, BetOnline, Bookmaker, Heritage, Pinnacle Sports, Sportsbook.com as well as the Westgate Superbook in Las Vegas.\n\nI will look at 2 simple betting strategies to determine profitability:\n\n1. Bet 100 on every game where either the home or away team winning probability from my model is higher than the implied odds \n2. Bet to win 100 on every game where either the home or away team winning probability from my model is higher than the implied odds",
"_____no_output_____"
],
[
"# Data Cleaning and Merging",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport numpy as np\nimport pickle\nsns.set_style(\"darkgrid\")\nsns.set_context(\"poster\")\npd.set_option('display.max_columns', None)",
"_____no_output_____"
],
[
"odds = pd.read_excel('data/nhl odds 2021.xlsx')",
"_____no_output_____"
],
[
"odds.head()",
"_____no_output_____"
],
[
"team_conversion = { 'Anaheim': 'ANA', \n 'Arizona' :'ARI', \n 'Boston': 'BOS', \n 'Buffalo':'BUF',\n 'Calgary': 'CGY', \n 'Carolina': 'CAR', \n 'Chicago': 'CHI', \n 'Colorado': 'COL',\n 'Columbus': 'CBJ',\n 'Dallas': 'DAL',\n 'Detroit': 'DET',\n 'Edmonton': 'EDM',\n 'Florida': 'FLA',\n 'LosAngeles': 'L.A',\n 'Minnesota': 'MIN',\n 'Montreal': 'MTL',\n 'Nashville': 'NSH',\n 'NewJersey': 'N.J',\n 'NYIslanders': 'NYI',\n 'NYRangers': 'NYR',\n 'Ottawa': 'OTT',\n 'Philadelphia': 'PHI',\n 'Pittsburgh': 'PIT',\n 'SanJose': 'S.J',\n 'St.Louis': 'STL',\n 'TampaBay': 'T.B',\n 'Toronto': 'TOR',\n 'Vancouver': 'VAN',\n 'Vegas':'VGK',\n 'Washington': 'WSH',\n 'Winnipeg': 'WPG'}",
"_____no_output_____"
],
[
"#convert date to proper datestring and create team key\nodds = odds.replace({'Team': team_conversion})\nodds['Month'] = odds['Date'].apply(lambda x: '0'+str(x)[0])\nodds['Day'] = odds['Date'].apply(lambda x: str(x)[1:])\nodds['Year'] = 2021\nodds['Datestring'] = odds[['Year','Month','Day']].astype(str).apply('-'.join, 1) \nodds['Team_Key'] = odds['Team'].astype(str)+'_'+odds['Datestring'].astype(str)\n#calculate implied odds\nodds['Implied_odds'] = np.where(odds['Close'] < 0, (odds['Close']*-1)/((odds['Close']*-1)+100) , 100/(odds['Close']+100))",
"_____no_output_____"
],
[
"odds.head(5)",
"_____no_output_____"
],
[
"#import file with predictions\npredictions = pd.read_csv('data/Predictions_2021b')",
"_____no_output_____"
],
[
"#merge my predictions with odd df\ndf = predictions.merge(odds.loc[:,['Team_Key', 'Implied_odds', 'Close']].add_prefix('home_'), how = 'left', left_on = 'Home_Team_Key', right_on = 'home_Team_Key').drop(columns = 'home_Team_Key')\ndf = df.merge(odds.loc[:,['Team_Key', 'Implied_odds', 'Close']].add_prefix('away_'), how = 'left', left_on = 'Away_Team_Key', right_on = 'away_Team_Key').drop(columns = 'away_Team_Key')",
"_____no_output_____"
],
[
"#odds info only contains info for games up to 5/4. These are the 15 missing games below. \ndf.isna().sum()",
"_____no_output_____"
],
[
"#drop missing games from df\ndf = df.dropna()",
"_____no_output_____"
],
[
"conditions = [df['Home Win Probability'] > df['home_Implied_odds'],\n df['Away Win Probability'] > df['away_Implied_odds']\n ]\n \n\nchoices = ['Home', \n 'Away']\n\ndf['Bet'] = np.select(conditions, choices, default = 'No Bet')\n",
"_____no_output_____"
],
[
"df['Favorites'] = np.where(df['home_Implied_odds'] >df['away_Implied_odds'], 'Home', 'Away' )\nconditions = [df['Bet'] == 'No Bet',\n df['Bet'] == df['Favorites'],\n df['Bet'] != df['Favorites']\n \n \n]\nchoices = ['No Bet',\n 'Favorite',\n 'Underdog'\n ]\n\ndf['Bet_For'] = np.select(conditions, choices)",
"_____no_output_____"
],
[
"#calculate profit for 100$ per game strat\nconditions = [((df['Bet'] == 'Home') & (df['Home_Team_Won'] == 1) & (df['home_Close'] <0)),\n ((df['Bet'] == 'Home') & (df['Home_Team_Won'] == 1) & (df['home_Close']>0)),\n ((df['Bet'] == 'Away') & (df['Home_Team_Won'] == 0) & (df['away_Close']<0)),\n ((df['Bet'] == 'Away') & (df['Home_Team_Won'] == 0) & (df['away_Close']>0)),\n df['Bet'] == 'No Bet'\n ]\n \n\nchoices = [-100 * (100/df['home_Close']), \n df['home_Close'],\n -100 * (100/df['away_Close']),\n df['away_Close'],\n 0]\n\ndf['Profit_Strat1'] = np.select(conditions, choices, default = -100)",
"_____no_output_____"
],
[
"#calculate profit for bet to win 100$ strat\nconditions = [((df['Bet'] == 'Home') & (df['Home_Team_Won'] == 1) & (df['home_Close'] <0)),\n ((df['Bet'] == 'Home') & (df['Home_Team_Won'] == 1) & (df['home_Close']>0)),\n ((df['Bet'] == 'Home') & (df['Home_Team_Won'] == 0) & (df['home_Close']>0)),\n ((df['Bet'] == 'Home') & (df['Home_Team_Won'] == 0) & (df['home_Close']<0)),\n ((df['Bet'] == 'Away') & (df['Home_Team_Won'] == 0) & (df['away_Close']<0)),\n ((df['Bet'] == 'Away') & (df['Home_Team_Won'] == 0) & (df['away_Close']>0)),\n ((df['Bet'] == 'Away') & (df['Home_Team_Won'] == 1) & (df['away_Close']>0)),\n ((df['Bet'] == 'Away') & (df['Home_Team_Won'] == 1) & (df['away_Close']<0)),\n df['Bet'] == 'No Bet'\n ]\n \n\nchoices = [100, \n 100,\n (100/df['home_Close'])*-100,\n df['home_Close'],\n 100,\n 100,\n (100/df['away_Close'])*-100,\n df['away_Close'],\n 0]\n\ndf['Profit_Strat2'] = np.select(conditions, choices)",
"_____no_output_____"
],
[
"#cost of bet to win 100$ strat\nconditions = [((df['Bet'] == 'Home') & (df['home_Close']>0)),\n ((df['Bet'] == 'Home') & (df['home_Close']<0)),\n ((df['Bet'] == 'Away') & (df['away_Close']>0)),\n ((df['Bet'] == 'Away') & (df['away_Close']<0)),\n df['Bet'] == 'No Bet'\n ]\n \n\nchoices = [(100/df['home_Close'])*100,\n df['home_Close']*-1,\n (100/df['away_Close'])*100,\n df['away_Close']*-1,\n 0]\n\ndf['Cost_Strat2'] = np.select(conditions, choices)",
"_____no_output_____"
],
[
"#convert date to pandas datetime\ndf['date'] = pd.to_datetime(df['date'])",
"_____no_output_____"
],
[
"#calculate cumulative profit for poth strategies\ndf['Profit_Strat2_cumsum'] = df['Profit_Strat2'].cumsum()\ndf['Profit_Strat1_cumsum'] = df['Profit_Strat1'].cumsum()",
"_____no_output_____"
],
[
"df['Won_Bet'] = np.where(df['Profit_Strat2'] > 0, 1, 0)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"# Evaluation\nLet's check the log loss from the implied odds. My model's log loss on the 2021 season was 0.655534. So the book implied odds are still performing slightly better with a log loss of 0.6529",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import log_loss, accuracy_score\n\nio_list = []\n\nfor index, row in df.iterrows():\n io_list.append([row['away_Implied_odds'], row['home_Implied_odds']])\n \n\nlog_loss(df['Home_Team_Won'], io_list)",
"_____no_output_____"
]
],
[
[
"How many bets would be for home vs away vs no bet? My model is definitley favoring the home team. From the EDA notebook, The home team had won 56.0% in the 17-18 season, 53.7% in 18-19, 53.1% in 19-20 and only 52.7% in 20-21 season. THe 20-21 season having no fans may be affecting this outcome for the home team and may have hurt the model slightly for the 20-21 season.",
"_____no_output_____"
]
],
[
[
"df['Bet'].value_counts()",
"_____no_output_____"
],
[
"df['Bet'].value_counts(normalize = True)",
"_____no_output_____"
]
],
[
[
"How many bets were for the favorite vs the underdrog? Interestingly the model liked underdogs more often. ",
"_____no_output_____"
]
],
[
[
"df['Bet_For'].value_counts(normalize = 'True')",
"_____no_output_____"
]
],
[
[
"The strategy of betting to win 100$ resulted in a per bet ROI of 2.04%",
"_____no_output_____"
]
],
[
[
"#ROI per bet\ndf['Profit_Strat2'].sum() / df['Cost_Strat2'].sum()",
"_____no_output_____"
]
],
[
[
"Total profit for this strategy would have been $1,473.69",
"_____no_output_____"
]
],
[
[
"#total profit\ndf['Profit_Strat2'].sum()",
"_____no_output_____"
]
],
[
[
"The strategy was profitable initially and dipped down into the red for short period in mid March. You would have only needed an initial bankroll of 325 to implement this and then would have needed to re-up 244 later for total out of pocket costs of 569",
"_____no_output_____"
]
],
[
[
"#initial bankroll needed \ndf[df['date'] == '2021-01-13']['Cost_Strat2'].sum()",
"_____no_output_____"
],
[
"df[df['Profit_Strat2_cumsum'] < 0]",
"_____no_output_____"
]
],
[
[
"I would have won only 49.6% of bets, but since the marjority of bets were for the underdog, the lower costs benefited profitability. ",
"_____no_output_____"
]
],
[
[
"df[df['Bet'] != 'No Bet']['Won_Bet'].value_counts(normalize = True)",
"_____no_output_____"
]
],
[
[
"Strategy 1, bet 100$ every bettable game was slightly profitable.",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize = (16,12))\n\nax = sns.lineplot(x = df['date'], y = df['Profit_Strat1_cumsum'], color = 'green')\nax.set_title('Cumulative Profit', fontsize = 24)\nax.set_ylabel('Cumulative Profit', fontsize =16, )\nax.set_xlabel('Date', fontsize =16)\nplt.xticks(rotation=45, fontsize = 16)\nax.axhline(0, linestyle = 'dashed', color = 'black')\nax.set_ylim(-2000,4000)\n\n\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Strategy 2, bet to win 100$ on every bettable game, was profitbale",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize = (16,12))\n\nax = sns.lineplot(x = df['date'], y = df['Profit_Strat2_cumsum'], color = 'green')\nax.set_title('Cumulative Profit', fontsize = 24)\nax.set_ylabel('Cumulative Profit', fontsize =18, )\nax.set_xlabel('Date', fontsize =18)\nplt.xticks(rotation=45, fontsize = 18)\nax.axhline(0, linestyle = 'dashed', color = 'black')\nax.set_ylim(-1000,4000)\n\n\n\nplt.show()",
"_____no_output_____"
],
[
"strat2 = pd.DataFrame(df.groupby('date').agg({'Profit_Strat2': 'sum'})).reset_index()\n\nstrat2['Cumulative Profit'] = strat2['Profit_Strat2'].cumsum()\n\nstrat2['date'] = pd.to_datetime(strat2['date'])",
"_____no_output_____"
],
[
"strat2.head()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize = (16,12))\n\nax = sns.lineplot(x = strat2['date'], y = strat2['Profit_Strat2'], palette = 'Blues')\nax.set_title('Daily Profit', fontsize = 18)\nax.set_ylabel('Daily Profit', fontsize =12, )\nax.set_xlabel('Date', fontsize =12)\nplt.xticks(rotation='vertical', fontsize = 12)\nax.axhline(0, color = 'black', linestyle = 'dashed')\nax.set_ylim(-900,900)\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7f590fdd1ac484374752b538b4fbbca1551c3b4 | 71,663 | ipynb | Jupyter Notebook | P2_Explore_Movie_Dataset/Explore Movie Dataset.ipynb | CCCCCaO/My_AIPND_Projects | 6904d6af6aef8a666392dfe5dce438fa8187defe | [
"MIT"
] | 2 | 2019-01-16T11:36:42.000Z | 2019-01-16T11:36:48.000Z | P3_Linear_Algebra/P2_Explore_Movie_Dataset/Explore Movie Dataset.ipynb | CCCCCaO/My_AIPND_Projects | 6904d6af6aef8a666392dfe5dce438fa8187defe | [
"MIT"
] | null | null | null | P3_Linear_Algebra/P2_Explore_Movie_Dataset/Explore Movie Dataset.ipynb | CCCCCaO/My_AIPND_Projects | 6904d6af6aef8a666392dfe5dce438fa8187defe | [
"MIT"
] | null | null | null | 50.32514 | 18,500 | 0.631246 | [
[
[
"## 探索电影数据集\n\n在这个项目中,你将尝试使用所学的知识,使用 `NumPy`、`Pandas`、`matplotlib`、`seaborn` 库中的函数,来对电影数据集进行探索。\n\n下载数据集:\n[TMDb电影数据](https://s3.cn-north-1.amazonaws.com.cn/static-documents/nd101/explore+dataset/tmdb-movies.csv)\n",
"_____no_output_____"
],
[
"\n数据集各列名称的含义:\n<table>\n<thead><tr><th>列名称</th><th>id</th><th>imdb_id</th><th>popularity</th><th>budget</th><th>revenue</th><th>original_title</th><th>cast</th><th>homepage</th><th>director</th><th>tagline</th><th>keywords</th><th>overview</th><th>runtime</th><th>genres</th><th>production_companies</th><th>release_date</th><th>vote_count</th><th>vote_average</th><th>release_year</th><th>budget_adj</th><th>revenue_adj</th></tr></thead><tbody>\n <tr><td>含义</td><td>编号</td><td>IMDB 编号</td><td>知名度</td><td>预算</td><td>票房</td><td>名称</td><td>主演</td><td>网站</td><td>导演</td><td>宣传词</td><td>关键词</td><td>简介</td><td>时常</td><td>类别</td><td>发行公司</td><td>发行日期</td><td>投票总数</td><td>投票均值</td><td>发行年份</td><td>预算(调整后)</td><td>票房(调整后)</td></tr>\n</tbody></table>\n",
"_____no_output_____"
],
[
"**请注意,你需要提交该报告导出的 `.html`、`.ipynb` 以及 `.py` 文件。**",
"_____no_output_____"
],
[
"\n\n---\n\n---\n\n## 第一节 数据的导入与处理\n\n在这一部分,你需要编写代码,使用 Pandas 读取数据,并进行预处理。",
"_____no_output_____"
],
[
"\n**任务1.1:** 导入库以及数据\n\n1. 载入需要的库 `NumPy`、`Pandas`、`matplotlib`、`seaborn`。\n2. 利用 `Pandas` 库,读取 `tmdb-movies.csv` 中的数据,保存为 `movie_data`。\n\n提示:记得使用 notebook 中的魔法指令 `%matplotlib inline`,否则会导致你接下来无法打印出图像。",
"_____no_output_____"
]
],
[
[
"# 各库导入\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sb\n\n# 数据读取\nmovie_data = pd.read_csv('./tmdb-movies.csv')",
"_____no_output_____"
]
],
[
[
"---\n\n**任务1.2: ** 了解数据\n\n你会接触到各种各样的数据表,因此在读取之后,我们有必要通过一些简单的方法,来了解我们数据表是什么样子的。\n\n1. 获取数据表的行列,并打印。\n2. 使用 `.head()`、`.tail()`、`.sample()` 方法,观察、了解数据表的情况。\n3. 使用 `.dtypes` 属性,来查看各列数据的数据类型。\n4. 使用 `isnull()` 配合 `.any()` 等方法,来查看各列是否存在空值。\n5. 使用 `.describe()` 方法,看看数据表中数值型的数据是怎么分布的。\n\n",
"_____no_output_____"
]
],
[
[
"print('1.电影数据集的行列数为:',movie_data.shape)",
"1.电影数据集的行列数为: (10866, 21)\n"
],
[
"movie_data.head() # 2.1 电影数据集的前五行",
"_____no_output_____"
],
[
"movie_data.tail() # 2-2 电影数据集的末五行",
"_____no_output_____"
],
[
"movie_data.sample() # 2-3 随机抽取一个电影数据样本",
"_____no_output_____"
],
[
"movie_data.dtypes # 3 获取每列的数据类型",
"_____no_output_____"
],
[
"movie_data.isnull().any() # 4 检查各列是否有NaN值",
"_____no_output_____"
],
[
"movie_data['id'].describe() # 5-1 编号id列(int64)的描述性统计信息",
"_____no_output_____"
],
[
"movie_data['popularity'].describe() # 5-2 知名度popularity列(float64)",
"_____no_output_____"
],
[
"movie_data['budget'].describe() # 5-3 预算budget列 (int64)",
"_____no_output_____"
],
[
"movie_data['revenue'].describe() # 5-4 票房revenue列 (int64)",
"_____no_output_____"
],
[
"movie_data['runtime'].describe() # 5-5 时长runtime列(int64)",
"_____no_output_____"
],
[
"movie_data['vote_count'].describe() # 5-5 投票总数vote_count列(int64)",
"_____no_output_____"
],
[
"movie_data['vote_average'].describe() # 5-6 投票均值vote_average列(float64)",
"_____no_output_____"
],
[
"movie_data['release_year'].describe() # 5-7 发行年份release_year列(int64)",
"_____no_output_____"
],
[
"movie_data['budget_adj'].describe() # 5-8 预算(调整后)budget_adj列(float64)",
"_____no_output_____"
],
[
"movie_data['revenue_adj'].describe() # 5-9 票房(调整后)revenue_adj列(float64)",
"_____no_output_____"
]
],
[
[
"---\n\n**任务1.3: ** 清理数据\n\n在真实的工作场景中,数据处理往往是最为费时费力的环节。但是幸运的是,我们提供给大家的 tmdb 数据集非常的「干净」,不需要大家做特别多的数据清洗以及处理工作。在这一步中,你的核心的工作主要是对数据表中的空值进行处理。你可以使用 `.fillna()` 来填补空值,当然也可以使用 `.dropna()` 来丢弃数据表中包含空值的某些行或者列。\n\n任务:使用适当的方法来清理空值,并将得到的数据保存。",
"_____no_output_____"
]
],
[
[
"# 这里采用将NaN值都替换为0 并保存至movie_data_adj中\nprint(\"处理前NaN值有:\", movie_data.isnull().sum().sum(),\"个\")\nmovie_data_adj = movie_data.fillna(0)\nprint(\"处理前NaN值有:\", movie_data_adj.isnull().sum().sum(),\"个\")",
"处理前NaN值有: 13434 个\n处理前NaN值有: 0 个\n"
]
],
[
[
"---\n\n---\n\n## 第二节 根据指定要求读取数据\n\n\n相比 Excel 等数据分析软件,Pandas 的一大特长在于,能够轻松地基于复杂的逻辑选择合适的数据。因此,如何根据指定的要求,从数据表当获取适当的数据,是使用 Pandas 中非常重要的技能,也是本节重点考察大家的内容。\n\n",
"_____no_output_____"
],
[
"---\n\n**任务2.1: ** 简单读取\n\n1. 读取数据表中名为 `id`、`popularity`、`budget`、`runtime`、`vote_average` 列的数据。\n2. 读取数据表中前1~20行以及48、49行的数据。\n3. 读取数据表中第50~60行的 `popularity` 那一列的数据。\n\n要求:每一个语句只能用一行代码实现。",
"_____no_output_____"
]
],
[
[
"# 注:参考了笔记和https://blog.csdn.net/u011089523/article/details/60341016\n\n# 2.1.1.读取某列数据 \n# 各列分别读取 df[['列名']]来访问\nmovie_data_id = movie_data[['id']]\nmovie_data_pop = movie_data[['popularity']]\nmovie_data_bud = movie_data[['budget']]\nmovie_data_rt = movie_data[['runtime']]\nmovie_data_vote_avg = movie_data[['vote_average']]\n# 各列一起读取 df[['列名1','列名2'...列名的列表]]来访问\nmovie_data_sel = movie_data[['id', 'popularity', 'budget', 'runtime', \n 'vote_average']]\n\n# 2.1.2 读取x行数据\n# 读取前20行的两种方法 df.head(n) 或 df[m:n]\nmovie_data_rows_1to20_1 = movie_data.head(20)\nmovie_data_rows_1to20_2 = movie_data[0:20]\n# 读取48,49行数据 注意索引从0开始 前闭后开\nmovie_data_rows_48to49 = movie_data[47:49]",
"_____no_output_____"
]
],
[
[
"---\n\n**任务2.2: **逻辑读取(Logical Indexing)\n\n1. 读取数据表中 **`popularity` 大于5** 的所有数据。\n2. 读取数据表中 **`popularity` 大于5** 的所有数据且**发行年份在1996年之后**的所有数据。\n\n提示:Pandas 中的逻辑运算符如 `&`、`|`,分别代表`且`以及`或`。\n\n要求:请使用 Logical Indexing实现。",
"_____no_output_____"
]
],
[
[
"# 参考了https://blog.csdn.net/GeekLeee/article/details/75268762\n# 1.读取popularity>5的所有数据\nmovie_data_pop_morethan5 = movie_data.loc[movie_data['popularity']>5]\n\n# 2.读取popularity>5 且 发行年份>1996的所有数据\nmovie_data_pop5p_rls1996p = movie_data.loc[(movie_data['popularity']>5)&(movie_data['release_year']>1996) ]",
"_____no_output_____"
]
],
[
[
"---\n\n**任务2.3: **分组读取\n\n1. 对 `release_year` 进行分组,使用 [`.agg`](http://pandas.pydata.org/pandas-docs/version/0.22/generated/pandas.core.groupby.DataFrameGroupBy.agg.html) 获得 `revenue` 的均值。\n2. 对 `director` 进行分组,使用 [`.agg`](http://pandas.pydata.org/pandas-docs/version/0.22/generated/pandas.core.groupby.DataFrameGroupBy.agg.html) 获得 `popularity` 的均值,从高到低排列。\n\n要求:使用 `Groupby` 命令实现。",
"_____no_output_____"
]
],
[
[
"data = movie_data\n# 按release_year分组 获取revenue均值\nrevenue_mean_groupby_rlsyear = data.groupby(['release_year'])['revenue'].agg('mean')\n\n# 按director分组 获取popularity均值\npopularity_mean_groupby_director = data.groupby(['director'])['popularity'].agg('mean')",
"_____no_output_____"
]
],
[
[
"---\n\n---\n\n## 第三节 绘图与可视化\n\n接着你要尝试对你的数据进行图像的绘制以及可视化。这一节最重要的是,你能够选择合适的图像,对特定的可视化目标进行可视化。所谓可视化的目标,是你希望从可视化的过程中,观察到怎样的信息以及变化。例如,观察票房随着时间的变化、哪个导演最受欢迎等。\n\n<table>\n<thead><tr><th>可视化的目标</th><th>可以使用的图像</th></tr></thead><tbody>\n <tr><td>表示某一属性数据的分布</td><td>饼图、直方图、散点图</td></tr>\n <tr><td>表示某一属性数据随着某一个变量变化</td><td>条形图、折线图、热力图</td></tr>\n <tr><td>比较多个属性的数据之间的关系</td><td>散点图、小提琴图、堆积条形图、堆积折线图</td></tr>\n</tbody></table>\n\n在这个部分,你需要根据题目中问题,选择适当的可视化图像进行绘制,并进行相应的分析。对于选做题,他们具有一定的难度,你可以尝试挑战一下~",
"_____no_output_____"
],
[
"**任务3.1:**对 `popularity` 最高的20名电影绘制其 `popularity` 值。",
"_____no_output_____"
]
],
[
[
"base_color = sb.color_palette()[0] # 取第一个颜色\ny_count = movie_data_adj['popularity'][:20]\n\"\"\" \n这块有些搞不懂如何去绘制? 应该用条形图合适还是用直方图合适 感觉二者都不合适 \n饼图不适合20个扇形 直方图和条形图似乎x和y有些问题 这里用条形图勉强绘制出 感觉不合适\n另有一个问题即如何在sb.barplot中标注出某个条形图具体的数值 在countplot中可以有办法标注出频率 \n我猜测应该可以在barplot标注出数值,可是并没有相关资料或者示例....\n有些疑惑,请求解答,谢谢!!!\n\"\"\"\n# 绘图\nsb.barplot(x = y_count.index.values+1, y = y_count, color = base_color, orient = \"v\")\n\n\"\"\"\n可以从图表中得知:\n热度第1(数值达32.98)和热度第2(数值达28.41)的电影其流行程度远超第3以及之后所有的电影,差距达到了一倍以上。\n第3到第20的电影其热度相差不大 数值均在5-15范围之内 较为稳定\n\"\"\";",
"_____no_output_____"
]
],
[
[
"---\n**任务3.2:**分析电影净利润(票房-成本)随着年份变化的情况,并简单进行分析。",
"_____no_output_____"
]
],
[
[
"# 需要考虑净利润随时间变化的情况 所以选择 折线图 适宜\n# 调整分箱边缘和中心点\nxbin_edges = np.arange(1960, movie_data_adj['release_year'].max()+2,2)\nxbin_centers = (xbin_edges + 0.25/2)[:-1]\n# 计算每个分箱中的统计数值\ndata_xbins = pd.cut(movie_data_adj['release_year'], xbin_edges, right = False, include_lowest = True)\ny_means = movie_data_adj['revenue_adj'].groupby(data_xbins).mean()-movie_data_adj['budget_adj'].groupby(data_xbins).mean()\ny_sems = movie_data_adj['revenue_adj'].groupby(data_xbins).sem()-movie_data_adj['budget_adj'].groupby(data_xbins).sem()\n# 绘图\nplt.errorbar(x = xbin_centers, y = y_means, yerr = y_sems)\nplt.xlabel('release year');\nplt.ylabel('Net profit');\n\n\"\"\"\n可以从图中看出:\n随着年份的变化(这里选取的是电影的发行年份作参考)\n净利润本在1960-1970年段先下降后上升再下降,较不稳定;\n而后在1970-1980年段达到了一个净利润的峰值,可见当时的电影市场火爆;\n而后在1980之后,净利润整体呈逐年下降的趋势,趋于稳定,市场也逐渐成熟。\n净利润的波动(即误差线)再1960-1980年间较大,考虑到电影市场刚刚兴起,符合实际;\n在后来进入市场成熟期之后,1980年之后,波动较小,更加稳定。\nPS:不太清楚如何写分析,应该从哪些角度入手,哪些东西该讲,哪些不用讲....\n\"\"\";",
"_____no_output_____"
]
],
[
[
"---\n\n**[选做]任务3.3:**选择最多产的10位导演(电影数量最多的),绘制他们排行前3的三部电影的票房情况,并简要进行分析。",
"_____no_output_____"
],
[
"---\n\n**[选做]任务3.4:**分析1968年~2015年六月电影的数量的变化。",
"_____no_output_____"
],
[
"---\n\n**[选做]任务3.5:**分析1968年~2015年六月电影 `Comedy` 和 `Drama` 两类电影的数量的变化。",
"_____no_output_____"
],
[
"> 注意: 当你写完了所有的代码,并且回答了所有的问题。你就可以把你的 iPython Notebook 导出成 HTML 文件。你可以在菜单栏,这样导出**File -> Download as -> HTML (.html)、Python (.py)** 把导出的 HTML、python文件 和这个 iPython notebook 一起提交给审阅者。",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
e7f5960522c25831c290b7d4eefb8f6dc242416f | 67,139 | ipynb | Jupyter Notebook | focus_test_bert.ipynb | sudheer007/BERT-N-ALBERT | 60fe0d6987ed2999b7501ece0f03dae2bbe219f4 | [
"MIT"
] | null | null | null | focus_test_bert.ipynb | sudheer007/BERT-N-ALBERT | 60fe0d6987ed2999b7501ece0f03dae2bbe219f4 | [
"MIT"
] | null | null | null | focus_test_bert.ipynb | sudheer007/BERT-N-ALBERT | 60fe0d6987ed2999b7501ece0f03dae2bbe219f4 | [
"MIT"
] | null | null | null | 38.299487 | 1,054 | 0.498846 | [
[
[
"%tensorflow_version 2.x\nimport tensorflow as tf\nprint(\"Tensorflow version \" + tf.__version__)\n\ntry:\n tpu = tf.distribute.cluster_resolver.TPUClusterResolver() # TPU detection\n print('Running on TPU ', tpu.cluster_spec().as_dict()['worker'])\nexcept ValueError:\n raise BaseException('ERROR: Not connected to a TPU runtime; please see the previous cell in this notebook for instructions!')\n\ntf.config.experimental_connect_to_cluster(tpu)\ntf.tpu.experimental.initialize_tpu_system(tpu)\ntpu_strategy = tf.distribute.experimental.TPUStrategy(tpu)",
"Tensorflow version 2.2.0\nRunning on TPU ['10.111.74.170:8470']\nINFO:tensorflow:Initializing the TPU system: grpc://10.111.74.170:8470\n"
],
[
"!pip install tokenizer\n!pip install transformers",
"Collecting tokenizer\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/a8/7e/e68e6b91c13028b12aa0e7079f962888a453cf9240d168ae25ded612a3e1/tokenizer-2.1.0-py2.py3-none-any.whl (471kB)\n\u001b[K |████████████████████████████████| 481kB 3.4MB/s \n\u001b[?25hInstalling collected packages: tokenizer\nSuccessfully installed tokenizer-2.1.0\nCollecting transformers\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/27/3c/91ed8f5c4e7ef3227b4119200fc0ed4b4fd965b1f0172021c25701087825/transformers-3.0.2-py3-none-any.whl (769kB)\n\u001b[K |████████████████████████████████| 778kB 3.4MB/s \n\u001b[?25hCollecting sacremoses\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/7d/34/09d19aff26edcc8eb2a01bed8e98f13a1537005d31e95233fd48216eed10/sacremoses-0.0.43.tar.gz (883kB)\n\u001b[K |████████████████████████████████| 890kB 16.7MB/s \n\u001b[?25hRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from transformers) (2.23.0)\nCollecting tokenizers==0.8.1.rc1\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/40/d0/30d5f8d221a0ed981a186c8eb986ce1c94e3a6e87f994eae9f4aa5250217/tokenizers-0.8.1rc1-cp36-cp36m-manylinux1_x86_64.whl (3.0MB)\n\u001b[K |████████████████████████████████| 3.0MB 23.1MB/s \n\u001b[?25hRequirement already satisfied: tqdm>=4.27 in /usr/local/lib/python3.6/dist-packages (from transformers) (4.41.1)\nRequirement already satisfied: filelock in /usr/local/lib/python3.6/dist-packages (from transformers) (3.0.12)\nRequirement already satisfied: packaging in /usr/local/lib/python3.6/dist-packages (from transformers) (20.4)\nCollecting sentencepiece!=0.1.92\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/d4/a4/d0a884c4300004a78cca907a6ff9a5e9fe4f090f5d95ab341c53d28cbc58/sentencepiece-0.1.91-cp36-cp36m-manylinux1_x86_64.whl (1.1MB)\n\u001b[K |████████████████████████████████| 1.1MB 45.4MB/s \n\u001b[?25hRequirement already satisfied: dataclasses; python_version < \"3.7\" in /usr/local/lib/python3.6/dist-packages (from transformers) (0.7)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from transformers) (1.18.5)\nRequirement already satisfied: regex!=2019.12.17 in /usr/local/lib/python3.6/dist-packages (from transformers) (2019.12.20)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers) (1.15.0)\nRequirement already satisfied: click in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers) (7.1.2)\nRequirement already satisfied: joblib in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers) (0.16.0)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (2020.6.20)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (1.24.3)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->transformers) (2.10)\nRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.6/dist-packages (from packaging->transformers) (2.4.7)\nBuilding wheels for collected packages: sacremoses\n Building wheel for sacremoses (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for sacremoses: filename=sacremoses-0.0.43-cp36-none-any.whl size=893260 sha256=ecb51bc6752809c3f482cd8d8dcede626ce234bbb9af544e3b0fff19ed871555\n Stored in directory: /root/.cache/pip/wheels/29/3c/fd/7ce5c3f0666dab31a50123635e6fb5e19ceb42ce38d4e58f45\nSuccessfully built sacremoses\nInstalling collected packages: sacremoses, tokenizers, sentencepiece, transformers\nSuccessfully installed sacremoses-0.0.43 sentencepiece-0.1.91 tokenizers-0.8.1rc1 transformers-3.0.2\n"
],
[
"import os\nimport re\nimport json\nimport string\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\nfrom tokenizers import BertWordPieceTokenizer\nfrom transformers import BertTokenizer, TFBertModel, BertConfig\n\nmax_len = 384\nconfiguration = BertConfig() # default parameters and configuration for BERT",
"_____no_output_____"
],
[
"# Save the slow pretrained tokenizer\nslow_tokenizer = BertTokenizer.from_pretrained(\"bert-base-uncased\")\nsave_path = \"bert_base_uncased/\"\nif not os.path.exists(save_path):\n os.makedirs(save_path)\nslow_tokenizer.save_pretrained(save_path)\n\n# Load the fast tokenizer from saved file\ntokenizer = BertWordPieceTokenizer(\"bert_base_uncased/vocab.txt\", lowercase=True)",
"_____no_output_____"
],
[
"train_data_url = \"https://rajpurkar.github.io/SQuAD-explorer/dataset/train-v1.1.json\"\ntrain_path = keras.utils.get_file(\"train.json\", train_data_url)\neval_data_url = \"https://rajpurkar.github.io/SQuAD-explorer/dataset/dev-v1.1.json\"\neval_path = keras.utils.get_file(\"eval.json\", eval_data_url)",
"Downloading data from https://rajpurkar.github.io/SQuAD-explorer/dataset/train-v1.1.json\n30294016/30288272 [==============================] - 1s 0us/step\nDownloading data from https://rajpurkar.github.io/SQuAD-explorer/dataset/dev-v1.1.json\n4857856/4854279 [==============================] - 0s 0us/step\n"
],
[
"class SquadExample:\n def __init__(self, question, context, start_char_idx, answer_text, all_answers):\n self.question = question\n self.context = context\n self.start_char_idx = start_char_idx\n self.answer_text = answer_text\n self.all_answers = all_answers\n self.skip = False\n\n def preprocess(self):\n context = self.context\n question = self.question\n answer_text = self.answer_text\n start_char_idx = self.start_char_idx\n\n # Clean context, answer and question\n context = \" \".join(str(context).split())\n question = \" \".join(str(question).split())\n answer = \" \".join(str(answer_text).split())\n\n # Find end character index of answer in context\n end_char_idx = start_char_idx + len(answer)\n if end_char_idx >= len(context):\n self.skip = True\n return\n\n # Mark the character indexes in context that are in answer\n is_char_in_ans = [0] * len(context)\n for idx in range(start_char_idx, end_char_idx):\n is_char_in_ans[idx] = 1\n\n # Tokenize context\n tokenized_context = tokenizer.encode(context)\n\n # Find tokens that were created from answer characters\n ans_token_idx = []\n for idx, (start, end) in enumerate(tokenized_context.offsets):\n if sum(is_char_in_ans[start:end]) > 0:\n ans_token_idx.append(idx)\n\n if len(ans_token_idx) == 0:\n self.skip = True\n return\n\n # Find start and end token index for tokens from answer\n start_token_idx = ans_token_idx[0]\n end_token_idx = ans_token_idx[-1]\n\n # Tokenize question\n tokenized_question = tokenizer.encode(question)\n\n # Create inputs\n input_ids = tokenized_context.ids + tokenized_question.ids[1:]\n token_type_ids = [0] * len(tokenized_context.ids) + [1] * len(\n tokenized_question.ids[1:]\n )\n attention_mask = [1] * len(input_ids)\n\n # Pad and create attention masks.\n # Skip if truncation is needed\n padding_length = max_len - len(input_ids)\n if padding_length > 0: # pad\n input_ids = input_ids + ([0] * padding_length)\n attention_mask = attention_mask + ([0] * padding_length)\n token_type_ids = token_type_ids + ([0] * padding_length)\n elif padding_length < 0: # skip\n self.skip = True\n return\n\n self.input_ids = input_ids\n self.token_type_ids = token_type_ids\n self.attention_mask = attention_mask\n self.start_token_idx = start_token_idx\n self.end_token_idx = end_token_idx\n self.context_token_to_char = tokenized_context.offsets\n\n\nwith open(train_path) as f:\n raw_train_data = json.load(f)\n\nwith open(eval_path) as f:\n raw_eval_data = json.load(f)\n\n\ndef create_squad_examples(raw_data):\n squad_examples = []\n for item in raw_data[\"data\"]:\n for para in item[\"paragraphs\"]:\n context = para[\"context\"]\n for qa in para[\"qas\"]:\n question = qa[\"question\"]\n answer_text = qa[\"answers\"][0][\"text\"]\n all_answers = [_[\"text\"] for _ in qa[\"answers\"]]\n start_char_idx = qa[\"answers\"][0][\"answer_start\"]\n squad_eg = SquadExample(\n question, context, start_char_idx, answer_text, all_answers\n )\n squad_eg.preprocess()\n squad_examples.append(squad_eg)\n return squad_examples\n\n\ndef create_inputs_targets(squad_examples):\n dataset_dict = {\n \"input_ids\": [],\n \"token_type_ids\": [],\n \"attention_mask\": [],\n \"start_token_idx\": [],\n \"end_token_idx\": [],\n }\n for item in squad_examples:\n if item.skip == False:\n for key in dataset_dict:\n dataset_dict[key].append(getattr(item, key))\n for key in dataset_dict:\n dataset_dict[key] = np.array(dataset_dict[key])\n\n x = [\n dataset_dict[\"input_ids\"],\n dataset_dict[\"token_type_ids\"],\n dataset_dict[\"attention_mask\"],\n ]\n y = [dataset_dict[\"start_token_idx\"], dataset_dict[\"end_token_idx\"]]\n return x, y\n\n\ntrain_squad_examples = create_squad_examples(raw_train_data)\nx_train, y_train = create_inputs_targets(train_squad_examples)\nprint(f\"{len(train_squad_examples)} training points created.\")\n\neval_squad_examples = create_squad_examples(raw_eval_data)\nx_eval, y_eval = create_inputs_targets(eval_squad_examples)\nprint(f\"{len(eval_squad_examples)} evaluation points created.\")",
"87599 training points created.\n10570 evaluation points created.\n"
],
[
"def normalize_text(text):\n text = text.lower()\n\n # Remove punctuations\n exclude = set(string.punctuation)\n text = \"\".join(ch for ch in text if ch not in exclude)\n\n # Remove articles\n regex = re.compile(r\"\\b(a|an|the)\\b\", re.UNICODE)\n text = re.sub(regex, \" \", text)\n\n # Remove extra white space\n text = \" \".join(text.split())\n return text\ndef normalize_text(text):\n text = text.lower()\n\n # Remove punctuations\n exclude = set(string.punctuation)\n text = \"\".join(ch for ch in text if ch not in exclude)\n\n # Remove articles\n regex = re.compile(r\"\\b(a|an|the)\\b\", re.UNICODE)\n text = re.sub(regex, \" \", text)\n\n # Remove extra white space\n text = \" \".join(text.split())\n return text\n\n\nclass ExactMatch(keras.callbacks.Callback):\n \"\"\"\n Each `SquadExample` object contains the character level offsets for each token\n in its input paragraph. We use them to get back the span of text corresponding\n to the tokens between our predicted start and end tokens.\n All the ground-truth answers are also present in each `SquadExample` object.\n We calculate the percentage of data points where the span of text obtained\n from model predictions matches one of the ground-truth answers.\n \"\"\"\n\n def __init__(self, x_eval, y_eval):\n self.x_eval = x_eval\n self.y_eval = y_eval\n\n def on_epoch_end(self, epoch, logs=None):\n pred_start, pred_end = self.model.predict(self.x_eval)\n count = 0\n eval_examples_no_skip = [_ for _ in eval_squad_examples if _.skip == False]\n for idx, (start, end) in enumerate(zip(pred_start, pred_end)):\n squad_eg = eval_examples_no_skip[idx]\n offsets = squad_eg.context_token_to_char\n start = np.argmax(start)\n end = np.argmax(end)\n if start >= len(offsets):\n continue\n pred_char_start = offsets[start][0]\n if end < len(offsets):\n pred_char_end = offsets[end][1]\n pred_ans = squad_eg.context[pred_char_start:pred_char_end]\n else:\n pred_ans = squad_eg.context[pred_char_start:]\n\n normalized_pred_ans = normalize_text(pred_ans)\n normalized_true_ans = [normalize_text(_) for _ in squad_eg.all_answers]\n if normalized_pred_ans in normalized_true_ans:\n count += 1\n acc = count / len(self.y_eval[0])\n print(f\"\\nepoch={epoch+1}, exact match score={acc:.2f}\")\nexact_match_callback = ExactMatch(x_eval, y_eval)",
"_____no_output_____"
],
[
"# Create distribution strategy\ntpu = tf.distribute.cluster_resolver.TPUClusterResolver()\ntf.config.experimental_connect_to_cluster(tpu)\ntf.tpu.experimental.initialize_tpu_system(tpu)\nstrategy = tf.distribute.experimental.TPUStrategy(tpu)",
"WARNING:tensorflow:TPU system grpc://10.111.74.170:8470 has already been initialized. Reinitializing the TPU can cause previously created variables on TPU to be lost.\n"
],
[
"def create_model():\n ## BERT encoder\n encoder = TFBertModel.from_pretrained(\"bert-base-uncased\")\n\n ## QA Model\n input_ids = layers.Input(shape=(max_len,), dtype=tf.int32)\n token_type_ids = layers.Input(shape=(max_len,), dtype=tf.int32)\n attention_mask = layers.Input(shape=(max_len,), dtype=tf.int32)\n embedding = encoder(\n input_ids, token_type_ids=token_type_ids, attention_mask=attention_mask\n )[0]\n\n start_logits = layers.Dense(1, name=\"start_logit\", use_bias=False)(embedding)\n start_logits = layers.Flatten()(start_logits)\n\n end_logits = layers.Dense(1, name=\"end_logit\", use_bias=False)(embedding)\n end_logits = layers.Flatten()(end_logits)\n\n start_probs = layers.Activation(keras.activations.softmax)(start_logits)\n end_probs = layers.Activation(keras.activations.softmax)(end_logits)\n\n model = keras.Model(\n inputs=[input_ids, token_type_ids, attention_mask],\n outputs=[start_probs, end_probs],\n )\n loss = keras.losses.SparseCategoricalCrossentropy(from_logits=False)\n optimizer = keras.optimizers.Adam(lr=5e-5)\n model.compile(optimizer=optimizer, loss=[loss, loss])\n return model",
"_____no_output_____"
],
[
"# Create model\nwith strategy.scope():\n model = create_model()\n\nmodel.summary()",
"WARNING:transformers.modeling_tf_utils:Some weights of the model checkpoint at bert-base-uncased were not used when initializing TFBertModel: ['mlm___cls', 'nsp___cls']\n- This IS expected if you are initializing TFBertModel from the checkpoint of a model trained on another task or with another architecture (e.g. initializing a BertForSequenceClassification model from a BertForPretraining model).\n- This IS NOT expected if you are initializing TFBertModel from the checkpoint of a model that you expect to be exactly identical (initializing a BertForSequenceClassification model from a BertForSequenceClassification model).\nWARNING:transformers.modeling_tf_utils:All the weights of TFBertModel were initialized from the model checkpoint at bert-base-uncased.\nIf your task is similar to the task the model of the ckeckpoint was trained on, you can already use TFBertModel for predictions without further training.\n"
],
[
"model.fit(\n x_train,\n y_train,\n epochs=3,\n verbose=2,\n batch_size=64,\n callbacks=[exact_match_callback],\n)",
"Epoch 1/3\nWARNING:tensorflow:Gradients do not exist for variables ['tf_bert_model_1/bert/pooler/dense/kernel:0', 'tf_bert_model_1/bert/pooler/dense/bias:0'] when minimizing the loss.\n"
],
[
"class QnATestData():\n \n def __init__(self):\n self.input_ids = []\n self.token_type_ids = []\n self.attention_masks = []\n self.context_token_to_char = []\n \n def preprocess(self, context, questions):\n input_id = []\n\n for each_question in questions:\n\n # Clean context, answer and question\n context = \" \".join(str(context).split())\n question = \" \".join(str(each_question).split())\n\n # Tokenize context and question\n tokenized_context = tokenizer.encode(context)\n tokenized_question = tokenizer.encode(each_question)\n\n # Create inputs\n input_id = tokenized_context.ids + tokenized_question.ids[1:]\n token_type_id = [0] * len(tokenized_context.ids) + [1] * len(tokenized_question.ids[1:])\n attention_mask = [1] * len(input_id)\n\n # Pad and create attention masks.\n # Skip if truncation is needed\n padding_length = max_len - len(input_id)\n \n if padding_length > 0: # pad\n input_id = input_id + ([0] * padding_length)\n attention_mask = attention_mask + ([0] * padding_length)\n token_type_id = token_type_id + ([0] * padding_length)\n elif padding_length < 0: # skip\n self.skip = True\n continue\n \n\n self.input_ids.append(input_id)\n self.token_type_ids.append(token_type_id)\n self.attention_masks.append(attention_mask)\n self.context_token_to_char.append(tokenized_context.offsets)\n\n def get_test_result(self, context, questions):\n pred_answer_list = []\n self.preprocess(context, questions)\n x = [\n np.array(self.input_ids),\n np.array(self.token_type_ids),\n np.array(self.attention_masks),\n ]\n\n pred_start, pred_end = model.predict(x)\n for idx, (start, end) in enumerate(zip(pred_start, pred_end)):\n offsets = self.context_token_to_char[idx]\n start = np.argmax(start)\n end = np.argmax(end)\n if start >= len(offsets):\n print(\"start is greater the offsets\")\n continue\n pred_char_start = offsets[start][0]\n\n\n if end < len(offsets):\n pred_char_end = offsets[end][1]\n pred_ans = context[pred_char_start:pred_char_end]\n else:\n pred_ans = context[idx][pred_char_start:]\n pred_answer_list.append(pred_ans)\n return pred_answer_list",
"_____no_output_____"
],
[
"context = '''Mike and Morris lived in the same village. While Morris owned the largest jewelry shop in the village, Mike was a poor farmer. Both had large families with many sons, daughters-in-law and grandchildren. One fine day, Mike, tired of not being able to feed his family, decided to leave the village and move to the city where he was certain to earn enough to feed everyone. Along with his family, he left the village for the city. At night, they stopped under a large tree. There was a stream running nearby where they could freshen up themselves. He told his sons to clear the area below the tree, he told his wife to fetch water and he instructed his daughters-in-law to make up the fire and started cutting wood from the tree himself. They didn’t know that in the branches of the tree, there was a thief hiding. He watched as Mike’s family worked together and also noticed that they had nothing to cook. Mike’s wife also thought the same and asked her husband ” Everything is ready but what shall we eat?”. Mike raised his h '''\nquestions = [\"What did Morris owned?\", \"What did Mike do for a living?\", \"what the instruction he gave to daughters-in-law? \"]\nqna_test_obj = QnATestData()\nqna_test_obj.get_test_result(context, questions)\n",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f59f2619833704c17dfe4195327f99425026ec | 292,477 | ipynb | Jupyter Notebook | GeneralExemplars/MLExemplars/Classification_k_NN_Notebook.ipynb | jstix/Exemplars2020 | 801ffdf37c05a0d7edb9762f356924b07fa131ba | [
"BSD-3-Clause"
] | null | null | null | GeneralExemplars/MLExemplars/Classification_k_NN_Notebook.ipynb | jstix/Exemplars2020 | 801ffdf37c05a0d7edb9762f356924b07fa131ba | [
"BSD-3-Clause"
] | null | null | null | GeneralExemplars/MLExemplars/Classification_k_NN_Notebook.ipynb | jstix/Exemplars2020 | 801ffdf37c05a0d7edb9762f356924b07fa131ba | [
"BSD-3-Clause"
] | null | null | null | 468.713141 | 129,888 | 0.941701 | [
[
[
"## This Notebook - Goals - FOR EDINA\n\n**What?:**\n- Standard classification method example/tutorial\n\n**Who?:**\n- Researchers in ML\n- Students in computer science\n- Teachers in ML/STEM\n\n**Why?:**\n- Demonstrate capability/simplicity of core scipy stack. \n- Demonstrate common ML concept known to learners and used by researchers.\n\n**Noteable features to exploit:**\n- use of pre-installed libraries: <code>numpy</code>, <code>scikit-learn</code>, <code>matplotlib</code>\n\n**How?:**\n- clear to understand - minimise assumed knowledge\n- clear visualisations - concise explanations\n- recognisable/familiar - use standard methods\n- Effective use of core libraries\n\n<hr>",
"_____no_output_____"
],
[
"# Classification - K nearest neighbours\n\nK nearest neighbours is a simple and effective way to deal with classification problems. This method classifies each sample based on the class of the points that are closest to it.\n\nThis is a supervised learning method, meaning that data used contains information on some feature that the model should predict.\n\nThis notebook shows the process of classifying handwritten digits. ",
"_____no_output_____"
],
[
"<hr>\n\n### Import libraries\n\nOn Noteable, all the libaries required for this notebook are pre-installed, so they simply need to be imported:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nimport sklearn.datasets as ds\nimport sklearn.model_selection as ms \n\nfrom sklearn import decomposition\nfrom sklearn import neighbors\nfrom sklearn import metrics\n\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"<hr>\n\n# Data - Handwritten Digits\n\nIn terms of data, [scikit-learn](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_digits.html) has a loading function for some data regarding hand written digits.",
"_____no_output_____"
]
],
[
[
"# get the digits data from scikit into the notebook\ndigits = ds.load_digits()",
"_____no_output_____"
]
],
[
[
"The cell above loads the data as a [bunch object](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_digits.html), meaning that the data (in this case images of handwritten digits) and the target (the number that is written) can be split by accessing the attributes of the bunch object:",
"_____no_output_____"
]
],
[
[
"# store data and targets seperately\nX = digits.data\ny = digits.target\n\nprint(\"The data is of the shape\", X.shape)\nprint(\"The target data is of the shape\", y.shape)",
"The data is of the shape (1797, 64)\nThe target data is of the shape (1797,)\n"
]
],
[
[
"The individual samples in the <code>X</code> array each represent an image. In this representation, 64 numbers are used to represent a greyscale value on an 8\\*8 square. The images can be examined by using pyplot's [matshow](https://matplotlib.org/3.3.0/api/_as_gen/matplotlib.pyplot.matshow.html) function.\n\nThe next cell displays the 17th sample in the dataset as an 8\\*8 image.",
"_____no_output_____"
]
],
[
[
"# create figure to display the 17th sample\nfig = plt.matshow(digits.images[17], cmap=plt.cm.gray)\nfig.axes.get_xaxis().set_visible(False)\nfig.axes.get_yaxis().set_visible(False)",
"_____no_output_____"
]
],
[
[
"Suppose instead of viewing the 17th sample, we want to see the average of samples corresponding to a certain value.\n\nThis can be done as follows (using 0 as an example):\n- All samples where the target value is 0 are located\n- The mean of these samples is taken\n- The resulting 64 long array is reshaped to be 8\\*8 (for display)\n- The image is displayed",
"_____no_output_____"
]
],
[
[
"# take samples with target=0\nizeros = np.where(y == 0)\n# take average across samples, reshape to visualise\nzeros = np.mean(X[izeros], axis=0).reshape(8,8)\n\n# display\nfig = plt.matshow(zeros, cmap=plt.cm.gray)\nfig.axes.get_xaxis().set_visible(False)\nfig.axes.get_yaxis().set_visible(False)",
"_____no_output_____"
]
],
[
[
"<hr>\n\n# Fit and test the model\n\n## Split the data",
"_____no_output_____"
],
[
"Now that you have an understanding of the data, the model can be fitted.\n\nFitting the model involves setting some of the data aside for testing, and allowing the model to \"see\" the target values corresponding to the training samples.\n\nOnce the model has been fitted to the training data, the model will be tested on some data it has not seen before. \n\nThe next cell uses [train_test_split](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html) to shuffle all data, then set some data aside for testing later. \n\nFor this example, $\\frac{1}{4}$ of the data will be set aside for testing, and the model will be trained on the remaining training set.\n\nAs before, <code>X</code> corresponds to data samples, and <code>y</code> corresponds to labels.",
"_____no_output_____"
]
],
[
[
"# split data to train and test sets\nX_train, X_test, y_train, y_test = \\\n ms.train_test_split(X, y, test_size=0.25, shuffle=True,\n random_state=22)",
"_____no_output_____"
]
],
[
[
"The data can be examined - here you can see that 1347 samples have been put into the training set, and 450 have been set aside for testing.",
"_____no_output_____"
]
],
[
[
"# print shape of data\nprint(\"training samples:\", X_train.shape) \nprint(\"testing samples :\", X_test.shape)\nprint(\"training targets:\", y_train.shape) \nprint(\"testing targets :\", y_test.shape) ",
"training samples: (1347, 64)\ntesting samples : (450, 64)\ntraining targets: (1347,)\ntesting targets : (450,)\n"
]
],
[
[
"## Using PCA to visualise data\n\nBefore diving into classifying, it is useful to visualise the data.\n\nSince each sample has 64 dimensions, some dimensionality reduction is needed in order to visualise the samples as points on a 2D map.\n\nOne of the easiest ways of visualising high dimensional data is by principal component analysis (PCA). This maps the 64 dimensional image data onto a lower dimension map (here we will map to 2D) so it can be easily viewed on a screen.\n\nIn this case, the 2 most important \"components\" are maintained.",
"_____no_output_____"
]
],
[
[
"# create PCA model with 2 components\npca = decomposition.PCA(n_components=2)",
"_____no_output_____"
]
],
[
[
"The next step is to perform the PCA on the samples, and store the results.",
"_____no_output_____"
]
],
[
[
"# transform training data to 2 principal components\nX_pca = pca.fit_transform(X_train)\n\n# transform test data to 2 principal components\nT_pca = pca.transform(X_test)",
"_____no_output_____"
],
[
"# check shape of result\nprint(X_pca.shape) \nprint(T_pca.shape)",
"(1347, 2)\n(450, 2)\n"
]
],
[
[
"As you can see from the above cell, the <code>X_pca</code> and <code>T_pca</code> data is now represented by only 2 elements per sample. The number of samples has remained the same.\n\nNow that there is a 2D representation of the data, it can be plotted on a regular scatter graph. Since the labels corresponding to each point are stored in the <code>y_train</code> variable, the plot can be colour coded by target value!\n\nDifferent coloured dots have different target values.",
"_____no_output_____"
]
],
[
[
"# choose the colours for each digit\ncmap_digits = plt.cm.tab10\n\n# plot training data with labels\nplt.figure(figsize = (9,6))\nplt.scatter(X_pca[:,0], X_pca[:,1], s=7, c=y_train,\n cmap=cmap_digits, alpha=0.7)\nplt.title(\"Training data coloured by target value\")\nplt.colorbar();",
"_____no_output_____"
]
],
[
[
"## Create and fit the model\n\nThe scikit-learn library allows fitting of a k-NN model just as with PCA above.\n\nFirst, create the classifier:",
"_____no_output_____"
]
],
[
[
"# create model\nknn = neighbors.KNeighborsClassifier()",
"_____no_output_____"
]
],
[
[
"The next step fits the k-NN model using the training data.",
"_____no_output_____"
]
],
[
[
"# fit model to training data\nknn.fit(X_train,y_train);",
"_____no_output_____"
]
],
[
[
"## Test model\n\nNow use the data that was set aside earlier - this stage involves getting the model to \"guess\" the samples (this time without seeing their target values).\n\nOnce the model has predicted the sample's class, a score can be calculated by checking how many samples the model guessed correctly.",
"_____no_output_____"
]
],
[
[
"# predict test data\npreds = knn.predict(X_test)\n\n# test model on test data\nscore = round(knn.score(X_test,y_test)*100, 2)\nprint(\"Score on test data: \" + str(score) + \"%\")",
"Score on test data: 98.44%\n"
]
],
[
[
"98.44% is a really high score, one that would not likely be seen on real life applications of the method.\n\nIt can often be useful to visualise the results of your example. Below are plots showing:\n- The labels that the model predicted for the test data\n- The actual labels for the test data\n- The data points that were incorrectly labelled\n\nIn this case, the predicted and actual plots are very similar, so these plots are not very informative. In other cases, this kind of visualisation may reveal patterns for you to explore further.",
"_____no_output_____"
]
],
[
[
"# plot 3 axes\nfig, axes = plt.subplots(2,2,figsize=(12,12))\n\n# top left axis for predictions\naxes[0,0].scatter(T_pca[:,0], T_pca[:,1], s=5, \n c=preds, cmap=cmap_digits)\naxes[0,0].set_title(\"Predicted labels\")\n\n# top right axis for actual targets\naxes[0,1].scatter(T_pca[:,0], T_pca[:,1], s=5, \n c=y_test, cmap=cmap_digits)\naxes[0,1].set_title(\"Actual labels\")\n\n# bottom left axis coloured to show correct and incorrect\naxes[1,0].scatter(T_pca[:,0], T_pca[:,1], s=5, \n c=(preds==y_test))\naxes[1,0].set_title(\"Incorrect labels\")\n\n# bottom right axis not used\naxes[1,1].set_axis_off()",
"_____no_output_____"
]
],
[
[
"So which samples did the model get wrong?\n\nThere were 7 samples that were misclassified. These can be displayed alongside their actual and predicted labels using the cell below:",
"_____no_output_____"
]
],
[
[
"# find the misclassified samples\nmisclass = np.where(preds!=y_test)[0]\n\n# display misclassified samples\nr, c = 1, len(misclass)\nfig, axes = plt.subplots(r,c,figsize=(10,5))\n\nfor i in range(c):\n ax = axes[i]\n ax.matshow(X_test[misclass[i]].reshape(8,8),cmap=plt.cm.gray)\n ax.set_axis_off()\n act = y_test[misclass[i]]\n pre = preds[misclass[i]]\n strng = \"actual: {a:.0f} \\npredicted: {p:.0f}\".format(a=act, p=pre)\n ax.set_title(strng)",
"_____no_output_____"
]
],
[
[
"Additionally, a confusion matrix can be used to identify which samples are misclassified by the model. This can help you identify if their are samples that are commonly misidentified - for example you may identify that 8's are often mistook for 1's.",
"_____no_output_____"
]
],
[
[
"# confusion matrix\nconf = metrics.confusion_matrix(y_test,preds)\n\n# figure\nf, ax = plt.subplots(figsize=(9,5))\nim = ax.imshow(conf, cmap=plt.cm.RdBu)\n\n# set labels as ticks on axes\nax.set_xticks(np.arange(10))\nax.set_yticks(np.arange(10))\nax.set_xticklabels(list(range(0,10)))\nax.set_yticklabels(list(range(0,10)))\nax.set_ylim(9.5,-0.5)\n\n# axes labels\nax.set_ylabel(\"actual value\")\nax.set_xlabel(\"predicted value\")\nax.set_title(\"Digit classification confusion matrix\")\n\n# display\nplt.colorbar(im).set_label(label=\"number of classifications\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f5ae285bbb1a9df24142e4c5b6662d348e6966 | 3,733 | ipynb | Jupyter Notebook | examples/reference/widgets/FloatInput.ipynb | slamer59/panel | 963a63af57e786a8121f2f2bff1f6533fe7ddffe | [
"BSD-3-Clause"
] | 1 | 2020-10-17T17:00:14.000Z | 2020-10-17T17:00:14.000Z | examples/reference/widgets/FloatInput.ipynb | slamer59/panel | 963a63af57e786a8121f2f2bff1f6533fe7ddffe | [
"BSD-3-Clause"
] | null | null | null | examples/reference/widgets/FloatInput.ipynb | slamer59/panel | 963a63af57e786a8121f2f2bff1f6533fe7ddffe | [
"BSD-3-Clause"
] | null | null | null | 31.108333 | 474 | 0.600589 | [
[
[
"import panel as pn\n\npn.extension()",
"_____no_output_____"
]
],
[
[
"The ``FloatInput`` widget allows selecting a floating point value using a spinbox. It behaves like a slider except that lower and upper bounds are optional and a specific value can be entered. Value can be changed using keyboard (up, down, page up, page down), mouse wheel and arrow buttons.\n\nFor more information about listening to widget events and laying out widgets refer to the [widgets user guide](../../user_guide/Widgets.ipynb). Alternatively you can learn how to build GUIs by declaring parameters independently of any specific widgets in the [param user guide](../../user_guide/Param.ipynb). To express interactivity entirely using Javascript without the need for a Python server take a look at the [links user guide](../../user_guide/Param.ipynb).\n\n#### Parameters:\n\nFor layout and styling related parameters see the [customization user guide](../../user_guide/Customization.ipynb).\n\n##### Core\n\n* **``value``** (float): The initial value of the spinner\n* **``value_throttled``** (float): The initial value of the spinner\n* **``step``** (float): The step added or subtracted to the current value on each click\n* **``start``** (float): Optional minimum allowable value\n* **``end``** (float): Optional maximum allowable value\n* **``format``** (str): Optional format to convert the float value in string, see : http://numbrojs.com/old-format.html\n* **``page_step_multiplier``** (int): Defines the multiplication factor applied to step when the page up and page down keys are pressed.\n\n##### Display\n\n* **``disabled``** (boolean): Whether the widget is editable\n* **``name``** (str): The title of the widget\n* **``placeholder``** (str): A placeholder string displayed when no value is entered\n\n___",
"_____no_output_____"
]
],
[
[
"float_input = pn.widgets.FloatInput(name='FloatInput', value=5., step=1e-1, start=0, end=1000)\n\nfloat_input",
"_____no_output_____"
]
],
[
[
"``FloatInput.value`` returns a float value:",
"_____no_output_____"
]
],
[
[
"float_input.value",
"_____no_output_____"
]
],
[
[
"### Controls\n\nThe `FloatSpinner` widget exposes a number of options which can be changed from both Python and Javascript. Try out the effect of these parameters interactively:",
"_____no_output_____"
]
],
[
[
"pn.Row(float_input.controls(jslink=True), float_input)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f5c72ed596eb220fbd2760c877211b3175dbf3 | 72,714 | ipynb | Jupyter Notebook | d2l-en/pytorch/chapter_attention-mechanisms/bahdanau-attention.ipynb | gr8khan/d2lai | 7c10432f38c80e86978cd075d0024902b47842a0 | [
"MIT"
] | null | null | null | d2l-en/pytorch/chapter_attention-mechanisms/bahdanau-attention.ipynb | gr8khan/d2lai | 7c10432f38c80e86978cd075d0024902b47842a0 | [
"MIT"
] | null | null | null | d2l-en/pytorch/chapter_attention-mechanisms/bahdanau-attention.ipynb | gr8khan/d2lai | 7c10432f38c80e86978cd075d0024902b47842a0 | [
"MIT"
] | null | null | null | 40.942568 | 751 | 0.498363 | [
[
[
"# Bahdanau Attention\n:label:`sec_seq2seq_attention`\n\nWe studied the machine translation\nproblem in :numref:`sec_seq2seq`,\nwhere we designed\nan encoder-decoder architecture based on two RNNs\nfor sequence to sequence learning.\nSpecifically,\nthe RNN encoder \ntransforms\na variable-length sequence\ninto a fixed-shape context variable,\nthen\nthe RNN decoder\ngenerates the output (target) sequence token by token\nbased on the generated tokens and the context variable.\nHowever,\neven though not all the input (source) tokens\nare useful for decoding a certain token,\nthe *same* context variable\nthat encodes the entire input sequence\nis still used at each decoding step.\n\n\nIn a separate but related\nchallenge of handwriting generation for a given text sequence,\nGraves designed a differentiable attention model\nto align text characters with the much longer pen trace,\nwhere the alignment moves only in one direction :cite:`Graves.2013`.\nInspired by the idea of learning to align,\nBahdanau et al. proposed a differentiable attention model\nwithout the severe unidirectional alignment limitation :cite:`Bahdanau.Cho.Bengio.2014`.\nWhen predicting a token,\nif not all the input tokens are relevant,\nthe model aligns (or attends)\nonly to parts of the input sequence that are relevant to the current prediction.\nThis is achieved\nby treating the context variable as an output of attention pooling.\n\n## Model\n\nWhen describing \nBahdanau attention\nfor the RNN encoder-decoder below,\nwe will follow the same notation in\n:numref:`sec_seq2seq`.\nThe new attention-based model\nis the same as that\nin :numref:`sec_seq2seq`\nexcept that\nthe context variable\n$\\mathbf{c}$\nin \n:eqref:`eq_seq2seq_s_t`\nis replaced by\n$\\mathbf{c}_{t'}$\nat any decoding time step $t'$.\nSuppose that\nthere are $T$ tokens in the input sequence,\nthe context variable at the decoding time step $t'$\nis the output of attention pooling:\n\n$$\\mathbf{c}_{t'} = \\sum_{t=1}^T \\alpha(\\mathbf{s}_{t' - 1}, \\mathbf{h}_t) \\mathbf{h}_t,$$\n\nwhere the decoder hidden state\n$\\mathbf{s}_{t' - 1}$ at time step $t' - 1$\nis the query,\nand the encoder hidden states $\\mathbf{h}_t$\nare both the keys and values,\nand the attention weight $\\alpha$\nis computed as in\n:eqref:`eq_attn-scoring-alpha`\nusing the additive attention scoring function\ndefined by\n:eqref:`eq_additive-attn`.\n\n\nSlightly different from \nthe vanilla RNN encoder-decoder architecture \nin :numref:`fig_seq2seq_details`,\nthe same architecture\nwith Bahdanau attention is depicted in \n:numref:`fig_s2s_attention_details`.\n\n\n:label:`fig_s2s_attention_details`\n",
"_____no_output_____"
]
],
[
[
"from d2l import torch as d2l\nimport torch\nfrom torch import nn",
"_____no_output_____"
]
],
[
[
"## Defining the Decoder with Attention\n\nTo implement the RNN encoder-decoder\nwith Bahdanau attention,\nwe only need to redefine the decoder.\nTo visualize the learned attention weights more conveniently,\nthe following `AttentionDecoder` class\ndefines the base interface for \ndecoders with attention mechanisms.\n",
"_____no_output_____"
]
],
[
[
"#@save\nclass AttentionDecoder(d2l.Decoder):\n \"\"\"The base attention-based decoder interface.\"\"\"\n def __init__(self, **kwargs):\n super(AttentionDecoder, self).__init__(**kwargs)\n\n @property\n def attention_weights(self):\n raise NotImplementedError",
"_____no_output_____"
]
],
[
[
"Now let us implement\nthe RNN decoder with Bahdanau attention\nin the following `Seq2SeqAttentionDecoder` class.\nThe state of the decoder\nis initialized with \ni) the encoder final-layer hidden states at all the time steps (as keys and values of the attention);\nii) the encoder all-layer hidden state at the final time step (to initialize the hidden state of the decoder);\nand iii) the encoder valid length (to exclude the padding tokens in attention pooling).\nAt each decoding time step,\nthe decoder final-layer hidden state at the previous time step is used as the query of the attention.\nAs a result, both the attention output\nand the input embedding are concatenated\nas the input of the RNN decoder.\n",
"_____no_output_____"
]
],
[
[
"class Seq2SeqAttentionDecoder(AttentionDecoder):\n def __init__(self, vocab_size, embed_size, num_hiddens, num_layers,\n dropout=0, **kwargs):\n super(Seq2SeqAttentionDecoder, self).__init__(**kwargs)\n self.attention = d2l.AdditiveAttention(\n num_hiddens, num_hiddens, num_hiddens, dropout)\n self.embedding = nn.Embedding(vocab_size, embed_size)\n self.rnn = nn.GRU(\n embed_size + num_hiddens, num_hiddens, num_layers,\n dropout=dropout)\n self.dense = nn.Linear(num_hiddens, vocab_size)\n\n def init_state(self, enc_outputs, enc_valid_lens, *args):\n # Shape of `outputs`: (`num_steps`, `batch_size`, `num_hiddens`).\n # Shape of `hidden_state[0]`: (`num_layers`, `batch_size`,\n # `num_hiddens`)\n outputs, hidden_state = enc_outputs\n return (outputs.permute(1, 0, 2), hidden_state, enc_valid_lens)\n\n def forward(self, X, state):\n # Shape of `enc_outputs`: (`batch_size`, `num_steps`, `num_hiddens`).\n # Shape of `hidden_state[0]`: (`num_layers`, `batch_size`,\n # `num_hiddens`)\n enc_outputs, hidden_state, enc_valid_lens = state\n # Shape of the output `X`: (`num_steps`, `batch_size`, `embed_size`)\n X = self.embedding(X).permute(1, 0, 2)\n outputs, self._attention_weights = [], []\n for x in X:\n # Shape of `query`: (`batch_size`, 1, `num_hiddens`)\n query = torch.unsqueeze(hidden_state[-1], dim=1)\n # Shape of `context`: (`batch_size`, 1, `num_hiddens`)\n context = self.attention(\n query, enc_outputs, enc_outputs, enc_valid_lens)\n # Concatenate on the feature dimension\n x = torch.cat((context, torch.unsqueeze(x, dim=1)), dim=-1)\n # Reshape `x` as (1, `batch_size`, `embed_size` + `num_hiddens`)\n out, hidden_state = self.rnn(x.permute(1, 0, 2), hidden_state)\n outputs.append(out)\n self._attention_weights.append(self.attention.attention_weights)\n # After fully-connected layer transformation, shape of `outputs`:\n # (`num_steps`, `batch_size`, `vocab_size`)\n outputs = self.dense(torch.cat(outputs, dim=0))\n return outputs.permute(1, 0, 2), [enc_outputs, hidden_state,\n enc_valid_lens]\n \n @property\n def attention_weights(self):\n return self._attention_weights",
"_____no_output_____"
]
],
[
[
"In the following, we test the implemented \ndecoder with Bahdanau attention\nusing a minibatch of 4 sequence inputs\nof 7 time steps.\n",
"_____no_output_____"
]
],
[
[
"encoder = d2l.Seq2SeqEncoder(vocab_size=10, embed_size=8, num_hiddens=16,\n num_layers=2)\nencoder.eval()\ndecoder = Seq2SeqAttentionDecoder(vocab_size=10, embed_size=8, num_hiddens=16,\n num_layers=2)\ndecoder.eval()\nX = torch.zeros((4, 7), dtype=torch.long) # (`batch_size`, `num_steps`)\nstate = decoder.init_state(encoder(X), None)\noutput, state = decoder(X, state)\noutput.shape, len(state), state[0].shape, len(state[1]), state[1][0].shape",
"_____no_output_____"
]
],
[
[
"## Training\n\n\nSimilar to :numref:`sec_seq2seq_training`,\nhere we specify hyperparemeters,\ninstantiate\nan encoder and a decoder with Bahdanau attention,\nand train this model for machine translation.\nDue to the newly added attention mechanism,\nthis training is much slower than\nthat in :numref:`sec_seq2seq_training` without attention mechanisms.\n",
"_____no_output_____"
]
],
[
[
"embed_size, num_hiddens, num_layers, dropout = 32, 32, 2, 0.1\nbatch_size, num_steps = 64, 10\nlr, num_epochs, device = 0.005, 250, d2l.try_gpu()\n\ntrain_iter, src_vocab, tgt_vocab = d2l.load_data_nmt(batch_size, num_steps)\nencoder = d2l.Seq2SeqEncoder(\n len(src_vocab), embed_size, num_hiddens, num_layers, dropout)\ndecoder = Seq2SeqAttentionDecoder(\n len(tgt_vocab), embed_size, num_hiddens, num_layers, dropout)\nnet = d2l.EncoderDecoder(encoder, decoder)\nd2l.train_seq2seq(net, train_iter, lr, num_epochs, tgt_vocab, device)",
"loss 0.020, 4902.7 tokens/sec on cuda:0\n"
]
],
[
[
"After the model is trained,\nwe use it to translate a few English sentences\ninto French and compute their BLEU scores.\n",
"_____no_output_____"
]
],
[
[
"engs = ['go .', \"i lost .\", 'he\\'s calm .', 'i\\'m home .']\nfras = ['va !', 'j\\'ai perdu .', 'il est calme .', 'je suis chez moi .']\nfor eng, fra in zip(engs, fras):\n translation, dec_attention_weight_seq = d2l.predict_seq2seq(\n net, eng, src_vocab, tgt_vocab, num_steps, device, True)\n print(f'{eng} => {translation}, ',\n f'bleu {d2l.bleu(translation, fra, k=2):.3f}')",
"go . => va !, bleu 1.000\ni lost . => j'ai perdu ., bleu 1.000\nhe's calm . => il est paresseux ., bleu 0.658\ni'm home . => je suis chez moi ., bleu 1.000\n"
],
[
"attention_weights = torch.cat(\n [step[0][0][0] for step in dec_attention_weight_seq], 0).reshape(\n (1, 1, -1, num_steps))",
"_____no_output_____"
]
],
[
[
"By visualizing the attention weights\nwhen translating the last English sentence,\nwe can see that each query assigns non-uniform weights\nover key-value pairs.\nIt shows that at each decoding step,\ndifferent parts of the input sequences \nare selectively aggregated in the attention pooling.\n",
"_____no_output_____"
]
],
[
[
"# Plus one to include the end-of-sequence token\nd2l.show_heatmaps(\n attention_weights[:, :, :, :len(engs[-1].split()) + 1].cpu(),\n xlabel='Key posistions', ylabel='Query posistions')",
"_____no_output_____"
]
],
[
[
"## Summary\n\n* When predicting a token, if not all the input tokens are relevant, the RNN encoder-decoder with Bahdanau attention selectively aggregates different parts of the input sequence. This is achieved by treating the context variable as an output of additive attention pooling.\n* In the RNN encoder-decoder, Bahdanau attention treats the decoder hidden state at the previous time step as the query, and the encoder hidden states at all the time steps as both the keys and values.\n\n\n## Exercises\n\n1. Replace GRU with LSTM in the experiment.\n1. Modify the experiment to replace the additive attention scoring function with the scaled dot-product. How does it influence the training efficiency?\n",
"_____no_output_____"
],
[
"[Discussions](https://discuss.d2l.ai/t/1065)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e7f5d5b6261dc4d2bb50c2e0a52ffdc79f6b83f9 | 61,612 | ipynb | Jupyter Notebook | experiment.ipynb | JeyDi/Digits-ConvNeuralNet | c61f08168ad780fd6dc14085a1dbbdd06c30cb97 | [
"Apache-2.0"
] | null | null | null | experiment.ipynb | JeyDi/Digits-ConvNeuralNet | c61f08168ad780fd6dc14085a1dbbdd06c30cb97 | [
"Apache-2.0"
] | null | null | null | experiment.ipynb | JeyDi/Digits-ConvNeuralNet | c61f08168ad780fd6dc14085a1dbbdd06c30cb97 | [
"Apache-2.0"
] | null | null | null | 198.748387 | 51,196 | 0.893884 | [
[
[
"# Assignement 3: CNN Exercise",
"_____no_output_____"
],
[
"## Load libraries",
"_____no_output_____"
]
],
[
[
"import pickle\nimport numpy as np\nimport pandas as pd\n\nfrom keras.datasets import mnist\nfrom keras.utils import to_categorical\nfrom keras import layers\nfrom keras import models\n\nimport matplotlib.pyplot as plt\nfrom numpy.random import seed\nfrom keras.utils import plot_model\n\nimport csv\nimport json\n\nfrom keras.callbacks import EarlyStopping\n\n",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\h5py\\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
]
],
[
[
"## Load the dataset",
"_____no_output_____"
]
],
[
[
"#Load the dataset\ntry:\n #Load the MNIST data\n (train_images, train_labels), (test_images, test_labels) = mnist.load_data()\n \n #Reshape and trasform the input\n train_images = train_images.reshape((60000, 28, 28, 1))\n train_images = train_images.astype('float32') / 255\n test_images = test_images.reshape((10000, 28, 28, 1))\n test_images = test_images.astype('float32') / 255\n\n train_labels = to_categorical(train_labels)\n test_labels = to_categorical(test_labels)\n \n print(\"Data Loaded\")\n \nexcept:\n print(\"Error Loading Data\")\n",
"Data Loaded\n"
]
],
[
[
"## Create the network, train and evaluate",
"_____no_output_____"
]
],
[
[
"seed(42)\n\nmodel = models.Sequential()\n\n#Create the network\nmodel.add(layers.Conv2D(16, (3, 3), activation='relu', input_shape=(28, 28, 1)))\nmodel.add(layers.MaxPooling2D((2, 2)))\nmodel.add(layers.Conv2D(16, (3, 3), activation='relu'))\nmodel.add(layers.MaxPooling2D((2, 2)))\n\nmodel.add(layers.Flatten())\nmodel.add(layers.Dense(16, activation='relu'))\nmodel.add(layers.Dense(10, activation='softmax'))\n\n#Create early stop callback\nearlystop = EarlyStopping(monitor='loss', min_delta=0.0001, patience=5, \\\n verbose=1, mode='auto')\ncallbacks_list = [earlystop]\n\n#Compile the model\nmodel.compile(\n optimizer='rmsprop',\n loss='categorical_crossentropy',\n metrics=['accuracy'])\n\n#Fit the model to the data\nhistory = model.fit(\n train_images, \n train_labels, \n epochs=10, \n callbacks=callbacks_list,\n batch_size=254,\n validation_data=(test_images,test_labels))\n\nprint(\"\\nTraining completed\")\n\n#Evaluate the test set\nhistory_evaluation = model.evaluate(test_images, test_labels)\n\nprint(\"\\nEvaluation completed\")\n",
"Train on 60000 samples, validate on 10000 samples\nEpoch 1/10\n60000/60000 [==============================] - 19s 322us/step - loss: 0.6149 - acc: 0.8169 - val_loss: 0.3241 - val_acc: 0.8938\nEpoch 2/10\n60000/60000 [==============================] - 22s 361us/step - loss: 0.2056 - acc: 0.9380 - val_loss: 0.1750 - val_acc: 0.9438\nEpoch 3/10\n60000/60000 [==============================] - 19s 314us/step - loss: 0.1427 - acc: 0.9563 - val_loss: 0.1713 - val_acc: 0.9451\nEpoch 4/10\n60000/60000 [==============================] - 18s 296us/step - loss: 0.1115 - acc: 0.9659 - val_loss: 0.1271 - val_acc: 0.9624\nEpoch 5/10\n60000/60000 [==============================] - 18s 292us/step - loss: 0.0933 - acc: 0.9715 - val_loss: 0.1065 - val_acc: 0.9670\nEpoch 6/10\n60000/60000 [==============================] - 18s 292us/step - loss: 0.0807 - acc: 0.9753 - val_loss: 0.1044 - val_acc: 0.9650\nEpoch 7/10\n60000/60000 [==============================] - 18s 293us/step - loss: 0.0723 - acc: 0.9777 - val_loss: 0.0735 - val_acc: 0.9772\nEpoch 8/10\n60000/60000 [==============================] - 18s 295us/step - loss: 0.0649 - acc: 0.9803 - val_loss: 0.0742 - val_acc: 0.9777\nEpoch 9/10\n60000/60000 [==============================] - 18s 292us/step - loss: 0.0597 - acc: 0.9815 - val_loss: 0.0579 - val_acc: 0.9813\nEpoch 10/10\n60000/60000 [==============================] - 18s 292us/step - loss: 0.0544 - acc: 0.9835 - val_loss: 0.0572 - val_acc: 0.9825\n\nTraining completed\n10000/10000 [==============================] - 1s 124us/step\n\nEvaluation completed\n\nHistory\n\n<keras.callbacks.History object at 0x00000179D8980160>\n"
]
],
[
[
"## Visualize results",
"_____no_output_____"
]
],
[
[
"model.summary()\n\n# Get training and test loss histories\ntraining_loss = history.history['loss']\ntest_loss = history.history['val_loss']\n# Get training and test accuracy histories\ntraining_accuracy = history.history['acc']\ntest_accuracy = history.history['val_acc']\n\n#print(history_evaluation)\n\nprint(\"Training Accuracy \" + str(training_accuracy[-1]))\nprint(\"Training Loss: \" + str(training_loss[-1]))\nprint(\"Test Accuracy: \" + str(test_accuracy[-1]))\nprint(\"Test Loss: \" + str(test_loss[-1]))\n\nprint(\"Model Parameters: \" + str(model.count_params()))\n\n# Plot the accuracy and cost summaries \nf, (ax1, ax2) = plt.subplots(2, 1, sharex=False, figsize=(13,13))\n\n# Create count of the number of epochs\nepoch_count = range(1, len(training_loss) + 1)\n\n# Visualize loss history\nax1.plot(epoch_count, training_loss, 'r--')\nax1.plot(epoch_count, test_loss, 'b-')\nax1.legend(['Training Loss', 'Test Loss'])\nax1.set_ylabel('Loss')\nax1.set_xlabel('Epoch')\n\n# Create count of the number of epochs\nepoch_count = range(1, len(training_accuracy) + 1)\n\n# Visualize accuracy history\nax2.plot(epoch_count, training_accuracy, 'r--')\nax2.plot(epoch_count, test_accuracy, 'b-')\nax2.legend(['Training Accuracy', 'Test Accuracy'])\nax2.set_ylabel('Accuracy Score')\nax1.set_xlabel('Epoch')\n\nplt.xlabel('Epoch')\nplt.show();\n",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d_10 (Conv2D) (None, 26, 26, 16) 160 \n_________________________________________________________________\nmax_pooling2d_9 (MaxPooling2 (None, 13, 13, 16) 0 \n_________________________________________________________________\nconv2d_11 (Conv2D) (None, 11, 11, 16) 2320 \n_________________________________________________________________\nmax_pooling2d_10 (MaxPooling (None, 5, 5, 16) 0 \n_________________________________________________________________\nflatten_5 (Flatten) (None, 400) 0 \n_________________________________________________________________\ndense_9 (Dense) (None, 16) 6416 \n_________________________________________________________________\ndense_10 (Dense) (None, 10) 170 \n=================================================================\nTotal params: 9,066\nTrainable params: 9,066\nNon-trainable params: 0\n_________________________________________________________________\nTraining Accuracy 0.9834500078916549\nTraining Loss: 0.05444586953427642\nTest Accuracy: 0.9825000083446502\nTest Loss: 0.05715448258873075\nModel Parameters: 9066\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f5dcb510f0ed7ed3f4db2fba8a9e3cb1302cb6 | 10,835 | ipynb | Jupyter Notebook | mwdsbe/Notebooks/Matching Algorithms/TF-IDF/tf-idf.ipynb | BinnyDaBin/MWDSBE | aa0de50f2289e47f7c2e9134334b23c3b5594f0c | [
"MIT"
] | null | null | null | mwdsbe/Notebooks/Matching Algorithms/TF-IDF/tf-idf.ipynb | BinnyDaBin/MWDSBE | aa0de50f2289e47f7c2e9134334b23c3b5594f0c | [
"MIT"
] | 10 | 2021-03-10T01:06:45.000Z | 2022-02-26T21:02:40.000Z | mwdsbe/Notebooks/Matching Algorithms/TF-IDF/tf-idf.ipynb | BinnyDaBin/MWDSBE | aa0de50f2289e47f7c2e9134334b23c3b5594f0c | [
"MIT"
] | null | null | null | 22.432712 | 182 | 0.537425 | [
[
[
"# TF-IDF\nJoining registry and license data using TF-IDF string matching algorithm",
"_____no_output_____"
]
],
[
[
"import mwdsbe\nimport mwdsbe.datasets.licenses as licenses\nimport schuylkill as skool\nimport time",
"_____no_output_____"
],
[
"registry = mwdsbe.load_registry() # geopandas df\nlicense = licenses.CommercialActivityLicenses().download()",
"_____no_output_____"
],
[
"# clean data\nignore_words = ['inc', 'group', 'llc', 'corp', 'pc', 'incorporated', 'ltd', 'co', 'associates', 'services', 'company', 'enterprises', 'enterprise', 'service', 'corporation']\ncleaned_registry = skool.clean_strings(registry, ['company_name', 'dba_name'], True, ignore_words)\ncleaned_license = skool.clean_strings(license, ['company_name'], True, ignore_words)",
"_____no_output_____"
],
[
"print('Total number of cleaned registry:', len(cleaned_registry))",
"Total number of cleaned registry: 3119\n"
],
[
"print('Total number of cleaned license:', len(cleaned_license))",
"Total number of cleaned license: 203578\n"
]
],
[
[
"## 1. Score-cutoff 90",
"_____no_output_____"
]
],
[
[
"t1 = time.time()\nmerged = (\n skool.tf_idf_merge(cleaned_registry, cleaned_license, on=\"company_name\", score_cutoff=90)\n .pipe(skool.tf_idf_merge, cleaned_registry, cleaned_license, left_on=\"dba_name\", right_on=\"company_name\", score_cutoff=90)\n)\nt = time.time() - t1",
"_____no_output_____"
],
[
"print('Execution time:', t, 'sec')",
"Execution time: 186.29000186920166 sec\n"
],
[
"matched = merged.dropna(subset=['company_name_y'])",
"_____no_output_____"
],
[
"print('Match:', len(matched), 'out of', len(cleaned_registry))",
"Match: 1391 out of 3119\n"
],
[
"non_exact_match = matched[matched.match_probability < 0.999999]\nnon_exact_match = non_exact_match[['company_name_x', 'match_probability', 'company_name_y']]\nprint('Non-exact match above 90:', len(non_exact_match), 'out of', len(matched))",
"Non-exact match above 90: 88 out of 1391\n"
],
[
"# non_exact_match.to_excel (r'C:\\Users\\dabinlee\\Desktop\\mwdsbe\\data\\tf-idf\\tf-idf-90.xlsx', index = None, header=True)",
"_____no_output_____"
]
],
[
[
"## 2. Score-cutoff 85",
"_____no_output_____"
]
],
[
[
"t1 = time.time()\nmerged = (\n skool.tf_idf_merge(cleaned_registry, cleaned_license, on=\"company_name\", score_cutoff=85)\n .pipe(skool.tf_idf_merge, cleaned_registry, cleaned_license, left_on=\"dba_name\", right_on=\"company_name\", score_cutoff=85)\n)\nt = time.time() - t1",
"_____no_output_____"
],
[
"print('Execution time:', t, 'sec')",
"Execution time: 187.34773302078247 sec\n"
],
[
"matched = merged.dropna(subset=['company_name_y'])",
"_____no_output_____"
],
[
"print('Match:', len(matched), 'out of', len(cleaned_registry))",
"Match: 1499 out of 3119\n"
],
[
"match_to_check = matched[matched.match_probability < 0.9]\nmatch_to_check = match_to_check[['company_name_x', 'match_probability', 'company_name_y']]\nprint('Match between 85 and 90:', len(match_to_check), 'out of', len(matched))",
"Match between 85 and 90: 111 out of 1499\n"
],
[
"# match_to_check.to_excel (r'C:\\Users\\dabinlee\\Desktop\\mwdsbe\\data\\tf-idf\\tf-idf-85.xlsx', index = None, header=True)",
"_____no_output_____"
]
],
[
[
"## 3. Score-cutoff 80",
"_____no_output_____"
]
],
[
[
"t1 = time.time()\nmerged = (\n skool.tf_idf_merge(cleaned_registry, cleaned_license, on=\"company_name\", score_cutoff=80)\n .pipe(skool.tf_idf_merge, cleaned_registry, cleaned_license, left_on=\"dba_name\", right_on=\"company_name\", score_cutoff=80)\n)\nt = time.time() - t1",
"_____no_output_____"
],
[
"print('Execution time:', t, 'sec')",
"Execution time: 188.21181917190552 sec\n"
],
[
"matched = merged.dropna(subset=['company_name_y'])",
"_____no_output_____"
],
[
"print('Match:', len(matched), 'out of', len(cleaned_registry))",
"Match: 1666 out of 3119\n"
],
[
"match_to_check = matched[matched.match_probability < 0.85]\nmatch_to_check = match_to_check[['company_name_x', 'match_probability', 'company_name_y']]\nprint('Match between 80 and 85:', len(match_to_check), 'out of', len(matched))",
"Match between 80 and 85: 172 out of 1666\n"
],
[
"# match_to_check.to_excel (r'C:\\Users\\dabinlee\\Desktop\\mwdsbe\\data\\tf-idf\\tf-idf-80.xlsx', index = None, header=True)",
"_____no_output_____"
]
],
[
[
"## 4. Score-cutoff 75",
"_____no_output_____"
]
],
[
[
"t1 = time.time()\nmerged = (\n skool.tf_idf_merge(cleaned_registry, cleaned_license, on=\"company_name\", score_cutoff=75)\n .pipe(skool.tf_idf_merge, cleaned_registry, cleaned_license, left_on=\"dba_name\", right_on=\"company_name\", score_cutoff=75)\n)\nt = time.time() - t1",
"_____no_output_____"
],
[
"print('Execution time:', t, 'sec')",
"Execution time: 186.20661854743958 sec\n"
],
[
"matched = merged.dropna(subset=['company_name_y'])",
"_____no_output_____"
],
[
"print('Match:', len(matched), 'out of', len(cleaned_registry))",
"Match: 1868 out of 3119\n"
],
[
"match_to_check = matched[matched.match_probability < 0.8]\nmatch_to_check = match_to_check[['company_name_x', 'match_probability', 'company_name_y']]\nprint('Match between 75 and 80:', len(match_to_check), 'out of', len(matched))",
"Match between 75 and 80: 208 out of 1868\n"
],
[
"# match_to_check.to_excel (r'C:\\Users\\dabinlee\\Desktop\\mwdsbe\\data\\tf-idf\\tf-idf-75.xlsx', index = None, header=True)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f5ebe733b94f069e0ff336df490298b151bc55 | 9,741 | ipynb | Jupyter Notebook | 00_quickstart/09_Run_Data_Bias_Analysis_ProcessingJob.ipynb | NRauschmayr/workshop | c890e38a5f4a339540697206ebdea479e66534e5 | [
"Apache-2.0"
] | null | null | null | 00_quickstart/09_Run_Data_Bias_Analysis_ProcessingJob.ipynb | NRauschmayr/workshop | c890e38a5f4a339540697206ebdea479e66534e5 | [
"Apache-2.0"
] | null | null | null | 00_quickstart/09_Run_Data_Bias_Analysis_ProcessingJob.ipynb | NRauschmayr/workshop | c890e38a5f4a339540697206ebdea479e66534e5 | [
"Apache-2.0"
] | null | null | null | 26.398374 | 269 | 0.582281 | [
[
[
"# Run Data Bias Analysis with SageMaker Clarify (Pre-Training)\n\n## Using SageMaker Processing Jobs",
"_____no_output_____"
]
],
[
[
"import boto3\nimport sagemaker\nimport pandas as pd\nimport numpy as np\n\nsess = sagemaker.Session()\nbucket = sess.default_bucket()\nrole = sagemaker.get_execution_role()\nregion = boto3.Session().region_name\n\nsm = boto3.Session().client(service_name=\"sagemaker\", region_name=region)",
"_____no_output_____"
]
],
[
[
"# Get Data from S3",
"_____no_output_____"
]
],
[
[
"%store -r bias_data_s3_uri",
"_____no_output_____"
],
[
"print(bias_data_s3_uri)",
"_____no_output_____"
],
[
"!aws s3 cp $bias_data_s3_uri ./data-clarify",
"_____no_output_____"
],
[
"import pandas as pd\n\ndata = pd.read_csv(\"./data-clarify/amazon_reviews_us_giftcards_software_videogames.csv\")\ndata.head()",
"_____no_output_____"
]
],
[
[
"# Analyze Unbalanced Data\nPlotting histograms for the distribution of the different features is a good way to visualize the data. ",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\n\nsns.countplot(data=data, x=\"star_rating\", hue=\"product_category\")",
"_____no_output_____"
]
],
[
[
"# Calculate Bias Metrics on Unbalanced Data\n\nSageMaker Clarify helps you detect possible pre- and post-training biases using a variety of metrics.",
"_____no_output_____"
]
],
[
[
"from sagemaker import clarify\n\nclarify_processor = clarify.SageMakerClarifyProcessor(\n role=role, instance_count=1, instance_type=\"ml.c5.2xlarge\", sagemaker_session=sess\n)",
"_____no_output_____"
]
],
[
[
"# Pre-training Bias\nBias can be present in your data before any model training occurs. Inspecting your data for bias before training begins can help detect any data collection gaps, inform your feature engineering, and hep you understand what societal biases the data may reflect.\n\nComputing pre-training bias metrics does not require a trained model.",
"_____no_output_____"
],
[
"## Writing DataConfig\nA `DataConfig` object communicates some basic information about data I/O to Clarify. We specify where to find the input dataset, where to store the output, the target column (`label`), the header names, and the dataset type.",
"_____no_output_____"
]
],
[
[
"bias_report_output_path = \"s3://{}/clarify\".format(bucket)\n\nbias_data_config = clarify.DataConfig(\n s3_data_input_path=bias_data_s3_uri,\n s3_output_path=bias_report_output_path,\n label=\"star_rating\",\n headers=data.columns.to_list(),\n dataset_type=\"text/csv\",\n)",
"_____no_output_____"
]
],
[
[
"# Configure `BiasConfig`\nSageMaker Clarify also needs the sensitive columns (`facets`) and the desirable outcomes (`label_values_or_threshold`).\n\nWe specify this information in the `BiasConfig` API. Here that the positive outcome is either `star rating==5` or `star_rating==4` and `product_category` is the sensitive facet that we analyze in this run.",
"_____no_output_____"
]
],
[
[
"bias_config = clarify.BiasConfig(\n label_values_or_threshold=[5, 4], facet_name=\"product_category\", group_name=\"product_category\"\n)",
"_____no_output_____"
]
],
[
[
"## Detect Bias with a SageMaker Processing Job and Clarify",
"_____no_output_____"
]
],
[
[
"clarify_processor.run_pre_training_bias(\n data_config=bias_data_config, data_bias_config=bias_config, methods=\"all\", wait=False, logs=False\n)",
"_____no_output_____"
],
[
"run_pre_training_bias_processing_job_name = clarify_processor.latest_job.job_name\nrun_pre_training_bias_processing_job_name",
"_____no_output_____"
],
[
"from IPython.core.display import display, HTML\n\ndisplay(\n HTML(\n '<b>Review <a target=\"blank\" href=\"https://console.aws.amazon.com/sagemaker/home?region={}#/processing-jobs/{}\">Processing Job</a></b>'.format(\n region, run_pre_training_bias_processing_job_name\n )\n )\n)",
"_____no_output_____"
],
[
"from IPython.core.display import display, HTML\n\ndisplay(\n HTML(\n '<b>Review <a target=\"blank\" href=\"https://console.aws.amazon.com/cloudwatch/home?region={}#logStream:group=/aws/sagemaker/ProcessingJobs;prefix={};streamFilter=typeLogStreamPrefix\">CloudWatch Logs</a> After About 5 Minutes</b>'.format(\n region, run_pre_training_bias_processing_job_name\n )\n )\n)",
"_____no_output_____"
],
[
"from IPython.core.display import display, HTML\n\ndisplay(\n HTML(\n '<b>Review <a target=\"blank\" href=\"https://s3.console.aws.amazon.com/s3/buckets/{}/{}/?region={}&tab=overview\">S3 Output Data</a> After The Processing Job Has Completed</b>'.format(\n bucket, run_pre_training_bias_processing_job_name, region\n )\n )\n)",
"_____no_output_____"
],
[
"running_processor = sagemaker.processing.ProcessingJob.from_processing_name(\n processing_job_name=run_pre_training_bias_processing_job_name, sagemaker_session=sess\n)\n\nprocessing_job_description = running_processor.describe()\n\nprint(processing_job_description)",
"_____no_output_____"
],
[
"running_processor.wait(logs=False)",
"_____no_output_____"
]
],
[
[
"# Download Report From S3\nThe class-imbalance metric should match the value calculated for the unbalanced dataset using the open source version above.",
"_____no_output_____"
]
],
[
[
"!aws s3 ls $bias_report_output_path/",
"_____no_output_____"
],
[
"!aws s3 cp --recursive $bias_report_output_path ./generated_bias_report/",
"_____no_output_____"
],
[
"from IPython.core.display import display, HTML\n\ndisplay(HTML('<b>Review <a target=\"blank\" href=\"./generated_bias_report/report.html\">Bias Report</a></b>'))",
"_____no_output_____"
]
],
[
[
"# Release Resources",
"_____no_output_____"
]
],
[
[
"%%html\n\n<p><b>Shutting down your kernel for this notebook to release resources.</b></p>\n<button class=\"sm-command-button\" data-commandlinker-command=\"kernelmenu:shutdown\" style=\"display:none;\">Shutdown Kernel</button>\n \n<script>\ntry {\n els = document.getElementsByClassName(\"sm-command-button\");\n els[0].click();\n}\ncatch(err) {\n // NoOp\n} \n</script>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f5fa3b8233e9c00a10e4b5b366e15d6e10f424 | 12,304 | ipynb | Jupyter Notebook | content/lessons/09/Class-Coding-Lab/CCL-Lists.ipynb | jvrecca-su/ist256project | 255ee5ae91e6f1d0a56519804701633443b75bc6 | [
"MIT"
] | null | null | null | content/lessons/09/Class-Coding-Lab/CCL-Lists.ipynb | jvrecca-su/ist256project | 255ee5ae91e6f1d0a56519804701633443b75bc6 | [
"MIT"
] | null | null | null | content/lessons/09/Class-Coding-Lab/CCL-Lists.ipynb | jvrecca-su/ist256project | 255ee5ae91e6f1d0a56519804701633443b75bc6 | [
"MIT"
] | null | null | null | 26.982456 | 201 | 0.529178 | [
[
[
"# In-Class Coding Lab: Lists\n\nThe goals of this lab are to help you understand:\n\n - List indexing and slicing\n - List methods such as insert, append, find, delete\n - How to iterate over lists with loops\n \n## Python Lists work like Real-Life Lists\n \nIn real life, we make lists all the time. To-Do lists. Shopping lists. Reading lists. These lists are collections of items, for example here's my shopping list:\n \n ```\n Milk, Eggs, Bread, Beer\n ```\n\nThere are 4 items in this list.\n\nLikewise, we can make a similar list in Python, and count the number of items in the list using the `len()` function:",
"_____no_output_____"
]
],
[
[
"shopping_list = [ 'Milk', 'Eggs', 'Bread', 'Beer']\nitem_count = len(shopping_list)\nprint(\"List: %s has %d items\" % (shopping_list, item_count))",
"List: ['Milk', 'Eggs', 'Bread', 'Beer'] has 4 items\n"
]
],
[
[
"## Enumerating Your List Items\n\nIn real-life, we *enumerate* lists all the time. We go through the items on our list one at a time and make a decision, for example: \"Did I add that to my shopping cart yet?\"\n\nIn Python we go through items in our lists with the `for` loop. We use `for` because the number of items in pre-determined and thus a **definite** loop is the appropriate choice. \n\nHere's an example:",
"_____no_output_____"
]
],
[
[
"for item in shopping_list:\n print(\"I need to buy some %s \" % (item))",
"I need to buy some Milk \nI need to buy some Eggs \nI need to buy some Bread \nI need to buy some Beer \n"
]
],
[
[
"## Now You Try It!\n\nWrite code in the space below to print each stock on its own line.",
"_____no_output_____"
]
],
[
[
"stocks = [ 'IBM', 'AAPL', 'GOOG', 'MSFT', 'TWTR', 'FB']\n#TODO: Write code here\nfor item in stocks:\n print(item)",
"IBM\nAAPL\nGOOG\nMSFT\nTWTR\nFB\n"
]
],
[
[
"## Indexing Lists\n\nSometimes we refer to our items by their place in the list. For example \"Milk is the first item on the list\" or \"Beer is the last item on the list.\"\n\nWe can also do this in Python, and it is called *indexing* the list. \n\n**IMPORTANT** The first item in a Python lists starts at index **0**.",
"_____no_output_____"
]
],
[
[
"print(\"The first item in the list is:\", shopping_list[0]) \nprint(\"The last item in the list is:\", shopping_list[3]) \nprint(\"This is also the last item in the list:\", shopping_list[-1]) \nprint(\"This is the second to last item in the list:\", shopping_list[-2])\n",
"The first item in the list is: Milk\nThe last item in the list is: Beer\nThis is also the last item in the list: Beer\nThis is the second to last item in the list: Bread\n"
]
],
[
[
"## For Loop with Index\n\nYou can also loop through your Python list using an index. In this case we use the `range()` function to determine how many times we should loop:",
"_____no_output_____"
]
],
[
[
"for i in range(len(shopping_list)):\n print(\"I need to buy some %s \" % (shopping_list[i]))",
"I need to buy some Milk \nI need to buy some Eggs \nI need to buy some Bread \nI need to buy some Beer \n"
]
],
[
[
"## Now You Try It!\n\nWrite code to print the 2nd and 4th stocks in the list variable `stocks`. For example:\n\n`AAPL MSFT`",
"_____no_output_____"
]
],
[
[
"#TODO: Write code here\nprint(\"This is the second stock in the list:\", stocks[1])\nprint(\"This is the fourth stock in the list:\", stocks[3])\n",
"This is the second stock in the list: AAPL\nThis is the fourth stock in the list: MSFT\n"
]
],
[
[
"## Lists are Mutable\n\nUnlike strings, lists are mutable. This means we can change a value in the list.\n\nFor example, I want `'Craft Beer'` not just `'Beer'`:",
"_____no_output_____"
]
],
[
[
"print(shopping_list)\nshopping_list[-1] = 'Craft Beer'\nprint(shopping_list)",
"['Milk', 'Eggs', 'Bread', 'Beer']\n['Milk', 'Eggs', 'Bread', 'Craft Beer']\n"
]
],
[
[
"## List Methods\n\nIn your readings and class lecture, you encountered some list methods. These allow us to maniupulate the list by adding or removing items.",
"_____no_output_____"
]
],
[
[
"print(\"Shopping List: %s\" %(shopping_list))\n\nprint(\"Adding 'Cheese' to the end of the list...\")\nshopping_list.append('Cheese') #add to end of list\nprint(\"Shopping List: %s\" %(shopping_list))\n\nprint(\"Adding 'Cereal' to position 0 in the list...\")\nshopping_list.insert(0,'Cereal') # add to the beginning of the list (position 0)\nprint(\"Shopping List: %s\" %(shopping_list))\n\nprint(\"Removing 'Cheese' from the list...\")\nshopping_list.remove('Cheese') # remove 'Cheese' from the list\nprint(\"Shopping List: %s\" %(shopping_list))\n\nprint(\"Removing item from position 0 in the list...\")\ndel shopping_list[0] # remove item at position 0\nprint(\"Shopping List: %s\" %(shopping_list))\n",
"Shopping List: ['Milk', 'Eggs', 'Bread', 'Craft Beer']\nAdding 'Cheese' to the end of the list...\nShopping List: ['Milk', 'Eggs', 'Bread', 'Craft Beer', 'Cheese']\nAdding 'Cereal' to position 0 in the list...\nShopping List: ['Cereal', 'Milk', 'Eggs', 'Bread', 'Craft Beer', 'Cheese']\nRemoving 'Cheese' from the list...\nShopping List: ['Cereal', 'Milk', 'Eggs', 'Bread', 'Craft Beer']\nRemoving item from position 0 in the list...\nShopping List: ['Milk', 'Eggs', 'Bread', 'Craft Beer']\n"
]
],
[
[
"## Now You Try It!\n\nWrite a program to remove the following stocks: `IBM` and `TWTR`\n\nThen add this stock to the end `NFLX` and this stock to the beginning `TSLA`\n\nPrint your list when you are done. It should look like this:\n\n`['TSLA', 'AAPL', 'GOOG', 'MSFT', 'FB', 'NFLX']`\n",
"_____no_output_____"
]
],
[
[
"# TODO: Write Code here\nprint(\"Stocks: %s\" % (stocks))\n\nprint('Removing Stocks: IBM, TWTR')\n\nstocks.remove('IBM')\nstocks.remove('TWTR')\n\nprint(\"Stocks: %s\" % (stocks))\n\nprint('Adding Stock to End:NFLX')\n\nstocks.append('NFLX')\nprint(\"Stocks: %s\" % (stocks))\n\nprint('Adding Stock to Beginning:TSLA')\nstocks.insert(0, 'TSLA')\n\nprint(\"Final Stocks: %s\" % (stocks))",
"Stocks: ['IBM', 'AAPL', 'GOOG', 'MSFT', 'TWTR', 'FB']\nRemoving Stocks: IBM, TWTR\nStocks: ['AAPL', 'GOOG', 'MSFT', 'FB']\nAdding Stock to End:NFLX\nStocks: ['AAPL', 'GOOG', 'MSFT', 'FB', 'NFLX']\nAdding Stock to Beginning:TSLA\nFinal Stocks: ['TSLA', 'AAPL', 'GOOG', 'MSFT', 'FB', 'NFLX']\n"
]
],
[
[
"## Sorting\n\nSince Lists are mutable. You can use the `sort()` method to re-arrange the items in the list alphabetically (or numerically if it's a list of numbers)",
"_____no_output_____"
]
],
[
[
"print(\"Before Sort:\", shopping_list)\nshopping_list.sort() \nprint(\"After Sort:\", shopping_list)",
"Before Sort: ['Milk', 'Eggs', 'Bread', 'Craft Beer']\nAfter Sort: ['Bread', 'Craft Beer', 'Eggs', 'Milk']\n"
]
],
[
[
"# Putting it all together\n\nWinning Lotto numbers. When the lotto numbers are drawn, they are in any order, when they are presented they're allways sorted. Let's write a program to input 5 numbers then output them sorted\n\n```\n1. for i in range(5)\n2. input a number\n3. append the number you input to the lotto_numbers list\n4. sort the lotto_numbers list\n5. print the lotto_numbers list like this: \n 'today's winning numbers are [1, 5, 17, 34, 56]'\n```",
"_____no_output_____"
]
],
[
[
"## TODO: Write program here:\n\nlotto_numbers = [] # start with an empty list\nfor i in range(5):\n number = int(input(\"Enter a number: \"))\n lotto_numbers.append(number)\nlotto_numbers.sort()\nprint(\"Today's winning lotto numbers are\", lotto_numbers)\n",
"Enter a number: 12\nEnter a number: 15\nEnter a number: 22\nEnter a number: 9\nEnter a number: 4\nToday's winning lotto numbers are [4, 9, 12, 15, 22]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f5fb8896ab4e4ac1170aff4aa9a35110071ba6 | 27,909 | ipynb | Jupyter Notebook | samples/tutorials/mnist/04_Reusable_and_Pre-build_Components_as_Pipeline.ipynb | eedorenko/pipelines | 213bf328d156c9d598c147486993f65735b2fd39 | [
"Apache-2.0"
] | null | null | null | samples/tutorials/mnist/04_Reusable_and_Pre-build_Components_as_Pipeline.ipynb | eedorenko/pipelines | 213bf328d156c9d598c147486993f65735b2fd39 | [
"Apache-2.0"
] | null | null | null | samples/tutorials/mnist/04_Reusable_and_Pre-build_Components_as_Pipeline.ipynb | eedorenko/pipelines | 213bf328d156c9d598c147486993f65735b2fd39 | [
"Apache-2.0"
] | null | null | null | 35.283186 | 499 | 0.589416 | [
[
[
"# Copyright 2019 Google Inc. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n# ==============================================================================",
"_____no_output_____"
]
],
[
[
"# Composing a pipeline from reusable, pre-built, and lightweight components\n\nThis tutorial describes how to build a Kubeflow pipeline from reusable, pre-built, and lightweight components. The following provides a summary of the steps involved in creating and using a reusable component:\n\n- Write the program that contains your component’s logic. The program must use files and command-line arguments to pass data to and from the component.\n- Containerize the program.\n- Write a component specification in YAML format that describes the component for the Kubeflow Pipelines system.\n- Use the Kubeflow Pipelines SDK to load your component, use it in a pipeline and run that pipeline.\n\nThen, we will compose a pipeline from a reusable component, a pre-built component, and a lightweight component. The pipeline will perform the following steps:\n- Train an MNIST model and export it to Google Cloud Storage.\n- Deploy the exported TensorFlow model on AI Platform Prediction service.\n- Test the deployment by calling the endpoint with test data.",
"_____no_output_____"
],
[
"Note: Ensure that you have Docker installed, if you want to build the image locally, by running the following command:\n \n`which docker`\n \nThe result should be something like:\n\n`/usr/bin/docker`",
"_____no_output_____"
]
],
[
[
"import kfp\nimport kfp.gcp as gcp\nimport kfp.dsl as dsl\nimport kfp.compiler as compiler\nimport kfp.components as comp\nimport datetime\n\nimport kubernetes as k8s",
"_____no_output_____"
],
[
"# Required Parameters\nPROJECT_ID='<ADD GCP PROJECT HERE>'\nGCS_BUCKET='gs://<ADD STORAGE LOCATION HERE>'",
"_____no_output_____"
]
],
[
[
"## Create client\n\nIf you run this notebook **outside** of a Kubeflow cluster, run the following command:\n- `host`: The URL of your Kubeflow Pipelines instance, for example \"https://`<your-deployment>`.endpoints.`<your-project>`.cloud.goog/pipeline\"\n- `client_id`: The client ID used by Identity-Aware Proxy\n- `other_client_id`: The client ID used to obtain the auth codes and refresh tokens.\n- `other_client_secret`: The client secret used to obtain the auth codes and refresh tokens.\n\n```python\nclient = kfp.Client(host, client_id, other_client_id, other_client_secret)\n```\n\nIf you run this notebook **within** a Kubeflow cluster, run the following command:\n```python\nclient = kfp.Client()\n```\n\nYou'll need to create OAuth client ID credentials of type `Other` to get `other_client_id` and `other_client_secret`. Learn more about [creating OAuth credentials](\nhttps://cloud.google.com/iap/docs/authentication-howto#authenticating_from_a_desktop_app)",
"_____no_output_____"
]
],
[
[
"# Optional Parameters, but required for running outside Kubeflow cluster\n\n# The host for 'AI Platform Pipelines' ends with 'pipelines.googleusercontent.com'\n# The host for pipeline endpoint of 'full Kubeflow deployment' ends with '/pipeline'\n# Examples are:\n# https://7c021d0340d296aa-dot-us-central2.pipelines.googleusercontent.com\n# https://kubeflow.endpoints.kubeflow-pipeline.cloud.goog/pipeline\nHOST = '<ADD HOST NAME TO TALK TO KUBEFLOW PIPELINE HERE>'\n\n# For 'full Kubeflow deployment' on GCP, the endpoint is usually protected through IAP, therefore the following \n# will be needed to access the endpoint.\nCLIENT_ID = '<ADD OAuth CLIENT ID USED BY IAP HERE>'\nOTHER_CLIENT_ID = '<ADD OAuth CLIENT ID USED TO OBTAIN AUTH CODES HERE>'\nOTHER_CLIENT_SECRET = '<ADD OAuth CLIENT SECRET USED TO OBTAIN AUTH CODES HERE>'",
"_____no_output_____"
],
[
"# This is to ensure the proper access token is present to reach the end point for 'AI Platform Pipelines'\n# If you are not working with 'AI Platform Pipelines', this step is not necessary\n! gcloud auth print-access-token",
"_____no_output_____"
],
[
"# Create kfp client\nin_cluster = True\ntry:\n k8s.config.load_incluster_config()\nexcept:\n in_cluster = False\n pass\n\nif in_cluster:\n client = kfp.Client()\nelse:\n if HOST.endswith('googleusercontent.com'):\n CLIENT_ID = None\n OTHER_CLIENT_ID = None\n OTHER_CLIENT_SECRET = None\n\n client = kfp.Client(host=HOST, \n client_id=CLIENT_ID,\n other_client_id=OTHER_CLIENT_ID, \n other_client_secret=OTHER_CLIENT_SECRET)",
"_____no_output_____"
]
],
[
[
"# Build reusable components",
"_____no_output_____"
],
[
"## Writing the program code",
"_____no_output_____"
],
[
"The following cell creates a file `app.py` that contains a Python script. The script downloads MNIST dataset, trains a Neural Network based classification model, writes the training log and exports the trained model to Google Cloud Storage.\n\nYour component can create outputs that the downstream components can use as inputs. Each output must be a string and the container image must write each output to a separate local text file. For example, if a training component needs to output the path of the trained model, the component writes the path into a local file, such as `/output.txt`.",
"_____no_output_____"
]
],
[
[
"%%bash\n\n# Create folders if they don't exist.\nmkdir -p tmp/reuse_components_pipeline/mnist_training\n\n# Create the Python file that lists GCS blobs.\ncat > ./tmp/reuse_components_pipeline/mnist_training/app.py <<HERE\nimport argparse\nfrom datetime import datetime\nimport tensorflow as tf\n\nparser = argparse.ArgumentParser()\nparser.add_argument(\n '--model_path', type=str, required=True, help='Name of the model file.')\nparser.add_argument(\n '--bucket', type=str, required=True, help='GCS bucket name.')\nargs = parser.parse_args()\n\nbucket=args.bucket\nmodel_path=args.model_path\n\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Flatten(input_shape=(28, 28)),\n tf.keras.layers.Dense(512, activation=tf.nn.relu),\n tf.keras.layers.Dropout(0.2),\n tf.keras.layers.Dense(10, activation=tf.nn.softmax)\n])\n\nmodel.compile(optimizer='adam',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\n\nprint(model.summary()) \n\nmnist = tf.keras.datasets.mnist\n(x_train, y_train),(x_test, y_test) = mnist.load_data()\nx_train, x_test = x_train / 255.0, x_test / 255.0\n\ncallbacks = [\n tf.keras.callbacks.TensorBoard(log_dir=bucket + '/logs/' + datetime.now().date().__str__()),\n # Interrupt training if val_loss stops improving for over 2 epochs\n tf.keras.callbacks.EarlyStopping(patience=2, monitor='val_loss'),\n]\n\nmodel.fit(x_train, y_train, batch_size=32, epochs=5, callbacks=callbacks,\n validation_data=(x_test, y_test))\n\nfrom tensorflow import gfile\n\ngcs_path = bucket + \"/\" + model_path\n# The export require the folder is new\nif gfile.Exists(gcs_path):\n gfile.DeleteRecursively(gcs_path)\ntf.keras.experimental.export_saved_model(model, gcs_path)\n\nwith open('/output.txt', 'w') as f:\n f.write(gcs_path)\nHERE",
"_____no_output_____"
]
],
[
[
"## Create a Docker container\nCreate your own container image that includes your program. ",
"_____no_output_____"
],
[
"### Creating a Dockerfile",
"_____no_output_____"
],
[
"Now create a container that runs the script. Start by creating a Dockerfile. A Dockerfile contains the instructions to assemble a Docker image. The `FROM` statement specifies the Base Image from which you are building. `WORKDIR` sets the working directory. When you assemble the Docker image, `COPY` copies the required files and directories (for example, `app.py`) to the file system of the container. `RUN` executes a command (for example, install the dependencies) and commits the results. ",
"_____no_output_____"
]
],
[
[
"%%bash\n\n# Create Dockerfile.\n# AI platform only support tensorflow 1.14\ncat > ./tmp/reuse_components_pipeline/mnist_training/Dockerfile <<EOF\nFROM tensorflow/tensorflow:1.14.0-py3\nWORKDIR /app\nCOPY . /app\nEOF",
"_____no_output_____"
]
],
[
[
"### Build docker image",
"_____no_output_____"
],
[
"Now that we have created our Dockerfile for creating our Docker image. Then we need to build the image and push to a registry to host the image. There are three possible options:\n- Use the `kfp.containers.build_image_from_working_dir` to build the image and push to the Container Registry (GCR). This requires [kaniko](https://cloud.google.com/blog/products/gcp/introducing-kaniko-build-container-images-in-kubernetes-and-google-container-builder-even-without-root-access), which will be auto-installed with 'full Kubeflow deployment' but not 'AI Platform Pipelines'.\n- Use [Cloud Build](https://cloud.google.com/cloud-build), which would require the setup of GCP project and enablement of corresponding API. If you are working with GCP 'AI Platform Pipelines' with GCP project running, it is recommended to use Cloud Build.\n- Use [Docker](https://www.docker.com/get-started) installed locally and push to e.g. GCR.",
"_____no_output_____"
],
[
"**Note**:\nIf you run this notebook **within Kubeflow cluster**, **with Kubeflow version >= 0.7** and exploring **kaniko option**, you need to ensure that valid credentials are created within your notebook's namespace.\n- With Kubeflow version >= 0.7, the credential is supposed to be copied automatically while creating notebook through `Configurations`, which doesn't work properly at the time of creating this notebook. \n- You can also add credentials to the new namespace by either [copying credentials from an existing Kubeflow namespace, or by creating a new service account](https://www.kubeflow.org/docs/gke/authentication/#kubeflow-v0-6-and-before-gcp-service-account-key-as-secret).\n- The following cell demonstrates how to copy the default secret to your own namespace.\n\n```bash\n%%bash\n\nNAMESPACE=<your notebook name space>\nSOURCE=kubeflow\nNAME=user-gcp-sa\nSECRET=$(kubectl get secrets \\${NAME} -n \\${SOURCE} -o jsonpath=\"{.data.\\${NAME}\\.json}\" | base64 -D)\nkubectl create -n \\${NAMESPACE} secret generic \\${NAME} --from-literal=\"\\${NAME}.json=\\${SECRET}\"\n```",
"_____no_output_____"
]
],
[
[
"IMAGE_NAME=\"mnist_training_kf_pipeline\"\nTAG=\"latest\" # \"v_$(date +%Y%m%d_%H%M%S)\"\n\nGCR_IMAGE=\"gcr.io/{PROJECT_ID}/{IMAGE_NAME}:{TAG}\".format(\n PROJECT_ID=PROJECT_ID,\n IMAGE_NAME=IMAGE_NAME,\n TAG=TAG\n)\n\nAPP_FOLDER='./tmp/reuse_components_pipeline/mnist_training/'",
"_____no_output_____"
],
[
"# In the following, for the purpose of demonstration\n# Cloud Build is choosen for 'AI Platform Pipelines'\n# kaniko is choosen for 'full Kubeflow deployment'\n\nif HOST.endswith('googleusercontent.com'):\n # kaniko is not pre-installed with 'AI Platform Pipelines'\n import subprocess\n # ! gcloud builds submit --tag ${IMAGE_NAME} ${APP_FOLDER}\n cmd = ['gcloud', 'builds', 'submit', '--tag', GCR_IMAGE, APP_FOLDER]\n build_log = (subprocess.run(cmd, stdout=subprocess.PIPE).stdout[:-1].decode('utf-8'))\n print(build_log)\n \nelse:\n if kfp.__version__ <= '0.1.36':\n # kfp with version 0.1.36+ introduce broken change that will make the following code not working\n import subprocess\n \n builder = kfp.containers._container_builder.ContainerBuilder(\n gcs_staging=GCS_BUCKET + \"/kfp_container_build_staging\"\n )\n\n kfp.containers.build_image_from_working_dir(\n image_name=GCR_IMAGE,\n working_dir=APP_FOLDER,\n builder=builder\n )\n else:\n raise(\"Please build the docker image use either [Docker] or [Cloud Build]\")",
"_____no_output_____"
]
],
[
[
"#### If you want to use docker to build the image\nRun the following in a cell\n```bash\n%%bash -s \"{PROJECT_ID}\"\n\nIMAGE_NAME=\"mnist_training_kf_pipeline\"\nTAG=\"latest\" # \"v_$(date +%Y%m%d_%H%M%S)\"\n\n# Create script to build docker image and push it.\ncat > ./tmp/components/mnist_training/build_image.sh <<HERE\nPROJECT_ID=\"${1}\"\nIMAGE_NAME=\"${IMAGE_NAME}\"\nTAG=\"${TAG}\"\nGCR_IMAGE=\"gcr.io/\\${PROJECT_ID}/\\${IMAGE_NAME}:\\${TAG}\"\ndocker build -t \\${IMAGE_NAME} .\ndocker tag \\${IMAGE_NAME} \\${GCR_IMAGE}\ndocker push \\${GCR_IMAGE}\ndocker image rm \\${IMAGE_NAME}\ndocker image rm \\${GCR_IMAGE}\nHERE\n\ncd tmp/components/mnist_training\nbash build_image.sh\n```",
"_____no_output_____"
]
],
[
[
"image_name = GCR_IMAGE",
"_____no_output_____"
]
],
[
[
"## Writing your component definition file\nTo create a component from your containerized program, you must write a component specification in YAML that describes the component for the Kubeflow Pipelines system.\n\nFor the complete definition of a Kubeflow Pipelines component, see the [component specification](https://www.kubeflow.org/docs/pipelines/reference/component-spec/). However, for this tutorial you don’t need to know the full schema of the component specification. The notebook provides enough information to complete the tutorial.\n\nStart writing the component definition (component.yaml) by specifying your container image in the component’s implementation section:",
"_____no_output_____"
]
],
[
[
"%%bash -s \"{image_name}\"\n\nGCR_IMAGE=\"${1}\"\necho ${GCR_IMAGE}\n\n# Create Yaml\n# the image uri should be changed according to the above docker image push output\n\ncat > mnist_pipeline_component.yaml <<HERE\nname: Mnist training\ndescription: Train a mnist model and save to GCS\ninputs:\n - name: model_path\n description: 'Path of the tf model.'\n type: String\n - name: bucket\n description: 'GCS bucket name.'\n type: String\noutputs:\n - name: gcs_model_path\n description: 'Trained model path.'\n type: GCSPath\nimplementation:\n container:\n image: ${GCR_IMAGE}\n command: [\n python, /app/app.py,\n --model_path, {inputValue: model_path},\n --bucket, {inputValue: bucket},\n ]\n fileOutputs:\n gcs_model_path: /output.txt\nHERE",
"_____no_output_____"
],
[
"import os\nmnist_train_op = kfp.components.load_component_from_file(os.path.join('./', 'mnist_pipeline_component.yaml')) ",
"_____no_output_____"
],
[
"mnist_train_op.component_spec",
"_____no_output_____"
]
],
[
[
"# Define deployment operation on AI Platform",
"_____no_output_____"
]
],
[
[
"mlengine_deploy_op = comp.load_component_from_url(\n 'https://raw.githubusercontent.com/kubeflow/pipelines/2df775a28045bda15372d6dd4644f71dcfe41bfe/components/gcp/ml_engine/deploy/component.yaml')\n\ndef deploy(\n project_id,\n model_uri,\n model_id,\n runtime_version,\n python_version):\n \n return mlengine_deploy_op(\n model_uri=model_uri,\n project_id=project_id, \n model_id=model_id, \n runtime_version=runtime_version, \n python_version=python_version,\n replace_existing_version=True, \n set_default=True)",
"_____no_output_____"
]
],
[
[
"Kubeflow serving deployment component as an option. **Note that, the deployed Endppoint URI is not availabe as output of this component.**\n```python\nkubeflow_deploy_op = comp.load_component_from_url(\n 'https://raw.githubusercontent.com/kubeflow/pipelines/2df775a28045bda15372d6dd4644f71dcfe41bfe/components/gcp/ml_engine/deploy/component.yaml')\n\ndef deploy_kubeflow(\n model_dir,\n tf_server_name):\n return kubeflow_deploy_op(\n model_dir=model_dir,\n server_name=tf_server_name,\n cluster_name='kubeflow', \n namespace='kubeflow',\n pvc_name='', \n service_type='ClusterIP')\n```",
"_____no_output_____"
],
[
"# Create a lightweight component for testing the deployment",
"_____no_output_____"
]
],
[
[
"def deployment_test(project_id: str, model_name: str, version: str) -> str:\n\n model_name = model_name.split(\"/\")[-1]\n version = version.split(\"/\")[-1]\n \n import googleapiclient.discovery\n \n def predict(project, model, data, version=None):\n \"\"\"Run predictions on a list of instances.\n\n Args:\n project: (str), project where the Cloud ML Engine Model is deployed.\n model: (str), model name.\n data: ([[any]]), list of input instances, where each input instance is a\n list of attributes.\n version: str, version of the model to target.\n\n Returns:\n Mapping[str: any]: dictionary of prediction results defined by the model.\n \"\"\"\n\n service = googleapiclient.discovery.build('ml', 'v1')\n name = 'projects/{}/models/{}'.format(project, model)\n\n if version is not None:\n name += '/versions/{}'.format(version)\n\n response = service.projects().predict(\n name=name, body={\n 'instances': data\n }).execute()\n\n if 'error' in response:\n raise RuntimeError(response['error'])\n\n return response['predictions']\n\n import tensorflow as tf\n import json\n \n mnist = tf.keras.datasets.mnist\n (x_train, y_train),(x_test, y_test) = mnist.load_data()\n x_train, x_test = x_train / 255.0, x_test / 255.0\n\n result = predict(\n project=project_id,\n model=model_name,\n data=x_test[0:2].tolist(),\n version=version)\n print(result)\n \n return json.dumps(result)",
"_____no_output_____"
],
[
"# # Test the function with already deployed version\n# deployment_test(\n# project_id=PROJECT_ID,\n# model_name=\"mnist\",\n# version='ver_bb1ebd2a06ab7f321ad3db6b3b3d83e6' # previous deployed version for testing\n# )",
"_____no_output_____"
],
[
"deployment_test_op = comp.func_to_container_op(\n func=deployment_test, \n base_image=\"tensorflow/tensorflow:1.15.0-py3\",\n packages_to_install=[\"google-api-python-client==1.7.8\"])",
"_____no_output_____"
]
],
[
[
"# Create your workflow as a Python function",
"_____no_output_____"
],
[
"Define your pipeline as a Python function. ` @kfp.dsl.pipeline` is a required decoration, and must include `name` and `description` properties. Then compile the pipeline function. After the compilation is completed, a pipeline file is created.",
"_____no_output_____"
]
],
[
[
"# Define the pipeline\[email protected](\n name='Mnist pipeline',\n description='A toy pipeline that performs mnist model training.'\n)\ndef mnist_reuse_component_deploy_pipeline(\n project_id: str = PROJECT_ID,\n model_path: str = 'mnist_model', \n bucket: str = GCS_BUCKET\n):\n train_task = mnist_train_op(\n model_path=model_path, \n bucket=bucket\n ).apply(gcp.use_gcp_secret('user-gcp-sa'))\n \n deploy_task = deploy(\n project_id=project_id,\n model_uri=train_task.outputs['gcs_model_path'],\n model_id=\"mnist\", \n runtime_version=\"1.14\",\n python_version=\"3.5\"\n ).apply(gcp.use_gcp_secret('user-gcp-sa')) \n \n deploy_test_task = deployment_test_op(\n project_id=project_id,\n model_name=deploy_task.outputs[\"model_name\"], \n version=deploy_task.outputs[\"version_name\"],\n ).apply(gcp.use_gcp_secret('user-gcp-sa'))\n \n return True",
"_____no_output_____"
]
],
[
[
"### Submit a pipeline run",
"_____no_output_____"
]
],
[
[
"pipeline_func = mnist_reuse_component_deploy_pipeline",
"_____no_output_____"
],
[
"experiment_name = 'minist_kubeflow'\n\narguments = {\"model_path\":\"mnist_model\",\n \"bucket\":GCS_BUCKET}\n\nrun_name = pipeline_func.__name__ + ' run'\n\n# Submit pipeline directly from pipeline function\nrun_result = client.create_run_from_pipeline_func(pipeline_func, \n experiment_name=experiment_name, \n run_name=run_name, \n arguments=arguments)",
"_____no_output_____"
]
],
[
[
"**As an alternative, you can compile the pipeline into a package.** The compiled pipeline can be easily shared and reused by others to run the pipeline.\n\n```python\npipeline_filename = pipeline_func.__name__ + '.pipeline.zip'\ncompiler.Compiler().compile(pipeline_func, pipeline_filename)\n\nexperiment = client.create_experiment('python-functions-mnist')\n\nrun_result = client.run_pipeline(\n experiment_id=experiment.id, \n job_name=run_name, \n pipeline_package_path=pipeline_filename, \n params=arguments)\n```",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e7f6026df05e834793a78d76c1448053d2599005 | 6,723 | ipynb | Jupyter Notebook | python_notes/using_python_for_research_ph256x_harvard/week2/4randomness_n_time.ipynb | ZaynChen/notes | 37a71aac067ff86d89fc4654782d9a483472e91f | [
"MulanPSL-1.0"
] | null | null | null | python_notes/using_python_for_research_ph256x_harvard/week2/4randomness_n_time.ipynb | ZaynChen/notes | 37a71aac067ff86d89fc4654782d9a483472e91f | [
"MulanPSL-1.0"
] | null | null | null | python_notes/using_python_for_research_ph256x_harvard/week2/4randomness_n_time.ipynb | ZaynChen/notes | 37a71aac067ff86d89fc4654782d9a483472e91f | [
"MulanPSL-1.0"
] | null | null | null | 19.26361 | 77 | 0.481928 | [
[
[
"# Notes for Using Python for Research(HarvardX PH526x)",
"_____no_output_____"
],
[
"## Part 4: Randomness and Time\n\n### 1. Simulating Randomness(模拟随机性)\n\n### 2. Examples Involving Randomness\n\n### 3. Using the NumPy Random Module\n\n### 4. Measuring Time(测量时间)\n\n### 5. Random Walks(RW,随机游走)\n\n$$\nx(t=x) = x(t=0) + \\Delta x(t=1) + \\ldots + \\Delta x(t=k)\n$$",
"_____no_output_____"
],
[
"#### Simulating Randomness 部分的代码",
"_____no_output_____"
]
],
[
[
"import random\nrandom.choice([\"H\",\"T\"])",
"_____no_output_____"
],
[
"random.choice([0, 1])",
"_____no_output_____"
],
[
"random.choice([1,2,3,4,5,6])",
"_____no_output_____"
],
[
"random.choice(range(1, 7))",
"_____no_output_____"
],
[
"random.choice([range(1,7)])",
"_____no_output_____"
],
[
"random.choice(random.choice([range(1, 7), range(1, 9), range(1, 11)]))",
"_____no_output_____"
]
],
[
[
"#### Examples Involving Randomness 部分的代码",
"_____no_output_____"
]
],
[
[
"import random\nimport matplotlib.pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"rolls = []\nfor k in range(100000):\n rolls.append(random.choice([1,2,3,4,5,6]))\nplt.hist(rolls, bins = np.linspace(0.5, 6.5, 7));",
"_____no_output_____"
],
[
"ys = []\n\nfor rep in range(100000):\n y = 0\n for k in range(10):\n x = random.choice([1,2,3,4,5,6])\n y = y + x\n ys.append(y)\nplt.hist(ys);",
"_____no_output_____"
]
],
[
[
"#### Using the NumPy Random Module 部分的代码",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"np.random.random()",
"_____no_output_____"
],
[
"np.random.random(5)",
"_____no_output_____"
],
[
"np.random.random((5, 3))",
"_____no_output_____"
],
[
"np.random.normal(0, 1)",
"_____no_output_____"
],
[
"np.random.normal(0, 1, 5)",
"_____no_output_____"
],
[
"np.random.normal(0, 1, (2, 5))",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nX = np.random.randint(1, 7, (100000, 10))\nY = np.sum(X, axis=1)\nplt.hist(Y);",
"_____no_output_____"
]
],
[
[
"#### Measuring Time 部分的代码",
"_____no_output_____"
]
],
[
[
"import time\nimport random\nimport numpy as np",
"_____no_output_____"
],
[
"\nstart_time = time.time()\nys = []\nfor rep in range(1000000):\n y = 0\n for k in range(10):\n x = random.choice([1,2,3,4,5,6])\n y = y + x\nys.append(y)\nend_time = time.time()\nprint(end_time - start_time)",
"_____no_output_____"
],
[
"start_time = time.time()\nX = np.random.randint(1, 7, (1000000, 10))\nY = np.sum(X, axis=1)\nend_time = time.time()\nprint(end_time - start_time)",
"_____no_output_____"
]
],
[
[
"#### Random Walks 部分的代码",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"delta_X = np.random.normal(0,1,(2,5))\nplt.plot(delta_X[0], delta_X[1], \"go\")",
"_____no_output_____"
],
[
"X = np.cumsum(delta_X, axis=1)\nX",
"_____no_output_____"
],
[
"X_0 = np.array([[0], [0]])\ndelta_X = np.random.normal(0, 1, (2, 100))\nX = np.concatenate((X_0, np.cumsum(delta_X, axis=1)), axis=1)\nplt.plot(X[0], X[1], \"ro-\")\n# plt.savefig(\"rw.pdf\")",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7f62aa67f752684247dabe6f376d03073bfee0e | 630,035 | ipynb | Jupyter Notebook | samples/demo.ipynb | jaewon-jun9/Mask_RCNN | ea4c72c833c941bd47c70a3bb01bc93db60813eb | [
"MIT"
] | null | null | null | samples/demo.ipynb | jaewon-jun9/Mask_RCNN | ea4c72c833c941bd47c70a3bb01bc93db60813eb | [
"MIT"
] | null | null | null | samples/demo.ipynb | jaewon-jun9/Mask_RCNN | ea4c72c833c941bd47c70a3bb01bc93db60813eb | [
"MIT"
] | null | null | null | 1,853.044118 | 618,608 | 0.95908 | [
[
[
"# Mask R-CNN Demo\n\nA quick intro to using the pre-trained model to detect and segment objects.",
"_____no_output_____"
]
],
[
[
"import os\nos.environ[\"CUDA_VISIBLE_DEVICES\"] = \"-1\"\nimport sys\nimport random\nimport math\nimport numpy as np\nimport skimage.io\nimport matplotlib\nimport matplotlib.pyplot as plt\n\n# Root directory of the project\nROOT_DIR = os.path.abspath(\"../\")\n\n# Import Mask RCNN\nsys.path.append(ROOT_DIR) # To find local version of the library\nfrom mrcnn import utils\nimport mrcnn.model as modellib\nfrom mrcnn import visualize\n# Import COCO config\nsys.path.append(os.path.join(ROOT_DIR, \"samples/coco/\")) # To find local version\nimport coco\n\n%matplotlib inline \n\n# Directory to save logs and trained model\nMODEL_DIR = os.path.join(ROOT_DIR, \"logs\")\n\n# Local path to trained weights file\nCOCO_MODEL_PATH = os.path.join(ROOT_DIR, \"mask_rcnn_coco.h5\")\n# Download COCO trained weights from Releases if needed\nif not os.path.exists(COCO_MODEL_PATH):\n utils.download_trained_weights(COCO_MODEL_PATH)\n\n# Directory of images to run detection on\nIMAGE_DIR = os.path.join(ROOT_DIR, \"images\")",
"Using TensorFlow backend.\n"
]
],
[
[
"## Configurations\n\nWe'll be using a model trained on the MS-COCO dataset. The configurations of this model are in the ```CocoConfig``` class in ```coco.py```.\n\nFor inferencing, modify the configurations a bit to fit the task. To do so, sub-class the ```CocoConfig``` class and override the attributes you need to change.",
"_____no_output_____"
]
],
[
[
"class InferenceConfig(coco.CocoConfig):\n # Set batch size to 1 since we'll be running inference on\n # one image at a time. Batch size = GPU_COUNT * IMAGES_PER_GPU\n GPU_COUNT = 1\n IMAGES_PER_GPU = 1\n\nconfig = InferenceConfig()\nconfig.display()",
"\nConfigurations:\nBACKBONE resnet101\nBACKBONE_STRIDES [4, 8, 16, 32, 64]\nBATCH_SIZE 1\nBBOX_STD_DEV [0.1 0.1 0.2 0.2]\nCOMPUTE_BACKBONE_SHAPE None\nDETECTION_MAX_INSTANCES 100\nDETECTION_MIN_CONFIDENCE 0.7\nDETECTION_NMS_THRESHOLD 0.3\nFPN_CLASSIF_FC_LAYERS_SIZE 1024\nGPU_COUNT 1\nGRADIENT_CLIP_NORM 5.0\nIMAGES_PER_GPU 1\nIMAGE_CHANNEL_COUNT 3\nIMAGE_MAX_DIM 1024\nIMAGE_META_SIZE 93\nIMAGE_MIN_DIM 800\nIMAGE_MIN_SCALE 0\nIMAGE_RESIZE_MODE square\nIMAGE_SHAPE [1024 1024 3]\nLEARNING_MOMENTUM 0.9\nLEARNING_RATE 0.001\nLOSS_WEIGHTS {'rpn_class_loss': 1.0, 'rpn_bbox_loss': 1.0, 'mrcnn_class_loss': 1.0, 'mrcnn_bbox_loss': 1.0, 'mrcnn_mask_loss': 1.0}\nMASK_POOL_SIZE 14\nMASK_SHAPE [28, 28]\nMAX_GT_INSTANCES 100\nMEAN_PIXEL [123.7 116.8 103.9]\nMINI_MASK_SHAPE (56, 56)\nNAME coco\nNUM_CLASSES 81\nPOOL_SIZE 7\nPOST_NMS_ROIS_INFERENCE 1000\nPOST_NMS_ROIS_TRAINING 2000\nPRE_NMS_LIMIT 6000\nROI_POSITIVE_RATIO 0.33\nRPN_ANCHOR_RATIOS [0.5, 1, 2]\nRPN_ANCHOR_SCALES (32, 64, 128, 256, 512)\nRPN_ANCHOR_STRIDE 1\nRPN_BBOX_STD_DEV [0.1 0.1 0.2 0.2]\nRPN_NMS_THRESHOLD 0.7\nRPN_TRAIN_ANCHORS_PER_IMAGE 256\nSTEPS_PER_EPOCH 1000\nTOP_DOWN_PYRAMID_SIZE 256\nTRAIN_BN False\nTRAIN_ROIS_PER_IMAGE 200\nUSE_MINI_MASK True\nUSE_RPN_ROIS True\nVALIDATION_STEPS 50\nWEIGHT_DECAY 0.0001\n\n\n"
]
],
[
[
"## Create Model and Load Trained Weights",
"_____no_output_____"
]
],
[
[
"# Create model object in inference mode.\nmodel = modellib.MaskRCNN(mode=\"inference\", model_dir=MODEL_DIR, config=config)\n\n# Load weights trained on MS-COCO\nmodel.load_weights(COCO_MODEL_PATH, by_name=True)",
"_____no_output_____"
]
],
[
[
"## Class Names\n\nThe model classifies objects and returns class IDs, which are integer value that identify each class. Some datasets assign integer values to their classes and some don't. For example, in the MS-COCO dataset, the 'person' class is 1 and 'teddy bear' is 88. The IDs are often sequential, but not always. The COCO dataset, for example, has classes associated with class IDs 70 and 72, but not 71.\n\nTo improve consistency, and to support training on data from multiple sources at the same time, our ```Dataset``` class assigns it's own sequential integer IDs to each class. For example, if you load the COCO dataset using our ```Dataset``` class, the 'person' class would get class ID = 1 (just like COCO) and the 'teddy bear' class is 78 (different from COCO). Keep that in mind when mapping class IDs to class names.\n\nTo get the list of class names, you'd load the dataset and then use the ```class_names``` property like this.\n```\n# Load COCO dataset\ndataset = coco.CocoDataset()\ndataset.load_coco(COCO_DIR, \"train\")\ndataset.prepare()\n\n# Print class names\nprint(dataset.class_names)\n```\n\nWe don't want to require you to download the COCO dataset just to run this demo, so we're including the list of class names below. The index of the class name in the list represent its ID (first class is 0, second is 1, third is 2, ...etc.)",
"_____no_output_____"
]
],
[
[
"# COCO Class names\n# Index of the class in the list is its ID. For example, to get ID of\n# the teddy bear class, use: class_names.index('teddy bear')\nclass_names = ['BG', 'person', 'bicycle', 'car', 'motorcycle', 'airplane',\n 'bus', 'train', 'truck', 'boat', 'traffic light',\n 'fire hydrant', 'stop sign', 'parking meter', 'bench', 'bird',\n 'cat', 'dog', 'horse', 'sheep', 'cow', 'elephant', 'bear',\n 'zebra', 'giraffe', 'backpack', 'umbrella', 'handbag', 'tie',\n 'suitcase', 'frisbee', 'skis', 'snowboard', 'sports ball',\n 'kite', 'baseball bat', 'baseball glove', 'skateboard',\n 'surfboard', 'tennis racket', 'bottle', 'wine glass', 'cup',\n 'fork', 'knife', 'spoon', 'bowl', 'banana', 'apple',\n 'sandwich', 'orange', 'broccoli', 'carrot', 'hot dog', 'pizza',\n 'donut', 'cake', 'chair', 'couch', 'potted plant', 'bed',\n 'dining table', 'toilet', 'tv', 'laptop', 'mouse', 'remote',\n 'keyboard', 'cell phone', 'microwave', 'oven', 'toaster',\n 'sink', 'refrigerator', 'book', 'clock', 'vase', 'scissors',\n 'teddy bear', 'hair drier', 'toothbrush']",
"_____no_output_____"
]
],
[
[
"## Run Object Detection",
"_____no_output_____"
]
],
[
[
"# Load a random image from the images folder\nfile_names = next(os.walk(IMAGE_DIR))[2]\nimage = skimage.io.imread(os.path.join(IMAGE_DIR, random.choice(file_names)))",
"_____no_output_____"
],
[
"# Run detection\nresults = model.detect([image], verbose=1)",
"Processing 1 images\nimage shape: (700, 700, 3) min: 0.00000 max: 255.00000 uint8\nmolded_images shape: (1, 1024, 1024, 3) min: -123.70000 max: 150.10000 float64\nimage_metas shape: (1, 93) min: 0.00000 max: 1024.00000 float64\nanchors shape: (1, 261888, 4) min: -0.35390 max: 1.29134 float32\n"
],
[
"# Visualize results\nr = results[0]\nvisualize.display_instances(image, r['rois'], r['masks'], r['class_ids'], \n class_names, r['scores'])",
"_____no_output_____"
],
[
"r.get",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7f631a793d834728e38974944e3e9f758441a3c | 22,573 | ipynb | Jupyter Notebook | NLP/Sequences/1/NLP_C3_W1_lecture_nb_02_classes.ipynb | verneh/DataSci | cbbecbd780368c2a4567aaf9ec8d66f7c7cdfa06 | [
"MIT"
] | 116 | 2020-07-22T15:47:17.000Z | 2022-03-25T17:36:30.000Z | NLP/Sequences/1/NLP_C3_W1_lecture_nb_02_classes.ipynb | verneh/DataSci | cbbecbd780368c2a4567aaf9ec8d66f7c7cdfa06 | [
"MIT"
] | null | null | null | NLP/Sequences/1/NLP_C3_W1_lecture_nb_02_classes.ipynb | verneh/DataSci | cbbecbd780368c2a4567aaf9ec8d66f7c7cdfa06 | [
"MIT"
] | 183 | 2020-08-27T10:24:21.000Z | 2022-03-18T17:05:26.000Z | 28.865729 | 531 | 0.565897 | [
[
[
"# Classes and subclasses \n\nIn this notebook, I will show you the basics of classes and subclasses in Python. As you've seen in the lectures from this week, `Trax` uses layer classes as building blocks for deep learning models, so it is important to understand how classes and subclasses behave in order to be able to build custom layers when needed. \n\nBy completing this notebook, you will:\n\n- Be able to define classes and subclasses in Python\n- Understand how inheritance works in subclasses\n- Be able to work with instances",
"_____no_output_____"
],
[
"# Part 1: Parameters, methods and instances",
"_____no_output_____"
],
[
"First, let's define a class `My_Class`. ",
"_____no_output_____"
]
],
[
[
"class My_Class: #Definition of My_class\n x = None ",
"_____no_output_____"
]
],
[
[
"`My_Class` has one parameter `x` without any value. You can think of parameters as the variables that every object assigned to a class will have. So, at this point, any object of class `My_Class` would have a variable `x` equal to `None`. To check this, I'll create two instances of that class and get the value of `x` for both of them.",
"_____no_output_____"
]
],
[
[
"instance_a= My_Class() #To create an instance from class \"My_Class\" you have to call \"My_Class\"\ninstance_b= My_Class()\nprint('Parameter x of instance_a: ' + str(instance_a.x)) #To get a parameter 'x' from an instance 'a', write 'a.x'\nprint('Parameter x of instance_b: ' + str(instance_b.x))",
"Parameter x of instance_a: None\nParameter x of instance_b: None\n"
]
],
[
[
"For an existing instance you can assign new values for any of its parameters. In the next cell, assign a value of `5` to the parameter `x` of `instance_a`.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE (1 line) ### \ninstance_a.x = 5\n### END CODE HERE ###\nprint('Parameter x of instance_a: ' + str(instance_a.x))",
"Parameter x of instance_a: 5\n"
]
],
[
[
"## 1.1 The `__init__` method",
"_____no_output_____"
],
[
"When you want to assign values to the parameters of your class when an instance is created, it is necessary to define a special method: `__init__`. The `__init__` method is called when you create an instance of a class. It can have multiple arguments to initialize the paramenters of your instance. In the next cell I will define `My_Class` with an `__init__` method that takes the instance (`self`) and an argument `y` as inputs.",
"_____no_output_____"
]
],
[
[
"class My_Class: \n def __init__(self, y): # The __init__ method takes as input the instance to be initialized and a variable y\n self.x = y # Sets parameter x to be equal to y",
"_____no_output_____"
]
],
[
[
"In this case, the parameter `x` of an instance from `My_Class` would take the value of an argument `y`. \nThe argument `self` is used to pass information from the instance being created to the method `__init__`. In the next cell, create an instance `instance_c`, with `x` equal to `10`.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE (1 line) ### \ninstance_c = My_Class(10)\n### END CODE HERE ###\nprint('Parameter x of instance_c: ' + str(instance_c.x))",
"Parameter x of instance_c: 10\n"
]
],
[
[
"Note that in this case, you had to pass the argument `y` from the `__init__` method to create an instance of `My_Class`.",
"_____no_output_____"
],
[
"## 1.2 The `__call__` method",
"_____no_output_____"
],
[
"Another important method is the `__call__` method. It is performed whenever you call an initialized instance of a class. It can have multiple arguments and you can define it to do whatever you want like\n\n- Change a parameter, \n- Print a message,\n- Create new variables, etc.\n\nIn the next cell, I'll define `My_Class` with the same `__init__` method as before and with a `__call__` method that adds `z` to parameter `x` and prints the result.",
"_____no_output_____"
]
],
[
[
"class My_Class: \n def __init__(self, y): # The __init__ method takes as input the instance to be initialized and a variable y\n self.x = y # Sets parameter x to be equal to y\n def __call__(self, z): # __call__ method with self and z as arguments\n self.x += z # Adds z to parameter x when called \n print(self.x)",
"_____no_output_____"
]
],
[
[
"Let’s create `instance_d` with `x` equal to 5.",
"_____no_output_____"
]
],
[
[
"instance_d = My_Class(5)",
"_____no_output_____"
]
],
[
[
"And now, see what happens when `instance_d` is called with argument `10`.",
"_____no_output_____"
]
],
[
[
"instance_d(10)",
"15\n"
]
],
[
[
"Now, you are ready to complete the following cell so any instance from `My_Class`:\n\n- Is initialized taking two arguments `y` and `z` and assigns them to `x_1` and `x_2`, respectively. And, \n- When called, takes the values of the parameters `x_1` and `x_2`, sums them, prints and returns the result.",
"_____no_output_____"
]
],
[
[
"class My_Class: \n def __init__(self, y, z): #Initialization of x_1 and x_2 with arguments y and z\n ### START CODE HERE (2 lines) ### \n self.x_1 = y\n self.x_2 = z\n ### END CODE HERE ###\n def __call__(self): #When called, adds the values of parameters x_1 and x_2, prints and returns the result \n ### START CODE HERE (1 line) ### \n result = self.x_1 + self.x_2 \n ### END CODE HERE ### \n print(\"Addition of {} and {} is {}\".format(self.x_1,self.x_2,result))\n return result",
"_____no_output_____"
]
],
[
[
"Run the next cell to check your implementation. If everything is correct, you shouldn't get any errors.",
"_____no_output_____"
]
],
[
[
"instance_e = My_Class(10,15)\ndef test_class_definition():\n \n assert instance_e.x_1 == 10, \"Check the value assigned to x_1\"\n assert instance_e.x_2 == 15, \"Check the value assigned to x_2\"\n assert instance_e() == 25, \"Check the __call__ method\"\n \n print(\"\\033[92mAll tests passed!\")\n \ntest_class_definition()",
"Addition of 10 and 15 is 25\n\u001b[92mAll tests passed!\n"
]
],
[
[
"## 1.3 Custom methods",
"_____no_output_____"
],
[
"In addition to the `__init__` and `__call__` methods, your classes can have custom-built methods to do whatever you want when called. To define a custom method, you have to indicate its input arguments, the instructions that you want it to perform and the values to return (if any). In the next cell, `My_Class` is defined with `my_method` that multiplies the values of `x_1` and `x_2`, sums that product with an input `w`, and returns the result.",
"_____no_output_____"
]
],
[
[
"class My_Class: \n def __init__(self, y, z): #Initialization of x_1 and x_2 with arguments y and z\n self.x_1 = y\n self.x_2 = z\n def __call__(self): #Performs an operation with x_1 and x_2, and returns the result\n a = self.x_1 - 2*self.x_2 \n return a\n def my_method(self, w): #Multiplies x_1 and x_2, adds argument w and returns the result\n result = self.x_1*self.x_2 + w\n return result",
"_____no_output_____"
]
],
[
[
"Create an instance `instance_f` of `My_Class` with any integer values that you want for `x_1` and `x_2`. For that instance, see the result of calling `My_method`, with an argument `w` equal to `16`.",
"_____no_output_____"
]
],
[
[
"### START CODE HERE (1 line) ### \ninstance_f = My_Class(1,10)\n### END CODE HERE ### \nprint(\"Output of my_method:\",instance_f.my_method(16))",
"Output of my_method: 26\n"
]
],
[
[
"As you can corroborate in the previous cell, to call a custom method `m`, with arguments `args`, for an instance `i` you must write `i.m(args)`. With that in mind, methods can call others within a class. In the following cell, try to define `new_method` which calls `my_method` with `v` as input argument. Try to do this on your own in the cell given below.\n\n",
"_____no_output_____"
]
],
[
[
"class My_Class: \n def __init__(self, y, z): #Initialization of x_1 and x_2 with arguments y and z\n self.x_1 = None\n self.x_2 = None\n def __call__(self): #Performs an operation with x_1 and x_2, and returns the result\n a = None \n return a\n def my_method(self, w): #Multiplies x_1 and x_2, adds argument w and returns the result\n b = None\n return b\n def new_method(self, v): #Calls My_method with argument v\n ### START CODE HERE (1 line) ### \n result = None\n ### END CODE HERE ### \n return result",
"_____no_output_____"
]
],
[
[
"<b>SPOILER ALERT</b> Solution:",
"_____no_output_____"
]
],
[
[
"# hidden-cell\nclass My_Class: \n def __init__(self, y, z): #Initialization of x_1 and x_2 with arguments y and z\n self.x_1 = y\n self.x_2 = z\n def __call__(self): #Performs an operation with x_1 and x_2, and returns the result\n a = self.x_1 - 2*self.x_2 \n return a\n def my_method(self, w): #Multiplies x_1 and x_2, adds argument w and returns the result\n b = self.x_1*self.x_2 + w\n return b\n def new_method(self, v): #Calls My_method with argument v\n result = self.my_method(v)\n return result",
"_____no_output_____"
],
[
"instance_g = My_Class(1,10)\nprint(\"Output of my_method:\",instance_g.my_method(16))\nprint(\"Output of new_method:\",instance_g.new_method(16))",
"Output of my_method: 26\nOutput of new_method: 26\n"
]
],
[
[
"# Part 2: Subclasses and Inheritance",
"_____no_output_____"
],
[
"`Trax` uses classes and subclasses to define layers. The base class in `Trax` is `layer`, which means that every layer from a deep learning model is defined as a subclass of the `layer` class. In this part of the notebook, you are going to see how subclasses work. To define a subclass `sub` from class `super`, you have to write `class sub(super):` and define any method and parameter that you want for your subclass. In the next cell, I define `sub_c` as a subclass of `My_Class` with only one method (`additional_method`).",
"_____no_output_____"
]
],
[
[
"class sub_c(My_Class): #Subclass sub_c from My_class\n def additional_method(self): #Prints the value of parameter x_1\n print(self.x_1)",
"_____no_output_____"
]
],
[
[
"## 2.1 Inheritance",
"_____no_output_____"
],
[
"When you define a subclass `sub`, every method and parameter is inherited from `super` class, including the `__init__` and `__call__` methods. This means that any instance from `sub` can use the methods defined in `super`. Run the following cell and see for yourself.",
"_____no_output_____"
]
],
[
[
"instance_sub_a = sub_c(1,10)\nprint('Parameter x_1 of instance_sub_a: ' + str(instance_sub_a.x_1))\nprint('Parameter x_2 of instance_sub_a: ' + str(instance_sub_a.x_2))\nprint(\"Output of my_method of instance_sub_a:\",instance_sub_a.my_method(16))\n",
"Parameter x_1 of instance_sub_a: 1\nParameter x_2 of instance_sub_a: 10\nOutput of my_method of instance_sub_a: 26\n"
]
],
[
[
"As you can see, `sub_c` does not have an initialization method `__init__`, it is inherited from `My_class`. However, you can overwrite any method you want by defining it again in the subclass. For instance, in the next cell define a class `sub_c` with a redefined `my_Method` that multiplies `x_1` and `x_2` but does not add any additional argument.",
"_____no_output_____"
]
],
[
[
"class sub_c(My_Class): #Subclass sub_c from My_class\n def my_method(self): #Multiplies x_1 and x_2 and returns the result\n ### START CODE HERE (1 line) ###\n b = self.x_1*self.x_2 \n ### END CODE HERE ###\n return b",
"_____no_output_____"
]
],
[
[
"To check your implementation run the following cell.",
"_____no_output_____"
]
],
[
[
"test = sub_c(3,10)\nassert test.my_method() == 30, \"The method my_method should return the product between x_1 and x_2\"\n\nprint(\"Output of overridden my_method of test:\",test.my_method()) #notice we didn't pass any parameter to call my_method\n#print(\"Output of overridden my_method of test:\",test.my_method(16)) #try to see what happens if you call it with 1 argument",
"Output of overridden my_method of test: 30\n"
]
],
[
[
"In the next cell, two instances are created, one of `My_Class` and another one of `sub_c`. The instances are initialized with equal `x_1` and `x_2` parameters.",
"_____no_output_____"
]
],
[
[
"y,z= 1,10\ninstance_sub_a = sub_c(y,z)\ninstance_a = My_Class(y,z)\nprint('My_method for an instance of sub_c returns: ' + str(instance_sub_a.my_method()))\nprint('My_method for an instance of My_Class returns: ' + str(instance_a.my_method(10)))",
"My_method for an instance of sub_c returns: 10\nMy_method for an instance of My_Class returns: 20\n"
]
],
[
[
"As you can see, even though `sub_c` is a subclass from `My_Class` and both instances are initialized with the same values, `My_method` returns different results for each instance because you overwrote `My_method` for `sub_c`.",
"_____no_output_____"
],
[
"<b>Congratulations!</b> You just reviewed the basics behind classes and subclasses. Now you can define your own classes and subclasses, work with instances and overwrite inherited methods. The concepts within this notebook are more than enough to understand how layers in `Trax` work.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e7f644d2e6d9a37867ad36abdc240a698bc0cf7a | 22,102 | ipynb | Jupyter Notebook | book/tutorials/lis/2_interactive_data_exploration.ipynb | zachghiaccio/website | b6f3f760ecba8700a5d989d94389ad044a59e214 | [
"MIT"
] | 1 | 2021-07-12T18:30:47.000Z | 2021-07-12T18:30:47.000Z | book/tutorials/lis/2_interactive_data_exploration.ipynb | slopezon/website | 50d47b7977fb5f8ac14f367ff806cf9187dfb268 | [
"MIT"
] | null | null | null | book/tutorials/lis/2_interactive_data_exploration.ipynb | slopezon/website | 50d47b7977fb5f8ac14f367ff806cf9187dfb268 | [
"MIT"
] | null | null | null | 34.861199 | 441 | 0.568546 | [
[
[
"# Interactive Data Exploration\n\nThis notebook demonstrates how the functions and techniques we covered in the first notebook can be combined to build interactive data exploration tools. The code in the cells below will generate two interactive panels. The The first panel enables comparison of LIS output, SNODAS, and SNOTEL snow depth and snow water equivalent at SNOTEL site locations. The second panel enables exploration of LIS output using an interactive map.\n\n**Note: some cells below take several minutes to run.**",
"_____no_output_____"
],
[
"## Import Libraries",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport geopandas\nimport xarray as xr\nimport fsspec\nimport s3fs\nfrom datetime import datetime as dt\nfrom scipy.spatial import distance\n\nimport holoviews as hv, geoviews as gv\nfrom geoviews import opts\nfrom geoviews import tile_sources as gvts\n\nfrom datashader.colors import viridis\nimport datashader\nfrom holoviews.operation.datashader import datashade, shade, dynspread, spread, rasterize\n\nfrom holoviews.streams import Selection1D, Params\nimport panel as pn\nimport param as pm\nimport hvplot.pandas \nimport hvplot.xarray",
"_____no_output_____"
],
[
"# create S3 filesystem object\ns3 = s3fs.S3FileSystem()\n\n# define S3 bucket name\nbucket = \"s3://eis-dh-hydro/SNOWEX-HACKWEEK\"\n\n# set holoviews backend to Bokeh\ngv.extension('bokeh')",
"_____no_output_____"
]
],
[
[
"## Load Data",
"_____no_output_____"
],
[
"### SNOTEL Sites info",
"_____no_output_____"
]
],
[
[
"# create dictionary linking state names and abbreviations\nsnotel = {\"AZ\" : \"arizona\",\n \"CO\" : \"colorado\",\n \"ID\" : \"idaho\",\n \"MT\" : \"montana\", \n \"NM\" : \"newmexico\",\n \"UT\" : \"utah\",\n \"WY\" : \"wyoming\"}",
"_____no_output_____"
],
[
"# load SNOTEL site metadata for sites in the given state\ndef load_site(state):\n \n # define path to file\n key = f\"SNOTEL/snotel_{state}.csv\"\n \n # load csv into pandas DataFrame\n df = pd.read_csv(s3.open(f'{bucket}/{key}', mode='r'))\n \n return df ",
"_____no_output_____"
]
],
[
[
"### SNOTEL Depth & SWE",
"_____no_output_____"
]
],
[
[
"def load_snotel_txt(state, var):\n \n # define path to file\n key = f\"SNOTEL/snotel_{state}{var}_20162020.txt\"\n\n # open text file\n fh = s3.open(f\"{bucket}/{key}\")\n \n # read each line and note those that begin with '#'\n lines = fh.readlines()\n skips = sum(1 for ln in lines if ln.decode('ascii').startswith('#'))\n \n # load txt file into pandas DataFrame (skipping lines beginning with '#')\n df = pd.read_csv(s3.open(f\"{bucket}/{key}\"), skiprows=skips)\n \n # convert Date column from str to pandas datetime objects\n df['Date'] = pd.to_datetime(df['Date'])\n return df",
"_____no_output_____"
],
[
"# load SNOTEL depth & swe into dictionaries\n\n# define empty dicts\nsnotel_depth = {}\nsnotel_swe = {}\n\n# loop over states and load SNOTEL data\nfor state in snotel.keys():\n print(f\"Loading state {state}\")\n snotel_depth[state] = load_snotel_txt(state, 'depth')\n snotel_swe[state] = load_snotel_txt(state, 'swe')",
"_____no_output_____"
]
],
[
[
"### SNODAS Depth & SWE\n\nLike the LIS output we have been working with, a sample of SNODAS data is available on our S3 bucket in Zarr format. We can therefore load the SNODAS just as we load the LIS data.",
"_____no_output_____"
]
],
[
[
"# load snodas depth data\nkey = \"SNODAS/snodas_snowdepth_20161001_20200930.zarr\"\nsnodas_depth = xr.open_zarr(s3.get_mapper(f\"{bucket}/{key}\"), consolidated=True)\n\n# load snodas swe data\nkey = \"SNODAS/snodas_swe_20161001_20200930.zarr\"\nsnodas_swe = xr.open_zarr(s3.get_mapper(f\"{bucket}/{key}\"), consolidated=True)",
"_____no_output_____"
]
],
[
[
"### LIS Outputs\n\nNext we'll load the LIS outputs. First, we'll define the helper function we saw in the previous notebook that adds `lat` and `lon` as coordinate variables. We'll use this immediately upon loading the data.",
"_____no_output_____"
]
],
[
[
"def add_latlon_coords(dataset: xr.Dataset)->xr.Dataset:\n \"\"\"Adds lat/lon as dimensions and coordinates to an xarray.Dataset object.\"\"\"\n \n # get attributes from dataset\n attrs = dataset.attrs\n \n # get x, y resolutions\n dx = round(float(attrs['DX']), 3)\n dy = round(float(attrs['DY']), 3)\n \n # get grid cells in x, y dimensions\n ew_len = len(dataset['east_west'])\n ns_len = len(dataset['north_south'])\n \n # get lower-left lat and lon\n ll_lat = round(float(attrs['SOUTH_WEST_CORNER_LAT']), 3)\n ll_lon = round(float(attrs['SOUTH_WEST_CORNER_LON']), 3)\n \n # calculate upper-right lat and lon\n ur_lat = ll_lat + (dy * ns_len)\n ur_lon = ll_lon + (dx * ew_len)\n \n # define the new coordinates\n coords = {\n # create an arrays containing the lat/lon at each gridcell\n 'lat': np.linspace(ll_lat, ur_lat, ns_len, dtype=np.float32, endpoint=False),\n 'lon': np.linspace(ll_lon, ur_lon, ew_len, dtype=np.float32, endpoint=False)\n }\n \n \n # drop the original lat and lon variables\n dataset = dataset.rename({'lon': 'orig_lon', 'lat': 'orig_lat'})\n # rename the grid dimensions to lat and lon\n dataset = dataset.rename({'north_south': 'lat', 'east_west': 'lon'})\n # assign the coords above as coordinates\n dataset = dataset.assign_coords(coords)\n # reassign variable attributes\n dataset.lon.attrs = dataset.orig_lon.attrs\n dataset.lat.attrs = dataset.orig_lat.attrs\n \n return dataset",
"_____no_output_____"
]
],
[
[
"Load the LIS data and apply `add_latlon_coords()`:",
"_____no_output_____"
]
],
[
[
"# LIS surfacemodel DA_10km\nkey = \"DA_SNODAS/SURFACEMODEL/LIS_HIST.d01.zarr\"\n\nlis_sf = xr.open_zarr(s3.get_mapper(f\"{bucket}/{key}\"), consolidated=True)\n\n# (optional for 10km simulation?)\nlis_sf = add_latlon_coords(lis_sf)\n\n# drop off irrelevant variables\ndrop_vars = ['_history', '_eis_source_path', 'orig_lat', 'orig_lon']\nlis_sf = lis_sf.drop(drop_vars)\nlis_sf",
"_____no_output_____"
]
],
[
[
"Working with the full LIS output dataset can be slow and consume lots of memory. Here we temporally subset the data to a shorter window of time. The full dataset contains daily values from 10/1/2016 to 9/30/2018. Feel free to explore the full dataset by modifying the `time_range` variable below and re-running all cells that follow.",
"_____no_output_____"
]
],
[
[
"# subset LIS data for two years \ntime_range = slice('2016-10-01', '2017-04-30')\nlis_sf = lis_sf.sel(time=time_range)",
"_____no_output_____"
]
],
[
[
"In the next cell, we extract the data variable names and timesteps from the LIS outputs. These will be used to define the widget options.",
"_____no_output_____"
]
],
[
[
"# gather metadata from LIS\n\n# get variable names:string\nvnames = list(lis_sf.data_vars)\nprint(vnames)\n\n# get time-stamps:string\ntstamps = list(np.datetime_as_string(lis_sf.time.values, 'D'))\nprint(len(tstamps), tstamps[0], tstamps[-1])",
"_____no_output_____"
]
],
[
[
"By default, the `holoviews` plotting library automatically adjusts the range of plot colorbars based on the range of values in the data being plotted. This may not be ideal when comparing data on different timesteps. In the next cell we extract the upper and lower bounds for each data variable which we'll later use to set a static colorbar range.\n\n**Note: this cell will take ~1m40s to run**",
"_____no_output_____"
]
],
[
[
"%%time\n# pre-load min/max range for LIS variables\ndef get_cmap_range(vns):\n vals = [(lis_sf[x].sel(time='2016-12').min(skipna=True).values.item(),\n lis_sf[x].sel(time='2016-12').max(skipna=True).values.item()) for x in vns]\n return dict(zip(vns, vals))\n\ncmap_lims = get_cmap_range(vnames)",
"_____no_output_____"
]
],
[
[
"## Interactive Widgets\n\n### SNOTEL Site Map and Timeseries\n\nThe two cells that follow will create an interactive panel for comparing LIS, SNODAS, and SNOTEL snow depth and snow water equivalent. The SNOTEL site locations are plotted as points on an interactive map. Hover over the sites to view metadata and click on a site to generate a timeseries!\n\n**Note: it will take some time for the timeseries to display.**",
"_____no_output_____"
]
],
[
[
"# get snotel depth\ndef get_depth(state, site, ts, te):\n df = snotel_depth[state]\n \n # subset between time range\n mask = (df['Date'] >= ts) & (df['Date'] <= te)\n df = df.loc[mask]\n \n # extract timeseries for the site\n return pd.concat([df.Date, df.filter(like=site)], axis=1).set_index('Date')\n\n# get snotel swe\ndef get_swe(state, site, ts, te):\n df = snotel_swe[state]\n \n # subset between time range\n mask = (df['Date'] >= ts) & (df['Date'] <= te)\n df = df.loc[mask]\n \n # extract timeseries for the site\n return pd.concat([df.Date, df.filter(like=site)], axis=1).set_index('Date')\n\n# co-locate site & LIS model cell\ndef nearest_grid(pt):\n # pt : input point, tuple (longtitude, latitude)\n # output:\n # x_idx, y_idx \n loc_valid = df_loc.dropna()\n pts = loc_valid[['lon', 'lat']].to_numpy()\n idx = distance.cdist([pt], pts).argmin()\n\n return loc_valid['east_west'].iloc[idx], loc_valid['north_south'].iloc[idx]\n\n# get LIS variable \ndef var_subset(dset, v, lon, lat, ts, te):\n return dset[v].sel(lon=lon, lat=lat, method=\"nearest\").sel(time=slice(ts, te)).load()\n\n# line plots\ndef line_callback(index, state, vname, ts_tag, te_tag):\n sites = load_site(snotel[state])\n row = sites.iloc[0]\n \n tmp = var_subset(lis_sf, vname, row.lon, row.lat, ts_tag, te_tag) \n xr_sf = xr.zeros_like(tmp)\n \n xr_snodas = xr_sf\n \n ck = get_depth(state, row.site_name, ts_tag, te_tag).to_xarray().rename({'Date': 'time'})\n xr_snotel = xr.zeros_like(ck)\n \n if not index:\n title='Var: -- Lon: -- Lat: --'\n return (xr_sf.hvplot(title=title, color='blue', label='LIS') \\\n * xr_snotel.hvplot(color='red', label='SNOTEL') \\\n * xr_snodas.hvplot(color='green', label='SNODAS')).opts(legend_position='right')\n \n\n else:\n sites = load_site(snotel[state])\n first_index = index[0]\n row = sites.iloc[first_index]\n \n \n xr_sf = var_subset(lis_sf, vname, row.lon, row.lat, ts_tag, te_tag)\n \n vs = vname.split('_')[0]\n title=f'Var: {vs} Lon: {row.lon} Lat: {row.lat}'\n\n \n # update snotel data \n if 'depth' in vname.lower():\n xr_snotel = get_depth(state, row.site_name, ts_tag, te_tag).to_xarray().rename({'Date': 'time'})*0.01\n xr_snodas = var_subset(snodas_depth, 'SNOWDEPTH', row.lon, row.lat, ts_tag, te_tag)*0.001\n \n if 'swe' in vname.lower():\n xr_snotel = get_swe(state, row.site_name, ts_tag, te_tag).to_xarray().rename({'Date': 'time'})\n xr_snodas = var_subset(snodas_swe, 'SWE', row.lon, row.lat, ts_tag, te_tag)\n\n \n return xr_sf.hvplot(title=title, color='blue', label='LIS') \\\n * xr_snotel.hvplot(color='red', label='SNOTEL') \\\n * xr_snodas.hvplot(color='green', label='SNODAS')\n\n",
"_____no_output_____"
],
[
"# sites on map\ndef plot_points(state): \n # dataframe to hvplot obj Points\n sites=load_site(snotel[state])\n pts_opts=dict(size=12, nonselection_alpha=0.4,tools=['tap', 'hover'])\n site_points=sites.hvplot.points(x='lon', y='lat', c='elev', cmap='fire', geo=True, hover_cols=['site_name', 'ntwk', 'state', 'lon', 'lat']).opts(**pts_opts) \n return site_points\n\n# base map\ntiles = gvts.OSM()\n\n# state widget\nstate_select = pn.widgets.Select(options=list(snotel.keys()), name=\"State\")\nstate_stream = Params(state_select, ['value'], rename={'value':'state'})\n\n# variable widget\nvar_select = pn.widgets.Select(options=['SnowDepth_tavg', 'SWE_tavg'], name=\"LIS Variable List\")\nvar_stream = Params(var_select, ['value'], rename={'value':'vname'})\n\n# date range widget\ndate_fmt = '%Y-%m-%d'\nsdate_input = pn.widgets.DatetimeInput(name='Start date', value=dt(2016,10,1),start=dt.strptime(tstamps[0], date_fmt), end=dt.strptime(tstamps[-1], date_fmt), format=date_fmt)\nsdate_stream = Params(sdate_input, ['value'], rename={'value':'ts_tag'})\nedate_input = pn.widgets.DatetimeInput(name='End date', value=dt(2017,3,31),start=dt.strptime(tstamps[0], date_fmt), end=dt.strptime(tstamps[-1], date_fmt),format=date_fmt)\nedate_stream = Params(edate_input, ['value'], rename={'value':'te_tag'})\n\n# generate site points as dynamic map\n# plots points and calls plot_points() when user selects a site\nsite_dmap = hv.DynamicMap(plot_points, streams=[state_stream]).opts(height=400, width=600)\n# pick site\nselect_stream = Selection1D(source=site_dmap)\n\n# link widgets to callback function\nline = hv.DynamicMap(line_callback, streams=[select_stream, state_stream, var_stream, sdate_stream, edate_stream])\n\n# create panel layout\npn.Row(site_dmap*tiles, pn.Column(state_select, var_select, pn.Row(sdate_input, edate_input), line))",
"_____no_output_____"
]
],
[
[
"### Interactive LIS Output Explorer\n\nThe cell below creates a `panel` layout for exploring LIS output rasters. Select a variable using the drop down and then use the date slider to scrub back and forth in time!",
"_____no_output_____"
]
],
[
[
"# date widget (slider & key in)\n# start and end dates\ndate_fmt = '%Y-%m-%d'\nb = dt.strptime('2016-10-01', date_fmt)\ne = dt.strptime('2017-04-30', date_fmt)\n\n# define date widgets\ndate_slider = pn.widgets.DateSlider(start=b, end=e, value=b, name=\"LIS Model Date\")\ndt_input = pn.widgets.DatetimeInput(name='LIS Model Date Input', value=b, format=date_fmt)\ndate_stream = Params(date_slider, ['value'], rename={'value':'date'})\n\n# variable widget\nvar_select = pn.widgets.Select(options=vnames, name=\"LIS Variable List\")\nvar_stream = Params(var_select, ['value'], rename={'value':'vname'})\n\n\n# base map widget\nmap_layer= pn.widgets.RadioButtonGroup(\n name='Base map layer',\n options=['Open Street Map', 'Satellite Imagery'],\n value='Satellite Imagery',\n button_type='primary',\n background='#f307eb')\n\n# lis output display callback function\n# returns plot of LIS output when date/variable is changed\ndef var_layer(vname, date):\n t_stamp = dt.strftime(date, '%Y-%m-%d')\n dssm = lis_sf[vname].sel(time=t_stamp)\n\n image = dssm.hvplot(geo=True)\n clim = cmap_lims[vname]\n return image.opts(clim=clim)\n\n# watches date widget for updates\[email protected](dt_input.param.value, watch=True)\ndef _update_date(dt_input):\n date_slider.value=dt_input\n\n# updates basemap on widget change\ndef update_map(maps):\n tile = gvts.OSM if maps=='Open Street Map' else gvts.EsriImagery\n return tile.opts(alpha=0.7)\n\n# link widgets to callback functions\nstreams = dict(vname=var_select.param.value, date=date_slider.param.value) \ndmap = hv.DynamicMap(var_layer, streams=streams)\ndtile = hv.DynamicMap(update_map, streams=dict(maps=map_layer.param.value))\n\n# create panel layout of widgets and plot\npn.Column(var_select, date_slider, dt_input, map_layer,\n dtile*rasterize(dmap, aggregator=datashader.mean()).opts(cmap=viridis,colorbar=True,width=800, height=600))\n",
"_____no_output_____"
]
],
[
[
"## Fin\n\nThank you for joining us for this tutorial. We hope that you are now more familiar with [NASA's Land Information System](https://lis.gsfc.nasa.gov/) and how to use Python to explore and use the model simulation output LIS generates. For more information please see the links under the \"More information\" dropdown on the introduction page of this tutorial.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e7f651bbe645f891ec2f54932bdc67ad296ea057 | 31,102 | ipynb | Jupyter Notebook | DS_Unit_2_Sprint_Challenge_4_Model_Validation.ipynb | donw385/DS-Unit-2-Sprint-4-Model-Validation | 7ff08c5763c21d4687ec116cf14351b0a35418d2 | [
"MIT"
] | null | null | null | DS_Unit_2_Sprint_Challenge_4_Model_Validation.ipynb | donw385/DS-Unit-2-Sprint-4-Model-Validation | 7ff08c5763c21d4687ec116cf14351b0a35418d2 | [
"MIT"
] | null | null | null | DS_Unit_2_Sprint_Challenge_4_Model_Validation.ipynb | donw385/DS-Unit-2-Sprint-4-Model-Validation | 7ff08c5763c21d4687ec116cf14351b0a35418d2 | [
"MIT"
] | null | null | null | 31.575635 | 292 | 0.460517 | [
[
[
"<a href=\"https://colab.research.google.com/github/donw385/DS-Unit-2-Sprint-4-Model-Validation/blob/master/DS_Unit_2_Sprint_Challenge_4_Model_Validation.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
" # Data Science Unit 2 Sprint Challenge 4 — Model Validation",
"_____no_output_____"
],
[
"Follow the instructions for each numbered part to earn a score of 2. See the bottom of the notebook for a list of ways you can earn a score of 3.",
"_____no_output_____"
],
[
"## Predicting Blood Donations\n\nOur dataset is from a mobile blood donation vehicle in Taiwan. The Blood Transfusion Service Center drives to different universities and collects blood as part of a blood drive.\n\nThe goal is to predict the last column, whether the donor made a donation in March 2007, using information about each donor's history. We'll measure success using recall score as the model evaluation metric.\n\nGood data-driven systems for tracking and predicting donations and supply needs can improve the entire supply chain, making sure that more patients get the blood transfusions they need.\n\n#### Run this cell to load the data:",
"_____no_output_____"
]
],
[
[
"# all imports here\nimport pandas as pd\nfrom sklearn.metrics import accuracy_score\nimport numpy as np\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.feature_selection import f_regression, SelectKBest\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.preprocessing import RobustScaler\nfrom sklearn.metrics import recall_score\n",
"_____no_output_____"
],
[
"\ndf = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/blood-transfusion/transfusion.data')\n\ndf = df.rename(columns={\n 'Recency (months)': 'months_since_last_donation', \n 'Frequency (times)': 'number_of_donations', \n 'Monetary (c.c. blood)': 'total_volume_donated', \n 'Time (months)': 'months_since_first_donation', \n 'whether he/she donated blood in March 2007': 'made_donation_in_march_2007'\n})",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"## Part 1.1 — Begin with baselines\n\nWhat **accuracy score** would you get here with a **\"majority class baseline\"?** \n \n(You don't need to split the data into train and test sets yet. You can answer this question either with a scikit-learn function or with a pandas function.)",
"_____no_output_____"
]
],
[
[
"#determine majority class\ndf['made_donation_in_march_2007'].value_counts(normalize=True)",
"_____no_output_____"
],
[
"# Guess the majority class for every prediction:\n\nmajority_class = 0\ny_pred = [majority_class] * len(df['made_donation_in_march_2007'])",
"_____no_output_____"
],
[
"#accuracy score same as majority class, because dataset not split yet\n\naccuracy_score(df['made_donation_in_march_2007'], y_pred)",
"_____no_output_____"
]
],
[
[
"What **recall score** would you get here with a **majority class baseline?**\n\n(You can answer this question either with a scikit-learn function or with no code, just your understanding of recall.)",
"_____no_output_____"
]
],
[
[
"#when it is actually yes, how often do you predict yes? 0, because always predicting no\n\n# recall = true_positive / actual_positive\n",
"_____no_output_____"
]
],
[
[
"## Part 1.2 — Split data\n\nIn this Sprint Challenge, you will use \"Cross-Validation with Independent Test Set\" for your model evaluation protocol.\n\nFirst, **split the data into `X_train, X_test, y_train, y_test`**, with random shuffle. (You can include 75% of the data in the train set, and hold out 25% for the test set.)\n",
"_____no_output_____"
]
],
[
[
"#split data\n\nX = df.drop(columns='made_donation_in_march_2007')\ny = df['made_donation_in_march_2007']\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=42)",
"_____no_output_____"
],
[
"#validate 75% in train set\nX_train.shape",
"_____no_output_____"
],
[
"#validate 25% in test set\nX_test.shape",
"_____no_output_____"
]
],
[
[
"## Part 2.1 — Make a pipeline\n\nMake a **pipeline** which includes:\n- Preprocessing with any scikit-learn [**Scaler**](https://scikit-learn.org/stable/modules/classes.html#module-sklearn.preprocessing)\n- Feature selection with **[`SelectKBest`](https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.SelectKBest.html)([`f_classif`](https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.f_classif.html))**\n- Classification with [**`LogisticRegression`**](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html)",
"_____no_output_____"
]
],
[
[
"#make pipeline with 3 prerequisites\n\nkbest = SelectKBest(f_regression)\npipeline = Pipeline([('scale', StandardScaler()),('kbest', kbest), ('lr', LogisticRegression(solver='lbfgs'))])\n\npipe = make_pipeline(RobustScaler(),SelectKBest(),LogisticRegression(solver='lbfgs'))\n",
"_____no_output_____"
]
],
[
[
"## Part 2.2 — Do Grid Search Cross-Validation\n\nDo [**GridSearchCV**](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html) with your pipeline. Use **5 folds** and **recall score**.\n\nInclude these **parameters for your grid:**\n\n#### `SelectKBest`\n- `k : 1, 2, 3, 4`\n\n#### `LogisticRegression`\n- `class_weight : None, 'balanced'`\n- `C : .0001, .001, .01, .1, 1.0, 10.0, 100.00, 1000.0, 10000.0`\n\n\n**Fit** on the appropriate data.",
"_____no_output_____"
]
],
[
[
"param_grid = {'selectkbest__k':[1,2,3,4],'logisticregression__class_weight':[None,'balanced'],'logisticregression__C':[.0001, .001, .01, .1, 1.0, 10.0, 100.00, 1000.0, 10000.0]}\n\n\ngs = GridSearchCV(pipe,param_grid,cv=5,scoring='recall')\ngs.fit(X_train, y_train)\n\n# grid_search = GridSearchCV(pipeline, { 'lr__class_weight': [None,'balanced'],'kbest__k': [1,2,3,4], 'lr__C': [.0001, .001, .01, .1, 1.0, 10.0, 100.00, 1000.0, 10000.0]},scoring='recall', cv=5,verbose=1)\n\n# grid_search.fit(X_train, y_train)",
"/usr/local/lib/python3.6/dist-packages/sklearn/model_selection/_search.py:841: DeprecationWarning: The default of the `iid` parameter will change from True to False in version 0.22 and will be removed in 0.24. This will change numeric results when test-set sizes are unequal.\n DeprecationWarning)\n"
]
],
[
[
"## Part 3 — Show best score and parameters\n\nDisplay your **best cross-validation score**, and the **best parameters** (the values of `k, class_weight, C`) from the grid search.\n\n(You're not evaluated here on how good your score is, or which parameters you find. You're only evaluated on being able to display the information. There are several ways you can get the information, and any way is acceptable.)",
"_____no_output_____"
]
],
[
[
"validation_score = gs.best_score_\nprint()\nprint('Cross-Validation Score:', -validation_score)\nprint()\nprint('Best estimator:', gs.best_estimator_)\nprint()",
"\nCross-Validation Score: -0.784519402166461\n\nBest estimator: Pipeline(memory=None,\n steps=[('robustscaler', RobustScaler(copy=True, quantile_range=(25.0, 75.0), with_centering=True,\n with_scaling=True)), ('selectkbest', SelectKBest(k=1, score_func=<function f_classif at 0x7f91ffc05730>)), ('logisticregression', LogisticRegression(C=0.0001, class_weight='balanced', dual=False,...enalty='l2', random_state=None,\n solver='lbfgs', tol=0.0001, verbose=0, warm_start=False))])\n\n"
],
[
"gs.best_estimator_",
"_____no_output_____"
],
[
"# Cross-Validation Score: -0.784519402166461\n\n\n# best parameters: k=1,C=0.0001,class_weight=balanced",
"_____no_output_____"
]
],
[
[
"## Part 4 — Calculate classification metrics from a confusion matrix\n\nSuppose this is the confusion matrix for your binary classification model:\n\n<table>\n <tr>\n <th colspan=\"2\" rowspan=\"2\"></th>\n <th colspan=\"2\">Predicted</th>\n </tr>\n <tr>\n <th>Negative</th>\n <th>Positive</th>\n </tr>\n <tr>\n <th rowspan=\"2\">Actual</th>\n <th>Negative</th>\n <td>85</td>\n <td>58</td>\n </tr>\n <tr>\n <th>Positive</th>\n <td>8</td>\n <td>36</td>\n </tr>\n</table>",
"_____no_output_____"
]
],
[
[
"true_negative = 85\nfalse_positive = 58\nfalse_negative = 8\ntrue_positive = 36\npredicted_positive = 58+36\nactual_positive = 8 + 36",
"_____no_output_____"
]
],
[
[
"Calculate accuracy",
"_____no_output_____"
]
],
[
[
"accuracy = (true_negative + true_positive) / (true_negative + false_positive +false_negative + true_positive)\nprint ('Accuracy:', accuracy)",
"Accuracy: 0.6470588235294118\n"
]
],
[
[
"Calculate precision",
"_____no_output_____"
]
],
[
[
"precision = true_positive / predicted_positive \nprint ('Precision:', precision)",
"Precision: 0.3829787234042553\n"
]
],
[
[
"Calculate recall",
"_____no_output_____"
]
],
[
[
"recall = true_positive / actual_positive\nprint ('Recall:', recall)",
"Recall: 0.8181818181818182\n"
]
],
[
[
"## BONUS — How you can earn a score of 3\n\n### Part 1\nDo feature engineering, to try improving your cross-validation score.\n\n### Part 2\nAdd transformations in your pipeline and parameters in your grid, to try improving your cross-validation score.\n\n### Part 3\nShow names of selected features. Then do a final evaluation on the test set — what is the test score?\n\n### Part 4\nCalculate F1 score and False Positive Rate. ",
"_____no_output_____"
]
],
[
[
"# # Which features were selected?\nselector = gs.best_estimator_.named_steps['selectkbest']\nall_names = X_train.columns\nselected_mask = selector.get_support()\nselected_names = all_names[selected_mask]\nunselected_names = all_names[~selected_mask]\n\nprint('Features selected:')\nfor name in selected_names:\n print(name)\n\nprint()\nprint('Features not selected:')\nfor name in unselected_names:\n print(name)",
"Features selected:\nmonths_since_last_donation\n\nFeatures not selected:\nnumber_of_donations\ntotal_volume_donated\nmonths_since_first_donation\n"
],
[
"# Predict with X_test features\ny_pred = grid_search.predict(X_test)\n\n# Compare predictions to y_test labels\ntest_score = recall_score(y_test, y_pred)\nprint('Test Score:', test_score)",
"Test Score: 0.7708333333333334\n"
],
[
"f1 = 2*precision*recall/(precision+recall)\nprint('f1:', f1)",
"f1: 0.5217391304347826\n"
],
[
"false_positive_rate = false_positive / (false_positive+true_negative)\nprint('False Positive Rate:', false_positive_rate)",
"False Positive Rate: 0.40559440559440557\n"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7f653d155a9063f03b57373dc8d9770806089f2 | 25,730 | ipynb | Jupyter Notebook | 07_Visualization/Chipotle/Exercises.ipynb | duongv/pandas_exercises | ed574a87a5d4c3756046f15124755bfe865c91da | [
"BSD-3-Clause"
] | null | null | null | 07_Visualization/Chipotle/Exercises.ipynb | duongv/pandas_exercises | ed574a87a5d4c3756046f15124755bfe865c91da | [
"BSD-3-Clause"
] | null | null | null | 07_Visualization/Chipotle/Exercises.ipynb | duongv/pandas_exercises | ed574a87a5d4c3756046f15124755bfe865c91da | [
"BSD-3-Clause"
] | null | null | null | 82.467949 | 16,716 | 0.772639 | [
[
[
"# Visualizing Chipotle's Data",
"_____no_output_____"
],
[
"This time we are going to pull data directly from the internet.\nSpecial thanks to: https://github.com/justmarkham for sharing the dataset and materials.\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom collections import Counter\nimport matplotlib.pyplot as plt \n\n# set this so the graphs open internally\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv). ",
"_____no_output_____"
],
[
"### Step 3. Assign it to a variable called chipo.",
"_____no_output_____"
]
],
[
[
"url = 'https://raw.githubusercontent.com/justmarkham/DAT8/master/data/chipotle.tsv'\n \nchipo = pd.read_csv(url, sep = '\\t')",
"_____no_output_____"
]
],
[
[
"### Step 4. See the first 10 entries",
"_____no_output_____"
]
],
[
[
"chipo.head(10)",
"_____no_output_____"
]
],
[
[
"### Step 5. Create a histogram of the top 5 items bought",
"_____no_output_____"
]
],
[
[
"# Create a Series of Item_name\nx = chipo.item_name\n\n# use the Counter to count frequency with keys and frequency.\nletter_counts = Counter(x)\nnew_data = pd.DataFrame.from_dict(letter_counts, orient='index')\ndata = new_data.sort_values(0,ascending=False)[0:5]\ndata.plot(kind='bar')\nplt.xlabel('Item')\nplt.ylabel ('The number of orders')\nplt.title('Most ordered Chipotle')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Step 6. Create a scatterplot with the number of items orderered per order price\n#### Hint: Price should be in the X-axis and Items ordered in the Y-axis",
"_____no_output_____"
],
[
"### Step 7. BONUS: Create a question and a graph to answer your own question.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e7f669a01c6f6178001a5d8c758ed20f3104cfa8 | 38,397 | ipynb | Jupyter Notebook | graphicalmethod.ipynb | andim/transitions-paper | 538e30fa92517fbc43c9c3768acb1b5eddd2e86e | [
"MIT"
] | null | null | null | graphicalmethod.ipynb | andim/transitions-paper | 538e30fa92517fbc43c9c3768acb1b5eddd2e86e | [
"MIT"
] | null | null | null | graphicalmethod.ipynb | andim/transitions-paper | 538e30fa92517fbc43c9c3768acb1b5eddd2e86e | [
"MIT"
] | null | null | null | 156.722449 | 30,648 | 0.871032 | [
[
[
"# Figure 2: Illustration of graphical method for finding best adaptation strategy in uncorrelated environments\n\nGoal: illustration of the steps of the graphical method",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport scipy.spatial\n\n%matplotlib inline\nimport matplotlib.pyplot as plt\nplt.style.use(['transitions.mplstyle'])\nimport matplotlib\ncolors = matplotlib.rcParams['axes.prop_cycle'].by_key()['color']\nfrom matplotlib import patches\n\nimport sys\nsys.path.append('lib/')\nimport evolimmune, plotting",
"_____no_output_____"
],
[
"def paretofrontier(points):\n \"Naive Pareto frontier calculation of a set of points where along every axis larger is better\"\n paretopoints = []\n for point in points:\n if not np.any(np.all(points - point > 0, axis=1)):\n paretopoints.append(point)\n paretopoints.sort(key=lambda row: row[0])\n return np.asarray(paretopoints)",
"_____no_output_____"
],
[
"fs = []\nprng = np.random.RandomState(1234)\nwhile len(fs) < 20:\n f = prng.rand(2)\n a = 1.7\n if f[1] < (1.0-f[0]**(1.0/a))**a and np.amin(f) > 0.04:\n if not fs or (np.amin(np.sum((f - np.asarray(fs))**2, axis=1)**.5) > 0.05):\n fs.append(f)\nfs = np.asarray(fs)",
"_____no_output_____"
],
[
"pienvs = [0.3, 0.7]\nfig, axes = plt.subplots(figsize=(7, 2), ncols=4, subplot_kw=dict(aspect='equal'))\n\n# plot phenotype fitnesses\nfor ax in [axes[0], axes[1]]:\n ax.scatter(fs[:, 0], fs[:, 1], color=colors[1])\n\n# calculate and plot convex hull\nhull = scipy.spatial.ConvexHull(fs)\np = patches.Polygon(fs[hull.vertices], alpha=0.5, color=colors[1])\naxes[1].add_patch(p)\n\n# calc pareto\npareto = [f for f in fs[hull.vertices] if f in paretofrontier(fs)]\npareto.sort(key=lambda row: row[0])\npareto = np.asarray(pareto)\n\n# plot pareto boundaries\nfor ax in [axes[1], axes[2]]:\n ax.plot(pareto[:, 0], pareto[:, 1], '-', c=colors[0], lw=2.0)\nfor i in range(len(pareto)-1):\n N = 100\n x, y = pareto[i:i+2, 0], pareto[i:i+2, 1]\n axes[3].plot(np.linspace(x[0], x[1], N), np.linspace(y[0], y[1], N), '-', c=colors[0], lw=2.0) \nfor ax in [axes[1], axes[2], axes[3]]:\n ax.plot(pareto[:, 0], pareto[:, 1], 'o', c=colors[0], markeredgecolor=colors[0])\n\n# calc optimal fitnesses for different pienvs\ncopts = []\nopts = []\nfor pienv in pienvs:\n for i in range(len(pareto)-1):\n pih = evolimmune.pihat(pienv, pareto[i], pareto[i+1])\n if 0.0 < pih < 1.0:\n opt = pareto[i]*pih + pareto[i+1]*(1.0-pih)\n opts.append(opt)\n copts.append(pienv*np.log(opt[1]) + (1.0-pienv)*np.log(opt[0]))\n\n \n# plot isolines\nf0 = np.linspace(0.001, 0.999)\nhandles = [None, None]\nfor i, copt in enumerate(copts):\n pienv = pienvs[i]\n alpha = (1.0-pienv)/pienv\n for dc in [-0.2, 0.0, 0.2]:\n c = copt + dc\n for ax in [axes[2], axes[3]]:\n l, = ax.plot(f0, np.exp(c/pienv)/f0**alpha, '-', c=colors[i+2], lw=.75, alpha=.5)\n handles[i] = l\naxes[3].legend(handles, pienvs, title='$p(x=2)$')\n\n# plot opt\nfor i, opt in enumerate(opts):\n for ax in [axes[2], axes[3]]:\n ax.plot(opt[0], opt[1], '*', c=colors[i+2], markeredgecolor=colors[i+2])\n\n# axes limits, labels, etc.\nfor ax in [axes[0], axes[1], axes[2]]:\n ax.set_xlim(0.0, 0.9)\n ax.set_ylim(0.0, 0.9)\n ax.set_xlabel('fitness in env. 1,\\n$f(x=1)$')\n ax.set_ylabel('fitness in env. 2,\\n$f(x=2)$')\nax = axes[3]\nax.set_xlim(0.03, 1.5)\nax.set_ylim(0.03, 1.5)\nax.set_xscale('log')\nax.set_yscale('log')\nax.set_xlabel('log-fitness in env. 1,\\n$m(x=1)$')\nax.set_ylabel('log-fitness in env. 2,\\n$m(x=2)$')\nfor ax in axes:\n plotting.despine(ax)\n ax.set_xticks([])\n ax.set_yticks([])\nplotting.label_axes(axes, xy=(-0.15, 0.95))\nfig.tight_layout(pad=0.25)\nfig.savefig('svgs/graphicalmethod.svg')",
"_____no_output_____"
]
],
[
[
"**Illustration of the steps of a graphical method for finding the best adaptation strategy in uncorrelated environments.** (A) Fitness values of phenotypes across environments (orange dots). (B) Fitness values achievable by switching strategies (orange area) are those inside the convex hull of the fitness values of the different phenotypes. A necessary condition for optimality is to lie on the Pareto frontier (blue line). (C, D) The optimal strategy has the fitnesses (red/green star) at which the isolines of the long-term growth rate for given environmental frequencies (red lines for $p(2)=0.7$, green lines for $p(2)=0.3$) are tangential to the Pareto frontier. (C) In fitness space the isolines are curved. (D) To determine the optimal strategy it is more convenient to work in log-fitness space, where the isolines are straight lines.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e7f66ba48855963a755ea61697dbdd5b12c84c9a | 36,438 | ipynb | Jupyter Notebook | notebooks/junctions_testcases.ipynb | devyanijivani/pygraspi | ee72cf0755acfa5d2e92151953f43f2b63172bd0 | [
"MIT"
] | null | null | null | notebooks/junctions_testcases.ipynb | devyanijivani/pygraspi | ee72cf0755acfa5d2e92151953f43f2b63172bd0 | [
"MIT"
] | 4 | 2022-02-28T17:26:36.000Z | 2022-03-15T16:34:21.000Z | notebooks/junctions_testcases.ipynb | devyanijivani/pygraspi | ee72cf0755acfa5d2e92151953f43f2b63172bd0 | [
"MIT"
] | null | null | null | 33.927374 | 6,808 | 0.635134 | [
[
[
"import numpy as np\nfrom skimage.morphology import medial_axis, skeletonize\nimport matplotlib.pyplot as plt\nimport pandas\nimport networkx as nx\nimport sknw\nimport glob\nimport os",
"_____no_output_____"
],
[
"morph = np.array(pandas.read_csv('data/data_0.558_2.4_000180.txt', delimiter=' ', header=None)).swapaxes(0, 1)\nskel, distance = medial_axis(morph, return_distance=True)\nskel",
"_____no_output_____"
],
[
"graph = sknw.build_sknw(skel)\nfor (s,e) in graph.edges():\n ps = graph[s][e]['pts']\n plt.plot(ps[:,1], ps[:,0], 'green', zorder=-1)\n \n# draw node by o\nnodes = graph.nodes()\nps = np.array([nodes[i]['o'] for i in nodes], dtype = int)\nplt.scatter(ps[:,1], ps[:,0], s = 5, c ='r')\n\n# title and show\nplt.title('Build Graph')\n\nplt.gca().set_aspect('equal')\n#plt.savefig('my_plot.png',dpi=1200)\n",
"_____no_output_____"
],
[
"plt.show()",
"_____no_output_____"
],
[
"morph = np.array([[1,1,1],\\\n [1,1,1],\\\n [1,1,1]])\nskel, distance = medial_axis(morph, return_distance=True)",
"_____no_output_____"
],
[
"set(np.array([list(graph.nodes[n].keys()) for n in graph.nodes()]).flatten())\n",
"_____no_output_____"
],
[
"for n1 in graph.nodes():\n print(graph.nodes[n1]['o'])\n",
"[0. 2.]\n[2. 0.]\n"
],
[
"for node1, node2, data in graph.edges(data=True):\n #print(data['pts'])\n print(node1, \" \", node2)",
"0 1\n"
],
[
"def getEndJunction(graph):\n l = [graph.degree[n] for n in graph.nodes()]\n return l.count(1), l.count(3)\n ",
"_____no_output_____"
],
[
"[E, J] = getEndJunction(graph)\nB = graph.number_of_edges\nB",
"_____no_output_____"
],
[
"def getBranchLen(graph):\n b_l = [graph.edges[e]['weight'] for e in graph.edges()]\n return np.array([len(b_l), sum(b_l)/len(b_l)])",
"_____no_output_____"
],
[
"getBranchLen(graph)\n",
"_____no_output_____"
],
[
"dist_on_skel = distance * skel\n\nmax(map(max, dist_on_skel))\n#min(map(min, dist_on_skel))\nd = dist_on_skel[skel]\nlen(d)",
"_____no_output_____"
]
],
[
[
"## Test SKNW for Cahn-Hillard dataset. \n ",
"_____no_output_____"
]
],
[
[
"#os.chdir(r'/Users/devyanijivani/git/pygraspi/notebooks/data')\ndest = \"/Users/devyanijivani/git/pygraspi/notebooks/junctions\"\nmyFiles = glob.glob('*.txt')\nmyFiles.sort()\n\nfor i, file in enumerate(myFiles):\n morph = np.array(pandas.read_csv(file, delimiter=' ', header=None)).swapaxes(0, 1)\n skel, distance = medial_axis(morph, return_distance=True)\n graph = sknw.build_sknw(skel)\n for (s,e) in graph.edges():\n ps = graph[s][e]['pts']\n plt.plot(ps[:,1], ps[:,0], 'green', zorder=-1)\n\n # draw node by o\n nodes = graph.nodes()\n ps = np.array([nodes[i]['o'] for i in nodes], dtype = int)\n plt.scatter(ps[:,1], ps[:,0], s = 1, c ='r')\n\n # title and show\n plt.title('Build Graph')\n \n plt.gca().set_aspect('equal')\n print(os.path.splitext(file)[0])\n \n file_loc = os.path.join(dest, os.path.splitext(file)[0]+'.png')\n #print(file_loc)\n #plt.savefig(file_loc,dpi=1200)\n \n plt.close()\n",
"data_0.514_2.4_000080\ndata_0.514_2.4_000100\ndata_0.514_2.4_000140\ndata_0.514_2.4_000160\ndata_0.514_2.4_000180\ndata_0.514_2.4_000220\ndata_0.514_2.4_000280\ndata_0.514_2.4_000440\ndata_0.514_2.4_000480\ndata_0.514_2.4_000560\ndata_0.514_2.4_000620\ndata_0.514_2.4_000760\ndata_0.514_2.4_000880\ndata_0.514_2.4_001240\ndata_0.514_2.4_001800\ndata_0.514_2.4_002480\ndata_0.514_2.4_004660\ndata_0.514_2.4_006400\ndata_0.514_2.6_000080\ndata_0.514_2.6_000100\ndata_0.514_2.6_000140\ndata_0.514_2.6_000180\ndata_0.514_2.6_000220\ndata_0.514_2.6_000240\ndata_0.514_2.6_000260\ndata_0.514_2.6_000340\ndata_0.514_2.6_000420\ndata_0.514_2.6_000460\ndata_0.514_2.6_000540\ndata_0.514_2.6_000600\ndata_0.514_2.6_000680\ndata_0.514_2.6_000980\ndata_0.514_2.6_002660\ndata_0.514_2.6_004160\ndata_0.514_2.6_005600\ndata_0.514_2.6_006460\ndata_0.514_2.8_000100\ndata_0.514_2.8_000120\ndata_0.514_2.8_000140\ndata_0.514_2.8_000200\ndata_0.514_2.8_000240\ndata_0.514_2.8_000320\ndata_0.514_2.8_000400\ndata_0.514_2.8_000460\ndata_0.514_2.8_000600\ndata_0.514_2.8_001080\ndata_0.514_2.8_002000\ndata_0.514_2.8_005540\ndata_0.514_2.8_005980\ndata_0.514_3.2_000180\ndata_0.514_3.2_000200\ndata_0.514_3.2_000220\ndata_0.514_3.2_000240\ndata_0.514_3.2_000260\ndata_0.514_3.2_000280\ndata_0.514_3.2_000300\ndata_0.514_3.2_000340\ndata_0.514_3.2_000400\ndata_0.514_3.2_000440\ndata_0.514_3.2_000500\ndata_0.514_3.2_000580\ndata_0.514_3.2_000660\ndata_0.514_3.2_000740\ndata_0.514_3.2_000860\ndata_0.514_3.2_000960\ndata_0.514_3.2_001080\ndata_0.514_3.2_001300\ndata_0.514_3.2_001560\ndata_0.514_3.2_001820\ndata_0.514_3.2_002200\ndata_0.514_3.2_002800\ndata_0.514_3.2_003720\ndata_0.514_3.2_004780\ndata_0.514_3.2_005300\ndata_0.514_3.2_006860\ndata_0.514_3.4_000080\ndata_0.514_3.4_000100\ndata_0.514_3.4_000120\ndata_0.514_3.4_000160\ndata_0.514_3.4_000180\ndata_0.514_3.4_000200\ndata_0.514_3.4_000240\ndata_0.514_3.4_000360\ndata_0.514_3.4_000400\ndata_0.514_3.4_000440\ndata_0.514_3.4_000500\ndata_0.514_3.4_000560\ndata_0.514_3.4_000620\ndata_0.514_3.4_000700\ndata_0.514_3.4_000760\ndata_0.514_3.4_000820\ndata_0.514_3.4_000880\ndata_0.514_3.4_000960\ndata_0.514_3.4_001160\ndata_0.514_3.4_001400\ndata_0.514_3.4_001740\ndata_0.514_3.4_002200\ndata_0.514_3.4_007320\ndata_0.514_3.4_007500\ndata_0.514_3.6_000060\ndata_0.514_3.6_000080\ndata_0.514_3.6_000100\ndata_0.514_3.6_000120\ndata_0.514_3.6_000140\ndata_0.514_3.6_000160\ndata_0.514_3.6_000180\ndata_0.514_3.6_000200\ndata_0.514_3.6_000220\ndata_0.514_3.6_000280\ndata_0.514_3.6_000320\ndata_0.514_3.6_000360\ndata_0.514_3.6_000460\ndata_0.514_3.6_000500\ndata_0.514_3.6_000560\ndata_0.514_3.6_000640\ndata_0.514_3.6_000720\ndata_0.514_3.6_000840\ndata_0.514_3.6_000980\ndata_0.514_3.6_001180\ndata_0.514_3.6_001360\ndata_0.514_3.6_001480\ndata_0.514_3.6_001660\ndata_0.514_3.6_001920\ndata_0.514_3.6_002100\ndata_0.514_3.6_002520\ndata_0.514_3.6_003320\ndata_0.514_3.6_005000\ndata_0.514_3.6_006980\ndata_0.514_3.6_007020\ndata_0.514_3.8_000100\ndata_0.514_3.8_000120\ndata_0.514_3.8_000140\ndata_0.514_3.8_000160\ndata_0.514_3.8_000180\ndata_0.514_3.8_000200\ndata_0.514_3.8_000240\ndata_0.514_3.8_000280\ndata_0.514_3.8_000300\ndata_0.514_3.8_000340\ndata_0.514_3.8_000380\ndata_0.514_3.8_000400\ndata_0.514_3.8_000460\ndata_0.514_3.8_000520\ndata_0.514_3.8_000660\ndata_0.514_3.8_000740\ndata_0.514_3.8_000860\ndata_0.514_3.8_001080\ndata_0.514_3.8_001360\ndata_0.514_3.8_001640\ndata_0.514_3.8_001900\ndata_0.514_3.8_002100\ndata_0.514_3.8_003240\ndata_0.514_3.8_003840\ndata_0.514_3.8_004240\ndata_0.514_3.8_006040\ndata_0.514_3.8_007040\ndata_0.514_4.0_000040\ndata_0.514_4.0_000060\ndata_0.514_4.0_000080\ndata_0.514_4.0_000100\ndata_0.514_4.0_000120\ndata_0.514_4.0_000140\ndata_0.514_4.0_000160\ndata_0.514_4.0_000180\ndata_0.514_4.0_000200\ndata_0.514_4.0_000220\ndata_0.514_4.0_000260\ndata_0.514_4.0_000280\ndata_0.514_4.0_000320\ndata_0.514_4.0_000340\ndata_0.514_4.0_000380\ndata_0.514_4.0_000440\ndata_0.514_4.0_000500\ndata_0.514_4.0_000560\ndata_0.514_4.0_000680\ndata_0.514_4.0_000880\ndata_0.514_4.0_001080\ndata_0.514_4.0_001700\ndata_0.514_4.0_002020\ndata_0.514_4.0_002320\ndata_0.514_4.0_005200\ndata_0.514_4.0_007020\ndata_0.528_2.4_000040\ndata_0.528_2.4_000060\ndata_0.528_2.4_000120\ndata_0.528_2.4_000200\ndata_0.528_2.4_000240\ndata_0.528_2.4_000340\ndata_0.528_2.4_000400\ndata_0.528_2.4_000520\ndata_0.528_2.4_000600\ndata_0.528_2.4_000660\ndata_0.528_2.4_000760\ndata_0.528_2.4_000940\ndata_0.528_2.4_001200\ndata_0.528_2.4_001820\ndata_0.528_2.4_002480\ndata_0.528_2.4_003740\ndata_0.528_2.4_006460\ndata_0.528_2.6_000040\ndata_0.528_2.6_000080\ndata_0.528_2.6_000100\ndata_0.528_2.6_000120\ndata_0.528_2.6_000140\ndata_0.528_2.6_000180\ndata_0.528_2.6_000240\ndata_0.528_2.6_000260\ndata_0.528_2.6_000300\ndata_0.528_2.6_000380\ndata_0.528_2.6_000460\ndata_0.528_2.6_000520\ndata_0.528_2.6_000580\ndata_0.528_2.6_000660\ndata_0.528_2.6_000940\ndata_0.528_2.6_002660\ndata_0.528_2.6_003940\ndata_0.528_2.6_005460\ndata_0.528_2.6_006300\ndata_0.528_2.8_000040\ndata_0.528_2.8_000080\ndata_0.528_2.8_000100\ndata_0.528_2.8_000140\ndata_0.528_2.8_000160\ndata_0.528_2.8_000180\ndata_0.528_2.8_000440\ndata_0.528_2.8_000500\ndata_0.528_2.8_000660\ndata_0.528_2.8_001220\ndata_0.528_2.8_001560\ndata_0.528_2.8_003680\ndata_0.528_2.8_004880\ndata_0.528_2.8_006380\ndata_0.528_3.2_000080\ndata_0.528_3.2_000100\ndata_0.528_3.2_000120\ndata_0.528_3.2_000140\ndata_0.528_3.2_000160\ndata_0.528_3.2_000180\ndata_0.528_3.2_000200\ndata_0.528_3.2_000220\ndata_0.528_3.2_000240\ndata_0.528_3.2_000260\ndata_0.528_3.2_000280\ndata_0.528_3.2_000320\ndata_0.528_3.2_000360\ndata_0.528_3.2_000420\ndata_0.528_3.2_000480\ndata_0.528_3.2_000560\ndata_0.528_3.2_000640\ndata_0.528_3.2_000720\ndata_0.528_3.2_000780\ndata_0.528_3.2_000880\ndata_0.528_3.2_001860\ndata_0.528_3.2_002320\ndata_0.528_3.2_002820\ndata_0.528_3.2_003500\ndata_0.528_3.2_005380\ndata_0.528_3.2_007540\ndata_0.528_3.2_007560\ndata_0.528_3.4_000080\ndata_0.528_3.4_000100\ndata_0.528_3.4_000120\ndata_0.528_3.4_000140\ndata_0.528_3.4_000160\ndata_0.528_3.4_000180\ndata_0.528_3.4_000200\ndata_0.528_3.4_000240\ndata_0.528_3.4_000260\ndata_0.528_3.4_000300\ndata_0.528_3.4_000340\ndata_0.528_3.4_000380\ndata_0.528_3.4_000440\ndata_0.528_3.4_000500\ndata_0.528_3.4_000560\ndata_0.528_3.4_000640\ndata_0.528_3.4_000740\ndata_0.528_3.4_000840\ndata_0.528_3.4_000980\ndata_0.528_3.4_001200\ndata_0.528_3.4_001380\ndata_0.528_3.4_001580\ndata_0.528_3.4_001780\ndata_0.528_3.4_001980\ndata_0.528_3.4_002380\ndata_0.528_3.4_003280\ndata_0.528_3.4_004620\ndata_0.528_3.4_005740\ndata_0.528_3.4_006440\ndata_0.528_3.4_007500\ndata_0.528_3.6_000080\ndata_0.528_3.6_000120\ndata_0.528_3.6_000260\ndata_0.528_3.6_000280\ndata_0.528_3.6_000320\ndata_0.528_3.6_000360\ndata_0.528_3.6_000380\ndata_0.528_3.6_000440\ndata_0.528_3.6_000480\ndata_0.528_3.6_000540\ndata_0.528_3.6_000620\ndata_0.528_3.6_000700\ndata_0.528_3.6_000800\ndata_0.528_3.6_000900\ndata_0.528_3.6_001060\ndata_0.528_3.6_001240\ndata_0.528_3.6_001400\ndata_0.528_3.6_001640\ndata_0.528_3.6_001940\ndata_0.528_3.6_002120\ndata_0.528_3.6_003200\ndata_0.528_3.6_004040\ndata_0.528_3.6_005180\ndata_0.528_3.6_006780\ndata_0.528_3.6_007080\ndata_0.528_3.8_000080\ndata_0.528_3.8_000100\ndata_0.528_3.8_000120\ndata_0.528_3.8_000140\ndata_0.528_3.8_000160\ndata_0.528_3.8_000180\ndata_0.528_3.8_000220\ndata_0.528_3.8_000240\ndata_0.528_3.8_000280\ndata_0.528_3.8_000300\ndata_0.528_3.8_000340\ndata_0.528_3.8_000400\ndata_0.528_3.8_000460\ndata_0.528_3.8_000520\ndata_0.528_3.8_000580\ndata_0.528_3.8_000640\ndata_0.528_3.8_000740\ndata_0.528_3.8_000900\ndata_0.528_3.8_001260\ndata_0.528_3.8_001500\ndata_0.528_3.8_001700\ndata_0.528_3.8_001920\ndata_0.528_3.8_002300\ndata_0.528_3.8_002740\ndata_0.528_3.8_003360\ndata_0.528_3.8_003960\ndata_0.528_3.8_004420\ndata_0.528_3.8_005900\ndata_0.528_3.8_007080\ndata_0.528_4.0_000040\ndata_0.528_4.0_000060\ndata_0.528_4.0_000080\ndata_0.528_4.0_000100\ndata_0.528_4.0_000140\ndata_0.528_4.0_000160\ndata_0.528_4.0_000180\ndata_0.528_4.0_000200\ndata_0.528_4.0_000220\ndata_0.528_4.0_000240\ndata_0.528_4.0_000280\ndata_0.528_4.0_000320\ndata_0.528_4.0_000340\ndata_0.528_4.0_000380\ndata_0.528_4.0_000440\ndata_0.528_4.0_000520\ndata_0.528_4.0_000920\ndata_0.528_4.0_001100\ndata_0.528_4.0_001260\ndata_0.528_4.0_001460\ndata_0.528_4.0_001720\ndata_0.528_4.0_001980\ndata_0.528_4.0_002240\ndata_0.528_4.0_002660\ndata_0.528_4.0_003300\ndata_0.528_4.0_003820\ndata_0.528_4.0_005000\ndata_0.528_4.0_006960\ndata_0.543_2.4_000040\ndata_0.543_2.4_000080\n"
],
[
"pwd",
"_____no_output_____"
],
[
"def skeletonize(morph):\n skel, distance = medial_axis(morph, return_distance=True)\n return skel, distance ",
"_____no_output_____"
],
[
"morph = np.array([[1,1,1],\\\n [1,1,1],\\\n [1,1,1]])\nskel = skeletonize(morph)[0]",
"_____no_output_____"
],
[
"skel",
"_____no_output_____"
],
[
"def getEndJunction(graph):\n l = [graph.degree[n] for n in graph.nodes()]\n return np.array([l.count(1), l.count(3)])\n",
"_____no_output_____"
],
[
"graph = sknw.build_sknw(skel)",
"_____no_output_____"
],
[
"getEndJunction(graph)",
"_____no_output_____"
],
[
"def getBranchLen(graph):\n b_l = [graph.edges[e]['weight'] for e in graph.edges()]\n return np.array([len(b_l), round(sum(b_l)/len(b_l), 2)]) \n",
"_____no_output_____"
],
[
"getBranchLen(graph)",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f67508236263122170eff97b047f342831db32 | 59,795 | ipynb | Jupyter Notebook | BCNcode/0_vibratioon_signal/1450/BCN/1450-011-512-x.ipynb | Decaili98/BCN-code-2022 | ab0ce085cb29fbf12b6d773861953cb2cef23e20 | [
"MulanPSL-1.0"
] | null | null | null | BCNcode/0_vibratioon_signal/1450/BCN/1450-011-512-x.ipynb | Decaili98/BCN-code-2022 | ab0ce085cb29fbf12b6d773861953cb2cef23e20 | [
"MulanPSL-1.0"
] | null | null | null | BCNcode/0_vibratioon_signal/1450/BCN/1450-011-512-x.ipynb | Decaili98/BCN-code-2022 | ab0ce085cb29fbf12b6d773861953cb2cef23e20 | [
"MulanPSL-1.0"
] | null | null | null | 96.755663 | 18,004 | 0.7611 | [
[
[
"import tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers\nfrom keras import initializers\nimport keras.backend as K\nimport numpy as np\nimport pandas as pd\nfrom tensorflow.keras.layers import *\nfrom keras.regularizers import l2#正则化",
"Using TensorFlow backend.\n"
],
[
"import pandas as pd\nimport numpy as np\nnormal = np.loadtxt(r'F:\\张老师课题学习内容\\code\\数据集\\试验数据(包括压力脉动和振动)\\2013.9.12-未发生缠绕前\\2013-9.12振动\\2013-9-12振动-1450rmin-mat\\1450r_normalvibx.txt', delimiter=',')\nchanrao = np.loadtxt(r'F:\\张老师课题学习内容\\code\\数据集\\试验数据(包括压力脉动和振动)\\2013.9.17-发生缠绕后\\振动\\9-17下午振动1450rmin-mat\\1450r_chanraovibx.txt', delimiter=',')\nprint(normal.shape,chanrao.shape,\"***************************************************\")\ndata_normal=normal[20:22] #提取前两行\ndata_chanrao=chanrao[20:22] #提取前两行\nprint(data_normal.shape,data_chanrao.shape)\nprint(data_normal,\"\\r\\n\",data_chanrao,\"***************************************************\")\ndata_normal=data_normal.reshape(1,-1)\ndata_chanrao=data_chanrao.reshape(1,-1)\nprint(data_normal.shape,data_chanrao.shape)\nprint(data_normal,\"\\r\\n\",data_chanrao,\"***************************************************\")",
"(22, 32768) (22, 32768) ***************************************************\n(2, 32768) (2, 32768)\n[[-0.48398 0.52771 0.81684 ... 0.10754 -0.16297 -0.14819 ]\n [-0.76699 0.060077 -1.3187 ... 0.41728 -0.19369 0.13473 ]] \r\n [[ 1.4783 -3.0477 0.4864 ... -0.32556 0.081582 0.75064 ]\n [-0.87572 0.45935 -0.91375 ... 1.6293 -0.60795 -0.26352 ]] ***************************************************\n(1, 65536) (1, 65536)\n[[-0.48398 0.52771 0.81684 ... 0.41728 -0.19369 0.13473]] \r\n [[ 1.4783 -3.0477 0.4864 ... 1.6293 -0.60795 -0.26352]] ***************************************************\n"
],
[
"#水泵的两种故障类型信号normal正常,chanrao故障\ndata_normal=data_normal.reshape(-1, 512)#(65536,1)-(128, 515)\ndata_chanrao=data_chanrao.reshape(-1,512)\nprint(data_normal.shape,data_chanrao.shape)\n",
"(128, 512) (128, 512)\n"
],
[
"import numpy as np\ndef yuchuli(data,label):#(4:1)(51:13)\n #打乱数据顺序\n np.random.shuffle(data)\n train = data[0:102,:]\n test = data[102:128,:]\n label_train = np.array([label for i in range(0,102)])\n label_test =np.array([label for i in range(0,26)])\n return train,test ,label_train ,label_test\ndef stackkk(a,b,c,d,e,f,g,h):\n aa = np.vstack((a, e))\n bb = np.vstack((b, f))\n cc = np.hstack((c, g))\n dd = np.hstack((d, h))\n return aa,bb,cc,dd\nx_tra0,x_tes0,y_tra0,y_tes0 = yuchuli(data_normal,0)\nx_tra1,x_tes1,y_tra1,y_tes1 = yuchuli(data_chanrao,1)\ntr1,te1,yr1,ye1=stackkk(x_tra0,x_tes0,y_tra0,y_tes0 ,x_tra1,x_tes1,y_tra1,y_tes1)\n\nx_train=tr1\nx_test=te1\ny_train = yr1\ny_test = ye1\n\n#打乱数据\nstate = np.random.get_state()\nnp.random.shuffle(x_train)\nnp.random.set_state(state)\nnp.random.shuffle(y_train)\n\nstate = np.random.get_state()\nnp.random.shuffle(x_test)\nnp.random.set_state(state)\nnp.random.shuffle(y_test)\n\n\n#对训练集和测试集标准化\ndef ZscoreNormalization(x):\n \"\"\"Z-score normaliaztion\"\"\"\n x = (x - np.mean(x)) / np.std(x)\n return x\nx_train=ZscoreNormalization(x_train)\nx_test=ZscoreNormalization(x_test)\n# print(x_test[0])\n\n\n#转化为一维序列\nx_train = x_train.reshape(-1,512,1)\nx_test = x_test.reshape(-1,512,1)\nprint(x_train.shape,x_test.shape)\n\ndef to_one_hot(labels,dimension=2):\n results = np.zeros((len(labels),dimension))\n for i,label in enumerate(labels):\n results[i,label] = 1\n return results\none_hot_train_labels = to_one_hot(y_train)\none_hot_test_labels = to_one_hot(y_test)",
"(204, 512, 1) (52, 512, 1)\n"
],
[
"#定义挤压函数\ndef squash(vectors, axis=-1):\n \"\"\"\n 对向量的非线性激活函数\n ## vectors: some vectors to be squashed, N-dim tensor\n ## axis: the axis to squash\n :return: a Tensor with same shape as input vectors\n \"\"\"\n s_squared_norm = K.sum(K.square(vectors), axis, keepdims=True)\n scale = s_squared_norm / (1 + s_squared_norm) / K.sqrt(s_squared_norm + K.epsilon())\n return scale * vectors\n\nclass Length(layers.Layer):\n \"\"\"\n 计算向量的长度。它用于计算与margin_loss中的y_true具有相同形状的张量\n Compute the length of vectors. This is used to compute a Tensor that has the same shape with y_true in margin_loss\n inputs: shape=[dim_1, ..., dim_{n-1}, dim_n]\n output: shape=[dim_1, ..., dim_{n-1}]\n \"\"\"\n def call(self, inputs, **kwargs):\n return K.sqrt(K.sum(K.square(inputs), -1))\n\n def compute_output_shape(self, input_shape):\n return input_shape[:-1]\n \n def get_config(self):\n config = super(Length, self).get_config()\n return config\n#定义预胶囊层\ndef PrimaryCap(inputs, dim_capsule, n_channels, kernel_size, strides, padding):\n \"\"\"\n 进行普通二维卷积 `n_channels` 次, 然后将所有的胶囊重叠起来\n :param inputs: 4D tensor, shape=[None, width, height, channels]\n :param dim_capsule: the dim of the output vector of capsule\n :param n_channels: the number of types of capsules\n :return: output tensor, shape=[None, num_capsule, dim_capsule]\n \"\"\"\n output = layers.Conv2D(filters=dim_capsule*n_channels, kernel_size=kernel_size, strides=strides,\n padding=padding,name='primarycap_conv2d')(inputs)\n outputs = layers.Reshape(target_shape=[-1, dim_capsule], name='primarycap_reshape')(output)\n return layers.Lambda(squash, name='primarycap_squash')(outputs)\n\nclass DenseCapsule(layers.Layer):\n \"\"\"\n 胶囊层. 输入输出都为向量. \n ## num_capsule: 本层包含的胶囊数量\n ## dim_capsule: 输出的每一个胶囊向量的维度\n ## routings: routing 算法的迭代次数\n \"\"\"\n def __init__(self, num_capsule, dim_capsule, routings=3, kernel_initializer='glorot_uniform',**kwargs):\n super(DenseCapsule, self).__init__(**kwargs)\n self.num_capsule = num_capsule\n self.dim_capsule = dim_capsule\n self.routings = routings\n self.kernel_initializer = kernel_initializer\n\n def build(self, input_shape):\n assert len(input_shape) >= 3, '输入的 Tensor 的形状[None, input_num_capsule, input_dim_capsule]'#(None,1152,8)\n self.input_num_capsule = input_shape[1]\n self.input_dim_capsule = input_shape[2]\n\n #转换矩阵\n self.W = self.add_weight(shape=[self.num_capsule, self.input_num_capsule,\n self.dim_capsule, self.input_dim_capsule],\n initializer=self.kernel_initializer,name='W')\n self.built = True\n\n def call(self, inputs, training=None):\n # inputs.shape=[None, input_num_capsuie, input_dim_capsule]\n # inputs_expand.shape=[None, 1, input_num_capsule, input_dim_capsule]\n inputs_expand = K.expand_dims(inputs, 1)\n # 运算优化:将inputs_expand重复num_capsule 次,用于快速和W相乘\n # inputs_tiled.shape=[None, num_capsule, input_num_capsule, input_dim_capsule]\n inputs_tiled = K.tile(inputs_expand, [1, self.num_capsule, 1, 1])\n\n # 将inputs_tiled的batch中的每一条数据,计算inputs+W\n # x.shape = [num_capsule, input_num_capsule, input_dim_capsule]\n # W.shape = [num_capsule, input_num_capsule, dim_capsule, input_dim_capsule]\n # 将x和W的前两个维度看作'batch'维度,向量和矩阵相乘:\n # [input_dim_capsule] x [dim_capsule, input_dim_capsule]^T -> [dim_capsule].\n # inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsutel\n inputs_hat = K.map_fn(lambda x: K.batch_dot(x, self.W, [2, 3]),elems=inputs_tiled)\n\n # Begin: Routing算法\n # 将系数b初始化为0.\n # b.shape = [None, self.num_capsule, self, input_num_capsule].\n b = tf.zeros(shape=[K.shape(inputs_hat)[0], self.num_capsule, self.input_num_capsule])\n \n assert self.routings > 0, 'The routings should be > 0.'\n for i in range(self.routings):\n # c.shape=[None, num_capsule, input_num_capsule]\n C = tf.nn.softmax(b ,axis=1)\n # c.shape = [None, num_capsule, input_num_capsule]\n # inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsule]\n # 将c与inputs_hat的前两个维度看作'batch'维度,向量和矩阵相乘:\n # [input_num_capsule] x [input_num_capsule, dim_capsule] -> [dim_capsule],\n # outputs.shape= [None, num_capsule, dim_capsule]\n outputs = squash(K. batch_dot(C, inputs_hat, [2, 2])) # [None, 10, 16]\n \n if i < self.routings - 1:\n # outputs.shape = [None, num_capsule, dim_capsule]\n # inputs_hat.shape = [None, num_capsule, input_num_capsule, dim_capsule]\n # 将outputs和inρuts_hat的前两个维度看作‘batch’ 维度,向量和矩阵相乘:\n # [dim_capsule] x [imput_num_capsule, dim_capsule]^T -> [input_num_capsule]\n # b.shape = [batch_size. num_capsule, input_nom_capsule]\n# b += K.batch_dot(outputs, inputs_hat, [2, 3]) to this b += tf.matmul(self.W, x)\n b += K.batch_dot(outputs, inputs_hat, [2, 3])\n\n # End: Routing 算法\n return outputs\n\n def compute_output_shape(self, input_shape):\n return tuple([None, self.num_capsule, self.dim_capsule])\n\n def get_config(self):\n config = {\n 'num_capsule': self.num_capsule,\n 'dim_capsule': self.dim_capsule,\n 'routings': self.routings\n }\n base_config = super(DenseCapsule, self).get_config()\n return dict(list(base_config.items()) + list(config.items()))",
"_____no_output_____"
],
[
"from tensorflow import keras\nfrom keras.regularizers import l2#正则化\nx = layers.Input(shape=[512,1, 1])\n#普通卷积层\nconv1 = layers.Conv2D(filters=16, kernel_size=(2, 1),activation='relu',padding='valid',name='conv1')(x)\n#池化层\nPOOL1 = MaxPooling2D((2,1))(conv1)\n#普通卷积层\nconv2 = layers.Conv2D(filters=32, kernel_size=(2, 1),activation='relu',padding='valid',name='conv2')(POOL1)\n#池化层\n# POOL2 = MaxPooling2D((2,1))(conv2)\n#Dropout层\nDropout=layers.Dropout(0.1)(conv2)\n\n# Layer 3: 使用“squash”激活的Conv2D层, 然后重塑 [None, num_capsule, dim_vector]\nprimarycaps = PrimaryCap(Dropout, dim_capsule=8, n_channels=12, kernel_size=(4, 1), strides=2, padding='valid')\n# Layer 4: 数字胶囊层,动态路由算法在这里工作。\ndigitcaps = DenseCapsule(num_capsule=2, dim_capsule=16, routings=3, name='digit_caps')(primarycaps)\n# Layer 5:这是一个辅助层,用它的长度代替每个胶囊。只是为了符合标签的形状。\nout_caps = Length(name='out_caps')(digitcaps)\n\nmodel = keras.Model(x, out_caps) \nmodel.summary() ",
"WARNING:tensorflow:From C:\\ProgramData\\Anaconda3\\envs\\tf2\\lib\\site-packages\\tensorflow\\python\\util\\deprecation.py:605: calling map_fn_v2 (from tensorflow.python.ops.map_fn) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nUse fn_output_signature instead\nModel: \"model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 512, 1, 1)] 0 \n_________________________________________________________________\nconv1 (Conv2D) (None, 511, 1, 16) 48 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 255, 1, 16) 0 \n_________________________________________________________________\nconv2 (Conv2D) (None, 254, 1, 32) 1056 \n_________________________________________________________________\ndropout (Dropout) (None, 254, 1, 32) 0 \n_________________________________________________________________\nprimarycap_conv2d (Conv2D) (None, 126, 1, 96) 12384 \n_________________________________________________________________\nprimarycap_reshape (Reshape) (None, 1512, 8) 0 \n_________________________________________________________________\nprimarycap_squash (Lambda) (None, 1512, 8) 0 \n_________________________________________________________________\ndigit_caps (DenseCapsule) (None, 2, 16) 387072 \n_________________________________________________________________\nout_caps (Length) (None, 2) 0 \n=================================================================\nTotal params: 400,560\nTrainable params: 400,560\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"#定义优化\nmodel.compile(metrics=['accuracy'],\n optimizer='adam',\n loss=lambda y_true,y_pred: y_true*K.relu(0.9-y_pred)**2 + 0.25*(1-y_true)*K.relu(y_pred-0.1)**2 \n )\nimport time\ntime_begin = time.time()\nhistory = model.fit(x_train,one_hot_train_labels,\n validation_split=0.1,\n epochs=50,batch_size=10,\n shuffle=True)\ntime_end = time.time()\ntime = time_end - time_begin\nprint('time:', time)\n",
"Epoch 1/50\n19/19 [==============================] - 4s 87ms/step - loss: 0.1533 - accuracy: 0.4660 - val_loss: 0.0937 - val_accuracy: 0.6667\nEpoch 2/50\n19/19 [==============================] - 1s 29ms/step - loss: 0.0889 - accuracy: 0.6230 - val_loss: 0.0729 - val_accuracy: 0.6667\nEpoch 3/50\n19/19 [==============================] - 1s 27ms/step - loss: 0.0634 - accuracy: 0.6625 - val_loss: 0.0691 - val_accuracy: 0.5238\nEpoch 4/50\n19/19 [==============================] - 1s 30ms/step - loss: 0.0502 - accuracy: 0.8526 - val_loss: 0.0471 - val_accuracy: 0.7143\nEpoch 5/50\n19/19 [==============================] - 1s 29ms/step - loss: 0.0175 - accuracy: 0.9850 - val_loss: 0.0296 - val_accuracy: 0.9048\nEpoch 6/50\n19/19 [==============================] - 1s 30ms/step - loss: 0.0074 - accuracy: 1.0000 - val_loss: 0.0136 - val_accuracy: 1.0000\nEpoch 7/50\n19/19 [==============================] - 1s 30ms/step - loss: 0.0036 - accuracy: 1.0000 - val_loss: 0.0117 - val_accuracy: 1.0000\nEpoch 8/50\n19/19 [==============================] - 1s 27ms/step - loss: 0.0014 - accuracy: 1.0000 - val_loss: 0.0098 - val_accuracy: 1.0000\nEpoch 9/50\n19/19 [==============================] - 1s 27ms/step - loss: 6.0852e-04 - accuracy: 1.0000 - val_loss: 0.0080 - val_accuracy: 1.0000\nEpoch 10/50\n19/19 [==============================] - 1s 28ms/step - loss: 5.3153e-04 - accuracy: 1.0000 - val_loss: 0.0098 - val_accuracy: 1.0000\nEpoch 11/50\n19/19 [==============================] - 1s 28ms/step - loss: 6.0077e-04 - accuracy: 1.0000 - val_loss: 0.0089 - val_accuracy: 1.0000\nEpoch 12/50\n19/19 [==============================] - 1s 28ms/step - loss: 1.6323e-04 - accuracy: 1.0000 - val_loss: 0.0065 - val_accuracy: 1.0000\nEpoch 13/50\n19/19 [==============================] - 0s 26ms/step - loss: 2.8485e-04 - accuracy: 1.0000 - val_loss: 0.0094 - val_accuracy: 1.0000\nEpoch 14/50\n19/19 [==============================] - 1s 30ms/step - loss: 2.8164e-04 - accuracy: 1.0000 - val_loss: 0.0072 - val_accuracy: 1.0000\nEpoch 15/50\n19/19 [==============================] - 1s 28ms/step - loss: 1.0347e-04 - accuracy: 1.0000 - val_loss: 0.0066 - val_accuracy: 1.0000\nEpoch 16/50\n19/19 [==============================] - 1s 27ms/step - loss: 9.9590e-05 - accuracy: 1.0000 - val_loss: 0.0062 - val_accuracy: 1.0000\nEpoch 17/50\n19/19 [==============================] - 1s 28ms/step - loss: 3.6152e-05 - accuracy: 1.0000 - val_loss: 0.0090 - val_accuracy: 1.0000\nEpoch 18/50\n19/19 [==============================] - 1s 28ms/step - loss: 6.0549e-05 - accuracy: 1.0000 - val_loss: 0.0075 - val_accuracy: 1.0000\nEpoch 19/50\n19/19 [==============================] - 1s 27ms/step - loss: 9.5141e-05 - accuracy: 1.0000 - val_loss: 0.0068 - val_accuracy: 1.0000\nEpoch 20/50\n19/19 [==============================] - 1s 27ms/step - loss: 1.9215e-05 - accuracy: 1.0000 - val_loss: 0.0072 - val_accuracy: 1.0000\nEpoch 21/50\n19/19 [==============================] - 1s 29ms/step - loss: 3.4054e-05 - accuracy: 1.0000 - val_loss: 0.0074 - val_accuracy: 1.0000\nEpoch 22/50\n19/19 [==============================] - 1s 28ms/step - loss: 1.6229e-05 - accuracy: 1.0000 - val_loss: 0.0074 - val_accuracy: 1.0000\nEpoch 23/50\n19/19 [==============================] - 1s 29ms/step - loss: 7.5350e-06 - accuracy: 1.0000 - val_loss: 0.0068 - val_accuracy: 1.0000\nEpoch 24/50\n19/19 [==============================] - 1s 28ms/step - loss: 4.3493e-05 - accuracy: 1.0000 - val_loss: 0.0095 - val_accuracy: 1.0000\nEpoch 25/50\n19/19 [==============================] - 1s 28ms/step - loss: 1.5976e-04 - accuracy: 1.0000 - val_loss: 0.0048 - val_accuracy: 1.0000\nEpoch 26/50\n19/19 [==============================] - 1s 34ms/step - loss: 5.1855e-04 - accuracy: 1.0000 - val_loss: 0.0099 - val_accuracy: 1.0000\nEpoch 27/50\n19/19 [==============================] - 1s 31ms/step - loss: 6.1930e-04 - accuracy: 1.0000 - val_loss: 0.0095 - val_accuracy: 1.0000\nEpoch 28/50\n19/19 [==============================] - 1s 28ms/step - loss: 6.0895e-04 - accuracy: 1.0000 - val_loss: 0.0074 - val_accuracy: 1.0000\nEpoch 29/50\n19/19 [==============================] - 1s 27ms/step - loss: 2.3088e-04 - accuracy: 1.0000 - val_loss: 0.0067 - val_accuracy: 1.0000\nEpoch 30/50\n19/19 [==============================] - 1s 28ms/step - loss: 3.8923e-04 - accuracy: 1.0000 - val_loss: 0.0055 - val_accuracy: 1.0000\nEpoch 31/50\n19/19 [==============================] - 1s 29ms/step - loss: 2.0402e-04 - accuracy: 1.0000 - val_loss: 0.0049 - val_accuracy: 1.0000\nEpoch 32/50\n19/19 [==============================] - 1s 27ms/step - loss: 1.2660e-04 - accuracy: 1.0000 - val_loss: 0.0059 - val_accuracy: 1.0000\nEpoch 33/50\n19/19 [==============================] - 1s 27ms/step - loss: 6.8963e-05 - accuracy: 1.0000 - val_loss: 0.0047 - val_accuracy: 1.0000\nEpoch 34/50\n19/19 [==============================] - 1s 27ms/step - loss: 5.8897e-05 - accuracy: 1.0000 - val_loss: 0.0051 - val_accuracy: 1.0000\nEpoch 35/50\n19/19 [==============================] - 1s 27ms/step - loss: 1.5125e-04 - accuracy: 1.0000 - val_loss: 0.0093 - val_accuracy: 1.0000\nEpoch 36/50\n19/19 [==============================] - 1s 28ms/step - loss: 1.6335e-04 - accuracy: 1.0000 - val_loss: 0.0052 - val_accuracy: 1.0000\nEpoch 37/50\n19/19 [==============================] - 1s 27ms/step - loss: 2.0596e-04 - accuracy: 1.0000 - val_loss: 0.0071 - val_accuracy: 1.0000\nEpoch 38/50\n19/19 [==============================] - 1s 31ms/step - loss: 7.1038e-05 - accuracy: 1.0000 - val_loss: 0.0060 - val_accuracy: 1.0000\nEpoch 39/50\n19/19 [==============================] - 1s 36ms/step - loss: 3.2049e-05 - accuracy: 1.0000 - val_loss: 0.0067 - val_accuracy: 1.0000\nEpoch 40/50\n19/19 [==============================] - 1s 35ms/step - loss: 8.6913e-05 - accuracy: 1.0000 - val_loss: 0.0069 - val_accuracy: 1.0000\nEpoch 41/50\n19/19 [==============================] - 1s 31ms/step - loss: 2.2522e-04 - accuracy: 1.0000 - val_loss: 0.0044 - val_accuracy: 1.0000\nEpoch 42/50\n19/19 [==============================] - 1s 30ms/step - loss: 1.1322e-04 - accuracy: 1.0000 - val_loss: 0.0053 - val_accuracy: 1.0000\nEpoch 43/50\n19/19 [==============================] - 1s 35ms/step - loss: 8.7830e-05 - accuracy: 1.0000 - val_loss: 0.0046 - val_accuracy: 1.0000\nEpoch 44/50\n19/19 [==============================] - 1s 33ms/step - loss: 3.2628e-04 - accuracy: 1.0000 - val_loss: 0.0046 - val_accuracy: 1.0000\nEpoch 45/50\n19/19 [==============================] - 1s 32ms/step - loss: 2.7120e-04 - accuracy: 1.0000 - val_loss: 0.0054 - val_accuracy: 1.0000\nEpoch 46/50\n19/19 [==============================] - 1s 33ms/step - loss: 0.0010 - accuracy: 1.0000 - val_loss: 0.0105 - val_accuracy: 1.0000\nEpoch 47/50\n19/19 [==============================] - 1s 30ms/step - loss: 4.7005e-04 - accuracy: 1.0000 - val_loss: 0.0070 - val_accuracy: 1.0000\nEpoch 48/50\n19/19 [==============================] - 1s 33ms/step - loss: 3.9388e-04 - accuracy: 1.0000 - val_loss: 0.0059 - val_accuracy: 1.0000\nEpoch 49/50\n19/19 [==============================] - 1s 32ms/step - loss: 4.7152e-04 - accuracy: 1.0000 - val_loss: 0.0050 - val_accuracy: 1.0000\nEpoch 50/50\n19/19 [==============================] - 1s 33ms/step - loss: 7.5303e-05 - accuracy: 1.0000 - val_loss: 0.0066 - val_accuracy: 1.0000\ntime: 31.37974715232849\n"
],
[
"score = model.evaluate(x_test,one_hot_test_labels, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.002860977780073881\nTest accuracy: 1.0\n"
],
[
"#绘制acc-loss曲线\nimport matplotlib.pyplot as plt\n\nplt.plot(history.history['loss'],color='r')\nplt.plot(history.history['val_loss'],color='g')\nplt.plot(history.history['accuracy'],color='b')\nplt.plot(history.history['val_accuracy'],color='k')\nplt.title('model loss and acc')\nplt.ylabel('Accuracy')\nplt.xlabel('epoch')\nplt.legend(['train_loss', 'test_loss','train_acc', 'test_acc'], loc='upper left')\n# plt.legend(['train_loss','train_acc'], loc='upper left')\n#plt.savefig('1.png')\nplt.show()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nplt.plot(history.history['loss'],color='r')\nplt.plot(history.history['accuracy'],color='b')\nplt.title('model loss and sccuracy ')\nplt.ylabel('loss/sccuracy')\nplt.xlabel('epoch')\nplt.legend(['train_loss', 'train_sccuracy'], loc='upper left')\nplt.show()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f67ecec348fd50629f79550b123255c9c29911 | 223,900 | ipynb | Jupyter Notebook | Pandas Lesson.ipynb | nsoontie/pythonPandasLesson | 1c33fd58c9648720e43c20415e462f3e1b96402c | [
"MIT"
] | 2 | 2016-02-29T11:29:05.000Z | 2017-01-19T03:55:24.000Z | Pandas Lesson.ipynb | nsoontie/pythonPandasLesson | 1c33fd58c9648720e43c20415e462f3e1b96402c | [
"MIT"
] | 2 | 2015-10-14T22:55:23.000Z | 2016-05-15T14:26:30.000Z | Pandas Lesson.ipynb | nsoontie/pythonPandasLesson | 1c33fd58c9648720e43c20415e462f3e1b96402c | [
"MIT"
] | 5 | 2015-08-28T23:14:33.000Z | 2020-01-02T20:09:03.000Z | 122.617744 | 45,336 | 0.837347 | [
[
[
"#Exploring tabular data with pandas\n\nIn this notebook, we will explore a time series of water levels at the Point Atkinson lighthouse using pandas. This is a basic introduction to pandas and we touch on the following topics:\n\n* Reading a csv file\n* Simple plots\n* Indexing and subsetting\n* DatetimeIndex\n* Grouping\n* Time series methods\n\n \n##Getting started\n\nYou will need to have the python libraries pandas, numpy and matplotlib installed. These are all available through the Anaconda distribution of python.\n\n* https://store.continuum.io/cshop/anaconda/\n\n##Resources\n\nThere is a wealth of information in the pandas documentation.\n\n* http://pandas.pydata.org/pandas-docs/stable/\n\nWater level data (7795-01-JAN-2000_slev.csv) is from Fisheries and Oceans Canada and is available at this website:\n* http://www.isdm-gdsi.gc.ca/isdm-gdsi/twl-mne/index-eng.htm",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\nimport matplotlib.pyplot as plt\nimport datetime\nimport numpy as np\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"##Read the data",
"_____no_output_____"
],
[
"It is helpful to understand the structure of your dataset before attempting to read it with pandas.",
"_____no_output_____"
]
],
[
[
"!head 7795-01-JAN-2000_slev.csv",
"Station_Name,Point Atkinson, B.C.\nStation_Number,7795\nLatitude_Decimal_Degrees,49.337\nLongitude_Decimal_Degrees,123.253\nDatum,CD\nTime_zone,UTC\nSLEV=Observed Water Level\nObs_date,SLEV(metres)\n2000/01/01 08:00,2.95,\n2000/01/01 09:00,3.34,\n"
]
],
[
[
"This dataset contains comma separated values. It has a few rows of metadata (station name, longitude, latitude, etc).The actual data begins with timestamps and water level records at row 9. We can read this data with a pandas function read_csv().\n\nread_csv() has many arguments to help customize the reading of many different csv files. For this file, we will\n* skip the first 8 rows\n* use index_col=False so that the first column is treated as data and not an index\n* tell pandas to read the first column as dates (parse_dates=[0])\n* name the columns as 'date' and 'wlev'.",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('7795-01-JAN-2000_slev.csv', skiprows = 8,\n index_col=False, parse_dates=[0], names=['date','wlev'])",
"_____no_output_____"
]
],
[
[
"data is a DataFrame object",
"_____no_output_____"
]
],
[
[
"type(data)",
"_____no_output_____"
]
],
[
[
"Let's take a quick peak at the dataset.",
"_____no_output_____"
]
],
[
[
"data.head()",
"_____no_output_____"
],
[
"data.tail()",
"_____no_output_____"
],
[
"data.describe()",
"_____no_output_____"
]
],
[
[
"Notice that pandas did not apply the summary statistics to the date column.",
"_____no_output_____"
],
[
"##Simple Plots",
"_____no_output_____"
],
[
"pandas has support for some simple plotting features, like line plots, scatter plots, box plots, etc. For full overview of plots visit http://pandas.pydata.org/pandas-docs/stable/visualization.html\n\nPlotting is really easy. pandas even takes care of labels and legends.",
"_____no_output_____"
]
],
[
[
"data.plot('date','wlev')",
"_____no_output_____"
],
[
"data.plot(kind='hist')",
"_____no_output_____"
],
[
"data.plot(kind='box')",
"_____no_output_____"
]
],
[
[
"##Indexing and Subsetting",
"_____no_output_____"
],
[
"We can index and subset the data in different ways.\n\n###By row number\n\nFor example, grab the first two rows.",
"_____no_output_____"
]
],
[
[
"data[0:2]",
"_____no_output_____"
]
],
[
[
"Note that accessing a single row by the row number doesn't work!",
"_____no_output_____"
]
],
[
[
"data[0]",
"_____no_output_____"
]
],
[
[
"In that case, I would recommend using .iloc or slice for one row. ",
"_____no_output_____"
]
],
[
[
"data.iloc[0]",
"_____no_output_____"
],
[
"data[0:1]",
"_____no_output_____"
]
],
[
[
"### By column\n\nFor example, print the first few lines of the wlev column.",
"_____no_output_____"
]
],
[
[
"data['wlev'].head()",
"_____no_output_____"
]
],
[
[
"### By a condition\n\nFor example, subset the data with date greater than Jan 1, 2008. We pass our condition into the square brackets of data.",
"_____no_output_____"
]
],
[
[
"data_20082009 = data[data['date']>datetime.datetime(2008,1,1)]\ndata_20082009.plot('date','wlev')",
"_____no_output_____"
]
],
[
[
"###Mulitple conditions\n\nFor example, look for extreme water level events. That is, instances where the water level is above 5 m or below 0 m.\n\nDon't forget to put brackets () around each part of the condition.",
"_____no_output_____"
]
],
[
[
"data_extreme = data[(data['wlev']>5) | (data['wlev']<0)]\ndata_extreme.head()",
"_____no_output_____"
]
],
[
[
"### Exercise\n\nWhat was the maximum water level in 2006? \n\nBonus: When?\n\n####Solution",
"_____no_output_____"
],
[
"Isolate the year 2006. Use describe to look up the max water level.",
"_____no_output_____"
]
],
[
[
"data_2006 = data[(data['date']>=datetime.datetime(2006,1,1)) & (data['date'] < datetime.datetime(2007,1,1))]\ndata_2006.describe()",
"_____no_output_____"
]
],
[
[
"The max water level is 5.49m. Use a condition to determine the date.",
"_____no_output_____"
]
],
[
[
"date_max = data_2006[data_2006['wlev']==5.49]['date']\nprint date_max",
"53399 2006-02-04 17:00:00\nName: date, dtype: datetime64[ns]\n"
]
],
[
[
"##Manipulating dates",
"_____no_output_____"
],
[
"In the above example, it would have been convenient if we could access only the year part of the time stamp. But this doesn't work:",
"_____no_output_____"
]
],
[
[
"data['date'].year",
"_____no_output_____"
]
],
[
[
"We can use the pandas DatetimeIndex class to make this work. The DatetimeIndex allows us to easily access properties, like year, month, and day of each timestamp. We will use this to add new Year, Month, Day, Hour and DayOfYear columns to the dataframe.",
"_____no_output_____"
]
],
[
[
"date_index = pd.DatetimeIndex(data['date'])\nprint date_index",
"DatetimeIndex(['2000-01-01 08:00:00', '2000-01-01 09:00:00',\n '2000-01-01 10:00:00', '2000-01-01 11:00:00',\n '2000-01-01 12:00:00', '2000-01-01 13:00:00',\n '2000-01-01 14:00:00', '2000-01-01 15:00:00',\n '2000-01-01 16:00:00', '2000-01-01 17:00:00', \n ...\n '2009-12-30 23:00:00', '2009-12-31 00:00:00',\n '2009-12-31 01:00:00', '2009-12-31 02:00:00',\n '2009-12-31 03:00:00', '2009-12-31 04:00:00',\n '2009-12-31 05:00:00', '2009-12-31 06:00:00',\n '2009-12-31 07:00:00', '2009-12-31 08:00:00'],\n dtype='datetime64[ns]', length=87608, freq=None, tz=None)\n"
],
[
"data['Day'] = date_index.day\ndata['Month'] = date_index.month\ndata['Year'] = date_index.year\ndata['Hour'] = date_index.hour\ndata['DayOfYear'] = date_index.dayofyear",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"data.describe()",
"_____no_output_____"
]
],
[
[
"Notice that now pandas applies the describe function to these new columns because it sees them as numerical data.\n\nNow, we can access a single year with a simpler conditional.",
"_____no_output_____"
]
],
[
[
"data_2006 = data[data['Year']==2006]\ndata_2006.head()",
"_____no_output_____"
]
],
[
[
"##Grouping",
"_____no_output_____"
],
[
"Sometimes, it is convenient to group data with similar characteristics. We can do this with the groupby() method.\n\nFor example, we might want to group by year.",
"_____no_output_____"
]
],
[
[
"data_annual = data.groupby(['Year'])\ndata_annual['wlev'].describe().head(20)",
"_____no_output_____"
]
],
[
[
"Now the data is organized into groups based on the year of the observation.\n\n###Aggregating\n\nOnce the data is grouped, we may want to summarize it in some way. We can do this with the apply() function. The argument of apply() is a function that we want to apply to each group. For example, we may want to calculate the mean sea level of each year.",
"_____no_output_____"
]
],
[
[
"annual_means = data_annual['wlev'].apply(np.mean)\nprint annual_means",
"Year\n2000 3.067434\n2001 3.057653\n2002 3.078112\n2003 3.112990\n2004 3.104097\n2005 3.127036\n2006 3.142052\n2007 3.095614\n2008 3.070757\n2009 3.080533\nName: wlev, dtype: float64\n"
]
],
[
[
"It is also really easy to plot the aggregated data.",
"_____no_output_____"
]
],
[
[
"annual_means.plot()",
"_____no_output_____"
]
],
[
[
"### Multiple aggregations\n\nWe may also want to apply multiple aggregations, like the mean, max, and min. We can do this with the agg() method and pass a list of aggregation functions as the argument.",
"_____no_output_____"
]
],
[
[
"annual_summary = data_annual['wlev'].agg([np.mean,np.max,np.min])\nprint annual_summary",
" mean amax amin\nYear \n2000 3.067434 5.11 0.07\n2001 3.057653 5.24 -0.07\n2002 3.078112 5.25 0.14\n2003 3.112990 5.45 -0.09\n2004 3.104097 5.14 -0.06\n2005 3.127036 5.43 0.00\n2006 3.142052 5.49 -0.01\n2007 3.095614 5.20 -0.14\n2008 3.070757 5.19 -0.12\n2009 3.080533 5.20 -0.20\n"
],
[
"annual_summary.plot()",
"_____no_output_____"
]
],
[
[
"### Iterating over groups\nIn some instances, we may want to iterate over each group. Each group is identifed by a key. If we know the group's key, then we can access that group with the get_group() method. \n\nFor example, for each year print the mean sea level.",
"_____no_output_____"
]
],
[
[
"for year in data_annual.groups.keys():\n data_year = data_annual.get_group(year)\n print year, data_year['wlev'].mean()",
"2000 3.06743417303\n2001 3.05765296804\n2002 3.07811187215\n2003 3.11298972603\n2004 3.1040974832\n2005 3.12703618873\n2006 3.14205230699\n2007 3.0956142955\n2008 3.07075714448\n2009 3.08053287593\n"
]
],
[
[
"We had calculated the annual mean sea level earlier, but this is another way to achieve a similar result.\n\n###Exercise\n\nFor each year, plot the monthly mean water level.\n\n####Solution",
"_____no_output_____"
]
],
[
[
"for year in data_annual.groups.keys():\n data_year = data_annual.get_group(year)\n month_mean = data_year.groupby('Month')['wlev'].apply(np.mean)\n month_mean.plot(label=year)\nplt.legend()\n",
"_____no_output_____"
]
],
[
[
"###Multiple groups\n\nWe can also group by multiple columns. For example, we might want to group by year and month. That is, a year/month combo defines the group.",
"_____no_output_____"
]
],
[
[
"data_yearmonth = data.groupby(['Year','Month'])\n\nmeans = data_yearmonth['wlev'].apply(np.mean)\nmeans.plot()\n",
"_____no_output_____"
]
],
[
[
"##Time Series\n\nThe x-labels on the plot above are a little bit awkward. A different approach would be to resample the data at a monthly freqeuncy. This can be accomplished by setting the date column as an index. Then we can resample the data at a desired frequency. The resampling method is flexible but a common choice is the average.\n\nFirst, we will need to set the index as a DatetimeIndex. Recall, the date_index variable we had assigned earlier. We will add this to the dataframe and make it into the dataframe index.",
"_____no_output_____"
]
],
[
[
"data['date_index'] = date_index\ndata.set_index('date_index', inplace=True)",
"_____no_output_____"
]
],
[
[
"Now we can resample at a monthly frequency and plot.",
"_____no_output_____"
]
],
[
[
"data_monthly = data['wlev'].resample('M', how='mean')\ndata_monthly.plot()",
"_____no_output_____"
]
],
[
[
"##Summary\n\npandas is a poweful tool for manipulating tabular data. There are many, many other features that were not discussed here. See the documentation for more features.\n\nhttp://pandas.pydata.org/pandas-docs/stable/index.html",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e7f680d8fc1f35bf7503a862df28a5d6abcd5d2f | 4,961 | ipynb | Jupyter Notebook | doc0/Exercise09/Exercise09.ipynb | nsingh216/edu | 95cbd3dc25c9b15160ba4b146b52eeeb2fa54ecb | [
"Apache-2.0"
] | null | null | null | doc0/Exercise09/Exercise09.ipynb | nsingh216/edu | 95cbd3dc25c9b15160ba4b146b52eeeb2fa54ecb | [
"Apache-2.0"
] | null | null | null | doc0/Exercise09/Exercise09.ipynb | nsingh216/edu | 95cbd3dc25c9b15160ba4b146b52eeeb2fa54ecb | [
"Apache-2.0"
] | null | null | null | 20.585062 | 177 | 0.50131 | [
[
[
"# Docker Exercise 09\n\n### Getting started with Docker Swarms\n\nMake sure that Swarm is enabled on your Docker Desktop by typing `docker system info`, and looking for a message `Swarm: active` (you might have to scroll up a little).\n\nIf Swarm isn't running, simply type `docker swarm init` in a shell prompt to set it up.\n\n\n### Create the networks:",
"_____no_output_____"
]
],
[
[
"docker network create --driver overlay --subnet=172.10.1.0/24 ex09-frontend\n\ndocker network create --driver overlay --subnet=172.10.2.0/23 ex09-backend",
"_____no_output_____"
]
],
[
[
"### Save the MySQL configuration\n\nSave the following to your `development.env` file.",
"_____no_output_____"
]
],
[
[
"MYSQL_USER=sys_admin\nMYSQL_PASSWORD=sys_password\nMYSQL_ROOT_PASSWORD=root_password",
"_____no_output_____"
]
],
[
[
"### Create your Docker Swarm configuration",
"_____no_output_____"
]
],
[
[
"version: \"3\"\n\nnetworks:\n ex09-frontend:\n external: true\n ex09-backend:\n external: true\n\nservices:\n\n ex09-db:\n image: mysql:8.0\n command: --default-authentication-plugin=mysql_native_password\n ports:\n - \"3306:3306\"\n networks:\n - ex09-backend\n env_file:\n - ./development.env\n\n ex09-www:\n image: dockerjames85/php-mysqli-apache:1.1\n ports:\n - \"8080:80\"\n networks:\n - ex09-backend\n - ex09-frontend\n depends_on:\n - ex09-db\n env_file:\n - ./development.env\n deploy:\n replicas: 5\n resources:\n limits:\n cpus: \"0.1\"\n memory: 100M\n restart_policy:\n condition: on-failure\n``` \n\n### Deploy the stack\n",
"_____no_output_____"
]
],
[
[
"docker stack deploy -c php-mysqli-apache.yml php-mysqli-apache",
"_____no_output_____"
]
],
[
[
"\n### Veify the stack has been deployed\n",
"_____no_output_____"
]
],
[
[
"docker stack ls",
"_____no_output_____"
]
],
[
[
"\n### Verify all the containers have been deployed\n",
"_____no_output_____"
]
],
[
[
"docker stack ps php-mysqli-apache",
"_____no_output_____"
]
],
[
[
"\n### Verify the load balancers have all the replicas and mapped the ports\n",
"_____no_output_____"
]
],
[
[
"docker stack services php-mysqli-apache",
"_____no_output_____"
]
],
[
[
"\n### See what containers are on the nodemanager in the swarm\n",
"_____no_output_____"
]
],
[
[
"docker ps",
"_____no_output_____"
]
],
[
[
"\n### Verify that the stack is working correctly\n",
"_____no_output_____"
]
],
[
[
"# local node master\ncurl http://localhost:8080",
"_____no_output_____"
]
],
[
[
"\n### Destory and remove the stack",
"_____no_output_____"
]
],
[
[
"docker stack rm php-mysqli-apache\n```",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e7f6893ee0d9596dc33cd046bd937cd731f3bf13 | 36,664 | ipynb | Jupyter Notebook | notebooks/0.2.0-whs-YOLO_etl.ipynb | whs2k/ImageComparison | 32d6f72680306cf376ff59975fa0349ae2d75b26 | [
"MIT"
] | null | null | null | notebooks/0.2.0-whs-YOLO_etl.ipynb | whs2k/ImageComparison | 32d6f72680306cf376ff59975fa0349ae2d75b26 | [
"MIT"
] | null | null | null | notebooks/0.2.0-whs-YOLO_etl.ipynb | whs2k/ImageComparison | 32d6f72680306cf376ff59975fa0349ae2d75b26 | [
"MIT"
] | null | null | null | 92.352645 | 3,280 | 0.636074 | [
[
[
"import glob2\nimport mxnet as mx\nfrom gluoncv import model_zoo, data, utils\nimport os\nfrom matplotlib import pyplot as plt\nfrom tqdm import tqdm \nimport traceback\nimport pandas as pd",
"_____no_output_____"
],
[
"def dogCrop(fn_to_crop):\n x, img = data.transforms.presets.yolo.load_test(fn_to_crop, short=512)\n class_IDs, scores, bounding_boxs = net(x)\n for index, bbox in enumerate(bounding_boxs[0]):\n class_ID = int(class_IDs[0][index].asnumpy()[0])\n class_name = net.classes[class_ID]\n class_score = scores[0][index].asnumpy()\n if (class_name == 'dog') & (class_score > 0.9):\n #print('index: ', index)\n #print('class_ID: ', class_ID)\n #print('class_name: ', class_name)\n #print('class_score: ',class_score)\n #print('bbox: ', bbox.asnumpy())\n xmin, ymin, xmax, ymax = [int(x) for x in bbox.asnumpy()]\n xmin = max(0, xmin)\n xmax = min(x.shape[3], xmax)\n ymin = max(0, ymin)\n ymax = min(x.shape[2], ymax)\n im_fname_save = fn.replace('.jpg','_dogCrop.jpg') \\\n .replace('.jpeg','_dogCrop.jpeg') \\\n .replace('.png','_dogCrop.png')\n plt.imsave(im_fname_save, img[ymin:ymax,xmin:xmax,:])",
"_____no_output_____"
],
[
"%time net = model_zoo.get_model('yolo3_darknet53_voc', pretrained=True)",
"CPU times: user 461 ms, sys: 299 ms, total: 760 ms\nWall time: 920 ms\n"
],
[
"fns = glob2.glob(os.path.join(os.path.dirname(os.getcwd()), 'data', 'raw','**/*.*'))\nfor fn in fns:\n if 'png' in fn:\n print(fn)\ndf = pd.DataFrame()\ndf['fn_dog'] = fns\ndf.tail()",
"/Users/officialbiznas/Documents/GitHub/ImageSimilarity/data/raw/img.png\n"
],
[
"%time df['fn_dog'].apply(lambda x: dogCrop(x))",
"_____no_output_____"
],
[
"for fn in tqdm(fns):\n try:\n if ('_dogCrop.jpg' in fn) | (fn.replace('.jpg','_dogCrop.jpg') in fns):\n continue\n x, img = data.transforms.presets.yolo.load_test(fn, short=512)\n class_IDs, scores, bounding_boxs = net(x)\n for index, bbox in enumerate(bounding_boxs[0]):\n class_ID = int(class_IDs[0][index].asnumpy()[0])\n class_name = net.classes[class_ID]\n class_score = scores[0][index].asnumpy()\n if (class_name == 'dog') & (class_score > 0.9):\n #print('index: ', index)\n #print('class_ID: ', class_ID)\n #print('class_name: ', class_name)\n #print('class_score: ',class_score)\n #print('bbox: ', bbox.asnumpy())\n xmin, ymin, xmax, ymax = [int(x) for x in bbox.asnumpy()]\n xmin = max(0, xmin)\n xmax = min(x.shape[3], xmax)\n ymin = max(0, ymin)\n ymax = min(x.shape[2], ymax)\n im_fname_save = fn.replace('.jpg','_dogCrop.jpg')\n plt.imsave(im_fname_save, img[ymin:ymax,xmin:xmax,:])\n break\n except Exception as e:\n print(fn)\n print(x.shape)\n print(xmin, ymin, xmax, ymax)\n print(traceback.print_exc())",
" 17%|█▋ | 3206/18855 [1:20:26<12:26:34, 2.86s/it]Traceback (most recent call last):\n File \"<ipython-input-6-3b683c17df7d>\", line 5, in <module>\n x, img = data.transforms.presets.yolo.load_test(fn, short=512)\n File \"/Users/officialbiznas/anaconda3/envs/dog/lib/python3.6/site-packages/gluoncv/data/transforms/presets/yolo.py\", line 99, in load_test\n imgs = [mx.image.imread(f) for f in filenames]\n File \"/Users/officialbiznas/anaconda3/envs/dog/lib/python3.6/site-packages/gluoncv/data/transforms/presets/yolo.py\", line 99, in <listcomp>\n imgs = [mx.image.imread(f) for f in filenames]\n File \"/Users/officialbiznas/anaconda3/envs/dog/lib/python3.6/site-packages/mxnet/image/image.py\", line 85, in imread\n return _internal._cvimread(filename, *args, **kwargs)\n File \"<string>\", line 35, in _cvimread\n File \"/Users/officialbiznas/anaconda3/envs/dog/lib/python3.6/site-packages/mxnet/_ctypes/ndarray.py\", line 92, in _imperative_invoke\n ctypes.byref(out_stypes)))\n File \"/Users/officialbiznas/anaconda3/envs/dog/lib/python3.6/site-packages/mxnet/base.py\", line 252, in check_call\n raise MXNetError(py_str(_LIB.MXGetLastError()))\nmxnet.base.MXNetError: [19:39:37] src/io/image_io.cc:146: Check failed: !res.empty() Decoding failed. Invalid image file.\n\nStack trace returned 8 entries:\n[bt] (0) 0 libmxnet.so 0x000000010dca0c90 std::__1::__tree<std::__1::__value_type<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, mxnet::NDArrayFunctionReg*>, std::__1::__map_value_compare<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, std::__1::__value_type<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, mxnet::NDArrayFunctionReg*>, std::__1::less<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> > >, true>, std::__1::allocator<std::__1::__value_type<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, mxnet::NDArrayFunctionReg*> > >::destroy(std::__1::__tree_node<std::__1::__value_type<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, mxnet::NDArrayFunctionReg*>, void*>*) + 2736\n[bt] (1) 1 libmxnet.so 0x000000010dca0a3f std::__1::__tree<std::__1::__value_type<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, mxnet::NDArrayFunctionReg*>, std::__1::__map_value_compare<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, std::__1::__value_type<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, mxnet::NDArrayFunctionReg*>, std::__1::less<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> > >, true>, std::__1::allocator<std::__1::__value_type<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, mxnet::NDArrayFunctionReg*> > >::destroy(std::__1::__tree_node<std::__1::__value_type<std::__1::basic_string<char, std::__1::char_traits<char>, std::__1::allocator<char> >, mxnet::NDArrayFunctionReg*>, void*>*) + 2143\n[bt] (2) 2 libmxnet.so 0x000000010f361296 mxnet::io::ImdecodeImpl(int, bool, void*, unsigned long, mxnet::NDArray*) + 3734\n[bt] (3) 3 libmxnet.so 0x000000010f36315b mxnet::io::Imread(nnvm::NodeAttrs const&, std::__1::vector<mxnet::NDArray, std::__1::allocator<mxnet::NDArray> > const&, std::__1::vector<mxnet::NDArray, std::__1::allocator<mxnet::NDArray> >*) + 2843\n[bt] (4) 4 libmxnet.so 0x000000010f339d60 mxnet::Imperative::Invoke(mxnet::Context const&, nnvm::NodeAttrs const&, std::__1::vector<mxnet::NDArray*, std::__1::allocator<mxnet::NDArray*> > const&, std::__1::vector<mxnet::NDArray*, std::__1::allocator<mxnet::NDArray*> > const&) + 320\n[bt] (5) 5 libmxnet.so 0x000000010f285d9e SetNDInputsOutputs(nnvm::Op const*, std::__1::vector<mxnet::NDArray*, std::__1::allocator<mxnet::NDArray*> >*, std::__1::vector<mxnet::NDArray*, std::__1::allocator<mxnet::NDArray*> >*, int, void* const*, int*, int, int, void***) + 1774\n[bt] (6) 6 libmxnet.so 0x000000010f286ac0 MXImperativeInvokeEx + 176\n[bt] (7) 7 libffi.6.dylib 0x0000000106769884 ffi_call_unix64 + 76\n\n\n"
],
[
"fns = glob2.glob(os.path.join(os.path.dirname(os.getcwd()), 'data', 'raw','**/*_dogCrop.jpg'))\nprint('_dogCropFns: ', len(fns))\nfor fn in fns[0:3]:\n print(fn)",
"_dogCropFns: 13732\n/Users/officialbiznas/Documents/GitHub/ImageSimilarity/data/raw/setter-irish/n02100877_8900_dogCrop.jpg\n/Users/officialbiznas/Documents/GitHub/ImageSimilarity/data/raw/setter-irish/n02100877_257_dogCrop.jpg\n/Users/officialbiznas/Documents/GitHub/ImageSimilarity/data/raw/setter-irish/n02100877_5229_dogCrop.jpg\n"
],
[
"fns = glob2.glob(os.path.join(os.path.dirname(os.getcwd()), 'data', 'raw','**/*.jpg'))\nprint('_dogCropFns: ', len(fns))\nfor fn in fns[0:3]:\n print(fn)",
"_dogCropFns: 31869\n/Users/officialbiznas/Documents/GitHub/ImageSimilarity/data/raw/setter-irish/n02100877_8900_dogCrop.jpg\n/Users/officialbiznas/Documents/GitHub/ImageSimilarity/data/raw/setter-irish/n02100877_257_dogCrop.jpg\n/Users/officialbiznas/Documents/GitHub/ImageSimilarity/data/raw/setter-irish/n02100877_1913.jpg\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f68a29cc48479b9696abfdc916406df528fb7a | 10,082 | ipynb | Jupyter Notebook | virion2replication.ipynb | pc4covid19/virion2replication | c8b72abb57d77a7a1a00ca41dde97eb88b61155b | [
"BSD-3-Clause"
] | null | null | null | virion2replication.ipynb | pc4covid19/virion2replication | c8b72abb57d77a7a1a00ca41dde97eb88b61155b | [
"BSD-3-Clause"
] | null | null | null | virion2replication.ipynb | pc4covid19/virion2replication | c8b72abb57d77a7a1a00ca41dde97eb88b61155b | [
"BSD-3-Clause"
] | null | null | null | 31.905063 | 140 | 0.403293 | [
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import sys, os\nsys.path.insert(0, os.path.abspath('bin'))\nimport virion2replication",
"_____no_output_____"
],
[
"virion2replication.gui",
"_____no_output_____"
],
[
"#from debug import debug_view\n#debug_view",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
e7f692c1b746c71b3b1a87824d486c1c8b779d6f | 295,541 | ipynb | Jupyter Notebook | notebooks/EDA.ipynb | wdy06/kaggle-data-science-bowl-2019 | 645d690595fccc4a130cd435aef536c3af2e9045 | [
"MIT"
] | null | null | null | notebooks/EDA.ipynb | wdy06/kaggle-data-science-bowl-2019 | 645d690595fccc4a130cd435aef536c3af2e9045 | [
"MIT"
] | null | null | null | notebooks/EDA.ipynb | wdy06/kaggle-data-science-bowl-2019 | 645d690595fccc4a130cd435aef536c3af2e9045 | [
"MIT"
] | null | null | null | 94.846277 | 54,096 | 0.761769 | [
[
[
"%load_ext autoreload\n%autoreload 2\nfrom IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity='all'",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nfrom pathlib import Path\nimport json\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\nimport pprint",
"_____no_output_____"
],
[
"DATA_DIR = Path('../data/original/')",
"_____no_output_____"
],
[
"specs_df = pd.read_csv(DATA_DIR / 'specs.csv')\ntrain_df = pd.read_csv(DATA_DIR / 'train.csv')\ntrain_labels_df = pd.read_csv(DATA_DIR / 'train_labels.csv')\ntest_df = pd.read_csv(DATA_DIR / 'test.csv')",
"_____no_output_____"
]
],
[
[
"## specs",
"_____no_output_____"
]
],
[
[
"spces_df.head()",
"_____no_output_____"
],
[
"specs_df.shape",
"_____no_output_____"
],
[
"specs_df.describe()",
"_____no_output_____"
],
[
"specs_df['info'][0]",
"_____no_output_____"
],
[
"json.loads(specs_df['args'][0])",
"_____no_output_____"
],
[
"specs_df['info'][3]",
"_____no_output_____"
],
[
"json.loads(specs_df['args'][3])",
"_____no_output_____"
]
],
[
[
"## train",
"_____no_output_____"
]
],
[
[
"train_df.head()",
"_____no_output_____"
],
[
"train_df.shape",
"_____no_output_____"
],
[
"train_df.describe()",
"_____no_output_____"
],
[
"train_df.event_id.nunique()",
"_____no_output_____"
],
[
"train_df.game_session.nunique()",
"_____no_output_____"
],
[
"train_df.timestamp.min()\ntrain_df.timestamp.max()",
"_____no_output_____"
],
[
"train_df.installation_id.nunique()",
"_____no_output_____"
],
[
"train_df.event_count.nunique()",
"_____no_output_____"
],
[
"sns.distplot(train_df.event_count, )",
"_____no_output_____"
],
[
"sns.distplot(np.log(train_df.event_count))",
"_____no_output_____"
],
[
"sns.distplot(train_df.game_time)",
"_____no_output_____"
],
[
"sns.distplot(np.log1p(train_df.game_time))",
"_____no_output_____"
],
[
"train_df.title.value_counts().plot(kind='bar')",
"_____no_output_____"
],
[
"sns.countplot(y='title', data=train_df, order=train_df.title.value_counts().index)",
"_____no_output_____"
],
[
"sns.countplot(x='type', data=train_df)",
"_____no_output_____"
],
[
"sns.countplot(x='world', data=train_df)",
"_____no_output_____"
],
[
"train_df.groupby(['title', 'type', 'world'])['event_id'].count().sort_values(ascending=False)",
"_____no_output_____"
],
[
"train_df.query('game_session==\"901acc108f55a5a1\" & event_code==4100')",
"_____no_output_____"
],
[
"train_df.query('game_session == \"0848ef14a8dc6892\"')",
"_____no_output_____"
]
],
[
[
"## train labels\n",
"_____no_output_____"
]
],
[
[
"train_labels_df.head()",
"_____no_output_____"
],
[
"train_labels_df.shape",
"_____no_output_____"
],
[
"train_labels_df.game_session.nunique()",
"_____no_output_____"
],
[
"train_labels_df.installation_id.nunique()",
"_____no_output_____"
],
[
"train_labels_df.query('game_session == \"0848ef14a8dc6892\"')",
"_____no_output_____"
]
],
[
[
"## test",
"_____no_output_____"
]
],
[
[
"test_df.head()",
"_____no_output_____"
],
[
"test_df.shape",
"_____no_output_____"
],
[
"test_df.event_id.nunique()",
"_____no_output_____"
],
[
"test_df.game_session.nunique()",
"_____no_output_____"
],
[
"test_df.installation_id.nunique()",
"_____no_output_____"
],
[
"test_df.title.unique()",
"_____no_output_____"
],
[
"len(test_df.query('~(title==\"Bird Measurer (Assessment)\") & event_code==4100'))",
"_____no_output_____"
],
[
"len(test_df.query('title==\"Bird Measurer (Assessment)\" & event_code==4110'))",
"_____no_output_____"
],
[
"test_df.query('installation_id == \"00abaee7\" & event_code==4100')",
"_____no_output_____"
]
],
[
[
"## sample submission",
"_____no_output_____"
]
],
[
[
"sample_submission = pd.read_csv(DATA_DIR / 'sample_submission.csv')",
"_____no_output_____"
],
[
"sample_submission.head()",
"_____no_output_____"
],
[
"sample_submission.shape",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e7f69978942816038e67d45d8687834e04908ed3 | 31,998 | ipynb | Jupyter Notebook | udacity-project.ipynb | abhiojha8/Optimizing_ML_Pipeline_Azure | 2c1fe146f9b547818fe762f469626b55aeece614 | [
"MIT"
] | null | null | null | udacity-project.ipynb | abhiojha8/Optimizing_ML_Pipeline_Azure | 2c1fe146f9b547818fe762f469626b55aeece614 | [
"MIT"
] | null | null | null | udacity-project.ipynb | abhiojha8/Optimizing_ML_Pipeline_Azure | 2c1fe146f9b547818fe762f469626b55aeece614 | [
"MIT"
] | null | null | null | 61.064885 | 7,114 | 0.618726 | [
[
[
"from azureml.core import Workspace, Experiment\n\nws = Workspace.from_config()\nexp = Experiment(workspace=ws, name=\"p1\")\n\nprint('Workspace name: ' + ws.name, \n 'Azure region: ' + ws.location, \n 'Subscription id: ' + ws.subscription_id, \n 'Resource group: ' + ws.resource_group, sep = '\\n')\n\nrun = exp.start_logging()",
"Workspace name: quick-starts-ws-142925\nAzure region: southcentralus\nSubscription id: f9d5a085-54dc-4215-9ba6-dad5d86e60a0\nResource group: aml-quickstarts-142925\n"
]
],
[
[
"### (Optional) Cancel existing runs",
"_____no_output_____"
]
],
[
[
"for run in exp.get_runs():\n print(run.id)\n if run.status==\"Running\":\n run.cancel()",
"HD_93afb055-83d9-41a7-9c50-e9a7e14cd79d\nHD_a78b0e0e-008f-4804-9296-85be82ed1739\n7dcaf971-c29d-4f68-b72e-0815db91a92d\n"
],
[
"from azureml.core.compute import ComputeTarget, AmlCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\ncluster_name = \"udacityAzureML\"\n\ntry:\n compute_target = ComputeTarget(workspace=ws, name=cluster_name)\n print('Found existing compute target')\nexcept ComputeTargetException:\n print('Creating a new compute target...')\n compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D2_V2', \n max_nodes=4)\n\n # create the cluster\n compute_target = ComputeTarget.create(ws, cluster_name, compute_config)\n\ncompute_target.wait_for_completion(show_output=True)\n\nprint(compute_target.get_status().serialize())",
"Found existing compute target\n\nRunning\n{'errors': [], 'creationTime': '2021-04-17T10:57:19.241877+00:00', 'createdBy': {'userObjectId': 'f3c9ec0e-655a-4344-8b94-baf1544d5426', 'userTenantId': '660b3398-b80e-49d2-bc5b-ac1dc93b5254', 'userName': 'ODL_User 142925'}, 'modifiedTime': '2021-04-17T10:59:36.333182+00:00', 'state': 'Running', 'vmSize': 'STANDARD_DS2_V2'}\n"
],
[
"from azureml.widgets import RunDetails\nfrom azureml.train.sklearn import SKLearn\nfrom azureml.train.hyperdrive.run import PrimaryMetricGoal\nfrom azureml.train.hyperdrive.policy import BanditPolicy\nfrom azureml.train.hyperdrive.sampling import RandomParameterSampling\nfrom azureml.train.hyperdrive.runconfig import HyperDriveConfig\nfrom azureml.train.hyperdrive.parameter_expressions import uniform\nfrom azureml.train.hyperdrive.parameter_expressions import choice, uniform\nimport os\n\nps = RandomParameterSampling(\n {\n '--C' : choice(0.001,0.01,0.1,1,10,100),\n '--max_iter': choice(50,100,200)\n }\n)\n\n# Specify a Policy\npolicy = BanditPolicy(evaluation_interval=2, slack_factor=0.1)\n\nif \"training\" not in os.listdir():\n os.mkdir(\"./training\")\n\n# Create a SKLearn estimator for use with train.py\nest = SKLearn(source_directory = \"./\",\n compute_target=compute_target,\n vm_size='STANDARD_D2_V2',\n entry_script=\"train.py\")\n\n# Create a HyperDriveConfig using the estimator, hyperparameter sampler, and policy.\nhyperdrive_config = HyperDriveConfig(hyperparameter_sampling=ps, \n primary_metric_name='Accuracy',\n primary_metric_goal=PrimaryMetricGoal.MAXIMIZE,\n policy=policy,\n estimator=est,\n max_total_runs=20,\n max_concurrent_runs=4)",
"'SKLearn' estimator is deprecated. Please use 'ScriptRunConfig' from 'azureml.core.script_run_config' with your own defined environment or the AzureML-Tutorial curated environment.\n'enabled' is deprecated. Please use the azureml.core.runconfig.DockerConfiguration object with the 'use_docker' param instead.\n"
],
[
"# Submit your hyperdrive run to the experiment and show run details with the widget.\n\nhyperdrive_run = exp.submit(hyperdrive_config)\nhyperdrive_run.wait_for_completion(show_output=True)\nassert(hyperdrive_run.get_status() == \"Completed\")",
"RunId: HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074\nWeb View: https://ml.azure.com/runs/HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074?wsid=/subscriptions/f9d5a085-54dc-4215-9ba6-dad5d86e60a0/resourcegroups/aml-quickstarts-142925/workspaces/quick-starts-ws-142925&tid=660b3398-b80e-49d2-bc5b-ac1dc93b5254\n\nStreaming azureml-logs/hyperdrive.txt\n=====================================\n\n\"<START>[2021-04-17T11:37:19.465848][API][INFO]Experiment created<END>\\n\"\"<START>[2021-04-17T11:37:20.345133][GENERATOR][INFO]Trying to sample '4' jobs from the hyperparameter space<END>\\n\"\"<START>[2021-04-17T11:37:20.678429][GENERATOR][INFO]Successfully sampled '4' jobs, they will soon be submitted to the execution target.<END>\\n\"<START>[2021-04-17T11:37:20.8164067Z][SCHEDULER][INFO]The execution environment is being prepared. Please be patient as it can take a few minutes.<END>\n\nExecution Summary\n=================\nRunId: HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074\nWeb View: https://ml.azure.com/runs/HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074?wsid=/subscriptions/f9d5a085-54dc-4215-9ba6-dad5d86e60a0/resourcegroups/aml-quickstarts-142925/workspaces/quick-starts-ws-142925&tid=660b3398-b80e-49d2-bc5b-ac1dc93b5254\n\n"
],
[
"import joblib\nbest_run = hyperdrive_run.get_best_run_by_primary_metric()\n\nprint(\"Best run metrics :\",best_run.get_metrics())\nprint(\"Best run details :\",best_run.get_details())\nprint(\"Best run file names :\",best_run.get_file_names())\n\nmodel = best_run.register_model(model_name='best_model', \n model_path='outputs/model.joblib')",
"Best run metrics : {'Regularization Strength:': 0.01, 'Max iterations:': 200, 'Accuracy': 0.9121396054628225}\nBest run details : {'runId': 'HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8', 'target': 'udacityAzureML', 'status': 'Completed', 'startTimeUtc': '2021-04-17T11:41:34.499205Z', 'endTimeUtc': '2021-04-17T11:42:23.823768Z', 'properties': {'_azureml.ComputeTargetType': 'amlcompute', 'ContentSnapshotId': 'ca7ce0c5-eaa5-407a-8367-18a958fd6a74', 'ProcessInfoFile': 'azureml-logs/process_info.json', 'ProcessStatusFile': 'azureml-logs/process_status.json'}, 'inputDatasets': [], 'outputDatasets': [], 'runDefinition': {'script': 'train.py', 'command': '', 'useAbsolutePath': False, 'arguments': ['--C', '0.01', '--max_iter', '200'], 'sourceDirectoryDataStore': None, 'framework': 'Python', 'communicator': 'None', 'target': 'udacityAzureML', 'dataReferences': {}, 'data': {}, 'outputData': {}, 'jobName': None, 'maxRunDurationSeconds': None, 'nodeCount': 1, 'priority': None, 'credentialPassthrough': False, 'identity': None, 'environment': {'name': 'Experiment p1 Environment', 'version': 'Autosave_2021-04-17T11:16:10Z_56ce6ab9', 'python': {'interpreterPath': 'python', 'userManagedDependencies': False, 'condaDependencies': {'channels': ['anaconda', 'conda-forge'], 'dependencies': ['python=3.6.2', {'pip': ['azureml-defaults', 'scikit-learn==0.20.3', 'scipy==1.2.1', 'joblib==0.13.2']}], 'name': 'azureml_ba9520bf386d662001eeb9523395794e'}, 'baseCondaEnvironment': None}, 'environmentVariables': {'EXAMPLE_ENV_VAR': 'EXAMPLE_VALUE'}, 'docker': {'baseImage': 'mcr.microsoft.com/azureml/intelmpi2018.3-ubuntu16.04:20200423.v1', 'platform': {'os': 'Linux', 'architecture': 'amd64'}, 'baseDockerfile': None, 'baseImageRegistry': {'address': None, 'username': None, 'password': None}, 'enabled': True, 'arguments': []}, 'spark': {'repositories': [], 'packages': [], 'precachePackages': False}, 'inferencingStackVersion': None}, 'history': {'outputCollection': True, 'directoriesToWatch': ['logs'], 'enableMLflowTracking': True, 'snapshotProject': True}, 'spark': {'configuration': {'spark.app.name': 'Azure ML Experiment', 'spark.yarn.maxAppAttempts': '1'}}, 'parallelTask': {'maxRetriesPerWorker': 0, 'workerCountPerNode': 1, 'terminalExitCodes': None, 'configuration': {}}, 'amlCompute': {'name': None, 'vmSize': 'STANDARD_D2_V2', 'retainCluster': False, 'clusterMaxNodeCount': 1}, 'aiSuperComputer': {'instanceType': None, 'imageVersion': None, 'location': None, 'aiSuperComputerStorageData': None, 'interactive': False, 'scalePolicy': None, 'virtualClusterArmId': None, 'tensorboardLogDirectory': None}, 'tensorflow': {'workerCount': 1, 'parameterServerCount': 1}, 'mpi': {'processCountPerNode': 1}, 'pyTorch': {'communicationBackend': 'nccl', 'processCount': None}, 'hdi': {'yarnDeployMode': 'Cluster'}, 'containerInstance': {'region': None, 'cpuCores': 2.0, 'memoryGb': 3.5}, 'exposedPorts': None, 'docker': {'useDocker': False, 'sharedVolumes': True, 'shmSize': '2g', 'arguments': []}, 'cmk8sCompute': {'configuration': {}}, 'commandReturnCodeConfig': {'returnCode': 'Zero', 'successfulReturnCodes': []}, 'environmentVariables': {}}, 'logFiles': {'azureml-logs/55_azureml-execution-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/azureml-logs/55_azureml-execution-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt?sv=2019-02-02&sr=b&sig=Q4LYoRgdHpevhrsppAFT0pdr1rOMSoLR9m26Zr6%2BAQI%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'azureml-logs/65_job_prep-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/azureml-logs/65_job_prep-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt?sv=2019-02-02&sr=b&sig=dZ9jo7ozoLbAWrgjcWZ%2BJRIDMFYpzm%2BjrsEAlZMouDU%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'azureml-logs/70_driver_log.txt': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/azureml-logs/70_driver_log.txt?sv=2019-02-02&sr=b&sig=9DXh%2Bb749L3zGOsbeIRvbG8epOevx2uQxkFNpeUyFRk%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'azureml-logs/75_job_post-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/azureml-logs/75_job_post-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt?sv=2019-02-02&sr=b&sig=HydjEcqwjb0rR6BSi3zcvPzuQNi97%2Fk7vqkTTo899Lc%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'azureml-logs/process_info.json': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/azureml-logs/process_info.json?sv=2019-02-02&sr=b&sig=HtnCBYFG76ziXQGmOqz23nTd7tEu%2BTLYq2nBtyhPDKQ%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'azureml-logs/process_status.json': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/azureml-logs/process_status.json?sv=2019-02-02&sr=b&sig=Lwxd3nbF13ErWS9pjz%2FNPlohbh0bjavjtzR0CEVPbvQ%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'logs/azureml/99_azureml.log': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/logs/azureml/99_azureml.log?sv=2019-02-02&sr=b&sig=Tnh1FU9gnmNWano8UmgU%2BV%2BOHHVyjLR8cqZd6cfjY8Q%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'logs/azureml/dataprep/backgroundProcess.log': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/logs/azureml/dataprep/backgroundProcess.log?sv=2019-02-02&sr=b&sig=FoaD75w%2FrsQmh3revejsf7nNXt95qv3yJ333rMQWk38%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'logs/azureml/dataprep/backgroundProcess_Telemetry.log': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/logs/azureml/dataprep/backgroundProcess_Telemetry.log?sv=2019-02-02&sr=b&sig=Qni4ZzgDW44p0tEOMRfa6wD6Q8sUNyfz1mXtQBpC1Vs%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'logs/azureml/job_prep_azureml.log': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/logs/azureml/job_prep_azureml.log?sv=2019-02-02&sr=b&sig=TOv5TGgL9LikrZehl42O44IaxYDsQcHvIroaBgunFBk%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r', 'logs/azureml/job_release_azureml.log': 'https://mlstrg142925.blob.core.windows.net/azureml/ExperimentRun/dcid.HD_4b351f6b-88c0-46ea-abf3-3a48ed4bf074_8/logs/azureml/job_release_azureml.log?sv=2019-02-02&sr=b&sig=9cWDAMUDRLdON%2BQ2D8pGpfR8dbMk8yeAnipFu0Bfoc0%3D&st=2021-04-17T11%3A47%3A56Z&se=2021-04-17T19%3A57%3A56Z&sp=r'}, 'submittedBy': 'ODL_User 142925'}\nBest run file names : ['azureml-logs/55_azureml-execution-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt', 'azureml-logs/65_job_prep-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt', 'azureml-logs/70_driver_log.txt', 'azureml-logs/75_job_post-tvmps_35a524d0de03b2def21f61357c47209d9165ba907a670cda89dd2c93cbbff911_d.txt', 'azureml-logs/process_info.json', 'azureml-logs/process_status.json', 'logs/azureml/99_azureml.log', 'logs/azureml/dataprep/backgroundProcess.log', 'logs/azureml/dataprep/backgroundProcess_Telemetry.log', 'logs/azureml/job_prep_azureml.log', 'logs/azureml/job_release_azureml.log', 'outputs/model.joblib']\n"
],
[
"from azureml.data.dataset_factory import TabularDatasetFactory\n\n# Create TabularDataset using TabularDatasetFactory\ndata_uri = \"https://automlsamplenotebookdata.blob.core.windows.net/automl-sample-notebook-data/bankmarketing_train.csv\"\nds = TabularDatasetFactory.from_delimited_files(path=data_uri)",
"_____no_output_____"
],
[
"from train import clean_data\n\n# Use the clean_data function to clean your data.\nx, y = clean_data(ds)",
"_____no_output_____"
],
[
"from azureml.train.automl import AutoMLConfig\n\n# Set parameters for AutoMLConfig\n# NOTE: DO NOT CHANGE THE experiment_timeout_minutes PARAMETER OR YOUR INSTANCE WILL TIME OUT.\n# If you wish to run the experiment longer, you will need to run this notebook in your own\n# Azure tenant, which will incur personal costs.\n\nautoml_config = AutoMLConfig(\n compute_target = compute_target,\n experiment_timeout_minutes=15,\n task='classification',\n primary_metric='accuracy',\n training_data=ds,\n label_column_name='y',\n enable_onnx_compatible_models=True,\n n_cross_validations=2)",
"_____no_output_____"
],
[
"# Submit your automl run\n\nautoml_run = exp.submit(automl_config, show_output = False)\nautoml_run.wait_for_completion()",
"Submitting remote run.\n"
],
[
"# Retrieve and save your best automl model.\n\nautoml_best_run, automl_best_model = automl_run.get_output()\n\nprint(\"Best run metrics :\", automl_best_run)\n# print(\"Best run details :\",automl_run.get_details())\n# print(\"Best run file names :\",best_run.get_file_names())\n\nbest_automl_model = automl_run.register_model(model_name='best_automl_model')",
"Best run metrics : Run(Experiment: p1,\nId: AutoML_320fd1f5-f07a-417d-82df-27d00606516e_11,\nType: azureml.scriptrun,\nStatus: Completed)\n"
],
[
"print(os.getcwd())",
"/mnt/batch/tasks/shared/LS_root/mounts/clusters/udacityazureml/code/Users/odl_user_142925/Optimizing_ML_Pipeline_Azure\n"
],
[
"# Delete cluster\ncompute_target.delete()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f6a0d014ddcd7dfb01babca69ca88a4a594455 | 148,381 | ipynb | Jupyter Notebook | ch8/lstm_extensions.ipynb | PacktPublishing/Natural-Language-Processing-with-TensorFlow | 54653384ff2d0bab356e3e0877bd86c3ba3a80a3 | [
"MIT"
] | 259 | 2018-06-07T02:46:53.000Z | 2022-03-29T03:31:26.000Z | ch8/lstm_extensions.ipynb | Chunlinx/Natural-Language-Processing-with-TensorFlow | 097b59a2f085379bf9a53b8285701cf3a0cb1d5e | [
"MIT"
] | 3 | 2018-07-21T01:41:01.000Z | 2020-10-06T06:47:00.000Z | ch8/lstm_extensions.ipynb | Chunlinx/Natural-Language-Processing-with-TensorFlow | 097b59a2f085379bf9a53b8285701cf3a0cb1d5e | [
"MIT"
] | 170 | 2018-06-01T23:56:47.000Z | 2022-03-30T06:27:22.000Z | 61.314463 | 1,298 | 0.518847 | [
[
[
"# Extending LSTMs: LSTMs with Peepholes and GRUs",
"_____no_output_____"
]
],
[
[
"# These are all the modules we'll be using later. Make sure you can import them\n# before proceeding further.\n%matplotlib inline\nfrom __future__ import print_function\nimport collections\nimport math\nimport numpy as np\nimport os\nimport random\nimport tensorflow as tf\nimport zipfile\nfrom matplotlib import pylab\nfrom six.moves import range\nfrom six.moves.urllib.request import urlretrieve\nimport tensorflow as tf\nimport csv",
"_____no_output_____"
]
],
[
[
"## Downloading Stories\nStories are automatically downloaded from https://www.cs.cmu.edu/~spok/grimmtmp/, if not detected in the disk. The total size of stories is around ~500KB. The dataset consists of 100 stories.",
"_____no_output_____"
]
],
[
[
"url = 'https://www.cs.cmu.edu/~spok/grimmtmp/'\n\n# Create a directory if needed\ndir_name = 'stories'\nif not os.path.exists(dir_name):\n os.mkdir(dir_name)\n \ndef maybe_download(filename):\n \"\"\"Download a file if not present\"\"\"\n print('Downloading file: ', dir_name+ os.sep+filename)\n \n if not os.path.exists(dir_name+os.sep+filename):\n filename, _ = urlretrieve(url + filename, dir_name+os.sep+filename)\n else:\n print('File ',filename, ' already exists.')\n \n return filename\n\nnum_files = 100\nfilenames = [format(i, '03d')+'.txt' for i in range(1,num_files+1)]\n\nfor fn in filenames:\n maybe_download(fn)",
"Downloading file: stories\\001.txt\nFile 001.txt already exists.\nDownloading file: stories\\002.txt\nFile 002.txt already exists.\nDownloading file: stories\\003.txt\nFile 003.txt already exists.\nDownloading file: stories\\004.txt\nFile 004.txt already exists.\nDownloading file: stories\\005.txt\nFile 005.txt already exists.\nDownloading file: stories\\006.txt\nFile 006.txt already exists.\nDownloading file: stories\\007.txt\nFile 007.txt already exists.\nDownloading file: stories\\008.txt\nFile 008.txt already exists.\nDownloading file: stories\\009.txt\nFile 009.txt already exists.\nDownloading file: stories\\010.txt\nFile 010.txt already exists.\nDownloading file: stories\\011.txt\nFile 011.txt already exists.\nDownloading file: stories\\012.txt\nFile 012.txt already exists.\nDownloading file: stories\\013.txt\nFile 013.txt already exists.\nDownloading file: stories\\014.txt\nFile 014.txt already exists.\nDownloading file: stories\\015.txt\nFile 015.txt already exists.\nDownloading file: stories\\016.txt\nFile 016.txt already exists.\nDownloading file: stories\\017.txt\nFile 017.txt already exists.\nDownloading file: stories\\018.txt\nFile 018.txt already exists.\nDownloading file: stories\\019.txt\nFile 019.txt already exists.\nDownloading file: stories\\020.txt\nFile 020.txt already exists.\nDownloading file: stories\\021.txt\nFile 021.txt already exists.\nDownloading file: stories\\022.txt\nFile 022.txt already exists.\nDownloading file: stories\\023.txt\nFile 023.txt already exists.\nDownloading file: stories\\024.txt\nFile 024.txt already exists.\nDownloading file: stories\\025.txt\nFile 025.txt already exists.\nDownloading file: stories\\026.txt\nFile 026.txt already exists.\nDownloading file: stories\\027.txt\nFile 027.txt already exists.\nDownloading file: stories\\028.txt\nFile 028.txt already exists.\nDownloading file: stories\\029.txt\nFile 029.txt already exists.\nDownloading file: stories\\030.txt\nFile 030.txt already exists.\nDownloading file: stories\\031.txt\nFile 031.txt already exists.\nDownloading file: stories\\032.txt\nFile 032.txt already exists.\nDownloading file: stories\\033.txt\nFile 033.txt already exists.\nDownloading file: stories\\034.txt\nFile 034.txt already exists.\nDownloading file: stories\\035.txt\nFile 035.txt already exists.\nDownloading file: stories\\036.txt\nFile 036.txt already exists.\nDownloading file: stories\\037.txt\nFile 037.txt already exists.\nDownloading file: stories\\038.txt\nFile 038.txt already exists.\nDownloading file: stories\\039.txt\nFile 039.txt already exists.\nDownloading file: stories\\040.txt\nFile 040.txt already exists.\nDownloading file: stories\\041.txt\nFile 041.txt already exists.\nDownloading file: stories\\042.txt\nFile 042.txt already exists.\nDownloading file: stories\\043.txt\nFile 043.txt already exists.\nDownloading file: stories\\044.txt\nFile 044.txt already exists.\nDownloading file: stories\\045.txt\nFile 045.txt already exists.\nDownloading file: stories\\046.txt\nFile 046.txt already exists.\nDownloading file: stories\\047.txt\nFile 047.txt already exists.\nDownloading file: stories\\048.txt\nFile 048.txt already exists.\nDownloading file: stories\\049.txt\nFile 049.txt already exists.\nDownloading file: stories\\050.txt\nFile 050.txt already exists.\nDownloading file: stories\\051.txt\nFile 051.txt already exists.\nDownloading file: stories\\052.txt\nFile 052.txt already exists.\nDownloading file: stories\\053.txt\nFile 053.txt already exists.\nDownloading file: stories\\054.txt\nFile 054.txt already exists.\nDownloading file: stories\\055.txt\nFile 055.txt already exists.\nDownloading file: stories\\056.txt\nFile 056.txt already exists.\nDownloading file: stories\\057.txt\nFile 057.txt already exists.\nDownloading file: stories\\058.txt\nFile 058.txt already exists.\nDownloading file: stories\\059.txt\nFile 059.txt already exists.\nDownloading file: stories\\060.txt\nFile 060.txt already exists.\nDownloading file: stories\\061.txt\nFile 061.txt already exists.\nDownloading file: stories\\062.txt\nFile 062.txt already exists.\nDownloading file: stories\\063.txt\nFile 063.txt already exists.\nDownloading file: stories\\064.txt\nFile 064.txt already exists.\nDownloading file: stories\\065.txt\nFile 065.txt already exists.\nDownloading file: stories\\066.txt\nFile 066.txt already exists.\nDownloading file: stories\\067.txt\nFile 067.txt already exists.\nDownloading file: stories\\068.txt\nFile 068.txt already exists.\nDownloading file: stories\\069.txt\nFile 069.txt already exists.\nDownloading file: stories\\070.txt\nFile 070.txt already exists.\nDownloading file: stories\\071.txt\nFile 071.txt already exists.\nDownloading file: stories\\072.txt\nFile 072.txt already exists.\nDownloading file: stories\\073.txt\nFile 073.txt already exists.\nDownloading file: stories\\074.txt\nFile 074.txt already exists.\nDownloading file: stories\\075.txt\nFile 075.txt already exists.\nDownloading file: stories\\076.txt\nFile 076.txt already exists.\nDownloading file: stories\\077.txt\nFile 077.txt already exists.\nDownloading file: stories\\078.txt\nFile 078.txt already exists.\nDownloading file: stories\\079.txt\nFile 079.txt already exists.\nDownloading file: stories\\080.txt\nFile 080.txt already exists.\nDownloading file: stories\\081.txt\nFile 081.txt already exists.\nDownloading file: stories\\082.txt\nFile 082.txt already exists.\nDownloading file: stories\\083.txt\nFile 083.txt already exists.\nDownloading file: stories\\084.txt\nFile 084.txt already exists.\nDownloading file: stories\\085.txt\nFile 085.txt already exists.\nDownloading file: stories\\086.txt\nFile 086.txt already exists.\nDownloading file: stories\\087.txt\nFile 087.txt already exists.\nDownloading file: stories\\088.txt\nFile 088.txt already exists.\nDownloading file: stories\\089.txt\nFile 089.txt already exists.\nDownloading file: stories\\090.txt\nFile 090.txt already exists.\nDownloading file: stories\\091.txt\nFile 091.txt already exists.\nDownloading file: stories\\092.txt\nFile 092.txt already exists.\nDownloading file: stories\\093.txt\nFile 093.txt already exists.\nDownloading file: stories\\094.txt\nFile 094.txt already exists.\nDownloading file: stories\\095.txt\nFile 095.txt already exists.\nDownloading file: stories\\096.txt\nFile 096.txt already exists.\nDownloading file: stories\\097.txt\nFile 097.txt already exists.\nDownloading file: stories\\098.txt\nFile 098.txt already exists.\nDownloading file: stories\\099.txt\nFile 099.txt already exists.\nDownloading file: stories\\100.txt\nFile 100.txt already exists.\n"
],
[
"for i in range(len(filenames)):\n file_exists = os.path.isfile(os.path.join(dir_name,filenames[i]))\n assert file_exists\nprint('%d files found.'%len(filenames))",
"100 files found.\n"
]
],
[
[
"## Reading data\nData will be stored in a list of lists where the each list represents a document and document is a list of words. We will then break the text into bigrams",
"_____no_output_____"
]
],
[
[
"def read_data(filename):\n \n with open(filename) as f:\n data = tf.compat.as_str(f.read())\n # make all the text lowercase\n data = data.lower()\n data = list(data)\n return data\n\ndocuments = []\nglobal documents\nfor i in range(num_files): \n print('\\nProcessing file %s'%os.path.join(dir_name,filenames[i]))\n chars = read_data(os.path.join(dir_name,filenames[i]))\n \n # Breaking the text into bigrams\n two_grams = [''.join(chars[ch_i:ch_i+2]) for ch_i in range(0,len(chars)-2,2)]\n # Creates a list of lists with the bigrams (outer loop different stories)\n documents.append(two_grams)\n print('Data size (Characters) (Document %d) %d' %(i,len(two_grams)))\n print('Sample string (Document %d) %s'%(i,two_grams[:50]))",
"\nProcessing file stories\\001.txt\nData size (Characters) (Document 0) 3667\nSample string (Document 0) ['in', ' o', 'ld', 'en', ' t', 'im', 'es', ' w', 'he', 'n ', 'wi', 'sh', 'in', 'g ', 'st', 'il', 'l ', 'he', 'lp', 'ed', ' o', 'ne', ', ', 'th', 'er', 'e ', 'li', 've', 'd ', 'a ', 'ki', 'ng', '\\nw', 'ho', 'se', ' d', 'au', 'gh', 'te', 'rs', ' w', 'er', 'e ', 'al', 'l ', 'be', 'au', 'ti', 'fu', 'l,']\n\nProcessing file stories\\002.txt\nData size (Characters) (Document 1) 4928\nSample string (Document 1) ['ha', 'rd', ' b', 'y ', 'a ', 'gr', 'ea', 't ', 'fo', 're', 'st', ' d', 'we', 'lt', ' a', ' w', 'oo', 'd-', 'cu', 'tt', 'er', ' w', 'it', 'h ', 'hi', 's ', 'wi', 'fe', ', ', 'wh', 'o ', 'ha', 'd ', 'an', '\\no', 'nl', 'y ', 'ch', 'il', 'd,', ' a', ' l', 'it', 'tl', 'e ', 'gi', 'rl', ' t', 'hr', 'ee']\n\nProcessing file stories\\003.txt\nData size (Characters) (Document 2) 9745\nSample string (Document 2) ['a ', 'ce', 'rt', 'ai', 'n ', 'fa', 'th', 'er', ' h', 'ad', ' t', 'wo', ' s', 'on', 's,', ' t', 'he', ' e', 'ld', 'er', ' o', 'f ', 'wh', 'om', ' w', 'as', ' s', 'ma', 'rt', ' a', 'nd', '\\ns', 'en', 'si', 'bl', 'e,', ' a', 'nd', ' c', 'ou', 'ld', ' d', 'o ', 'ev', 'er', 'yt', 'hi', 'ng', ', ', 'bu']\n\nProcessing file stories\\004.txt\nData size (Characters) (Document 3) 2852\nSample string (Document 3) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' u', 'po', 'n ', 'a ', 'ti', 'me', ' a', 'n ', 'ol', 'd ', 'go', 'at', ' w', 'ho', ' h', 'ad', ' s', 'ev', 'en', ' l', 'it', 'tl', 'e ', 'ki', 'ds', ', ', 'an', 'd\\n', 'lo', 've', 'd ', 'th', 'em', ' w', 'it', 'h ', 'al', 'l ', 'th', 'e ', 'lo', 've', ' o']\n\nProcessing file stories\\005.txt\nData size (Characters) (Document 4) 8189\nSample string (Document 4) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' u', 'po', 'n ', 'a ', 'ti', 'me', ' a', 'n ', 'ol', 'd ', 'ki', 'ng', ' w', 'ho', ' w', 'as', ' i', 'll', ' a', 'nd', ' t', 'ho', 'ug', 'ht', ' t', 'o\\n', 'hi', 'ms', 'el', 'f ', \"'i\", ' a', 'm ', 'ly', 'in', 'g ', 'on', ' w', 'ha', 't ', 'mu', 'st', ' b']\n\nProcessing file stories\\006.txt\nData size (Characters) (Document 5) 4369\nSample string (Document 5) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' a', ' p', 'ea', 'sa', 'nt', ' w', 'ho', ' h', 'ad', ' d', 'ri', 've', 'n ', 'hi', 's ', 'co', 'w ', 'to', ' t', 'he', ' f', 'ai', 'r,', ' a', 'nd', ' s', 'ol', 'd\\n', 'he', 'r ', 'fo', 'r ', 'se', 've', 'n ', 'ta', 'le', 'rs', '. ', ' o', 'n ', 'th', 'e ']\n\nProcessing file stories\\007.txt\nData size (Characters) (Document 6) 5216\nSample string (Document 6) ['th', 'er', 'e ', 'we', 're', ' o', 'nc', 'e ', 'up', 'on', ' a', ' t', 'im', 'e ', 'a ', 'ki', 'ng', ' a', 'nd', ' a', ' q', 'ue', 'en', ' w', 'ho', ' l', 'iv', 'ed', '\\nh', 'ap', 'pi', 'ly', ' t', 'og', 'et', 'he', 'r ', 'an', 'd ', 'ha', 'd ', 'tw', 'el', 've', ' c', 'hi', 'ld', 're', 'n,', ' b']\n\nProcessing file stories\\008.txt\nData size (Characters) (Document 7) 6097\nSample string (Document 7) ['li', 'tt', 'le', ' b', 'ro', 'th', 'er', ' t', 'oo', 'k ', 'hi', 's ', 'li', 'tt', 'le', ' s', 'is', 'te', 'r ', 'by', ' t', 'he', ' h', 'an', 'd ', 'an', 'd ', 'sa', 'id', ', ', 'si', 'nc', 'e\\n', 'ou', 'r ', 'mo', 'th', 'er', ' d', 'ie', 'd ', 'we', ' h', 'av', 'e ', 'ha', 'd ', 'no', ' h', 'ap']\n\nProcessing file stories\\009.txt\nData size (Characters) (Document 8) 3699\nSample string (Document 8) ['th', 'er', 'e ', 'we', 're', ' o', 'nc', 'e ', 'a ', 'ma', 'n ', 'an', 'd ', 'a ', 'wo', 'ma', 'n ', 'wh', 'o ', 'ha', 'd ', 'lo', 'ng', ' i', 'n ', 'va', 'in', '\\nw', 'is', 'he', 'd ', 'fo', 'r ', 'a ', 'ch', 'il', 'd.', ' ', 'at', ' l', 'en', 'gt', 'h ', 'th', 'e ', 'wo', 'ma', 'n ', 'ho', 'pe']\n\nProcessing file stories\\010.txt\nData size (Characters) (Document 9) 5268\nSample string (Document 9) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' a', ' m', 'an', ' w', 'ho', 'se', ' w', 'if', 'e ', 'di', 'ed', ', ', 'an', 'd ', 'a ', 'wo', 'ma', 'n ', 'wh', 'os', 'e ', 'hu', 'sb', 'an', 'd\\n', 'di', 'ed', ', ', 'an', 'd ', 'th', 'e ', 'ma', 'n ', 'ha', 'd ', 'a ', 'da', 'ug', 'ht', 'er', ', ', 'an']\n\nProcessing file stories\\011.txt\nData size (Characters) (Document 10) 2377\nSample string (Document 10) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' a', ' g', 'ir', 'l ', 'wh', 'o ', 'wa', 's ', 'id', 'le', ' a', 'nd', ' w', 'ou', 'ld', ' n', 'ot', ' s', 'pi', 'n,', ' a', 'nd', '\\nl', 'et', ' h', 'er', ' m', 'ot', 'he', 'r ', 'sa', 'y ', 'wh', 'at', ' s', 'he', ' w', 'ou', 'ld', ', ', 'sh', 'e ', 'co']\n\nProcessing file stories\\012.txt\nData size (Characters) (Document 11) 7695\nSample string (Document 11) ['ha', 'rd', ' b', 'y ', 'a ', 'gr', 'ea', 't ', 'fo', 're', 'st', ' d', 'we', 'lt', ' a', ' p', 'oo', 'r ', 'wo', 'od', '-c', 'ut', 'te', 'r ', 'wi', 'th', ' h', 'is', ' w', 'if', 'e\\n', 'an', 'd ', 'hi', 's ', 'tw', 'o ', 'ch', 'il', 'dr', 'en', '. ', ' t', 'he', ' b', 'oy', ' w', 'as', ' c', 'al']\n\nProcessing file stories\\013.txt\nData size (Characters) (Document 12) 3665\nSample string (Document 12) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' o', 'n ', 'a ', 'ti', 'me', ' a', ' p', 'oo', 'r ', 'ma', 'n,', ' w', 'ho', ' c', 'ou', 'ld', ' n', 'o ', 'lo', 'ng', 'er', '\\ns', 'up', 'po', 'rt', ' h', 'is', ' o', 'nl', 'y ', 'so', 'n.', ' ', 'th', 'en', ' s', 'ai', 'd ', 'th', 'e ', 'so', 'n,', ' d']\n\nProcessing file stories\\014.txt\nData size (Characters) (Document 13) 4178\nSample string (Document 13) ['a ', 'lo', 'ng', ' t', 'im', 'e ', 'ag', 'o ', 'th', 'er', 'e ', 'li', 've', 'd ', 'a ', 'ki', 'ng', ' w', 'ho', ' w', 'as', ' f', 'am', 'ed', ' f', 'or', ' h', 'is', ' w', 'is', 'do', 'm\\n', 'th', 'ro', 'ug', 'h ', 'al', 'l ', 'th', 'e ', 'la', 'nd', '. ', ' n', 'ot', 'hi', 'ng', ' w', 'as', ' h']\n\nProcessing file stories\\015.txt\nData size (Characters) (Document 14) 8674\nSample string (Document 14) ['on', 'e ', 'su', 'mm', 'er', \"'s\", ' m', 'or', 'ni', 'ng', ' a', ' l', 'it', 'tl', 'e ', 'ta', 'il', 'or', ' w', 'as', ' s', 'it', 'ti', 'ng', ' o', 'n ', 'hi', 's ', 'ta', 'bl', 'e\\n', 'by', ' t', 'he', ' w', 'in', 'do', 'w,', ' h', 'e ', 'wa', 's ', 'in', ' g', 'oo', 'd ', 'sp', 'ir', 'it', 's,']\n\nProcessing file stories\\016.txt\nData size (Characters) (Document 15) 7018\nSample string (Document 15) ['\\tc', 'in', 'de', 're', 'll', 'a\\n', 'th', 'e ', 'wi', 'fe', ' o', 'f ', 'a ', 'ri', 'ch', ' m', 'an', ' f', 'el', 'l ', 'si', 'ck', ', ', 'an', 'd ', 'as', ' s', 'he', ' f', 'el', 't ', 'th', 'at', ' h', 'er', ' e', 'nd', '\\nw', 'as', ' d', 'ra', 'wi', 'ng', ' n', 'ea', 'r,', ' s', 'he', ' c', 'al']\n\nProcessing file stories\\017.txt\nData size (Characters) (Document 16) 3039\nSample string (Document 16) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' a', ' k', 'in', \"g'\", 's ', 'so', 'n ', 'wh', 'o ', 'wa', 's ', 'se', 'iz', 'ed', ' w', 'it', 'h ', 'a ', 'de', 'si', 're', ' t', 'o ', 'tr', 'av', 'el', '\\na', 'bo', 'ut', ' t', 'he', ' w', 'or', 'ld', ', ', 'an', 'd ', 'to', 'ok', ' n', 'o ', 'on', 'e ']\n\nProcessing file stories\\018.txt\nData size (Characters) (Document 17) 3020\nSample string (Document 17) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' a', ' w', 'id', 'ow', ' w', 'ho', ' h', 'ad', ' t', 'wo', ' d', 'au', 'gh', 'te', 'rs', ' -', ' o', 'ne', ' o', 'f\\n', 'wh', 'om', ' w', 'as', ' p', 're', 'tt', 'y ', 'an', 'd ', 'in', 'du', 'st', 'ri', 'ou', 's,', ' w', 'hi', 'ls', 't ', 'th', 'e ', 'ot']\n\nProcessing file stories\\019.txt\nData size (Characters) (Document 18) 2465\nSample string (Document 18) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' a', ' m', 'an', ' w', 'ho', ' h', 'ad', ' s', 'ev', 'en', ' s', 'on', 's,', ' a', 'nd', ' s', 'ti', 'll', ' h', 'e ', 'ha', 'd\\n', 'no', ' d', 'au', 'gh', 'te', 'r,', ' h', 'ow', 'ev', 'er', ' m', 'uc', 'h ', 'he', ' w', 'is', 'he', 'd ', 'fo', 'r ', 'on']\n\nProcessing file stories\\020.txt\nData size (Characters) (Document 19) 3703\nSample string (Document 19) ['\\tl', 'it', 'tl', 'e ', 're', 'd-', 'ca', 'p\\n', '\\no', 'nc', 'e ', 'up', 'on', ' a', ' t', 'im', 'e ', 'th', 'er', 'e ', 'wa', 's ', 'a ', 'de', 'ar', ' l', 'it', 'tl', 'e ', 'gi', 'rl', ' w', 'ho', ' w', 'as', ' l', 'ov', 'ed', '\\nb', 'y ', 'ev', 'er', 'y ', 'on', 'e ', 'wh', 'o ', 'lo', 'ok', 'ed']\n\nProcessing file stories\\021.txt\nData size (Characters) (Document 20) 1924\nSample string (Document 20) ['in', ' a', ' c', 'er', 'ta', 'in', ' c', 'ou', 'nt', 'ry', ' t', 'he', 're', ' w', 'as', ' o', 'nc', 'e ', 'gr', 'ea', 't ', 'la', 'me', 'nt', 'at', 'io', 'n ', 'ov', 'er', ' a', '\\nw', 'il', 'd ', 'bo', 'ar', ' t', 'ha', 't ', 'la', 'id', ' w', 'as', 'te', ' t', 'he', ' f', 'ar', 'me', \"r'\", 's ']\n\nProcessing file stories\\022.txt\nData size (Characters) (Document 21) 6561\nSample string (Document 21) ['th', 'er', 'e ', 'wa', 's ', 'on', 'ce', ' a', ' p', 'oo', 'r ', 'wo', 'ma', 'n ', 'wh', 'o ', 'ga', 've', ' b', 'ir', 'th', ' t', 'o ', 'a ', 'li', 'tt', 'le', ' s', 'on', ',\\n', 'an', 'd ', 'as', ' h', 'e ', 'ca', 'me', ' i', 'nt', 'o ', 'th', 'e ', 'wo', 'rl', 'd ', 'wi', 'th', ' a', ' c', 'au']\n\nProcessing file stories\\023.txt\n"
]
],
[
[
"## Building the Dictionaries (Bigrams)\nBuilds the following. To understand each of these elements, let us also assume the text \"I like to go to school\"\n\n* `dictionary`: maps a string word to an ID (e.g. {I:0, like:1, to:2, go:3, school:4})\n* `reverse_dictionary`: maps an ID to a string word (e.g. {0:I, 1:like, 2:to, 3:go, 4:school}\n* `count`: List of list of (word, frequency) elements (e.g. [(I,1),(like,1),(to,2),(go,1),(school,1)]\n* `data` : Contain the string of text we read, where string words are replaced with word IDs (e.g. [0, 1, 2, 3, 2, 4])\n\nIt also introduces an additional special token `UNK` to denote rare words to are too rare to make use of.",
"_____no_output_____"
]
],
[
[
"def build_dataset(documents):\n chars = []\n # This is going to be a list of lists\n # Where the outer list denote each document\n # and the inner lists denote words in a given document\n data_list = []\n \n for d in documents:\n chars.extend(d)\n print('%d Characters found.'%len(chars))\n count = []\n # Get the bigram sorted by their frequency (Highest comes first)\n count.extend(collections.Counter(chars).most_common())\n \n # Create an ID for each bigram by giving the current length of the dictionary\n # And adding that item to the dictionary\n # Start with 'UNK' that is assigned to too rare words\n dictionary = dict({'UNK':0})\n for char, c in count:\n # Only add a bigram to dictionary if its frequency is more than 10\n if c > 10:\n dictionary[char] = len(dictionary) \n \n unk_count = 0\n # Traverse through all the text we have\n # to replace each string word with the ID of the word\n for d in documents:\n data = list()\n for char in d:\n # If word is in the dictionary use the word ID,\n # else use the ID of the special token \"UNK\"\n if char in dictionary:\n index = dictionary[char] \n else:\n index = dictionary['UNK']\n unk_count += 1\n data.append(index)\n \n data_list.append(data)\n \n reverse_dictionary = dict(zip(dictionary.values(), dictionary.keys())) \n return data_list, count, dictionary, reverse_dictionary\n\nglobal data_list, count, dictionary, reverse_dictionary,vocabulary_size\n\n# Print some statistics about data\ndata_list, count, dictionary, reverse_dictionary = build_dataset(documents)\nprint('Most common words (+UNK)', count[:5])\nprint('Least common words (+UNK)', count[-15:])\nprint('Sample data', data_list[0][:10])\nprint('Sample data', data_list[1][:10])\nprint('Vocabulary: ',len(dictionary))\nvocabulary_size = len(dictionary)\ndel documents # To reduce memory.",
"449177 Characters found.\nMost common words (+UNK) [('e ', 15229), ('he', 15164), (' t', 13443), ('th', 13076), ('d ', 10687)]\nLeast common words (+UNK) [('rz', 1), ('zi', 1), ('i?', 1), ('\\ts', 1), ('\".', 1), ('hc', 1), ('sd', 1), ('z ', 1), ('m?', 1), ('\\tc', 1), ('oz', 1), ('iq', 1), ('pw', 1), ('tz', 1), ('yr', 1)]\nSample data [15, 28, 86, 23, 3, 95, 74, 11, 2, 16]\nSample data [22, 156, 25, 37, 82, 185, 43, 9, 90, 19]\nVocabulary: 544\n"
]
],
[
[
"## Generating Batches of Data\nThe following object generates a batch of data which will be used to train the RNN. More specifically the generator breaks a given sequence of words into `batch_size` segments. We also maintain a cursor for each segment. So whenever we create a batch of data, we sample one item from each segment and update the cursor of each segment. ",
"_____no_output_____"
]
],
[
[
"class DataGeneratorOHE(object):\n \n def __init__(self,text,batch_size,num_unroll):\n # Text where a bigram is denoted by its ID\n self._text = text\n # Number of bigrams in the text\n self._text_size = len(self._text)\n # Number of datapoints in a batch of data\n self._batch_size = batch_size\n # Num unroll is the number of steps we unroll the RNN in a single training step\n # This relates to the truncated backpropagation we discuss in Chapter 6 text\n self._num_unroll = num_unroll\n # We break the text in to several segments and the batch of data is sampled by\n # sampling a single item from a single segment\n self._segments = self._text_size//self._batch_size\n self._cursor = [offset * self._segments for offset in range(self._batch_size)]\n \n def next_batch(self):\n '''\n Generates a single batch of data\n '''\n # Train inputs (one-hot-encoded) and train outputs (one-hot-encoded)\n batch_data = np.zeros((self._batch_size,vocabulary_size),dtype=np.float32)\n batch_labels = np.zeros((self._batch_size,vocabulary_size),dtype=np.float32)\n \n # Fill in the batch datapoint by datapoint\n for b in range(self._batch_size):\n # If the cursor of a given segment exceeds the segment length\n # we reset the cursor back to the beginning of that segment\n if self._cursor[b]+1>=self._text_size:\n self._cursor[b] = b * self._segments\n \n # Add the text at the cursor as the input\n batch_data[b,self._text[self._cursor[b]]] = 1.0\n # Add the preceding bigram as the label to be predicted\n batch_labels[b,self._text[self._cursor[b]+1]]= 1.0 \n # Update the cursor\n self._cursor[b] = (self._cursor[b]+1)%self._text_size\n \n return batch_data,batch_labels\n \n def unroll_batches(self):\n '''\n This produces a list of num_unroll batches\n as required by a single step of training of the RNN\n '''\n unroll_data,unroll_labels = [],[]\n for ui in range(self._num_unroll):\n data, labels = self.next_batch() \n unroll_data.append(data)\n unroll_labels.append(labels)\n \n return unroll_data, unroll_labels\n \n def reset_indices(self):\n '''\n Used to reset all the cursors if needed\n '''\n self._cursor = [offset * self._segments for offset in range(self._batch_size)]\n \n# Running a tiny set to see if things are correct\ndg = DataGeneratorOHE(data_list[0][25:50],5,5)\nu_data, u_labels = dg.unroll_batches()\n\n# Iterate through each data batch in the unrolled set of batches\nfor ui,(dat,lbl) in enumerate(zip(u_data,u_labels)): \n print('\\n\\nUnrolled index %d'%ui)\n dat_ind = np.argmax(dat,axis=1)\n lbl_ind = np.argmax(lbl,axis=1)\n print('\\tInputs:')\n for sing_dat in dat_ind:\n print('\\t%s (%d)'%(reverse_dictionary[sing_dat],sing_dat),end=\", \")\n print('\\n\\tOutput:')\n for sing_lbl in lbl_ind: \n print('\\t%s (%d)'%(reverse_dictionary[sing_lbl],sing_lbl),end=\", \")",
"\n\nUnrolled index 0\n\tInputs:\n\te (1), \tki (131), \t d (48), \t w (11), \tbe (70), \n\tOutput:\n\tli (98), \tng (33), \tau (195), \ter (14), \tau (195), \n\nUnrolled index 1\n\tInputs:\n\tli (98), \tng (33), \tau (195), \ter (14), \tau (195), \n\tOutput:\n\tve (41), \t\nw (169), \tgh (106), \te (1), \tti (112), \n\nUnrolled index 2\n\tInputs:\n\tve (41), \t\nw (169), \tgh (106), \te (1), \tti (112), \n\tOutput:\n\td (5), \tho (62), \tte (61), \tal (84), \tfu (229), \n\nUnrolled index 3\n\tInputs:\n\td (5), \tho (62), \tte (61), \tal (84), \tfu (229), \n\tOutput:\n\ta (82), \tse (58), \trs (137), \tl (57), \tl, (257), \n\nUnrolled index 4\n\tInputs:\n\ta (82), \tse (58), \trs (137), \tl (57), \tbe (70), \n\tOutput:\n\tki (131), \t d (48), \t w (11), \tbe (70), \tau (195), "
]
],
[
[
"## Defining the LSTM, LSTM with Peepholes and GRUs\n\n* A LSTM has 5 main components\n * Cell state, Hidden state, Input gate, Forget gate, Output gate\n* A LSTM with peephole connections\n * Introduces several new sets of weights that connects the cell state to the gates\n* A GRU has 3 main components\n * Hidden state, Reset gate and a Update gate\n",
"_____no_output_____"
],
[
"## Defining hyperparameters\n\nHere we define several hyperparameters and are very similar to the ones we defined in Chapter 6. However additionally we use dropout; a technique that helps to avoid overfitting.",
"_____no_output_____"
]
],
[
[
"num_nodes = 128\nbatch_size = 64\nnum_unrollings = 50\ndropout = 0.2\n\n# Use this in the CSV filename when saving\n# when using dropout\nfilename_extension = ''\nif dropout>0.0:\n filename_extension = '_dropout'\n ",
"_____no_output_____"
]
],
[
[
"## Defining Inputs and Outputs\n\nIn the code we define two different types of inputs. \n* Training inputs (The stories we downloaded) (batch_size > 1 with unrolling)\n* Validation inputs (An unseen validation dataset) (bach_size =1, no unrolling)\n* Test input (New story we are going to generate) (batch_size=1, no unrolling)",
"_____no_output_____"
]
],
[
[
"tf.reset_default_graph()\n\n# Training Input data.\ntrain_inputs, train_labels = [],[]\n\n# Defining unrolled training inputs\nfor ui in range(num_unrollings):\n train_inputs.append(tf.placeholder(tf.float32, shape=[batch_size,vocabulary_size],name='train_inputs_%d'%ui))\n train_labels.append(tf.placeholder(tf.float32, shape=[batch_size,vocabulary_size], name = 'train_labels_%d'%ui))\n\nvalid_inputs = tf.placeholder(tf.float32, shape=[1, vocabulary_size])\nvalid_labels = tf.placeholder(tf.float32, shape=[1, vocabulary_size])\n\n# Text generation: batch 1, no unrolling.\ntest_input = tf.placeholder(tf.float32, shape=[1, vocabulary_size])\n",
"_____no_output_____"
]
],
[
[
"## Defining Model Parameters and Cell Computation\n\nWe define parameters and cell computation functions for all the different variants (LSTM, LSTM with peepholes and GRUs). **Make sure you only run a single cell withing this section (either the LSTM/ LSTM with peepholes or GRUs)",
"_____no_output_____"
],
[
"### Standard LSTM\n\nHere we define the parameters and the cell computation function for a standard LSTM",
"_____no_output_____"
]
],
[
[
"# Input gate (i_t) - How much memory to write to cell state\n# Connects the current input to the input gate\nix = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.02))\n# Connects the previous hidden state to the input gate\nim = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.02))\n# Bias of the input gate\nib = tf.Variable(tf.random_uniform([1, num_nodes],-0.02, 0.02))\n\n# Forget gate (f_t) - How much memory to discard from cell state\n# Connects the current input to the forget gate\nfx = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.02))\n# Connects the previous hidden state to the forget gate\nfm = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.02))\n# Bias of the forget gate\nfb = tf.Variable(tf.random_uniform([1, num_nodes],-0.02, 0.02))\n\n# Candidate value (c~_t) - Used to compute the current cell state\n# Connects the current input to the candidate\ncx = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.02))\n# Connects the previous hidden state to the candidate\ncm = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.02))\n# Bias of the candidate\ncb = tf.Variable(tf.random_uniform([1, num_nodes],-0.02,0.02))\n\n# Output gate - How much memory to output from the cell state\n# Connects the current input to the output gate\nox = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.02))\n# Connects the previous hidden state to the output gate\nom = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.02))\n# Bias of the output gate\nob = tf.Variable(tf.random_uniform([1, num_nodes],-0.02,0.02))\n\n\n# Softmax Classifier weights and biases.\nw = tf.Variable(tf.truncated_normal([num_nodes, vocabulary_size], stddev=0.02))\nb = tf.Variable(tf.random_uniform([vocabulary_size],-0.02,0.02))\n\n# Variables saving state across unrollings.\n# Hidden state\nsaved_output = tf.Variable(tf.zeros([batch_size, num_nodes]), trainable=False)\n# Cell state\nsaved_state = tf.Variable(tf.zeros([batch_size, num_nodes]), trainable=False)\n\nsaved_valid_output = tf.Variable(tf.zeros([1, num_nodes]), trainable=False)\nsaved_valid_state = tf.Variable(tf.zeros([1, num_nodes]), trainable=False)\n\n# Same variables for testing phase\nsaved_test_output = tf.Variable(tf.zeros([1, num_nodes]),trainable=False)\nsaved_test_state = tf.Variable(tf.zeros([1, num_nodes]),trainable=False)\n\nalgorithm = 'lstm'\nfilename_to_save = algorithm + filename_extension +'.csv'\n# Definition of the cell computation.\ndef lstm_cell(i, o, state):\n \"\"\"Create an LSTM cell\"\"\"\n input_gate = tf.sigmoid(tf.matmul(i, ix) + tf.matmul(o, im) + ib)\n forget_gate = tf.sigmoid(tf.matmul(i, fx) + tf.matmul(o, fm) + fb)\n update = tf.matmul(i, cx) + tf.matmul(o, cm) + cb\n state = forget_gate * state + input_gate * tf.tanh(update)\n output_gate = tf.sigmoid(tf.matmul(i, ox) + tf.matmul(o, om) + ob)\n return output_gate * tf.tanh(state), state\n",
"_____no_output_____"
]
],
[
[
"### LSTMs with Peephole Connections\n\nWe define the parameters and cell computation for a LSTM with peepholes. Note that we are using diagonal peephole connections (for more details refer the text).",
"_____no_output_____"
]
],
[
[
"# Parameters:\n# Input gate: input, previous output, and bias.\nix = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.01))\nim = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.01))\nic = tf.Variable(tf.truncated_normal([1,num_nodes], stddev=0.01))\nib = tf.Variable(tf.random_uniform([1, num_nodes],0.0, 0.01))\n# Forget gate: input, previous output, and bias.\nfx = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.01))\nfm = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.01))\nfc = tf.Variable(tf.truncated_normal([1,num_nodes], stddev=0.01))\nfb = tf.Variable(tf.random_uniform([1, num_nodes],0.0, 0.01))\n# Memory cell: input, state and bias. \ncx = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.01))\ncm = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.01))\ncb = tf.Variable(tf.random_uniform([1, num_nodes],0.0,0.01))\n# Output gate: input, previous output, and bias.\nox = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.01))\nom = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.01))\noc = tf.Variable(tf.truncated_normal([1,num_nodes], stddev=0.01))\nob = tf.Variable(tf.random_uniform([1, num_nodes],0.0,0.01))\n\n# Softmax Classifier weights and biases.\nw = tf.Variable(tf.truncated_normal([num_nodes, vocabulary_size], stddev=0.01))\nb = tf.Variable(tf.random_uniform([vocabulary_size],0.0,0.01))\n\n# Variables saving state across unrollings.\nsaved_output = tf.Variable(tf.zeros([batch_size, num_nodes]), trainable=False)\nsaved_state = tf.Variable(tf.zeros([batch_size, num_nodes]), trainable=False)\n\nsaved_valid_output = tf.Variable(tf.zeros([1, num_nodes]), trainable=False)\nsaved_valid_state = tf.Variable(tf.zeros([1, num_nodes]), trainable=False)\n\nsaved_test_output = tf.Variable(tf.zeros([1, num_nodes]), trainable=False)\nsaved_test_state = tf.Variable(tf.zeros([1, num_nodes]), trainable=False)\n\nalgorithm = 'lstm_peephole'\nfilename_to_save = algorithm + filename_extension +'.csv'\n# Definition of the cell computation.\ndef lstm_with_peephole_cell(i, o, state):\n '''\n LSTM with peephole connections\n Our implementation for peepholes is based on \n https://static.googleusercontent.com/media/research.google.com/en//pubs/archive/43905.pdf \n '''\n input_gate = tf.sigmoid(tf.matmul(i, ix) + state*ic + tf.matmul(o, im) + ib)\n forget_gate = tf.sigmoid(tf.matmul(i, fx) + state*fc + tf.matmul(o, fm) + fb)\n update = tf.matmul(i, cx) + tf.matmul(o, cm) + cb\n state = forget_gate * state + input_gate * tf.tanh(update)\n output_gate = tf.sigmoid(tf.matmul(i, ox) + state*oc + tf.matmul(o, om) + ob)\n\n return output_gate * tf.tanh(state), state",
"_____no_output_____"
]
],
[
[
"### Gated Recurrent Units (GRUs)\n\nFinally we define the parameters and cell computations for the GRU cell.",
"_____no_output_____"
]
],
[
[
"# Parameters:\n# Reset gate: input, previous output, and bias.\nrx = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.01))\nrh = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.01))\nrb = tf.Variable(tf.random_uniform([1, num_nodes],0.0, 0.01))\n\n# Hidden State: input, previous output, and bias.\nhx = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.01))\nhh = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.01))\nhb = tf.Variable(tf.random_uniform([1, num_nodes],0.0, 0.01))\n\n# Update gate: input, previous output, and bias.\nzx = tf.Variable(tf.truncated_normal([vocabulary_size, num_nodes], stddev=0.01))\nzh = tf.Variable(tf.truncated_normal([num_nodes, num_nodes], stddev=0.01))\nzb = tf.Variable(tf.random_uniform([1, num_nodes],0.0, 0.01))\n\n# Softmax Classifier weights and biases.\nw = tf.Variable(tf.truncated_normal([num_nodes, vocabulary_size], stddev=0.01))\nb = tf.Variable(tf.random_uniform([vocabulary_size],0.0,0.01))\n\n# Variables saving state across unrollings.\nsaved_output = tf.Variable(tf.zeros([batch_size, num_nodes]), trainable=False)\nsaved_valid_output = tf.Variable(tf.zeros([1, num_nodes]),trainable=False)\nsaved_test_output = tf.Variable(tf.zeros([1, num_nodes]),trainable=False)\n\nalgorithm = 'gru'\nfilename_to_save = algorithm + filename_extension +'.csv'\n\n# Definition of the cell computation.\ndef gru_cell(i, o):\n \"\"\"Create a GRU cell.\"\"\"\n reset_gate = tf.sigmoid(tf.matmul(i, rx) + tf.matmul(o, rh) + rb)\n h_tilde = tf.tanh(tf.matmul(i,hx) + tf.matmul(reset_gate * o, hh) + hb)\n z = tf.sigmoid(tf.matmul(i,zx) + tf.matmul(o, zh) + zb)\n h = (1-z)*o + z*h_tilde\n \n return h",
"_____no_output_____"
]
],
[
[
"## Defining LSTM/GRU/LSTM-Peephole Computations\nHere first we define the LSTM cell computations as a consice function. Then we use this function to define training and test-time inference logic.",
"_____no_output_____"
]
],
[
[
"# =========================================================\n#Training related inference logic\n\n# Keeps the calculated state outputs in all the unrollings\n# Used to calculate loss\noutputs = list()\n\n# These two python variables are iteratively updated\n# at each step of unrolling\noutput = saved_output\nif algorithm=='lstm' or algorithm=='lstm_peephole':\n state = saved_state\n\n# Compute the hidden state (output) and cell state (state)\n# recursively for all the steps in unrolling\n# Note: there is no cell state for GRUs\nfor i in train_inputs:\n if algorithm=='lstm':\n output, state = lstm_cell(i, output, state)\n train_state_update_ops = [saved_output.assign(output),\n saved_state.assign(state)]\n elif algorithm=='lstm_peephole':\n output, state = lstm_with_peephole_cell(i, output, state)\n train_state_update_ops = [saved_output.assign(output),\n saved_state.assign(state)]\n elif algorithm=='gru':\n output = gru_cell(i, output)\n train_state_update_ops = [saved_output.assign(output)]\n \n output = tf.nn.dropout(output,keep_prob=1.0-dropout)\n # Append each computed output value\n outputs.append(output)\n\n# calculate the score values\nlogits = tf.matmul(tf.concat(axis=0, values=outputs), w) + b\n \n# Compute predictions.\ntrain_prediction = tf.nn.softmax(logits)\n\n# Compute training perplexity\ntrain_perplexity_without_exp = tf.reduce_sum(tf.concat(train_labels,0)*-tf.log(tf.concat(train_prediction,0)+1e-10))/(num_unrollings*batch_size)\n\n# ========================================================================\n# Validation phase related inference logic\n\nvalid_output = saved_valid_output\nif algorithm=='lstm' or algorithm=='lstm_peephole':\n valid_state = saved_valid_state\n\n# Compute the LSTM cell output for validation data\nif algorithm=='lstm':\n valid_output, valid_state = lstm_cell(\n valid_inputs, saved_valid_output, saved_valid_state)\n valid_state_update_ops = [saved_valid_output.assign(valid_output),\n saved_valid_state.assign(valid_state)]\n \nelif algorithm=='lstm_peephole':\n valid_output, valid_state = lstm_with_peephole_cell(\n valid_inputs, saved_valid_output, saved_valid_state)\n valid_state_update_ops = [saved_valid_output.assign(valid_output),\n saved_valid_state.assign(valid_state)]\nelif algorithm=='gru':\n valid_output = gru_cell(valid_inputs, valid_output)\n valid_state_update_ops = [saved_valid_output.assign(valid_output)]\n\nvalid_logits = tf.nn.xw_plus_b(valid_output, w, b)\n# Make sure that the state variables are updated\n# before moving on to the next iteration of generation\nwith tf.control_dependencies(valid_state_update_ops):\n valid_prediction = tf.nn.softmax(valid_logits)\n\n# Compute validation perplexity\nvalid_perplexity_without_exp = tf.reduce_sum(valid_labels*-tf.log(valid_prediction+1e-10))\n\n# ========================================================================\n# Testing phase related inference logic\n\n# Compute the LSTM cell output for testing data\nif algorithm=='lstm':\n test_output, test_state = lstm_cell(test_input, saved_test_output, saved_test_state)\n test_state_update_ops = [saved_test_output.assign(test_output),\n saved_test_state.assign(test_state)]\nelif algorithm=='lstm_peephole':\n test_output, test_state = lstm_with_peephole_cell(test_input, saved_test_output, saved_test_state)\n test_state_update_ops = [saved_test_output.assign(test_output),\n saved_test_state.assign(test_state)]\nelif algorithm=='gru':\n test_output = gru_cell(test_input, saved_test_output)\n test_state_update_ops = [saved_test_output.assign(test_output)]\n\n# Make sure that the state variables are updated\n# before moving on to the next iteration of generation\nwith tf.control_dependencies(test_state_update_ops):\n test_prediction = tf.nn.softmax(tf.nn.xw_plus_b(test_output, w, b))",
"_____no_output_____"
]
],
[
[
"## Calculating LSTM Loss\nWe calculate the training loss of the LSTM here. It's a typical cross entropy loss calculated over all the scores we obtained for training data (`loss`).",
"_____no_output_____"
]
],
[
[
"# Before calcualting the training loss,\n# save the hidden state and the cell state to\n# their respective TensorFlow variables\nwith tf.control_dependencies(train_state_update_ops):\n\n # Calculate the training loss by\n # concatenating the results from all the unrolled time steps\n loss = tf.reduce_mean(\n tf.nn.softmax_cross_entropy_with_logits_v2(\n logits=logits, labels=tf.concat(axis=0, values=train_labels)))\n",
"_____no_output_____"
]
],
[
[
"## Resetting Operations for Resetting Hidden States\nSometimes the state variable needs to be reset (e.g. when starting predictions at a beginning of a new epoch). But since GRU doesn't have a cell state we have a conditioned reset_state ops",
"_____no_output_____"
]
],
[
[
"if algorithm=='lstm' or algorithm=='lstm_peephole':\n # Reset train state\n reset_train_state = tf.group(tf.assign(saved_state, tf.zeros([batch_size, num_nodes])),\n tf.assign(saved_output, tf.zeros([batch_size, num_nodes])))\n\n reset_valid_state = tf.group(tf.assign(saved_valid_state, tf.zeros([1, num_nodes])),\n tf.assign(saved_valid_output, tf.zeros([1, num_nodes])))\n \n # Reset test state. We use imputations in the test state reset\n reset_test_state = tf.group(\n saved_test_output.assign(tf.random_normal([1, num_nodes],stddev=0.01)),\n saved_test_state.assign(tf.random_normal([1, num_nodes],stddev=0.01)))\n \nelif algorithm=='gru':\n # Reset train state\n reset_train_state = [tf.assign(saved_output, tf.zeros([batch_size, num_nodes]))]\n\n # Reset valid state\n reset_valid_state = [tf.assign(saved_valid_output, tf.zeros([1, num_nodes]))]\n \n # Reset test state. We use imputations in the test state reset\n reset_test_state = [saved_test_output.assign(tf.random_normal([1, num_nodes],stddev=0.01))]\n\n",
"_____no_output_____"
]
],
[
[
"## Defining Learning Rate and the Optimizer with Gradient Clipping\nHere we define the learning rate and the optimizer we're going to use. We will be using the Adam optimizer as it is one of the best optimizers out there. Furthermore we use gradient clipping to prevent any gradient explosions.",
"_____no_output_____"
]
],
[
[
"# Used for decaying learning rate\ngstep = tf.Variable(0, trainable=False)\n\n# Running this operation will cause the value of gstep\n# to increase, while in turn reducing the learning rate\ninc_gstep = tf.assign(gstep, gstep+1)\n\n# Decays learning rate everytime the gstep increases\ntf_learning_rate = tf.train.exponential_decay(0.001,gstep,decay_steps=1, decay_rate=0.5)\n\n# Adam Optimizer. And gradient clipping.\noptimizer = tf.train.AdamOptimizer(tf_learning_rate)\n\ngradients, v = zip(*optimizer.compute_gradients(loss))\n# Clipping gradients\ngradients, _ = tf.clip_by_global_norm(gradients, 5.0)\n\noptimizer = optimizer.apply_gradients(\n zip(gradients, v))",
"_____no_output_____"
]
],
[
[
"## Greedy Sampling to Break the Repetition\nHere we write some simple logic to break the repetition in text. Specifically instead of always getting the word that gave this highest prediction probability, we sample randomly where the probability of being selected given by their prediction probabilities.",
"_____no_output_____"
]
],
[
[
"def sample(distribution):\n '''Greedy Sampling\n We pick the three best predictions given by the LSTM and sample\n one of them with very high probability of picking the best one'''\n best_inds = np.argsort(distribution)[-3:]\n best_probs = distribution[best_inds]/np.sum(distribution[best_inds])\n best_idx = np.random.choice(best_inds,p=best_probs)\n return best_idx",
"_____no_output_____"
]
],
[
[
"## Running the LSTM to Generate Text\n\nHere we train the model on the available data and generate text using the trained model for several steps. From each document we extract text for `steps_per_document` steps to train the model on. We also report the train perplexity at the end of each step. Finally we test the model by asking it to generate some new text starting from a randomly picked bigram.",
"_____no_output_____"
],
[
"### Learning rate Decay Logic\n\nHere we define the logic to decrease learning rate whenever the validation perplexity does not decrease",
"_____no_output_____"
]
],
[
[
"# Learning rate decay related\n# If valid perpelxity does not decrease\n# continuously for this many epochs\n# decrease the learning rate\ndecay_threshold = 5\n# Keep counting perplexity increases\ndecay_count = 0\n\nmin_perplexity = 1e10\n\n# Learning rate decay logic\ndef decay_learning_rate(session, v_perplexity):\n global decay_threshold, decay_count, min_perplexity \n # Decay learning rate\n if v_perplexity < min_perplexity:\n decay_count = 0\n min_perplexity= v_perplexity\n else:\n decay_count += 1\n\n if decay_count >= decay_threshold:\n print('\\t Reducing learning rate')\n decay_count = 0\n session.run(inc_gstep)",
"_____no_output_____"
]
],
[
[
"### Running Training, Validation and Generation\n\nWe traing the LSTM on existing training data, check the validaiton perplexity on an unseen chunk of text and generate a fresh segment of text",
"_____no_output_____"
]
],
[
[
"# Some hyperparameters needed for the training process\n\nnum_steps = 26\nsteps_per_document = 100\ndocs_per_step = 10\nvalid_summary = 1\ntrain_doc_count = num_files\n\nsession = tf.InteractiveSession()\n\n# Capture the behavior of train/valid perplexity over time\ntrain_perplexity_ot = []\nvalid_perplexity_ot = []\n\n# Initializing variables\ntf.global_variables_initializer().run()\nprint('Initialized Global Variables ')\n\naverage_loss = 0 # Calculates the average loss ever few steps\n\n# We use the first 10 documents that has \n# more than 10*steps_per_document bigrams for creating the validation dataset\n\n# Identify the first 10 documents following the above condition\nlong_doc_ids = []\nfor di in range(num_files):\n if len(data_list[di])>10*steps_per_document:\n long_doc_ids.append(di)\n if len(long_doc_ids)==10:\n break\n \n# Generating validation data\ndata_gens = []\nvalid_gens = []\nfor fi in range(num_files):\n # Get all the bigrams if the document id is not in the validation document ids\n if fi not in long_doc_ids:\n data_gens.append(DataGeneratorOHE(data_list[fi],batch_size,num_unrollings))\n # if the document is in the validation doc ids, only get up to the \n # last steps_per_document bigrams and use the last steps_per_document bigrams as validation data\n else:\n data_gens.append(DataGeneratorOHE(data_list[fi][:-steps_per_document],batch_size,num_unrollings))\n # Defining the validation data generator\n valid_gens.append(DataGeneratorOHE(data_list[fi][-steps_per_document:],1,1))\n\nfeed_dict = {}\nfor step in range(num_steps):\n \n for di in np.random.permutation(train_doc_count)[:docs_per_step]: \n doc_perplexity = 0\n for doc_step_id in range(steps_per_document):\n \n # Get a set of unrolled batches\n u_data, u_labels = data_gens[di].unroll_batches()\n \n # Populate the feed dict by using each of the data batches\n # present in the unrolled data\n for ui,(dat,lbl) in enumerate(zip(u_data,u_labels)): \n feed_dict[train_inputs[ui]] = dat\n feed_dict[train_labels[ui]] = lbl\n \n # Running the TensorFlow operations\n _, l, step_perplexity = session.run([optimizer, loss, train_perplexity_without_exp], \n feed_dict=feed_dict)\n \n # Update doc_perpelxity variable\n doc_perplexity += step_perplexity\n \n # Update the average_loss variable\n average_loss += step_perplexity\n \n # shows the training progress\n print('(%d).'%di,end='') \n \n # resetting hidden state after processing a single document\n # It's still questionable if this adds value in terms of learning\n # One one hand it's intuitive to reset the state when learning a new document\n # On the other hand this approach creates a bias for the state to be zero\n # We encourage the reader to investigate further the effect of resetting the state\n #session.run(reset_train_state) # resetting hidden state for each document\n session.run(reset_train_state) # resetting hidden state for each document\n \n print('')\n \n \n # Generate new samples\n if (step+1) % valid_summary == 0:\n \n # Compute average loss\n average_loss = average_loss / (valid_summary*docs_per_step*steps_per_document)\n \n # Print losses \n print('Average loss at step %d: %f' % (step+1, average_loss))\n print('\\tPerplexity at step %d: %f' %(step+1, np.exp(average_loss)))\n train_perplexity_ot.append(np.exp(average_loss))\n \n average_loss = 0 # reset loss\n \n valid_loss = 0 # reset loss\n \n # calculate valid perplexity\n for v_doc_id in range(10):\n # Remember we process things as bigrams\n # So need to divide by 2\n for v_step in range(steps_per_document//2):\n uvalid_data,uvalid_labels = valid_gens[v_doc_id].unroll_batches() \n\n # Run validation phase related TensorFlow operations \n v_perp = session.run(\n valid_perplexity_without_exp,\n feed_dict = {valid_inputs:uvalid_data[0],valid_labels: uvalid_labels[0]}\n )\n\n valid_loss += v_perp\n \n session.run(reset_valid_state)\n \n # Reset validation data generator cursor\n valid_gens[v_doc_id].reset_indices() \n \n print()\n v_perplexity = np.exp(valid_loss/(steps_per_document*10.0//2))\n print(\"Valid Perplexity: %.2f\\n\"%v_perplexity)\n valid_perplexity_ot.append(v_perplexity)\n \n decay_learning_rate(session, v_perplexity)\n\n # Generating new text ...\n # We will be generating one segment having 500 bigrams\n # Feel free to generate several segments by changing\n # the value of segments_to_generate\n print('Generated Text after epoch %d ... '%step) \n segments_to_generate = 1\n chars_in_segment = 500\n \n for _ in range(segments_to_generate):\n print('======================== New text Segment ==========================')\n \n # Start with a random word\n test_word = np.zeros((1,vocabulary_size),dtype=np.float32)\n test_word[0,data_list[np.random.randint(0,num_files)][np.random.randint(0,100)]] = 1.0\n print(\"\\t\",reverse_dictionary[np.argmax(test_word[0])],end='')\n \n # Generating words within a segment by feeding in the previous prediction\n # as the current input in a recursive manner\n for _ in range(chars_in_segment): \n sample_pred = session.run(test_prediction, feed_dict = {test_input:test_word}) \n next_ind = sample(sample_pred.ravel())\n test_word = np.zeros((1,vocabulary_size),dtype=np.float32)\n test_word[0,next_ind] = 1.0\n print(reverse_dictionary[next_ind],end='')\n print(\"\")\n \n # Reset train state\n session.run(reset_test_state)\n print('====================================================================')\n print(\"\")\n\nsession.close()\n\n# Write the perplexity data to a CSV\n\nwith open(filename_to_save, 'wt') as f:\n writer = csv.writer(f,delimiter=',')\n writer.writerow(train_perplexity_ot)\n writer.writerow(valid_perplexity_ot)",
"Initialized Global Variables \n(98).(25).(91).(5).(88).(49).(85).(96).(14).(73).\nAverage loss at step 1: 4.500272\n\tPerplexity at step 1: 90.041577\n\nValid Perplexity: 53.93\n\nGenerated Text after epoch 0 ... \n======================== New text Segment ==========================\n\t her, it the spirit, \"one his that to and said the money the spirit, and\nhere, and have, and\nthe gold all wile you that it the morester, and the spirit and had with ith the spirit, and hered hen hen that have the spirit, and the spiras, i will said the spirout. i will said, \"i will wout on that to and said, \"i wither in the bover, \"the spirit, \"one the father, \"i will said, \"the boy, and had to that it to the have\nto the father, \"and\nhere, that had come you came, and here, and here, and, \"the spirour wither as the money the spirler the spirit, i must the bected hen the boy that to you with the father to the first had come to the fore the monen the spneit, and have, answered and said, \"the gon that and he could hey the money you will sood that in ther, what have the spirit.\" the gold for to you the more his plaster, \"i will the fathen all had come, and wound in the boke, i will had come to the father, it you that then your had then your as you came, and have, and hen the boner to the had\n====================================================================\n\n(49).(87).(32).(14).(4).(51).(90).(16).(60).(43).\nAverage loss at step 2: 2.719010\n\tPerplexity at step 2: 15.165307\n\nValid Perplexity: 38.30\n\nGenerated Text after epoch 1 ... \n======================== New text Segment ==========================\n\t r, but the name, and the queen's allered that is name is the name, the queen was the name, and the queen was hease, and the little hands that he more himself in, and the man came the manikin two man, and the manikin was jumping, he pulled at his left leg so hard that is name in, and the little man whow to her leg were in his the deall the manikin his whole leg the queen's dever had told the names your name, the my the names that he plunged his right the little man came in, and the manikin she knew, that to\nthe\nname. but the name in the queen, what is my name in his whole his told you thatUNK\nthe devil has told yound the manikin said, is two my name.\n\non the little man came in, and foot so\nthe\nmanikin said, not the manikin said, is that is not not no the little man, and all the little man cantle the\nnauntribs, of the little man, and then in the little hands and the queen's dever's child, what is you thatUNK\nthe dever has hold. but\nshe had in, and\nthe little man, and in the night, that int\n====================================================================\n\n(48).(25).(81).(71).(45).(13).(0).(53).(28).(40).\nAverage loss at step 3: 2.477577\n\tPerplexity at step 3: 11.912361\n\nValid Perplexity: 32.62\n\nGenerated Text after epoch 2 ... \n======================== New text Segment ==========================\n\t asked his which she put of in two egg-should now will been the must splet down and said, i have done. when he said, i am to humble you can been to the king's heart, and she had driven her the most splendown the king throuhbeard of the king's daughter was too. i wish you will\nhappened to the corner of the king's evil danced, and that the most splet son her\nfor and the king's door this did now began in that her and will\nbe on which your promised that it down on this will down on the maid, i with you had to the\ncornest. i have been to the heart, and she was laughter and dide once with they down the maid to the comforted the pon the kindly, there\ntoo. and her and\nthen the prode, who she said to this days that when the maidUNKin-waiting came and put on her to the maidUNKin-waiting came and put on her the most splend and were of the hand that with your father and that the poor and\nbeen she was to this days wedding.\nthen the door, which your wife. but he said, be court sprangs the kind's ear\n====================================================================\n\n(78).(49).(12).(40).(27).(34).(89).(28).(66).(58).\nAverage loss at step 4: 2.076020\n\tPerplexity at step 4: 7.972671\n\nValid Perplexity: 50.50\n\nGenerated Text after epoch 3 ... \n======================== New text Segment ==========================\n\t out of which to the ground and broke. then they bought him his eyes for a while, and presently\nbegan to gather to the table, and\nhenceforth always let him eat with them, and likewise said nothing if\nhe did spill a little of anything. and they took the old grandfather to the table, and\nhenceforth always let him eat with them, and likewise said nothing if\nhe did spill a little of anything. i am making a little trough,\nanswered the child, for father and mother to eat with them, and likewise said nothing if\nhe did spill a little of anything. the old grandfather to the table while, and presently\nbegan to cry. then they took the old grandfather to the table, and\nhenceforth always let him eat with them, for a while, and presently\nbegan to cry. then they took the old grandfather to gather to the table, and\nhenceforth always let him eat with them, and likewise said nothing if\nhe did nothing if\nhe did spill a little of anything. i am making a little trough,\nanswered the child, for for a whi\n====================================================================\n\n(72).(5).(55).(2).(42).(75).(57).(80).(47).(14).\nAverage loss at step 5: 2.553451\n\tPerplexity at step 5: 12.851376\n\nValid Perplexity: 23.96\n\nGenerated Text after epoch 4 ... \n======================== New text Segment ==========================\n\t \nwither, as which,\nso the king, which should not he was came to the little said me that he tailor, and after her\nfather and bear, the little tailor, when he who had been\nboil, and they were\none of the king, and they were to be dought the little tailor was comforted the tree. then the king, who hans went one boy, and after her\nfather's death became to his have not the will tailor again the two\nother so low. i smote not\nliked the little tailor, who was at one of them to the board against the tailor. when the wild boy, and after her\nfather, and then the little tailor was and remadeed, and the little tailor and the little tailor, and tailor half of the tailor standing, and it was thouse, what he had heard the tailor so the little said, the tailor had faller asleep the treat with them, and then it, and they who who was no one of the king was thoubly and they will for\nhim, and then the king, who was caught, but they will forest again, who had heard the tree. the two giants and said than he\n====================================================================\n\n(25).(89).(52).(2).(63).(74).(61).(10).(56).(64).\nAverage loss at step 6: 2.086129\n\tPerplexity at step 6: 8.053676\n\nValid Perplexity: 24.38\n\nGenerated Text after epoch 5 ... \n======================== New text Segment ==========================\n\t e the bridegroom with the nut. immediately\ncame and said, nevery that they were she came to the bride she was ablew on the bride was on their deathe great before in the midst. and the bridegroom with the griffin the griffind there, but the bride was\nin sleep and said, i have been complaints, they were in the money she said, and the nut. immediately the chamber, and there in the chamber, and blood, and, and began to repating there she came the nut. and the bride had been as the bride was again led me, where they seated her where they for sease, but the princess there in the might of them and dragess, and there in the\nchamber there by that, and there in who had been the bird formed it, there went the\nprincess, who had been complace. then they sat down and said, i have been prepared\nthemselves and but on the chamber the prince went to the red of the princess, who was perfectly safe and they were in the\nchamber, and but on the chamber, and said, i have been complaints,\nand said, i will \n====================================================================\n\n(18).(96).(40).(95).(54).(2).(52).(37).(44).(55).\nAverage loss at step 7: 2.244664\n\tPerplexity at step 7: 9.437240\n\nValid Perplexity: 21.08\n\nGenerated Text after epoch 6 ... \n======================== New text Segment ==========================\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f6a0dc7be69fb2527c023359c4d08a11ea18a3 | 194,226 | ipynb | Jupyter Notebook | Bank_Fantasy/Golden_Bridge/recommendation_experiments/similarity_recommendations.ipynb | hanhanwu/Hanhan_Break_the_Limits | af6fd0bf333d4f7a8c96bf768a38ed8954a52222 | [
"MIT"
] | null | null | null | Bank_Fantasy/Golden_Bridge/recommendation_experiments/similarity_recommendations.ipynb | hanhanwu/Hanhan_Break_the_Limits | af6fd0bf333d4f7a8c96bf768a38ed8954a52222 | [
"MIT"
] | null | null | null | Bank_Fantasy/Golden_Bridge/recommendation_experiments/similarity_recommendations.ipynb | hanhanwu/Hanhan_Break_the_Limits | af6fd0bf333d4f7a8c96bf768a38ed8954a52222 | [
"MIT"
] | null | null | null | 73.794073 | 58,336 | 0.717834 | [
[
[
"# Similarity Recommendation\n\n* Collaborative Filtering\n * Similarity score is merchant similarity rank\n * Products list is most sold products in recent X weeks\n * Didn't choose most valuable products from `product_values` table is because they are largely overlapped with the top products in each merchant.\n * Also excluded the most sold products of the target merchant.\n * Avg daily purchase frequency is the count of each product in the list",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport datetime\nimport Levenshtein\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"import ray\n\nray.shutdown()",
"_____no_output_____"
],
[
"ray.init()",
"_____no_output_____"
],
[
"target_merchant = '49th Parallel Grocery'",
"_____no_output_____"
],
[
"all_order_train = pd.read_pickle('../all_order_train.pkl')\nall_order_test = pd.read_pickle('../all_order_test.pkl')\nprint(all_order_train.shape, all_order_test.shape)\n\nall_order_train.head()",
"(33720820, 12) (98286, 12)\n"
],
[
"target_train = all_order_train.loc[all_order_train['merchant'] == target_merchant]\ntarget_test = all_order_test.loc[all_order_test['merchant'] == target_merchant]\n\nprint(target_train.shape, target_test.shape)\ntarget_train.head()",
"(1365312, 12) (3850, 12)\n"
],
[
"all_order_train = all_order_train.loc[all_order_train['merchant'] != target_merchant]\nall_order_test = all_order_test.loc[all_order_test['merchant'] != target_merchant]\n\nprint(all_order_train.shape, all_order_test.shape)\nall_order_train.head()",
"(32355508, 12) (94436, 12)\n"
]
],
[
[
"## Merchant Similarity Score\n\n* Here, I converted the 3 similarity factors (top products, size, name) into 1 score, higher score represents higher similarity.\n* Commapring with sorting by 3 factors, 1 similarity score brings a bit different results.",
"_____no_output_____"
]
],
[
[
"@ray.remote\ndef get_merchant_data(merchant_df, top=10):\n merchant_size = merchant_df[['merchant', 'product_id']].astype('str').drop_duplicates()\\\n .groupby(['merchant'], as_index=False)['product_id']\\\n .agg('count').reset_index(drop=True).T.to_dict()\n merchant_data = merchant_size[0]\n merchant_data['product_ct'] = merchant_data.pop('product_id')\n \n top_prod_lst_df = merchant_df[['product_id', 'order_id']].astype('str').drop_duplicates()\\\n .groupby(['product_id'], as_index=False)['order_id']\\\n .agg('count').reset_index(drop=True)\\\n .sort_values(by='order_id', ascending=False)\\\n .head(n=top)\n top_prod_lst = list(top_prod_lst_df['product_id'].values)\n \n merchant_data['top_prod_lst'] = top_prod_lst\n \n return merchant_data\n\n\[email protected]\ndef get_merchant_similarity(target_merchant_dct, merchant_dct):\n prod_similarity = len(set(target_merchant_dct['top_prod_lst']).intersection(set(merchant_dct['top_prod_lst'])))\n size_similarity = abs(target_merchant_dct['product_ct'] - merchant_dct['product_ct'])\n name_similarity = Levenshtein.ratio(target_merchant_dct['merchant'], merchant_dct['merchant'])\n \n return {'merchant': merchant_dct['merchant'], 'prod_sim': prod_similarity, 'size_sim': size_similarity, 'name_sim': name_similarity}",
"_____no_output_____"
],
[
"target_merchant_train = get_merchant_data.remote(target_train[['merchant', 'product_id', 'order_id']], top=10)\ntarget_merchant_dct = ray.get(target_merchant_train)\n\nprint(target_merchant_dct)",
"{'merchant': '49th Parallel Grocery', 'product_ct': 37655, 'top_prod_lst': ['24852', '13176', '21137', '21903', '47209', '47766', '47626', '16797', '26209', '27845']}\n"
],
[
"merchant_lst = all_order_train['merchant'].unique()\n\nresults = [get_merchant_data.remote(all_order_train.loc[all_order_train['merchant']==merchant][['merchant', 'product_id', 'order_id']]) \n for merchant in merchant_lst]\nmerchant_data_lst = ray.get(results)\n\nprint(len(merchant_data_lst))\nmerchant_data_lst[7:9]",
"48\n"
],
[
"results = [get_merchant_similarity.remote(target_merchant_train, merchant_dct) for merchant_dct in merchant_data_lst]\nmerchant_similarity_lst = ray.get(results)\n\nmerchant_similarity_df = pd.DataFrame(merchant_similarity_lst)\nprint(merchant_similarity_df.shape)\n\nmerchant_similarity_df = merchant_similarity_df.sort_values(by=['prod_sim', 'size_sim', 'name_sim'], ascending=[False, True, False])\nmerchant_similarity_df.head()",
"(48, 4)\n"
],
[
"prod_sim_min = min(merchant_similarity_df['prod_sim'])\nprod_sim_max = max(merchant_similarity_df['prod_sim'])\n\nsize_sim_min = min(merchant_similarity_df['size_sim'])\nsize_sim_max = max(merchant_similarity_df['size_sim'])\n\nprint(prod_sim_min, prod_sim_max, size_sim_min, size_sim_max)",
"9 10 20 23859\n"
],
[
"def get_similarity_score(r):\n similarity = (r['prod_sim'] - prod_sim_min)/(prod_sim_max - prod_sim_min) * (size_sim_max - r['size_sim'])/(size_sim_max - size_sim_min) * r['name_sim']\n \n return round(similarity, 4)",
"_____no_output_____"
],
[
"merchant_similarity_df['similarity_score'] = merchant_similarity_df.apply(get_similarity_score, axis=1)\nmerchant_similarity_df = merchant_similarity_df.sort_values(by='similarity_score', ascending=False)\n\nmerchant_similarity_df.head()",
"_____no_output_____"
]
],
[
[
"## Recent Popular Products\n\nExcluding top products of the target merchant.",
"_____no_output_____"
]
],
[
[
"all_order_train.head()",
"_____no_output_____"
],
[
"latest_period = 2 # in weeks\nweek_lst = sorted(all_order_train['week_number'].unique())[-latest_period:]\nweek_lst",
"_____no_output_____"
],
[
"prod_ct_df = all_order_train.loc[all_order_train['week_number'].isin(week_lst)][['product_id', 'product_name', 'order_id']].astype('str').drop_duplicates()\\\n .groupby(['product_id', 'product_name'], as_index=False)['order_id']\\\n .agg('count').reset_index(drop=True)\\\n .sort_values(by='order_id', ascending=False)\n\n# remove product_id that's in target merchant's top popular products\nprod_ct_df = prod_ct_df.loc[~prod_ct_df['product_id'].isin(target_merchant_dct['top_prod_lst'])]\nprod_ct_df.head()",
"_____no_output_____"
],
[
"n = 20\nproduct_lst = prod_ct_df['product_id'].values[:n]\nprint(product_lst)\nprint()\nprint(prod_ct_df['product_name'].values[:n])",
"['49683' '24964' '27966' '22935' '39275' '45007' '28204' '4605' '42265'\n '44632' '5876' '4920' '40706' '30391' '30489' '8518' '27104' '45066'\n '5077' '17794']\n\n['Cucumber Kirby' 'Organic Garlic' 'Organic Raspberries'\n 'Organic Yellow Onion' 'Organic Blueberries' 'Organic Zucchini'\n 'Organic Fuji Apple' 'Yellow Onions' 'Organic Baby Carrots'\n 'Sparkling Water Grapefruit' 'Organic Lemon' 'Seedless Red Grapes'\n 'Organic Grape Tomatoes' 'Organic Cucumber' 'Original Hummus'\n 'Organic Red Onion' 'Fresh Cauliflower' 'Honeycrisp Apple'\n '100% Whole Wheat Bread' 'Carrots']\n"
]
],
[
[
"## Collaborative Filtering",
"_____no_output_____"
]
],
[
[
"merchant_similarity_df.head()",
"_____no_output_____"
],
[
"all_order_train.head()",
"_____no_output_____"
],
[
"n_merchant = 10\nsimilar_merchant_lst = merchant_similarity_df['merchant'].values[:n_merchant]\nmerchant_similarity_lst = merchant_similarity_df['similarity_score'].values[:n_merchant]\n\[email protected]\ndef get_product_score(prod_df, product_id, product_name):\n total_weighted_frequency = 0.0\n total_similarity = 0.0\n \n for i in range(len(similar_merchant_lst)):\n merchant = similar_merchant_lst[i]\n tmp_df = prod_df.loc[prod_df['merchant']==merchant]\n if tmp_df.shape[0] > 0:\n daily_avg = tmp_df['order_id'].nunique()/tmp_df['purchase_date'].nunique()\n similarity = merchant_similarity_lst[i]\n \n total_similarity += similarity\n total_weighted_frequency += similarity * daily_avg\n prod_score = total_weighted_frequency/total_similarity\n \n return {'product_id': product_id, 'product_name': product_name, 'prod_score': round(prod_score, 4)}",
"_____no_output_____"
],
[
"prod_score_lst = [get_product_score.remote(all_order_train.loc[all_order_train['product_id']==int(product_lst[i])][['merchant', 'order_id', 'purchase_date']],\n product_lst[i], prod_ct_df['product_name'].values[i])\n for i in range(len(product_lst))]",
"_____no_output_____"
],
[
"prod_score_df = pd.DataFrame(ray.get(prod_score_lst))\nprod_score_df = prod_score_df.sort_values(by='prod_score', ascending=False)\nprod_score_df",
"_____no_output_____"
]
],
[
[
"## Forecasting Recommendations",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.metrics import mean_squared_error\nfrom math import sqrt\nimport matplotlib.pyplot as plt\n\n# the logger here is to remove the warnings about plotly\nimport logging\nlogger = logging.getLogger('fbprophet.plot')\nlogger.setLevel(logging.CRITICAL)\nfrom fbprophet import Prophet\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"sample_train_df1 = pd.read_pickle('../sample_train_df1.pkl')\nsample_test_df1 = pd.read_pickle('../sample_test_df1.pkl')\nprint(sample_train_df1.shape, sample_test_df1.shape)",
"(100, 2) (14, 2)\n"
],
[
"train1_col = sample_train_df1['purchase_amount']\ntest1_col = sample_test_df1['purchase_amount']\n\n# Generate logged moving average for both time series sequences\nts_log_train1 = np.log(train1_col)\nts_moving_avg_train1 = ts_log_train1.rolling(window=4,center=False).mean()\nts_log_test1 = np.log(test1_col)\nts_moving_avg_test1 = ts_log_test1.rolling(window=4,center=False).mean()\n\nts_moving_avg_train1.head(n=10)",
"_____no_output_____"
],
[
"ts_ma_train1 = pd.DataFrame(ts_moving_avg_train1.copy())\nts_ma_train1['ds'] = ts_ma_train1.index\nts_ma_train1['y'] = ts_moving_avg_train1.values\nts_ma_train1.drop(['purchase_amount'], inplace=True, axis=1)\nprint(ts_ma_train1.shape)\n\nts_ma_test1 = pd.DataFrame(ts_moving_avg_test1.copy())\nts_ma_test1['ds'] = ts_ma_test1.index\nts_ma_test1['y'] = ts_moving_avg_test1.values\nts_ma_test1.drop(['purchase_amount'], inplace=True, axis=1)\nprint(ts_ma_test1.shape)\n\nts_ma_train1.head()",
"(100, 2)\n(14, 2)\n"
],
[
"latest_period = 14\nforecast_period = 7\n\ntrain = ts_ma_train1.tail(n=latest_period)\ntest = ts_ma_test1.head(n=forecast_period)\n\nprint(train.shape, test.shape)\n\ntrain.head()",
"(14, 2) (7, 2)\n"
],
[
"prophet_model = Prophet(daily_seasonality = True, yearly_seasonality=False, weekly_seasonality=False,\n seasonality_mode = 'multiplicative', n_changepoints=5,\n changepoint_prior_scale=0.05, seasonality_prior_scale=0.1)\nprophet_model.fit(train)\n \nperiods = len(test.index)\nfuture = prophet_model.make_future_dataframe(periods=periods)\nforecast = prophet_model.predict(future)\n\nprint(train.shape, test.shape, forecast.shape)\nall_ts = train.append(test).dropna()\nselected_forecast = forecast.loc[forecast['ds'].isin(all_ts.index)]\nrmse = round(sqrt(mean_squared_error(all_ts['y'].values, selected_forecast['yhat'].values)), 4)\nprint(rmse)\n\nforecast.head()",
"(14, 2) (7, 2) (21, 16)\n0.0391\n"
],
[
"exp_forecast = forecast[['ds', 'yhat']]\nexp_forecast['y_origin'] = np.exp(exp_forecast['yhat'])\nexp_forecast.head()",
"_____no_output_____"
],
[
"original_ts = sample_train_df1.iloc[sample_train_df1.index.isin(train.index)][['purchase_amount']]\noriginal_ts = original_ts.append(sample_test_df1.iloc[sample_test_df1.index.isin(test.index)][['purchase_amount']])\nprint(original_ts.shape)\n\nplt.figure(figsize=(16,7))\nplt.plot(original_ts.index, original_ts, label='Original Values', color='green')\nplt.plot(exp_forecast['ds'], exp_forecast['y_origin'].values, label='Forecasted Values', color='purple')\nplt.legend(loc='best')\nplt.title(\"Sample 1 - Original Values vs Forecasted Values (Without Recommended Products) - RMSE:\" + str(rmse))\nplt.show()",
"(21, 1)\n"
],
[
"product_values_df = pd.read_pickle('product_values.pkl')\nproduct_values_df.head()",
"_____no_output_____"
],
[
"product_values_df['product_id'] = product_values_df['product_id'].astype(str)\nprod_score_sales_df = prod_score_df.merge(product_values_df[['product_id', 'avg_daily_sales']], on='product_id')\n\nprod_score_sales_df.head()",
"_____no_output_____"
],
[
"test_ct = 20\ndaily_sales_increase = 0\n\noriginal_ts = sample_train_df1.iloc[sample_train_df1.index.isin(train.index)][['purchase_amount']]\noriginal_ts = original_ts.append(sample_test_df1.iloc[sample_test_df1.index.isin(test.index)][['purchase_amount']])\nprint(original_ts.shape)\n\nexp_forecast['y_forecast'] = exp_forecast['y_origin']\nforecast_ts_train = exp_forecast.head(n=latest_period)\nforecast_ts_test = exp_forecast.tail(n=forecast_period)\n\nfor idx, r in prod_score_sales_df.iterrows():\n added_daily_sales = r['avg_daily_sales']\n forecast_ts_test['y_forecast'] += added_daily_sales\n \n daily_sales_increase += added_daily_sales\n \n if idx >= test_ct:\n break",
"(21, 1)\n"
],
[
"forecast_ts = forecast_ts_train.append(forecast_ts_test)\nprint('Total sales increased: ' + str(daily_sales_increase * forecast_period))\n\nplt.figure(figsize=(16,7))\nplt.plot(original_ts.index, original_ts, label='Original Values', color='green')\nplt.plot(exp_forecast['ds'], exp_forecast['y_origin'].values, label='Forecasted Values No Recommendation', color='purple')\nplt.plot(forecast_ts['ds'], forecast_ts['y_forecast'].values, label='Forecasted Values With Recommendation', color='orange')\nplt.legend(loc='best')\nplt.title(\"Sample 1 - Original Values vs Forecasted Values (With Recommended Products) - Daily Sales Increased: \" + str(daily_sales_increase))\nplt.show()",
"Total sales increased: 2162.51\n"
]
],
[
[
"## Summary\n\n* If we recommend the top 7 products as popularity_recommendation method, the daily increase in this method is still higher.\n* If we want the forecasting curve obvious, mainly need to increase the number of recommended products.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e7f6a10d670424d4909840520e716eab8c5125ea | 13,674 | ipynb | Jupyter Notebook | semana-02/lista-exercicio/lista-3/poo2-lista3-larissa_justen.ipynb | larissajusten/ufsc-object-oriented-programming | 839e6abcc20580ea1a47479232c3ed3cb0153e4b | [
"MIT"
] | 6 | 2021-11-29T05:43:19.000Z | 2022-03-15T21:54:54.000Z | semana-02/lista-exercicio/lista-3/poo2-lista3-larissa_justen.ipynb | larissajusten/ufsc-object-oriented-programming | 839e6abcc20580ea1a47479232c3ed3cb0153e4b | [
"MIT"
] | 3 | 2021-11-21T03:44:03.000Z | 2021-11-21T03:44:05.000Z | semana-02/lista-exercicio/lista-3/poo2-lista3-larissa_justen.ipynb | larissajusten/ufsc-object-oriented-programming | 839e6abcc20580ea1a47479232c3ed3cb0153e4b | [
"MIT"
] | null | null | null | 33.930521 | 1,009 | 0.551192 | [
[
[
"1️⃣ **Exercício 1.** Escreva uma função que conta a frequência de ocorrência de cada palavra em um texto (arquivo txt) e armazena tal quantidade em um dicionário, onde a chave é a vogal considerada.",
"_____no_output_____"
],
[
"**Correção:** \"onde a chave é a PALAVRA considerada\"",
"_____no_output_____"
]
],
[
[
"from collections import Counter",
"_____no_output_____"
],
[
"def count_palavras(nome_arquivo: str):\n file = open(f'{nome_arquivo}.txt', 'rt')\n texto = file.read()\n\n palavras = [palavra for palavra in texto.split(' ')]\n dicionario = dict(Counter(palavras))\n # dicionario2 = {i: palavras.count(i) for i in list(set(palavras))}\n\n return dicionario",
"_____no_output_____"
],
[
"nome_arquivo = input('Digite o nome do arquivo de texto: ')\ndicionario = count_palavras(nome_arquivo)\nprint(dicionario)",
"Digite o nome do arquivo de texto: teste\n{'Gostaria': 1, 'de': 2, 'enfatizar': 1, 'que': 1, 'a': 2, 'hegemonia': 1, 'do': 3, 'ambiente': 2, 'político': 1, 'obstaculiza': 1, 'apreciação': 1, 'fluxo': 1, 'informações.': 1}\n"
]
],
[
[
"2️⃣ **Exercício 2.** Escreva uma função que apaga do dicionário anterior, todas as palavras que sejam ‘stopwords’.\nVer https://gist.github.com/alopes/5358189",
"_____no_output_____"
]
],
[
[
"stopwords = ['de', 'a', 'o', 'que', 'e', 'do', 'da', 'em', 'um', 'para', 'é', 'com', 'não', 'uma', 'os', 'no', 'se', 'na', 'por', 'mais', 'as', 'dos', 'como', 'mas', 'foi', 'ao', 'ele', 'das', 'tem', 'à', 'seu', 'sua', 'ou', 'ser', 'quando', 'muito', 'há', 'nos', 'já', 'está', 'eu', 'também', 'só', 'pelo', 'pela', 'até', 'isso', 'ela', 'entre', 'era', 'depois', 'sem', 'mesmo', 'aos', 'ter', 'seus', 'quem', 'nas', 'me', 'esse', 'eles', 'estão', 'você', 'tinha', 'foram', 'essa', 'num', 'nem', 'suas', 'meu', 'às', 'minha', 'têm', 'numa', 'pelos', 'elas', 'havia', 'seja', 'qual', 'será', 'nós', 'tenho', 'lhe', 'deles', 'essas', 'esses', 'pelas', 'este', 'fosse', 'dele', 'tu', 'te', 'vocês', 'vos', 'lhes', 'meus', 'minhas', 'teu', 'tua', 'teus', 'tuas', 'nosso', 'nossa', 'nossos', 'nossas', 'dela', 'delas', 'esta', 'estes', 'estas', 'aquele', 'aquela', 'aqueles', 'aquelas', 'isto', 'aquilo', 'estou', 'está', 'estamos', 'estão', 'estive', 'esteve', 'estivemos', 'estiveram', 'estava',\n 'estávamos', 'estavam', 'estivera', 'estivéramos', 'esteja', 'estejamos', 'estejam', 'estivesse', 'estivéssemos', 'estivessem', 'estiver', 'estivermos', 'estiverem', 'hei', 'há', 'havemos', 'hão', 'houve', 'houvemos', 'houveram', 'houvera', 'houvéramos', 'haja', 'hajamos', 'hajam', 'houvesse', 'houvéssemos', 'houvessem', 'houver', 'houvermos', 'houverem', 'houverei', 'houverá', 'houveremos', 'houverão', 'houveria', 'houveríamos', 'houveriam', 'sou', 'somos', 'são', 'era', 'éramos', 'eram', 'fui', 'foi', 'fomos', 'foram', 'fora', 'fôramos', 'seja', 'sejamos', 'sejam', 'fosse', 'fôssemos', 'fossem', 'for', 'formos', 'forem', 'serei', 'será', 'seremos', 'serão', 'seria', 'seríamos', 'seriam', 'tenho', 'tem', 'temos', 'tém', 'tinha', 'tínhamos', 'tinham', 'tive', 'teve', 'tivemos', 'tiveram', 'tivera', 'tivéramos', 'tenha', 'tenhamos', 'tenham', 'tivesse', 'tivéssemos', 'tivessem', 'tiver', 'tivermos', 'tiverem', 'terei', 'terá', 'teremos', 'terão', 'teria', 'teríamos', 'teriam']",
"_____no_output_____"
],
[
"def delete_stopwords(dicionario):\n for stopword in stopwords:\n if stopword in dicionario.keys():\n dicionario.pop(stopword, None)\n\n return dicionario",
"_____no_output_____"
],
[
"nome_arquivo = input('Digite o nome do arquivo de texto: ')\ndicionario = count_palavras(nome_arquivo)\nprint(f'\\nDicionario: {dicionario}')\nnovo_dicionario = delete_stopwords(dicionario)\nprint(f'\\nApos apagar stopwords: {novo_dicionario}')",
"Digite o nome do arquivo de texto: teste\n\nDicionario: {'Gostaria': 1, 'de': 2, 'enfatizar': 1, 'que': 1, 'a': 2, 'hegemonia': 1, 'do': 3, 'ambiente': 2, 'político': 1, 'obstaculiza': 1, 'apreciação': 1, 'fluxo': 1, 'informações.': 1}\n\nApos apagar stopwords: {'Gostaria': 1, 'enfatizar': 1, 'hegemonia': 1, 'ambiente': 2, 'político': 1, 'obstaculiza': 1, 'apreciação': 1, 'fluxo': 1, 'informações.': 1}\n"
]
],
[
[
"3️⃣ **Exercício 3.** Escreva um programa que lê duas notas de vários alunos e armazena tais notas em um dicionário, onde a chave é o nome do aluno. A entrada de dados deve terminar quando for lida uma string vazia como nome. Escreva uma função que retorna a média do aluno, dado seu nome.",
"_____no_output_____"
]
],
[
[
"def le_notas(dicionario = {}):\n nome_aluno = input('Digite o nome do aluno: ')\n if nome_aluno.isalpha() and nome_aluno not in dicionario.keys():\n nota1 = float(input('Digite a primeira nota: (somente numeros) '))\n nota2 = float(input('Digite a segunda nota: (somente numeros) '))\n dicionario[nome_aluno] = [nota1, nota2]\n le_notas(dicionario)\n elif nome_aluno in dicionario.keys():\n print('Aluno ja adicionado!')\n le_notas(dicionario)\n\n return dicionario",
"_____no_output_____"
],
[
"def retorna_nota_aluno(dicionario, nome_aluno):\n return (dicionario[nome_aluno][0] + dicionario[nome_aluno][1]) / 2",
"_____no_output_____"
],
[
"dicionario = le_notas()\nnome_aluno = input('\\nDigite o nome do aluno que deseja saber a nota: ')\n\nif dicionario and nome_aluno in dicionario.keys():\n media = retorna_nota_aluno(dicionario, nome_aluno)\n print(f'{nome_aluno}: {media}')",
"Digite o nome do aluno: Larissa\nDigite a primeira nota: (somente numeros) 1\nDigite a segunda nota: (somente numeros) 2\nDigite o nome do aluno: Jesus\nDigite a primeira nota: (somente numeros) 0\nDigite a segunda nota: (somente numeros) 0\nDigite o nome do aluno: \n\nDigite o nome do aluno que deseja saber a nota: Jesus\nJesus: 0.0\n"
]
],
[
[
"4️⃣ **Exercício 4.** Uma pista de Kart permite 10 voltas para cada um de 6 corredores. Escreva um programa que leia todos os tempos em segundos e os guarde em um dicionário, onde a chave é o nome do corredor. Ao final diga de quem foi a melhor volta da prova e em que volta; e ainda a classificação final em ordem (1o o campeão). O campeão é o que tem a menor média de tempos.",
"_____no_output_____"
]
],
[
[
"def le_tempos_corridas(array_tempos=[], numero_voltas=0):\n if numero_voltas < 10:\n tempo_volta = float(\n input(f'[{numero_voltas+1}] Digite o tempo: (numerico/seg) '))\n if tempo_volta > 0:\n array_tempos.append(tempo_volta)\n le_tempos_corridas(array_tempos, numero_voltas+1)\n else:\n print('# Valor invalido no tempo da volta!')\n le_tempos_corridas(array_tempos, numero_voltas)\n return array_tempos",
"_____no_output_____"
],
[
"def le_corredores(dicionario={}, num_corredores=0):\n if num_corredores < 6:\n nome_corredor = input(\n f'[{num_corredores+1}] Digite o nome do corredor: ')\n if nome_corredor.isalpha():\n array_tempos = le_tempos_corridas(array_tempos=[])\n dicionario[nome_corredor] = sorted(array_tempos)\n le_corredores(dicionario, num_corredores+1)\n else:\n print('# Valor invalido no nome do corredor!')\n le_corredores(dicionario, num_corredores)\n return dicionario",
"_____no_output_____"
],
[
"def calc_media_tempos(dicionario):\n return {corredor: sum(array_tempos)/len(array_tempos) for corredor, array_tempos in dicionario.items()}",
"_____no_output_____"
],
[
"dicionario = le_corredores()\n\nfor i in sorted(dicionario, key=dicionario.get):\n print(\n f'# {i.capitalize()} teve a melhor volta com duracao de {dicionario[i][0]} segundos!')\n break\n\ndicionario_medias = calc_media_tempos(dicionario)\nfor index, i in enumerate(sorted(dicionario_medias, key=dicionario_medias.get)):\n print(\n f'[{index+1} Lugar] {i.capitalize()} com media de {dicionario_medias[i]} segundos!')\n if index == 2:\n break",
"[1] Digite o nome do corredor: Larissa\n[1] Digite o tempo: (numerico/seg) 10\n[2] Digite o tempo: (numerico/seg) 15\n[2] Digite o nome do corredor: Jesus\n[1] Digite o tempo: (numerico/seg) 0\n# Valor invalido no tempo da volta!\n[1] Digite o tempo: (numerico/seg) 1\n[2] Digite o tempo: (numerico/seg) 1\n# Jesus teve a melhor volta com duracao de 1.0 segundos!\n[1 Lugar] Jesus com media de 1.0 segundos!\n[2 Lugar] Larissa com media de 12.5 segundos!\n"
]
],
[
[
"6️⃣ **Exercício 6.** Criar 10 frozensets com 30 números aleatórios cada, e construir um dicionário que contenha a soma de cada um deles.",
"_____no_output_____"
]
],
[
[
"import random",
"_____no_output_____"
],
[
"def get_random_set(size):\n return frozenset(random.sample(range(1, 100), size))",
"_____no_output_____"
],
[
"def get_random_sets(size, num_sets):\n return [get_random_set(size) for _ in range(num_sets)]",
"_____no_output_____"
],
[
"def get_dict_from_sets_sum(sets):\n return {key: sum(value) for key, value in enumerate(sets)}",
"_____no_output_____"
],
[
"_sets = get_random_sets(30, 10)\n_dict = get_dict_from_sets_sum(_sets)\nprint(_dict)",
"{0: 1334, 1: 1552, 2: 1762, 3: 1387, 4: 1535, 5: 1672, 6: 1422, 7: 1572, 8: 1567, 9: 1562}\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7f6a163aa4c501ec65925ae316d6e0f2982794a | 18,152 | ipynb | Jupyter Notebook | decipherd/notebooks/deciphrd_v2_BCM.ipynb | berebolledo/sofia | f5f4e0f5891213c4548aad193ed32a2a911c8073 | [
"MIT"
] | null | null | null | decipherd/notebooks/deciphrd_v2_BCM.ipynb | berebolledo/sofia | f5f4e0f5891213c4548aad193ed32a2a911c8073 | [
"MIT"
] | null | null | null | decipherd/notebooks/deciphrd_v2_BCM.ipynb | berebolledo/sofia | f5f4e0f5891213c4548aad193ed32a2a911c8073 | [
"MIT"
] | null | null | null | 30.610455 | 190 | 0.515315 | [
[
[
"# Import python modules\nimport os\nimport datetime\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# Requires the following databses\n# phenotypes_db.txt : database of genes and known phenotypes associations\n# mim2gene_mod.txt : gene mim ID\n# pLI.gnomad.txt : gnomad pLI calculation per gene\nlocalfiles = \"/Users/boris/GoogleDrive/UDD/research/bioinformatics/SABIO/projects/01_DECIPHERD/00_run_pipeline/\"\nphenotypes_db = localfiles + \"/databases/phenotypes_db.txt\"\nmim2gene_db = localfiles + \"/databases/mim2gene_mod.txt\"\npLI_db = localfiles + \"/databases/pLI.gnomad.txt\"",
"_____no_output_____"
],
[
"# Inputs\ntimestamp = str(datetime.datetime.now().strftime(\"%Y-%m-%d_%H_%M\"))\nrun = 'run004'\nprefix = \"UDD034\"\ninputdir = localfiles + \"/\" + run + \"/\" + prefix + \"_inputfiles/\"\noutdir = localfiles + \"/\" + run + \"/\" + prefix + \"_\" + timestamp + \"/\"\nwannovar_file = inputdir + prefix + \"_annovar.txt\"\nintervar_file = inputdir + prefix + \"_intervar.txt\"\nphenotips_file = inputdir + prefix + \"_phenotips.tsv\"\nsophia_file = inputdir + prefix + \"_sophia.xls\"",
"_____no_output_____"
],
[
"# The function takes frequency values. If frequency values exist, they are compared to the AF<=0.01, otherwise, if\n# no info is available, keep the site anyway\ndef freq_check(values, NotANum):\n results = 0\n for freq in values:\n if freq is NotANum:\n freq2 = 0\n else:\n freq2 = freq\n try:\n f = float(freq2)\n if f <= 0.01:\n results+-1\n else:\n results+=1\n except:\n results+=0\n if results <= 0:\n return \"rare|unknown\"\n else:\n return \"common\"\n\n# The function takes impact values. If impact values exist, at least two criteria agree with functional impact, otherwise, if\n# no info is available, keep the site anyway\ndef impact_check(values, NotANum):\n results = 0\n for impact in values:\n if impact is NotANum:\n imp2 = \"D\"\n else:\n imp2 = impact\n try:\n i = str(imp2)\n if i in [\"D\", \"P\", \"H\", \"M\", \"A\"]:\n results+=-1\n else:\n results+=1\n except:\n results+=0\n if results <= 0:\n return \"affected|unknown\"\n else:\n return \"unaffected\"\n\n# This function check for DP of the variant call >=50\ndef check_DP(depth, cutoff=50):\n #dp = depth.strip().split(\":\")[2]\n dp = depth\n if float(dp) >= cutoff:\n return True\n else:\n return False\n\n# This function check for GT \ndef check_GT(genotype, gt_default='1/1'):\n gt = genotype.strip().split(\":\")[0]\n if gt == gt_default:\n return True\n else:\n return False\n\n# This function splits DP and GT \ndef split_format(fmt, n):\n out = fmt.strip().split(\":\")[n]\n if n in [2,3]:\n return int(out)\n else:\n return out\n\n# Determine zygosity\ndef zygosity(genotype):\n A1,A2 = genotype.split(\"/\")\n if A1==A2:\n return \"hom\"\n else:\n return \"het\"\n\ndef compound(genes, gene_occ):\n gene_list = str(genes).split(',')\n occurence = [gene_occ[gene] for gene in gene_list]\n if len([i for i in occurence if i>1]) !=0:\n return 'true'\n else:\n return 'false'\n\ndef pheno_dic(entry):\n genes = entry[0]\n pheno = entry[1]\n for gene in genes.split(','):\n return (gene,pheno)\n\ndef get_pheno(genes,phenodb):\n phenotypes = []\n for gene in str(genes).split(','):\n try:\n phenotypes.append(phenodb[gene])\n except:\n pass\n return ''.join(list(set(phenotypes)))\n\ndef get_info(info, val):\n info_values = info.split(';')\n tmp = [i for i in info_values if i.startswith(val)]\n try:\n out = float(tmp[0].split(\"=\")[1])\n except:\n out = ''\n return out\n\ndef check_gene(genes, db):\n genelist = str(genes).split(',')\n match = [g for g in genelist if g in db.keys()]\n if match:\n n, feat, hpo = pheno_db[match[0]]\n return pd.Series([\"yes\", n, feat, hpo])\n else:\n return pd.Series([\"no\",0,'-','-'])",
"_____no_output_____"
],
[
"% mkdir {outdir}",
"_____no_output_____"
],
[
"# Read genomic data table generated by uploading HC-GATK raw VCF to http://wannovar.wglab.org/ \ndata = pd.read_csv(wannovar_file, low_memory=False, sep='\\t')",
"_____no_output_____"
],
[
"inter = pd.read_csv(intervar_file, low_memory=False, sep='\\t')\ninter_min = inter[[list(inter.columns)[i] for i in [0,1,2,3,4,12,13]]]\ninter_min.columns = [\"Chr\", \"Start\", \"End\", \"Ref\", \"Alt\" , \"ClinVar\", \"InterVar\"]\nclinvar_col = list(inter_min['ClinVar'].apply(lambda x: x.split(':')[1].strip()))\nintervar_col = list(inter_min['InterVar'].apply(lambda x: x.split(\":\")[1].split(\"P\")[0].strip()))\n\ninter_def = inter_min.iloc[:,0:5]\ninter_def['ClinVar'] = clinvar_col\ninter_def['ACMG_InterVar'] = intervar_col\ndel inter\ndel inter_min\ndel clinvar_col\ndel intervar_col\n\nclinical = inter_def.drop_duplicates()\ndel inter_def",
"_____no_output_____"
],
[
"tmp = data.merge(clinical, how='left', on=list(clinical.columns)[:-2])\ndel data\ndel clinical\ndata = tmp",
"_____no_output_____"
],
[
"# Add MIM phenotypes info\npheno_0 = pd.read_csv(phenotypes_db, header=None, sep='\\t')\npheno_0.columns = ['Gene.refgene','Phenotypes']\npheno_1 = pheno_0.dropna()\nphenodbase = dict(list(pheno_1.apply(pheno_dic, axis=1)))\ndata[\"Phenotypes\"] = data['Gene.refGene'].apply(get_pheno, phenodb=phenodbase)\n\ndel pheno_0\ndel pheno_1\ndel phenotypes_db",
"_____no_output_____"
],
[
"#Add MIM gene info\nomim = pd.read_csv(mim2gene_db, low_memory=False, header=None, sep='\\t')\nomim.columns = ['mim', 'Gene.refGene']\nweblinks = omim.mim.apply(lambda x: '=HYPERLINK(\"https://www.omim.org/entry/%s\", \"%s\")' % (str(x), str(x)))\nomim['mim'] = weblinks\ndata_2 = pd.merge(data, omim, how = 'left')\n\ndel omim\ndel mim2gene_db",
"_____no_output_____"
],
[
"#Add pLI gene info\npli = pd.read_csv(pLI_db, low_memory=False, sep='\\t')\npli.columns = ['Gene.refGene', 'pLI']\ndata_3 = pd.merge(data_2, pli, how = 'left')\n\ndel pli\ndel pLI_db",
"_____no_output_____"
],
[
"# Add cleaner columns to data table \ndel data\ndel data_2\ndata = data_3\n\ndata[\"Genotype\"] = data[\"Otherinfo.12\"].apply(split_format, n=0)\ndata[\"Genotype.qual\"] = data[\"Otherinfo.12\"].apply(split_format, n=3)\ndata[\"Depth\"] = data[\"Otherinfo.12\"].apply(split_format, n=2)\ndata[\"Zygosity\"] = data['Genotype'].apply(zygosity)\ndata[\"Allele_counts\"] = data[\"Otherinfo.12\"].apply(split_format, n=1)\ndata[\"Site.qual\"] = data[\"Otherinfo.8\"]\ndata[\"strand.FS\"] = data['Otherinfo.10'].apply(get_info, val='FS')\ndata[\"strand.OR\"] = data['Otherinfo.10'].apply(get_info, val='SOR')\n\ndata[\"CHROM\"] = data[\"Otherinfo.3\"]\ndata[\"POS\"] = data[\"Otherinfo.4\"]\ndata[\"REF\"] = data[\"Otherinfo.6\"]\ndata[\"ALT\"] = data[\"Otherinfo.7\"]\n\ndata['Effect'] = data['GeneDetail.refGene'].str.cat(data['AAChange.refGene'], sep =\",\") ",
"_____no_output_____"
],
[
"Snp = []\nfor snp, chrom, pos, ref,alt in data[['avsnp147', 'CHROM','POS','REF','ALT']].values:\n if snp!='.':\n Snp.append('=HYPERLINK(\"https://www.ncbi.nlm.nih.gov/snp/%s\", \"%s\")' % (snp, snp))\n else:\n loc = \"%s:%s%s>%s\" % (chrom,pos,ref,alt)\n Snp.append('=HYPERLINK(\"https://gnomad.broadinstitute.org/region/%s-%s-%s\", \"%s\")' % (chrom,pos,pos,loc))\n\ndata['dbSNP147'] = Snp ",
"_____no_output_____"
],
[
"# transform dbSNP to weblink\n#snp = data['avsnp147'].apply(lambda x: '=HYPERLINK(\"https://www.ncbi.nlm.nih.gov/snp/%s\", \"%s\")' % (str(x), str(x)))\n#data['dbSNP147'] = snp",
"_____no_output_____"
],
[
"# Extract frequency databases column names\nfreq_cols_tmp = [col for col in list(data.columns) if col.endswith(\"_ALL\") or col.endswith(\"_all\")]\nfreq_cols = [freq_cols_tmp[i] for i in [0,2,3,4]]\n\n# Extract Clinvar column\n#clinVar_cols = [col for col in list(data.columns) if col.startswith(\"ClinVar\")][1:]\nclinVar_cols = [\"CLNSIG\"]\n\n# Define impact criterias to be considered\nimpact_cols = [\"SIFT_pred\", \"Polyphen2_HDIV_pred\", \"Polyphen2_HVAR_pred\", \n \"MutationTaster_pred\", \"MutationAssessor_pred\"]",
"_____no_output_____"
],
[
"filter_a = ~( data[\"ExonicFunc.refGene\"].isin([\"synonymous SNV\"]) \n | data[\"Func.refGene\"].isin([\"intergenic\"])\n | data[\"Func.refGene\"].str.contains(\"intronic\")\n | data[\"Func.refGene\"].str.contains(\"stream\"))\n\nfilter_b = data[\"Site.qual\"]>=30\nfilter_c = data[\"Depth\"]>=10\nfilter_d = data[\"strand.FS\"]<=200\nfilter_e = data[\"strand.OR\"]<=10\n\n#reject FS>60, SOR >3 for SNPs and FS>200, SOR>10 for indels.\n\nfilter_1 = filter_a & filter_b & filter_c & filter_d & filter_d\ndel filter_a\ndel filter_b\ndel filter_c\ndel filter_d\ndel filter_e",
"_____no_output_____"
],
[
"allele_freq = data[freq_cols].apply(freq_check, axis=1, NotANum=np.nan)",
"_____no_output_____"
],
[
"pred_effect = data[impact_cols].apply(impact_check, axis=1, NotANum=np.nan)",
"_____no_output_____"
],
[
"data['db_AF'] = allele_freq\ndata['pred_effect'] = pred_effect",
"_____no_output_____"
],
[
"# Was gene suggested by Phenotips?\nphenotips = pd.read_csv(phenotips_file, low_memory=False, sep='\\t')\nphenotips.columns = ['Gene.refGene', 'GeneID', 'Phenotips_Score', 'Features','HPOs']\n\nDB = [tuple(x) for x in phenotips[['Gene.refGene','Phenotips_Score', 'Features','HPOs']].values]\npheno_db = dict([(x[0], x[1:4]) for x in DB])\n\nphenotype = data['Gene.refGene'].apply(check_gene, db=pheno_db)\nphenotype.columns = ['in_phenotips', 'n_phenotips', 'feat_phenotips', 'HPO_terms']",
"_____no_output_____"
],
[
"data2 = pd.concat([data,phenotype], axis=1)",
"_____no_output_____"
],
[
"sophia = pd.read_excel(open(sophia_file))\nsophia_cols = ['chromosome','genome_position','ref','alt','Category','ACMG value']\nsophia_labs = ['CHROM','POS','REF','ALT','Category_sophia','ACMG_sophia']\nsophia_short = sophia[sophia_cols]\nsophia_short.columns = sophia_labs\ndel sophia\nsophia = sophia_short",
"_____no_output_____"
],
[
"data3 = pd.merge(data2.astype(str), sophia.astype(str), how='left').fillna('-')",
"_____no_output_____"
],
[
"out_cols = [\"dbSNP147\",\"Zygosity\", \"Allele_counts\", \"db_AF\", \"pred_effect\",\n \"Func.refGene\", \"ExonicFunc.refGene\", \"Gene.refGene\", \n \"Effect\", \"Phenotypes\", \"mim\", \"pLI\", \"CADD_phred\",\"ClinVar\", \"ACMG_InterVar\",\n \"Category_sophia\",\"ACMG_sophia\"] + list(phenotype.columns)",
"_____no_output_____"
],
[
"data4 = data3[filter_1][out_cols]\ndata4.drop_duplicates(inplace=True)",
"_____no_output_____"
],
[
"t = {'Pathogenic':1000, 'Likely pathogenic':500, 'Uncertain significance':100, 'A':1000, 'B':500, 'C':100, '3':100, '4':500, '5':1000}\nscore = []\nfor array in data4[['ACMG_InterVar', 'ACMG_sophia', 'Category_sophia', 'n_phenotips']].values:\n total = 0\n if int(array[3]) > 0:\n n = int(array[3])\n else:\n n = 1 \n for v in array[:3]:\n try:\n total += t[v]\n except:\n total += 0\n score.append(total*n)",
"_____no_output_____"
],
[
"data4['score'] = score\ndata4.sort_values(by=['score','n_phenotips'], ascending=False, inplace=True)",
"_____no_output_____"
],
[
"writer = pd.ExcelWriter(os.path.expanduser(outdir + prefix + \"_\" + timestamp + \".xlsx\"))\ndata4.to_excel(writer, prefix, index=False)\nwriter.save()",
"_____no_output_____"
],
[
"acmg = {'Benign': 1, 'Likely benign': 2, 'Uncertain significance': 3, 'Likely pathogenic' : 4, 'Pathogenic' : 5}",
"_____no_output_____"
],
[
"compare_acmg = data3[(data3['ACMG_sophia'] !='-') & (data3['ACMG_InterVar'] != '')][['ACMG_InterVar','ACMG_sophia']].apply(lambda x: pd.Series([ acmg[x[0]], x[1] ]),axis=1).astype(str)",
"_____no_output_____"
],
[
"# % Identity between Intervar y Sophia Genetics for ACMG classification\nsum(compare_acmg[0] == compare_acmg[1])/float(len(compare_acmg))",
"_____no_output_____"
],
[
"nb = os.path.expanduser(outdir + prefix + \"_\" + timestamp + \".ipynb\")",
"_____no_output_____"
],
[
"%notebook -e $nb",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f6a4957fd9a75e146ea72f3c769507267121f7 | 3,148 | ipynb | Jupyter Notebook | 5 - synthetic-data-applications/regular-tabular/credit_card_fraud-balancing/pipeline/sample_synth.ipynb | ydataai/Blog | 7994727deb1e1ca35e06c6fe4680482920b95855 | [
"MIT"
] | null | null | null | 5 - synthetic-data-applications/regular-tabular/credit_card_fraud-balancing/pipeline/sample_synth.ipynb | ydataai/Blog | 7994727deb1e1ca35e06c6fe4680482920b95855 | [
"MIT"
] | null | null | null | 5 - synthetic-data-applications/regular-tabular/credit_card_fraud-balancing/pipeline/sample_synth.ipynb | ydataai/Blog | 7994727deb1e1ca35e06c6fe4680482920b95855 | [
"MIT"
] | null | null | null | 23.848485 | 239 | 0.583863 | [
[
[
"# Creating synthetic samples\nAfter training the synthesizer on top of fraudulent events we are able to generate as many as desired synthetic samples, always having in mind there's a trade-off between the number of records used for the model training and privacy.",
"_____no_output_____"
]
],
[
[
"#Importing the required packages\nimport os \n\nfrom ydata.synthesizers.regular import RegularSynthesizer\n\ntry:\n os.mkdir('outputs')\nexcept FileExistsError as e:\n print('Directory already exists')",
"_____no_output_____"
]
],
[
[
"### Init the synth & Samples generation",
"_____no_output_____"
]
],
[
[
"n_samples = os.environ['NSAMPLES']",
"_____no_output_____"
],
[
"model = RegularSynthesizer.load('outputs/synth_model.pkl')\nsynth_data = model.sample(int(n_samples))",
"INFO: 2022-02-20 23:44:25,790 [SYNTHESIZER] - Start generating model samples.\n"
]
],
[
[
"### Sending the synthetic samples to the next pipeline stage",
"_____no_output_____"
]
],
[
[
"OUTPUT_PATH=os.environ['OUTPUT_PATH']",
"_____no_output_____"
],
[
"from ydata.connectors.filetype import FileType\nfrom ydata.connectors import LocalConnector\n\nconn = LocalConnector()",
"_____no_output_____"
],
[
"#Creating the output with the synthetic sample\nconn.write_file(synth_data, path=OUTPUT_PATH, file_type = FileType.CSV)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e7f6a76e47e659c1641982798a69edf9a18a32da | 1,755 | ipynb | Jupyter Notebook | 00_core.ipynb | akshaysynerzip/hello_nbdev | db0d637f036ca402976d63de73c8bebe3878f483 | [
"Apache-2.0"
] | null | null | null | 00_core.ipynb | akshaysynerzip/hello_nbdev | db0d637f036ca402976d63de73c8bebe3878f483 | [
"Apache-2.0"
] | 2 | 2021-09-28T05:33:43.000Z | 2022-02-26T09:53:06.000Z | 00_core.ipynb | akshaysynerzip/hello_nbdev | db0d637f036ca402976d63de73c8bebe3878f483 | [
"Apache-2.0"
] | null | null | null | 17.038835 | 55 | 0.480342 | [
[
[
"# default_exp core",
"_____no_output_____"
]
],
[
[
"# module name here\n\n> API details.",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.showdoc import *\nfrom fastcore.test import *",
"_____no_output_____"
]
],
[
[
"This is a function to say hello",
"_____no_output_____"
]
],
[
[
"#export\ndef say_hello(to):\n \"Say hello to somebody\"\n return f'Hello {to}!'",
"_____no_output_____"
],
[
"say_hello(\"Akshay\")",
"_____no_output_____"
],
[
"test_eq(say_hello(\"akshay\"), \"Hello akshay!\")",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e7f6a8fd0bf9e678cfb7cbad67d7ff8dc23d3492 | 12,541 | ipynb | Jupyter Notebook | notebooks/03_ShiftAmountActivity.ipynb | TUDelft-CITG/Hydraulic-Infrastructure-Realisation | c5888ef8f8bd1676536e268701dbb974e6f87c40 | [
"MIT"
] | 6 | 2019-11-14T08:12:08.000Z | 2021-04-08T11:13:35.000Z | notebooks/03_ShiftAmountActivity.ipynb | TUDelft-CITG/Hydraulic-Infrastructure-Realisation | c5888ef8f8bd1676536e268701dbb974e6f87c40 | [
"MIT"
] | 16 | 2019-06-25T16:44:13.000Z | 2022-02-15T18:05:28.000Z | notebooks/03_ShiftAmountActivity.ipynb | TUDelft-CITG/Hydraulic-Infrastructure-Realisation | c5888ef8f8bd1676536e268701dbb974e6f87c40 | [
"MIT"
] | 8 | 2019-07-03T08:28:26.000Z | 2021-07-12T08:11:53.000Z | 27.623348 | 420 | 0.463599 | [
[
[
"## Demo: ShiftAmountActivity\nThe basic steps to set up an OpenCLSim simulation are:\n* Import libraries\n* Initialise simpy environment\n* Define object classes\n* Create objects\n * Create sites\n * Create vessels\n * Create activities\n* Register processes and run simpy\n\n----\n\nThis notebook shows the workings of the ShiftAmountActivity. This activity uses a processor to transfer a specified number of objects from an origin resource, which must have a container, to a destination resource, which also must have a container. In this case it shifts payload from a from_site to vessel01.\n\nNB: The ShiftAmountActivity checks the possible amount of objects which can be transferred, based on the number of objects available in the origin, the number of objects which can be stored in the destination and the number of objects requested to be transferred. If the number of actually to be transferred objects is zero than an exception is raised. These cases have to be prevented by using appropriate events.",
"_____no_output_____"
],
[
"#### 0. Import libraries",
"_____no_output_____"
]
],
[
[
"import datetime, time\nimport simpy\n\nimport shapely.geometry\nimport pandas as pd\n\nimport openclsim.core as core\nimport openclsim.model as model\nimport openclsim.plot as plot",
"_____no_output_____"
]
],
[
[
"#### 1. Initialise simpy environment",
"_____no_output_____"
]
],
[
[
"# setup environment\nsimulation_start = 0\nmy_env = simpy.Environment(initial_time=simulation_start)",
"_____no_output_____"
]
],
[
[
"#### 2. Define object classes",
"_____no_output_____"
]
],
[
[
"# create a Site object based on desired mixin classes\nSite = type(\n \"Site\",\n (\n core.Identifiable,\n core.Log,\n core.Locatable,\n core.HasContainer,\n core.HasResource,\n ),\n {},\n)\n\n# create a TransportProcessingResource object based on desired mixin classes\nTransportProcessingResource = type(\n \"TransportProcessingResource\",\n (\n core.Identifiable,\n core.Log,\n core.ContainerDependentMovable,\n core.HasResource,\n core.Processor,\n ),\n {},\n)",
"_____no_output_____"
]
],
[
[
"#### 3. Create objects\n##### 3.1. Create site object(s)",
"_____no_output_____"
]
],
[
[
"# prepare input data for from_site\nlocation_from_site = shapely.geometry.Point(4.18055556, 52.18664444)\ndata_from_site = {\"env\": my_env,\n \"name\": \"from_site\",\n \"geometry\": location_from_site,\n \"capacity\": 100,\n \"level\": 100\n }\n# instantiate to_site \nfrom_site = Site(**data_from_site)",
"_____no_output_____"
]
],
[
[
"##### 3.2. Create vessel object(s)",
"_____no_output_____"
]
],
[
[
"# prepare input data for vessel_01\ndata_vessel01 = {\"env\": my_env,\n \"name\": \"vessel01\",\n \"geometry\": location_from_site, \n \"capacity\": 5,\n \"compute_v\": lambda x: 10\n }\n# instantiate vessel_01 \nvessel01 = TransportProcessingResource(**data_vessel01)",
"_____no_output_____"
]
],
[
[
"##### 3.3 Create activity/activities",
"_____no_output_____"
]
],
[
[
"# initialise registry\nregistry = {}",
"_____no_output_____"
],
[
"shift_amount_activity_data = model.ShiftAmountActivity(\n env=my_env,\n name=\"Shift amount activity\",\n registry=registry,\n processor=vessel01,\n origin=from_site,\n destination=vessel01,\n amount=100,\n duration=60,\n)",
"_____no_output_____"
]
],
[
[
"#### 4. Register processes and run simpy",
"_____no_output_____"
]
],
[
[
"model.register_processes([shift_amount_activity_data])\nmy_env.run()",
"_____no_output_____"
]
],
[
[
"#### 5. Inspect results\n##### 5.1 Inspect logs\nWe can now inspect the logs. The model now shifted cargo from the from_site onto vessel01.",
"_____no_output_____"
]
],
[
[
"display(plot.get_log_dataframe(shift_amount_activity_data, [shift_amount_activity_data]))",
"_____no_output_____"
],
[
"display(plot.get_log_dataframe(from_site, [shift_amount_activity_data]))",
"_____no_output_____"
],
[
"display(plot.get_log_dataframe(vessel01, [shift_amount_activity_data]))",
"_____no_output_____"
]
],
[
[
"Observe that an amount has been shifted from from_site to vessel01. There was no movement.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
e7f6ad7d37190e384c6c3e1702b981819d5797cf | 43,911 | ipynb | Jupyter Notebook | quests/rl/early_rl/early_rl.ipynb | mohakala/training-data-analyst | 2b0e2647d528a4d0fb5c589e8c549836590b60cf | [
"Apache-2.0"
] | 3 | 2021-09-26T00:11:36.000Z | 2021-12-06T05:55:25.000Z | quests/rl/early_rl/early_rl.ipynb | mohakala/training-data-analyst | 2b0e2647d528a4d0fb5c589e8c549836590b60cf | [
"Apache-2.0"
] | 2 | 2021-05-20T04:58:35.000Z | 2021-05-20T05:09:15.000Z | quests/rl/early_rl/early_rl.ipynb | mohakala/training-data-analyst | 2b0e2647d528a4d0fb5c589e8c549836590b60cf | [
"Apache-2.0"
] | 4 | 2021-08-28T22:18:03.000Z | 2021-09-05T22:27:05.000Z | 42.100671 | 787 | 0.618911 | [
[
[
"# Early Reinforcement Learning\n\nWith the advances of modern computing power, the study of Reinforcement Learning is having a heyday. Machines are now able to learn complex tasks once thought to be solely in the domain of humans, from controlling the [heating and cooling in massive data centers](https://www.technologyreview.com/s/611902/google-just-gave-control-over-data-center-cooling-to-an-ai/) to beating [grandmasters at Starcraft](https://storage.googleapis.com/deepmind-media/research/alphastar/AlphaStar_unformatted.pdf). As magnificent as it may seem today, it had humble roots many decades ago. Seeing how far it's come, it's a wonder to see how far it will go!\n\nLet's take a step back in time to see how these early algorithms developed. Many of these algorithms make sense given the context of when they were created. Challenge yourself and see if you can come up with the same strategies given the right problem. Ok! Time to cozy up for a story.\n\n<img src=\"images/hero.jpg\" width=\"488\" height=\"172\">\n\nThis is the hero of our story, the gumdrop emoji. It was enjoying a cool winter day building a snowman when suddenly, it slipped and fell on a frozen lake of death.\n\n\n<img src=\"images/lake.jpg\" width=\"900\" height=\"680\">\n\nThe lake can be thought of as a 4 x 4 grid where the gumdrop can move left (0), down (1), right (2) and up (3). Unfortunately, this frozen lake of death has holes of death where if the gumdrop enters that square, it will fall in and meet an untimely demise. To make matters worse, the lake is surrounded by icy boulders that if the gumdrop attempts to climb, will have it slip back into its original position. Thankfully, at the bottom right of the lake is a safe ramp that leads to a nice warm cup of hot cocoa.\n\n## Set Up\n\nWe can try and save the gumdrop ourselves! This is a common game people begin their Reinforcement Learning journey with, and is included in the OpenAI's python package [Gym](https://gym.openai.com/) and is aptly named [FrozenLake-v0](https://gym.openai.com/envs/FrozenLake-v0/) ([code](https://github.com/openai/gym/blob/master/gym/envs/toy_text/frozen_lake.py)). No time to waste, let's get the environment up and running. Run the below to install the needed libraries if they are not installed already.",
"_____no_output_____"
]
],
[
[
"# Ensure the right version of Tensorflow is installed.\n!pip install tensorflow==2.5 --user",
"_____no_output_____"
]
],
[
[
"**NOTE**: In the output of the above cell you may ignore any WARNINGS or ERRORS related to the dependency resolver.",
"_____no_output_____"
],
[
"If you get any related errors mentioned above please rerun the above cell.",
"_____no_output_____"
]
],
[
[
"!pip install gym==0.12.5 --user",
"_____no_output_____"
]
],
[
[
"There are [four methods from Gym](http://gym.openai.com/docs/) that are going to be useful to us in order to save the gumdrop.\n* `make` allows us to build the environment or game that we can pass actions to\n* `reset` will reset an environment to it's starting configuration and return the state of the player\n* `render` displays the environment for human eyes\n* `step` takes an action and returns the player's next state.\n\nLet's make, reset, and render the game. The output is an ANSI string with the following characters:\n* `S` for starting point\n* `F` for frozen\n* `H` for hole\n* `G` for goal\n* A red square indicates the current position",
"_____no_output_____"
],
[
"**Note**: Restart the kernel if the above libraries needed to be installed\n",
"_____no_output_____"
]
],
[
[
"import gym\nimport numpy as np\nimport random\nenv = gym.make('FrozenLake-v0', is_slippery=False)\nstate = env.reset()\nenv.render()",
"_____no_output_____"
]
],
[
[
"If we print the state we'll get `0`. This is telling us which square we're in. Each square is labeled from `0` to `15` from left to right, top to bottom, like this:\n\n| | | | |\n|-|-|-|-|\n|0|1|2|3|\n|4|5|6|7|\n|8|9|10|11|\n|12|13|14|15|\n",
"_____no_output_____"
]
],
[
[
"print(state)",
"_____no_output_____"
]
],
[
[
"We can make a simple print function to let us know whether it's game won, game over, or game on.",
"_____no_output_____"
]
],
[
[
"def print_state(state, done):\n statement = \"Still Alive!\"\n if done:\n statement = \"Cocoa Time!\" if state == 15 else \"Game Over!\" \n print(state, \"-\", statement)",
"_____no_output_____"
]
],
[
[
"We can control the gumdrop ourselves with the `step` method. Run the below cell over and over again trying to move from the starting position to the goal. Good luck!",
"_____no_output_____"
]
],
[
[
"#0 left\n#1 down\n#2 right\n#3 up\n\n# Uncomment to reset the game\n#env.reset()\naction = 2 # Change me, please!\nstate, _, done, _ = env.step(action)\nenv.render()\nprint_state(state, done)",
"_____no_output_____"
]
],
[
[
"Were you able to reach the hot chocolate? If so, great job! There are multiple paths through the maze. One solution is `[1, 1, 2, 2, 1, 2]`. Let's loop through our actions in order to get used to interacting with the environment programmatically.",
"_____no_output_____"
]
],
[
[
"def play_game(actions):\n state = env.reset()\n step = 0\n done = False\n\n while not done and step < len(actions):\n action = actions[step]\n state, _, done, _ = env.step(action)\n env.render()\n step += 1\n print_state(state, done)\n \nactions = [1, 1, 2, 2, 1, 2] # Replace with your favorite path.\nplay_game(actions)",
"_____no_output_____"
]
],
[
[
"Nice, so we know how to get through the maze, but how do we teach that to the gumdrop? It's just some bytes in an android phone. It doesn't have our human insight.\n\nWe could give it our list of actions directly, but then it would be copying us and not really learning. This was a tricky one to the mathematicians and computer scientists originally trying to solve this problem. How do we teach a machine to do this without human insight?\n\n## Value Iteration\n\nLet's turn the clock back on our time machines to 1957 to meet Mr. [Richard Bellman](https://en.wikipedia.org/wiki/Richard_E._Bellman). Bellman started his academic career in mathematics, but due to World War II, left his postgraduate studies at John Hopkins to teach electronics as part of the war effort (as chronicled by J. J. O'Connor and E. F. Robertson [here](https://www-history.mcs.st-andrews.ac.uk/Biographies/Bellman.html)). When the war was over, and it came time for him to focus on his next area of research, he became fascinated with [Dynamic Programming](https://en.wikipedia.org/wiki/Dynamic_programming): the idea of breaking a problem down into sub-problems and using recursion to solve the larger problem.\n\nEventually, his research landed him on [Markov Decision Processes](https://en.wikipedia.org/wiki/Markov_decision_process). These processes are a graphical way of representing how to make a decision based on a current state. States are connected to other states with positive and negative rewards that can be picked up along the way.\n\nSound familiar at all? Perhaps our Frozen Lake?\n\nIn the lake case, each cell is a state. The `H`s and the `G` are a special type of state called a \"Terminal State\", meaning they can be entered, but they have no leaving connections. What of rewards? Let's say the value of losing our life is the negative opposite of getting to the goal and staying alive. Thus, we can assign the reward of entering a death hole as -1, and the reward of escaping as +1.\n\nBellman's first breakthrough with this type of problem is now known as Value Iteration ([his original paper](http://www.iumj.indiana.edu/IUMJ/FULLTEXT/1957/6/56038)). He introduced a variable, gamma (γ), to represent discounted future rewards. He also introduced a function of policy (π) that takes a state (s), and outputs corresponding suggested action (a). The goal is to find the value of a state (V), given the rewards that occur when following an action in a particular state (R).\n\nGamma, the discount, is the key ingredient here. If my time steps were in days, and my gamma was .9, `$100` would be worth `$100` to me today, `$90` tomorrow, `$81` the day after, and so on. Putting this all together, we get the Bellman Equation\n\n<img src=\"images/bellman_equation.jpg\" width=\"500\">\n\nsource: [Wikipedia](https://en.wikipedia.org/wiki/Bellman_equation)\n\nIn other words, the value of our current state, `current_values`, is equal to the discount times the value of the next state, `next_values`, given the policy the agent will follow. For now, we'll have our agent assume a greedy policy: it will move towards the state with the highest calculated value. If you're wondering what P is, don't worry, we'll get to that later.\n\nLet's program it out and see it in action! We'll set up an array representing the lake with -1 as the holes, and 1 as the goal. Then, we'll set up an array of zeros to start our iteration.",
"_____no_output_____"
]
],
[
[
"LAKE = np.array([[0, 0, 0, 0],\n [0, -1, 0, -1],\n [0, 0, 0, -1],\n [-1, 0, 0, 1]])\nLAKE_WIDTH = len(LAKE[0])\nLAKE_HEIGHT = len(LAKE)\n\nDISCOUNT = .9 # Change me to be a value between 0 and 1.\ncurrent_values = np.zeros_like(LAKE)",
"_____no_output_____"
]
],
[
[
"The Gym environment class has a handy property for finding the number of states in an environment called `observation_space`. In our case, there a 16 integer states, so it will label it as \"Discrete\". Similarly, `action_space` will tell us how many actions are available to the agent.\n\nLet's take advantage of these to make our code portable between different lakes sizes.",
"_____no_output_____"
]
],
[
[
"print(\"env.observation_space -\", env.observation_space)\nprint(\"env.observation_space.n -\", env.observation_space.n)\nprint(\"env.action_space -\", env.action_space)\nprint(\"env.action_space.n -\", env.action_space.n)\n\nSTATE_SPACE = env.observation_space.n\nACTION_SPACE = env.action_space.n\nSTATE_RANGE = range(STATE_SPACE)\nACTION_RANGE = range(ACTION_SPACE)",
"_____no_output_____"
]
],
[
[
"We'll need some sort of function to figure out what the best neighboring cell is. The below function take's a cell of the lake, and looks at the current value mapping (to be called with `current_values`, and see's what the value of the adjacent state is corresponding to the given `action`.",
"_____no_output_____"
]
],
[
[
"def get_neighbor_value(state_x, state_y, values, action):\n \"\"\"Returns the value of a state's neighbor.\n \n Args:\n state_x (int): The state's horizontal position, 0 is the lake's left.\n state_y (int): The state's vertical position, 0 is the lake's top.\n values (float array): The current iteration's state values.\n policy (int): Which action to check the value for.\n \n Returns:\n The corresponding action's value.\n \"\"\"\n left = [state_y, state_x-1]\n down = [state_y+1, state_x]\n right = [state_y, state_x+1]\n up = [state_y-1, state_x]\n actions = [left, down, right, up]\n\n direction = actions[action]\n check_x = direction[1]\n check_y = direction[0]\n \n is_boulder = check_y < 0 or check_y >= LAKE_HEIGHT \\\n or check_x < 0 or check_x >= LAKE_WIDTH\n \n value = values[state_y, state_x]\n if not is_boulder:\n value = values[check_y, check_x]\n \n return value",
"_____no_output_____"
]
],
[
[
"But this doesn't find the best action, and the gumdrop is going to need that if it wants to greedily get off the lake. The `get_max_neighbor` function we've defined below takes a number corresponding to a cell as `state_number` and the same value mapping as `get_neighbor_value`.",
"_____no_output_____"
]
],
[
[
"def get_state_coordinates(state_number):\n state_x = state_number % LAKE_WIDTH\n state_y = state_number // LAKE_HEIGHT\n return state_x, state_y\n\ndef get_max_neighbor(state_number, values):\n \"\"\"Finds the maximum valued neighbor for a given state.\n \n Args:\n state_number (int): the state to find the max neighbor for\n state_values (float array): the respective value of each state for\n each cell of the lake.\n \n Returns:\n max_value (float): the value of the maximum neighbor.\n policy (int): the action to take to move towards the maximum neighbor.\n \"\"\"\n state_x, state_y = get_state_coordinates(state_number)\n \n # No policy or best value yet\n best_policy = -1\n max_value = -np.inf\n\n # If the cell has something other than 0, it's a terminal state.\n if LAKE[state_y, state_x]:\n return LAKE[state_y, state_x], best_policy\n \n for action in ACTION_RANGE:\n neighbor_value = get_neighbor_value(state_x, state_y, values, action)\n if neighbor_value > max_value:\n max_value = neighbor_value\n best_policy = action\n \n return max_value, best_policy",
"_____no_output_____"
]
],
[
[
"Now, let's write our value iteration code. We'll write a function that comes out one step of the iteration by checking each state and finding its maximum neighbor. The values will be reshaped so that it's in the form of the lake, but the policy will stay as a list of ints. This way, when Gym returns a state, all we need to do is look at the corresponding index in the policy list to tell our agent where to go.",
"_____no_output_____"
]
],
[
[
"def iterate_value(current_values):\n \"\"\"Finds the future state values for an array of current states.\n \n Args:\n current_values (int array): the value of current states.\n\n Returns:\n next_values (int array): The value of states based on future states.\n next_policies (int array): The recommended action to take in a state.\n \"\"\"\n next_values = []\n next_policies = []\n\n for state in STATE_RANGE:\n value, policy = get_max_neighbor(state, current_values)\n next_values.append(value)\n next_policies.append(policy)\n \n next_values = np.array(next_values).reshape((LAKE_HEIGHT, LAKE_WIDTH))\n return next_values, next_policies\n\nnext_values, next_policies = iterate_value(current_values)",
"_____no_output_____"
]
],
[
[
"This is what our values look like after one step. Right now, it just looks like the lake. That's because we started with an array of zeros for `current_values`, and the terminal states of the lake were loaded in.",
"_____no_output_____"
]
],
[
[
"next_values",
"_____no_output_____"
]
],
[
[
"And this is what our policy looks like reshaped into the form of the lake. The `-1`'s are terminal states. Right now, the agent will move left in any non-terminal state, because it sees all of those states as equal. Remember, if the gumdrop is along the leftmost side of the lake, and tries to move left, it will slip on a boulder and return to the same position.",
"_____no_output_____"
]
],
[
[
"np.array(next_policies).reshape((LAKE_HEIGHT ,LAKE_WIDTH))",
"_____no_output_____"
]
],
[
[
"There's one last step to apply the Bellman Equation, the `discount`! We'll multiply our next states by the `discount` and set that to our `current_values`. One loop done!",
"_____no_output_____"
]
],
[
[
"current_values = DISCOUNT * next_values\ncurrent_values",
"_____no_output_____"
]
],
[
[
"Run the below cell over and over again to see how our values change with each iteration. It should be complete after six iterations when the values no longer change. The policy will also change as the values are updated.",
"_____no_output_____"
]
],
[
[
"next_values, next_policies = iterate_value(current_values)\nprint(\"Value\")\nprint(next_values)\nprint(\"Policy\")\nprint(np.array(next_policies).reshape((4,4)))\ncurrent_values = DISCOUNT * next_values",
"_____no_output_____"
]
],
[
[
"Have a completed policy? Let's see it in action! We'll update our `play_game` function to instead take our list of policies. That way, we can start in a random position and still get to the end.",
"_____no_output_____"
]
],
[
[
"def play_game(policy):\n state = env.reset()\n step = 0\n done = False\n\n while not done:\n action = policy[state] # This line is new.\n state, _, done, _ = env.step(action)\n env.render()\n step += 1\n print_state(state, done)\n\nplay_game(next_policies)",
"_____no_output_____"
]
],
[
[
"Phew! Good job, team! The gumdrop made it out alive. So what became of our gumdrop hero? Well, the next day, it was making another snowman and fell onto an even more slippery and deadly lake. Doh! Turns out this story is part of a trilogy. Feel free to move onto the next section after your own sip of cocoa, coffee, tea, or poison of choice.\n\n## Policy Iteration\n\nYou may have noticed that the first lake was built with the parameter `is_slippery=False`. This time, we're going to switch it to `True`.",
"_____no_output_____"
]
],
[
[
"env = gym.make('FrozenLake-v0', is_slippery=True)\nstate = env.reset()\nenv.render()",
"_____no_output_____"
]
],
[
[
"Hmm, looks the same as before. Let's try applying our old policy and see what happens.",
"_____no_output_____"
]
],
[
[
"play_game(next_policies)",
"_____no_output_____"
]
],
[
[
"Was there a game over? There's a small chance that the gumdrop made it to the end, but it's much more likely that it accidentally slipped and fell into a hole. Oh no! We can try repeatedly testing the above code cell over and over again, but it might take a while. In fact, this is a similar roadblock Bellman and his colleagues faced.\n\nHow efficient is Value Iteration? On our modern machines, this algorithm ran fairly quickly, but back in 1960, that wasn't the case. Let's say our lake is a long straight line like this:\n\n| | | | | | | |\n|-|-|-|-|-|-|-|\n|S|F|F|F|F|F|H|\n\nThis is the worst case scenario for value iteration. In each iteration, we look at every state (s) and each action per state (a), so one step of value iteration is O(s*a). In the case of our lake line, each iteration only updates one cell. In other words, the value iteration step needs to be run `s` times. In total, that's O(s<sup>2</sup>a).\n\nBack in 1960, that was computationally heavy, and so [Ronald Howard](https://en.wikipedia.org/wiki/Ronald_A._Howard) developed an alteration of Value Iteration that mildly sacrificed mathematical accuracy for speed.\n\nHere's the strategy: it was observed that the optimal policy often converged before value iteration was complete. To take advantage of this, we'll start with random policy. When we iterate over our values, we'll use this policy instead of trying to find the maximum neighbor. This has been coded out in `find_future_values` below.",
"_____no_output_____"
]
],
[
[
"def find_future_values(current_values, current_policies):\n \"\"\"Finds the next set of future values based on the current policy.\"\"\"\n next_values = []\n\n for state in STATE_RANGE:\n current_policy = current_policies[state]\n state_x, state_y = get_state_coordinates(state)\n\n # If the cell has something other than 0, it's a terminal state.\n value = LAKE[state_y, state_x]\n if not value:\n value = get_neighbor_value(\n state_x, state_y, current_values, current_policy)\n next_values.append(value)\n\n return np.array(next_values).reshape((LAKE_HEIGHT, LAKE_WIDTH))",
"_____no_output_____"
]
],
[
[
"After we've calculated our new values, then we'll update the policy (and not the values) based on the maximum neighbor. If there's no change in the policy, then we're done. The below is very similar to our `get_max_neighbor` function. Can you see the differences?",
"_____no_output_____"
]
],
[
[
"def find_best_policy(next_values):\n \"\"\"Finds the best policy given a value mapping.\"\"\"\n next_policies = []\n for state in STATE_RANGE:\n state_x, state_y = get_state_coordinates(state)\n\n # No policy or best value yet\n max_value = -np.inf\n best_policy = -1\n\n if not LAKE[state_y, state_x]:\n for policy in ACTION_RANGE:\n neighbor_value = get_neighbor_value(\n state_x, state_y, next_values, policy)\n if neighbor_value > max_value:\n max_value = neighbor_value\n best_policy = policy\n \n next_policies.append(best_policy)\n return next_policies",
"_____no_output_____"
]
],
[
[
"To complete the Policy Iteration algorithm, we'll combine the two functions above. Conceptually, we'll be alternating between updating our value function and updating our policy function.",
"_____no_output_____"
]
],
[
[
"def iterate_policy(current_values, current_policies):\n \"\"\"Finds the future state values for an array of current states.\n \n Args:\n current_values (int array): the value of current states.\n current_policies (int array): a list where each cell is the recommended\n action for the state matching its index.\n\n Returns:\n next_values (int array): The value of states based on future states.\n next_policies (int array): The recommended action to take in a state.\n \"\"\"\n next_values = find_future_values(current_values, current_policies)\n next_policies = find_best_policy(next_values)\n return next_values, next_policies",
"_____no_output_____"
]
],
[
[
"Next, let's modify the `get_neighbor_value` function to now include the slippery ice. Remember the `P` in the Bellman Equation above? It stands for the probability of ending up in a new state given the current state and action taken. That is, we'll take a weighted sum of the values of all possible states based on our chances to be in those states.\n\nHow does the physics of the slippery ice work? For this lake, whenever the gumdrop tries to move in a particular direction, there are three possible positions that it could end up with. It could move where it was intending to go, but it could also end up to the left or right of the direction it was facing. For instance, if it wanted to move right, it could end up on the square above or below it! This is depicted below, with the yellow squares being potential positions after attempting to move right.\n\n<img src=\"images/slipping.jpg\" width=\"360\" height=\"270\">\n\nEach of these has an equal probability chance of happening. So since there are three outcomes, they each have about a 33% chance to happen. What happens if we slip in the direction of a boulder? No problem, we'll just end up not moving anywhere. Let's make a function to find what our possible locations could be given a policy and state coordinates.",
"_____no_output_____"
]
],
[
[
"def get_locations(state_x, state_y, policy):\n left = [state_y, state_x-1]\n down = [state_y+1, state_x]\n right = [state_y, state_x+1]\n up = [state_y-1, state_x]\n directions = [left, down, right, up]\n num_actions = len(directions)\n\n gumdrop_right = (policy - 1) % num_actions\n gumdrop_left = (policy + 1) % num_actions\n locations = [gumdrop_left, policy, gumdrop_right]\n return [directions[location] for location in locations]",
"_____no_output_____"
]
],
[
[
"Then, we can add it to `get_neighbor_value` to find the weighted value of all the possible states the gumdrop can end up in.",
"_____no_output_____"
]
],
[
[
"def get_neighbor_value(state_x, state_y, values, policy):\n \"\"\"Returns the value of a state's neighbor.\n \n Args:\n state_x (int): The state's horizontal position, 0 is the lake's left.\n state_y (int): The state's vertical position, 0 is the lake's top.\n values (float array): The current iteration's state values.\n policy (int): Which action to check the value for.\n \n Returns:\n The corresponding action's value.\n \"\"\"\n locations = get_locations(state_x, state_y, policy)\n location_chance = 1.0 / len(locations)\n total_value = 0\n\n for location in locations:\n check_x = location[1]\n check_y = location[0]\n\n is_boulder = check_y < 0 or check_y >= LAKE_HEIGHT \\\n or check_x < 0 or check_x >= LAKE_WIDTH\n \n value = values[state_y, state_x]\n if not is_boulder:\n value = values[check_y, check_x]\n total_value += location_chance * value\n\n return total_value",
"_____no_output_____"
]
],
[
[
"For Policy Iteration, we'll start off with a random policy if only because the Gumdrop doesn't know any better yet. We'll reset our current values while we're at it.",
"_____no_output_____"
]
],
[
[
"current_values = np.zeros_like(LAKE)\npolicies = np.random.choice(ACTION_RANGE, size=STATE_SPACE)\nnp.array(policies).reshape((4,4))",
"_____no_output_____"
]
],
[
[
"As before with Value Iteration, run the cell below multiple until the policy no longer changes. It should only take 2-3 clicks compared to Value Iteration's 6.",
"_____no_output_____"
]
],
[
[
"next_values, policies = iterate_policy(current_values, policies)\nprint(\"Value\")\nprint(next_values)\nprint(\"Policy\")\nprint(np.array(policies).reshape((4,4)))\ncurrent_values = DISCOUNT * next_values",
"_____no_output_____"
]
],
[
[
"Hmm, does this work? Let's see! Run the cell below to watch the gumdrop slip its way to victory.",
"_____no_output_____"
]
],
[
[
"play_game(policies)",
"_____no_output_____"
]
],
[
[
"So what was the learned strategy here? The gumdrop learned to hug the left wall of boulders until it was down far enough to make a break for the exit. Instead of heading directly for it though, it took advantage of actions that did not have a hole of death in them. Patience is a virtue!\n\nWe promised this story was a trilogy, and yes, the next day, the gumdrop fell upon a frozen lake yet again.\n\n## Q Learning\nValue Iteration and Policy Iteration are great techniques, but what if we don't know how big the lake is? With real world problems, not knowing how many potential states are can be a definite possibility.\n\nEnter [Chris Watkins](http://www.cs.rhul.ac.uk/~chrisw/). Inspired by how animals learn with delayed rewards, he came up with the idea of [Q Learning](http://www.cs.rhul.ac.uk/~chrisw/new_thesis.pdf) as an evolution of [Richard Sutton's](https://en.wikipedia.org/wiki/Richard_S._Sutton) [Temporal Difference Learning](https://en.wikipedia.org/wiki/Temporal_difference_learning). Watkins noticed that animals learn from positive and negative rewards, and that they often make mistakes in order to optimize a skill.\n\nFrom this emerged the idea of a Q table. In the lake case, it would look something like this.\n\n| |Left|Down|Right|Up|\n|-|-|-|-|-|\n|0| | | | |\n|1| | | | |\n|...| | | | |\n\nHere's the strategy: our agent will explore the environment. As the agent observes new states, we'll add more rows to our table. Whenever it moves from one state to the next, we'll update the cell corresponding to the old state based on the Bellman Equation. The agent doesn't need to know what the probabilities are between transitions. It'll learn the value of these as it experiments.\n\nFor Q learning, this works by looking at the row that corresponds to the agent's current state. Then, we'll select the action with the highest value. There are multiple ways to initialize the Q-table, but for us, we'll start with all zeros. In that case, when selecting the best action, we'll randomly select between tied max values. If we don't, the agent will favor certain actions which will limit its exploration.\n\nTo be able to handle an unknown number of states, we'll initialize our q_table as one row to represent our initial state. Then, we'll make a dictionary to map new states to rows in the table.",
"_____no_output_____"
]
],
[
[
"new_row = np.zeros((1, env.action_space.n))\nq_table = np.copy(new_row)\nq_map = {0: 0}\n\ndef print_q(q_table, q_map):\n print(\"mapping\")\n print(q_map)\n print(\"q_table\")\n print(q_table)\n\nprint_q(q_table, q_map)",
"_____no_output_____"
]
],
[
[
"Our new `get_action` function will help us read the `q_table` and find the best action.\n\nFirst, we'll give the agent the ability to act randomly as opposed to choosing the best known action. This gives it the ability to explore and find new situations. This is done with a random chance to act randomly. So random!\n\nWhen the Gumdrop chooses not to act randomly, it will instead act based on the best action recorded in the `q_table`. Numpy's [argwhere](https://docs.scipy.org/doc/numpy/reference/generated/numpy.argwhere.html) is used to find the indexes with the maximum value in the q-table row corresponding to our current state. Since numpy is often used with higher dimensional data, each index is returned as a list of ints. Our indexes are really one dimensional since we're just looking within a single row, so we'll use [np.squeeze](https://docs.scipy.org/doc/numpy/reference/generated/numpy.squeeze.html) to remove the extra brackets. To randomly select from the indexes, we'll use [np.random.choice](https://docs.scipy.org/doc/numpy-1.14.1/reference/generated/numpy.random.choice.html).",
"_____no_output_____"
]
],
[
[
"def get_action(q_map, q_table, state_row, random_rate):\n \"\"\"Find max-valued actions and randomly select from them.\"\"\"\n if random.random() < random_rate:\n return random.randint(0, ACTION_SPACE-1)\n\n action_values = q_table[state_row]\n max_indexes = np.argwhere(action_values == action_values.max())\n max_indexes = np.squeeze(max_indexes, axis=-1)\n action = np.random.choice(max_indexes)\n return action",
"_____no_output_____"
]
],
[
[
"Here, we'll define how the `q_table` gets updated. We'll apply the Bellman Equation as before, but since there is so much luck involved between slipping and random actions, we'll update our `q_table` as a weighted average between the `old_value` we're updating and the `future_value` based on the best action in the next state. That way, there's a little bit of memory between old and new experiences.",
"_____no_output_____"
]
],
[
[
"def update_q(q_table, new_state_row, reward, old_value):\n \"\"\"Returns an updated Q-value based on the Bellman Equation.\"\"\"\n learning_rate = .1 # Change to be between 0 and 1.\n future_value = reward + DISCOUNT * np.max(q_table[new_state_row])\n return old_value + learning_rate * (future_value - old_value)",
"_____no_output_____"
]
],
[
[
"We'll update our `play_game` function to take our table and mapping, and at the end, we'll return any updates to them. Once we observe new states, we'll check our mapping and add then to the table if space isn't allocated for them already.\n\nFinally, for every `state` - `action` - `new-state` transition, we'll update the cell in `q_table` that corresponds to the `state` and `action` with the Bellman Equation.\n\nThere's a little secret to solving this lake problem, and that's to have a small negative reward when moving between states. Otherwise, the gumdrop will become too afraid of slipping in a death hole to explore out of what is thought to be safe positions.",
"_____no_output_____"
]
],
[
[
"def play_game(q_table, q_map, random_rate, render=False):\n state = env.reset()\n step = 0\n done = False\n\n while not done:\n state_row = q_map[state]\n action = get_action(q_map, q_table, state_row, random_rate)\n new_state, _, done, _ = env.step(action)\n\n #Add new state to table and mapping if it isn't there already.\n if new_state not in q_map:\n q_map[new_state] = len(q_table)\n q_table = np.append(q_table, new_row, axis=0)\n new_state_row = q_map[new_state]\n\n reward = -.01 #Encourage exploration.\n if done:\n reward = 1 if new_state == 15 else -1\n current_q = q_table[state_row, action]\n q_table[state_row, action] = update_q(\n q_table, new_state_row, reward, current_q)\n\n step += 1\n if render:\n env.render()\n print_state(new_state, done)\n state = new_state\n \n return q_table, q_map",
"_____no_output_____"
]
],
[
[
"Ok, time to shine, gumdrop emoji! Let's do one simulation and see what happens.",
"_____no_output_____"
]
],
[
[
"# Run to refresh the q_table.\nrandom_rate = 1\nq_table = np.copy(new_row)\nq_map = {0: 0}",
"_____no_output_____"
],
[
"q_table, q_map = play_game(q_table, q_map, random_rate, render=True)\nprint_q(q_table, q_map)",
"_____no_output_____"
]
],
[
[
"Unless the gumdrop was incredibly lucky, chances were, it fell in some death water. Q-learning is markedly different from Value Iteration or Policy Iteration in that it attempts to simulate how an animal learns in unknown situations. Since the layout of the lake is unknown to the Gumdrop, it doesn't know which states are death holes, and which ones are safe. Because of this, it's going to make many mistakes before it can start making successes.\n\nFeel free to run the above cell multiple times to see how the gumdrop steps through trial and error. When you're ready, run the below cell to have the gumdrop play 1000 times.",
"_____no_output_____"
]
],
[
[
"for _ in range(1000):\n q_table, q_map = play_game(q_table, q_map, random_rate)\n random_rate = random_rate * .99\nprint_q(q_table, q_map)\nrandom_rate",
"_____no_output_____"
]
],
[
[
"Cats have nine lives, our Gumdrop lived a thousand! Moment of truth. Can it get out of the lake now that it matters?",
"_____no_output_____"
]
],
[
[
"q_table, q_map = play_game(q_table, q_map, 0, render=True)",
"_____no_output_____"
]
],
[
[
"Third time's the charm!\n\nEach of these techniques has its pros and cons. For instance, while Value Iteration is the mathematically correct solution, it's not as time efficient at Policy Iteration or as flexible as Q-Learning.\n\n| |Value Iteration|Policy Iteration|Q Tables|\n|-|-|-|-|\n|Avoids locally optimal routes|✓|x|x|\n|On-policy (greedy)|✓|✓|x|\n|Model Free|x|x|✓|\n|Most time efficient|x|✓|x|\n\nCongratulations on making it through to the end. Now if you ever fall on a Frozen Lake, you'll have many different ways to calculate your survival. The gumdrop thank you!\n\n<img src=\"images/end.jpg\" width=\"178\" height=\"234\">",
"_____no_output_____"
],
[
"Copyright 2020 Google Inc.\nLicensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at\nhttp://www.apache.org/licenses/LICENSE-2.0\nUnless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
e7f6b15d04972b19aa6dec8111218c422edf89aa | 18,498 | ipynb | Jupyter Notebook | 06_ui.ipynb | CMB-S4/s4_design_sim_tool | 660e3388d968f0bfd26e4539aa193feea5d32cc1 | [
"Apache-2.0"
] | 1 | 2020-03-24T18:16:28.000Z | 2020-03-24T18:16:28.000Z | 06_ui.ipynb | CMB-S4/s4_design_sim_tool | 660e3388d968f0bfd26e4539aa193feea5d32cc1 | [
"Apache-2.0"
] | 22 | 2020-03-26T21:48:15.000Z | 2022-02-26T06:48:24.000Z | 06_ui.ipynb | CMB-S4/s4_design_sim_tool | 660e3388d968f0bfd26e4539aa193feea5d32cc1 | [
"Apache-2.0"
] | null | null | null | 27.733133 | 200 | 0.513461 | [
[
[
"# Web interface `s4_design_sim_tool`\n> Interactive web-based user interface for the CMB-S4 reference simulation tool",
"_____no_output_____"
],
[
"See the [Documentation](https://cmb-s4.github.io/s4_design_sim_tool/)\n\nIf your browser doesn't visualize the widget input boxes, try reloading the page and **disable your adblocker**.\n\nFor support requests, [open an issue on the `s4_design_sim_tool` repository](https://github.com/CMB-S4/s4_design_sim_tool/issues)",
"_____no_output_____"
]
],
[
[
"# default_exp ui",
"_____no_output_____"
],
[
"import ipywidgets as widgets\nfrom IPython.display import display",
"_____no_output_____"
],
[
"w = {}",
"_____no_output_____"
],
[
"for emission in [\"foreground_emission\", \"CMB_unlensed\", \"CMB_lensing_signal\"]:\n w[emission] = widgets.BoundedFloatText(\n value=1,\n min=0,\n max=1,\n step=0.01,\n description='Weight:',\n disabled=False\n )",
"_____no_output_____"
],
[
"emission = \"CMB_tensor_to_scalar_ratio\"\nw[emission] = widgets.BoundedFloatText(\n value=3e-3,\n min=0,\n max=1,\n step=1e-5,\n description=f'r:',\n disabled=False\n )\n",
"_____no_output_____"
]
],
[
[
"## Sky emission weighting\n\nEach sky emission has a weighting factor between 0 and 1\n\n### Foreground emission\n\nSynchrotron, Dust, Free-free, AME\nWebsky CIB, tSZ, kSZ",
"_____no_output_____"
]
],
[
[
"display(w[\"foreground_emission\"])",
"_____no_output_____"
]
],
[
[
"### Unlensed CMB\nPlanck cosmological parameters, no tensor modes",
"_____no_output_____"
]
],
[
[
"display(w[\"CMB_unlensed\"])",
"_____no_output_____"
]
],
[
[
"### CMB lensing signal\n\nCMB lensed - CMB unlensed:\n* 1 for lensed CMB\n* 0 for unlensed CMB\n* `>0, <1` for residual after de-lensing\n\nFor the case of partial de-lensing, consider that lensing is a non-linear and this is a very rough approximation, still it could be useful in same cases, for example low-ell BB modes.",
"_____no_output_____"
]
],
[
[
"display(w[\"CMB_lensing_signal\"])",
"_____no_output_____"
]
],
[
[
"### CMB tensor to scalar ratio\n\nValue of the `r` cosmological parameter",
"_____no_output_____"
]
],
[
[
"display(w[\"CMB_tensor_to_scalar_ratio\"])",
"_____no_output_____"
]
],
[
[
"## Experiment parameters\n\n### Total experiment length\n\nIn years, supports decimals",
"_____no_output_____"
]
],
[
[
"w[\"total_experiment_length_years\"] = widgets.BoundedFloatText(\n value=7,\n min=0,\n max=15,\n step=0.1,\n description='Years:',\n disabled=False\n )",
"_____no_output_____"
],
[
"display(w[\"total_experiment_length_years\"])",
"_____no_output_____"
],
[
"w[\"observing_efficiency\"] = widgets.BoundedFloatText(\n value=0.2,\n min=0,\n max=1,\n step=0.01,\n description='Efficiency:',\n disabled=False\n )",
"_____no_output_____"
]
],
[
[
"### Observing efficiency\n\nTypically 20%, use decimal notation",
"_____no_output_____"
]
],
[
[
"display(w[\"observing_efficiency\"])",
"_____no_output_____"
],
[
"w[\"number_of_splits\"] = widgets.BoundedIntText(\n value=1,\n min=1,\n max=7,\n step=1,\n description='Splits:',\n disabled=False\n )",
"_____no_output_____"
]
],
[
[
"### Number of splits\n\nNumber of splits, 1 generates only full mission\n2-7 generates the full mission map and then the requested number\nof splits scaled accordingly. E.g. 7 generates the full mission\nmap and 7 equal (yearly) maps",
"_____no_output_____"
]
],
[
[
"display(w[\"number_of_splits\"])",
"_____no_output_____"
]
],
[
[
"## Telescope configuration\n\nCurrently we constraint to have a total of 6 SAT and 3 LAT,\neach SAT has a maximum of 3 tubes, each LAT of 19.\nThe checkbox on the right of each telescope checks that the amount of number of tubes is correct.",
"_____no_output_____"
]
],
[
[
"import toml\nconfig = toml.load(\"s4_design.toml\")",
"_____no_output_____"
],
[
"def define_check_sum(telescope_widgets, max_tubes):\n def check_sum(_):\n total_tubes = sum([w.value for w in telescope_widgets[1:1+4]])\n telescope_widgets[0].value = total_tubes == max_tubes\n return check_sum",
"_____no_output_____"
],
[
"telescopes = {\"SAT\":{}, \"LAT\":{}}\nfor telescope, tubes in config[\"telescopes\"][\"SAT\"].items():\n telescopes[\"SAT\"][telescope] = [widgets.Valid(\n value=True, description=telescope, layout=widgets.Layout(width='120px')\n )]\n telescope_check = define_check_sum(telescopes[\"SAT\"][telescope], 3)\n for k,v in tubes.items():\n if k == \"site\":\n wid = widgets.Dropdown(\n options=['Pole', 'Chile'],\n value=v,\n description=k,\n disabled=False, layout=widgets.Layout(width='150px')\n )\n elif k == \"years\":\n wid = widgets.BoundedFloatText(\n value=v,\n min=0,\n max=20,\n step=0.1,\n description='years',\n disabled=False, layout=widgets.Layout(width='130px')\n )\n else:\n \n wid = widgets.BoundedIntText(\n value=v,\n min=0,\n max=3,\n step=1,\n description=k,\n disabled=False, layout=widgets.Layout(width='130px')\n ) \n wid.observe(telescope_check)\n telescopes[\"SAT\"][telescope].append(wid)",
"_____no_output_____"
],
[
"for k, v in telescopes[\"SAT\"].items():\n display(widgets.HBox(v))",
"_____no_output_____"
],
[
"for telescope, tubes in config[\"telescopes\"][\"LAT\"].items():\n telescopes[\"LAT\"][telescope] = [widgets.Valid(\n value=True, description=telescope, layout=widgets.Layout(width='120px')\n )]\n telescope_check = define_check_sum(telescopes[\"LAT\"][telescope], 19)\n for k,v in tubes.items():\n if k == \"site\":\n wid = widgets.Dropdown(\n options=['Pole', 'Chile'],\n value=v,\n description=k,\n disabled=False, layout=widgets.Layout(width='150px')\n )\n elif k == \"years\":\n wid = widgets.BoundedFloatText(\n value=v,\n min=0,\n max=20,\n step=0.1,\n description='years',\n disabled=False, layout=widgets.Layout(width='130px')\n )\n else:\n \n wid = widgets.BoundedIntText(\n value=v,\n min=0,\n max=19,\n step=1,\n description=k,\n disabled=False, layout=widgets.Layout(width='130px')\n ) \n wid.observe(telescope_check)\n telescopes[\"LAT\"][telescope].append(wid)",
"_____no_output_____"
],
[
"for k, v in telescopes[\"LAT\"].items():\n display(widgets.HBox(v))",
"_____no_output_____"
],
[
"import toml",
"_____no_output_____"
],
[
"toml_decoder = toml.decoder.TomlDecoder()\ntoml_encoder = toml.TomlPreserveInlineDictEncoder()\n\ndef generate_toml():\n output_config = {}\n for section in [\"sky_emission\", \"experiment\"]:\n output_config[section] = {}\n for k in config[section]:\n output_config[section][k] = w[k].value\n output_config[\"telescopes\"] = {\"SAT\":{}, \"LAT\":{}}\n for t in [\"SAT\", \"LAT\"]:\n for telescope, tubes in telescopes[t].items():\n output_config[\"telescopes\"][t][telescope] = toml_decoder.get_empty_inline_table()\n for tube_type in tubes[1:]:\n output_config[\"telescopes\"][t][telescope][tube_type.description] = tube_type.value\n if tube_type.description == \"years\":\n output_config[\"telescopes\"][t][telescope][tube_type.description] = int(tube_type.value)\n return toml.dumps(output_config, encoder=toml_encoder)",
"_____no_output_____"
],
[
"from s4_design_sim_tool.cli import md5sum_string, S4RefSimTool\nfrom pathlib import Path",
"_____no_output_____"
]
],
[
[
"## Generate a TOML configuration file\n\nClick on the button to generate the TOML file and display it.",
"_____no_output_____"
]
],
[
[
"import os\noutput_location = os.environ.get(\"S4REFSIMTOOL_OUTPUT_URL\", \"\")",
"_____no_output_____"
],
[
"button = widgets.Button(\n description='Generate TOML',\n disabled=False,\n button_style='info', # 'success', 'info', 'warning', 'danger' or ''\n tooltip='Click me',\n icon='check'\n)\noutput_label = widgets.HTML(value=\"\")\noutput = widgets.Output(layout={'border': '1px solid black'})\n\ndisplay(button, output_label, output)\n\ndef on_button_clicked(b):\n output.clear_output()\n \n toml_string = generate_toml()\n md5sum = md5sum_string(toml_string)\n\n output_path = Path(\"output\") / md5sum\n output_label.value = \"\"\n \n if output_path.exists():\n output_label.value = \"This exact CMB-S4 configuration has <b>already been executed</b><br />\" + \\\n f\"<a href='{output_location}/output/{md5sum}' target='blank'><button class='p-Widget jupyter-widgets jupyter-button widget-button mod-success'>Access the maps </button></a>\"\n output_label.value += \"<p>TOML file preview:</p>\"\n \n with output:\n print(toml_string)\n\nbutton.on_click(on_button_clicked)",
"_____no_output_____"
],
[
"import ipywidgets as widgets\nimport logging\n\nclass OutputWidgetHandler(logging.Handler):\n \"\"\" Custom logging handler sending logs to an output widget \"\"\"\n\n def __init__(self, *args, **kwargs):\n super(OutputWidgetHandler, self).__init__(*args, **kwargs)\n layout = {\n 'width': '100%',\n 'height': '500px',\n 'border': '1px solid black'\n }\n self.out = widgets.Output(layout=layout)\n\n def emit(self, record):\n \"\"\" Overload of logging.Handler method \"\"\"\n formatted_record = self.format(record)\n new_output = {\n 'name': 'stdout',\n 'output_type': 'stream',\n 'text': formatted_record+'\\n'\n }\n self.out.outputs = (new_output, ) + self.out.outputs\n\n def show_logs(self):\n \"\"\" Show the logs \"\"\"\n display(self.out)\n\n def clear_logs(self):\n \"\"\" Clear the current logs \"\"\"\n self.out.clear_output()\n\n\nlogger = logging.root\nhandler = OutputWidgetHandler()\nhandler.setFormatter(logging.Formatter('%(asctime)s - [%(levelname)s] %(message)s'))\nlogger.addHandler(handler)\nlogger.setLevel(logging.INFO)",
"_____no_output_____"
]
],
[
[
"## Run the simulation\n\nGenerate the output maps",
"_____no_output_____"
]
],
[
[
"#export\n\n\ndef create_wget_script(folder, output_location):\n with open(folder / \"download_all.sh\", \"w\") as f:\n f.write(\"#!/bin/bash\\n\")\n for o in folder.iterdir():\n if not str(o).endswith(\"sh\"):\n f.write(f\"wget {output_location}/{o}\\n\")",
"_____no_output_____"
],
[
"def run_simulation(toml_filename, md5sum):\n output_path = toml_filename.parents[0]\n sim = S4RefSimTool(toml_filename, output_folder=output_path)\n sim.run(channels=\"all\", sites=[\"Pole\", \"Chile\"])\n \n logger.info(\"Create the wget script\")\n create_wget_script(output_path, output_location)",
"_____no_output_____"
],
[
"run_button = widgets.Button(\n description='Create the maps',\n disabled=False,\n button_style='danger', # 'success', 'info', 'warning', 'danger' or ''\n tooltip='Click me',\n icon='check'\n)\n\nrun_output_label = widgets.HTML(value=\"\")\n\nhandler.clear_logs()\ndisplay(run_button, run_output_label, handler.out)\n\ndef on_run_button_clicked(_):\n run_button.disabled = True\n\n toml_string = generate_toml()\n md5sum = md5sum_string(toml_string)\n\n output_path = Path(\"output\") / md5sum\n \n if output_path.exists():\n logger.error(\"This configuration has already been executed\")\n run_button.disabled = False\n return\n \n output_path.mkdir(parents=True, exist_ok=True)\n toml_filename = output_path / \"config.toml\"\n\n with open(toml_filename, \"w\") as f:\n f.write(toml_string)\n \n run_output_label.value = \"<p> The simulation has been launched, see the logs below, access the TOML configuration file and the maps as they are created clicking on the button </p>\" + \\\n f\"<a href='{output_location}/output/{md5sum}' target='blank'><button class='p-Widget jupyter-widgets jupyter-button widget-button mod-success'>Access the maps </button></a>\"\n \n run_simulation(toml_filename, md5sum)\n run_button.disabled = False\n \n\nrun_button.on_click(on_run_button_clicked)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e7f6d682b11eeeafeffd4ba597141afe05ecf549 | 4,679 | ipynb | Jupyter Notebook | labwork/lab1/tensorboard_mnsit.ipynb | YogeshHiremath/ml_lab_ecsc_306 | 985c20ea5340f760fc209b9eef84aa3dbd217d99 | [
"Apache-2.0"
] | null | null | null | labwork/lab1/tensorboard_mnsit.ipynb | YogeshHiremath/ml_lab_ecsc_306 | 985c20ea5340f760fc209b9eef84aa3dbd217d99 | [
"Apache-2.0"
] | null | null | null | labwork/lab1/tensorboard_mnsit.ipynb | YogeshHiremath/ml_lab_ecsc_306 | 985c20ea5340f760fc209b9eef84aa3dbd217d99 | [
"Apache-2.0"
] | 1 | 2018-01-18T05:50:11.000Z | 2018-01-18T05:50:11.000Z | 33.905797 | 107 | 0.549904 | [
[
[
"import tensorflow as tf\n\n# reset everything to rerun in jupyter\ntf.reset_default_graph()\n\n# config\nbatch_size = 100\nlearning_rate = 0.5\ntraining_epochs = 5\nlogs_path = \"/tmp/mnist/2\"\n\n# load mnist data set\nfrom tensorflow.examples.tutorials.mnist import input_data\nmnist = input_data.read_data_sets('MNIST_data', one_hot=True)\n\n# input images\nwith tf.name_scope('input'):\n # None -> batch size can be any size, 784 -> flattened mnist image\n x = tf.placeholder(tf.float32, shape=[None, 784], name=\"x-input\") \n # target 10 output classes\n y_ = tf.placeholder(tf.float32, shape=[None, 10], name=\"y-input\")\n\n# model parameters will change during training so we use tf.Variable\nwith tf.name_scope(\"weights\"):\n W = tf.Variable(tf.zeros([784, 10]))\n\n# bias\nwith tf.name_scope(\"biases\"):\n b = tf.Variable(tf.zeros([10]))\n\n# implement model\nwith tf.name_scope(\"softmax\"):\n # y is our prediction\n y = tf.nn.softmax(tf.matmul(x,W) + b)\n\n# specify cost function\nwith tf.name_scope('cross_entropy'):\n # this is our cost\n cross_entropy = tf.reduce_mean(-tf.reduce_sum(y_ * tf.log(y), reduction_indices=[1]))\n\n# specify optimizer\nwith tf.name_scope('train'):\n # optimizer is an \"operation\" which we can execute in a session\n train_op = tf.train.GradientDescentOptimizer(learning_rate).minimize(cross_entropy)\n\nwith tf.name_scope('Accuracy'):\n # Accuracy\n correct_prediction = tf.equal(tf.argmax(y,1), tf.argmax(y_,1))\n accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))\n \n# create a summary for our cost and accuracy\ntf.summary.scalar(\"cost\", cross_entropy)\ntf.summary.scalar(\"accuracy\", accuracy)\n\n# merge all summaries into a single \"operation\" which we can execute in a session \nsummary_op = tf.summary.merge_all()\n\nwith tf.Session() as sess:\n # variables need to be initialized before we can use them\n sess.run(tf.initialize_all_variables())\n\n # create log writer object\n writer = tf.summary.FileWriter('/tmp/tensorflow_logs', graph=tf.get_default_graph())\n \n \n # perform training cycles\n for epoch in range(training_epochs):\n \n # number of batches in one epoch\n batch_count = int(mnist.train.num_examples/batch_size)\n \n for i in range(batch_count):\n batch_x, batch_y = mnist.train.next_batch(batch_size)\n \n # perform the operations we defined earlier on batch\n _, summary = sess.run([train_op, summary_op], feed_dict={x: batch_x, y_: batch_y})\n \n # write log\n writer.add_summary(summary, epoch * batch_count + i)\n \n if epoch % 5 == 0: \n print (\"Epoch: \", epoch )\n print (\"Accuracy: \", accuracy.eval(feed_dict={x: mnist.test.images, y_: mnist.test.labels}))\n print (\"done\")\n \n \n \n ",
"_____no_output_____"
],
[
"## execute this command in terminal to see the visualization of increasing/decreasing accuracy\n## tensorboard --logdir=run1:/tmp/tensorflow_logs --port=6006\n## for multiple runs data visualization\n## tensorboard --logdir=run1:/tmp/mnist/1,run2:/tmp/mnist/2 --port=6006",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
e7f6db9af801a284121fc1271120e1a15131ac6a | 28,896 | ipynb | Jupyter Notebook | sagemaker_xgboost_hpo.ipynb | bbonik/sagemaker-xgboost-with-hpo | fa0abd1440ed9bf4a9c49b772d17f81e478efd01 | [
"MIT"
] | null | null | null | sagemaker_xgboost_hpo.ipynb | bbonik/sagemaker-xgboost-with-hpo | fa0abd1440ed9bf4a9c49b772d17f81e478efd01 | [
"MIT"
] | null | null | null | sagemaker_xgboost_hpo.ipynb | bbonik/sagemaker-xgboost-with-hpo | fa0abd1440ed9bf4a9c49b772d17f81e478efd01 | [
"MIT"
] | null | null | null | 46.086124 | 557 | 0.640746 | [
[
[
"# Amazon SageMaker with XGBoost and Hyperparameter Tuning for Direct Marketing predictions \n_**Supervised Learning with Gradient Boosted Trees: A Binary Prediction Problem With Unbalanced Classes**_\n\n---\n\n---\n\n## Contents\n\n1. [Objective](#Objective)\n1. [Background](#Background)\n1. [Environment Prepration](#Environment-preparation)\n1. [Data Downloading](#Data-downloading-and-exploration)\n1. [Data Transformation](#Data-Transformation)\n1. [SageMaker: Training](#Training)\n1. [SageMaker: Deploying and evaluating model](#Deploying-and-evaluating-model)\n1. [SageMaker: Hyperparameter Optimization (HPO)](#Hyperparameter-Optimization-(HPO))\n1. [Conclusions](#Conclusions)\n1. [Releasing cloud resources](#Releasing-cloud-resources)\n\n\n---\n\n## Objective\nThe goal of this workshop is to serve as a **Minimum Viable Example about SageMaker**, teaching you how to do a **basic ML training** and **Hyper-Parameter Optimization (HPO)** in AWS. Teaching an in-depth Data Science approach is out of the scope of this workshop. We hope that you can use it as a starting point and modify it according to your future projects. \n\n---\n\n## Background (problem description and approach)\n\n- **Direct marketing**: contacting potential new customers via mail, email, phone call etc. \n- **Challenge**: A) too many potential customers. B) limited resources of the approacher (time, money etc.).\n- **Problem: Which are the potential customers with the higher chance of becoming actual customers**? (so as to focus the effort only on them). \n- **Our setting**: A bank who wants to predict *whether a customer will enroll for a term deposit, after one or more phone calls*.\n- **Our approach**: Build a ML model to do this prediction, from readily available information e.g. demographics, past interactions etc. (features).\n- **Our tools**: We will be using the **XGBoost** algorithm in AWS **SageMaker**, followed by **Hyperparameter Optimization (HPO)** to produce the best model.\n\n\n\n---\n\n## Environment preparation\n\nSageMaker requires some minimal setup at the begining. This setup is standard and you can use it for any of your future projects. \nThings to specify:\n- The **S3 bucket** and **prefix** that you want to use for training and model data. **This should be within the same region as SageMaker training**!\n- The **IAM role** used to give training access to your data. See SageMaker documentation for how to create these.",
"_____no_output_____"
]
],
[
[
"import numpy as np # For matrix operations and numerical processing\nimport pandas as pd # For munging tabular data\nimport time\nimport os\nfrom util.ml_reporting_tools import generate_classification_report # helper function for classification reports\n\n# setting up SageMaker parameters\nimport sagemaker\nimport boto3\n\nsgmk_region = boto3.Session().region_name \nsgmk_client = boto3.Session().client(\"sagemaker\")\nsgmk_role = sagemaker.get_execution_role()\nsgmk_bucket = sagemaker.Session().default_bucket() # a default bucket has been created for this session\nsgmk_prefix = \"sagemaker/xgboost-hpo\"\n",
"_____no_output_____"
]
],
[
[
"---\n\n## Data downloading and exploration\nLet's start by downloading the [direct marketing dataset](https://archive.ics.uci.edu/ml/datasets/bank+marketing) from UCI's ML Repository. \nWe can run shell commands from Jupyter using the following code:",
"_____no_output_____"
]
],
[
[
"# (Running shell commands from Jupyter)\n!wget -P data/ -N https://archive.ics.uci.edu/ml/machine-learning-databases/00222/bank-additional.zip\n!unzip -o data/bank-additional.zip -d data/\n",
"_____no_output_____"
]
],
[
[
"Now lets read this into a Pandas data frame and take a look.",
"_____no_output_____"
]
],
[
[
"df_data = pd.read_csv(\"./data/bank-additional/bank-additional-full.csv\", sep=\";\")\ndf_data.head() # show part of the dataframe\n",
"_____no_output_____"
]
],
[
[
"_**Specifics on each of the features:**_\n\n*Demographics:*\n* `age`: Customer's age (numeric)\n* `job`: Type of job (categorical: 'admin.', 'services', ...)\n* `marital`: Marital status (categorical: 'married', 'single', ...)\n* `education`: Level of education (categorical: 'basic.4y', 'high.school', ...)\n\n*Past customer events:*\n* `default`: Has credit in default? (categorical: 'no', 'unknown', ...)\n* `housing`: Has housing loan? (categorical: 'no', 'yes', ...)\n* `loan`: Has personal loan? (categorical: 'no', 'yes', ...)\n\n*Past direct marketing contacts:*\n* `contact`: Contact communication type (categorical: 'cellular', 'telephone', ...)\n* `month`: Last contact month of year (categorical: 'may', 'nov', ...)\n* `day_of_week`: Last contact day of the week (categorical: 'mon', 'fri', ...)\n* `duration`: Last contact duration, in seconds (numeric). Important note: If duration = 0 then `y` = 'no'.\n \n*Campaign information:*\n* `campaign`: Number of contacts performed during this campaign and for this client (numeric, includes last contact)\n* `pdays`: Number of days that passed by after the client was last contacted from a previous campaign (numeric)\n* `previous`: Number of contacts performed before this campaign and for this client (numeric)\n* `poutcome`: Outcome of the previous marketing campaign (categorical: 'nonexistent','success', ...)\n\n*External environment factors:*\n* `emp.var.rate`: Employment variation rate - quarterly indicator (numeric)\n* `cons.price.idx`: Consumer price index - monthly indicator (numeric)\n* `cons.conf.idx`: Consumer confidence index - monthly indicator (numeric)\n* `euribor3m`: Euribor 3 month rate - daily indicator (numeric)\n* `nr.employed`: Number of employees - quarterly indicator (numeric)\n\n*Target variable* **(the one we want to eventually predict):**\n* `y`: Has the client subscribed to a term deposit? (binary: 'yes','no')",
"_____no_output_____"
],
[
"---\n\n## Data Transformation\nCleaning up data is part of nearly every ML project. Several common steps include:\n\n* **Handling missing values**: In our case there are no missing values.\n* **Handling weird/outlier values**: There are some values in the dataset that may require manipulation.\n* **Converting categorical to numeric**: There are a lot of categorical variables in our dataset. We need to address this.\n* **Oddly distributed data**: We will be using XGBoost, which is a non-linear method, and is minimally affected by the data distribution.\n* **Remove unnecessary data**: There are lots of columns representing general economic features that may not be available during inference time.\n\nTo summarise, we need to A) address some weird values, B) convert the categorical to numeric valriables and C) Remove unnecessary data:",
"_____no_output_____"
],
[
"1. Many records have the value of \"999\" for `pdays`. It is very likely to be a 'magic' number to represent that *no contact was made before*. Considering that, we will create a new column called \"no_previous_contact\", then grant it value of \"1\" when pdays is 999 and \"0\" otherwise.\n\n2. In the `job` column, there are more than one categories for people who don't work e.g., \"student\", \"retired\", and \"unemployed\". It is very likely the decision to enroll or not to a term deposit depends a lot on whether the customer is working or not. A such, we generate a new column to show whether the customer is working based on `job` column.\n\n3. We will remove the economic features and `duration` from our data as they would need to be forecasted with high precision to be used as features during inference time.\n\n4. We convert categorical variables to numeric using *one hot encoding*.",
"_____no_output_____"
]
],
[
[
"# Indicator variable to capture when pdays takes a value of 999\ndf_data[\"no_previous_contact\"] = np.where(df_data[\"pdays\"] == 999, 1, 0)\n\n# Indicator for individuals not actively employed\ndf_data[\"not_working\"] = np.where(np.in1d(df_data[\"job\"], [\"student\", \"retired\", \"unemployed\"]), 1, 0)\n\n# remove unnecessary data\ndf_model_data = df_data.drop(\n [\"duration\", \n \"emp.var.rate\", \n \"cons.price.idx\", \n \"cons.conf.idx\", \n \"euribor3m\", \n \"nr.employed\"], \n axis=1,\n)\n\ndf_model_data = pd.get_dummies(df_model_data) # Convert categorical variables to sets of indicators\n\ndf_model_data.head() # Show part of the new transformed dataframe (which will be used for training)\n",
"_____no_output_____"
]
],
[
[
"---\n\n## Training\n\nBefore initializing training, there are some things that need to be done:\n1. Suffle and split dataset. \n2. Convert the dataset to the right format the SageMaker algorithm expects (e.g. CSV).\n3. Copy the dataset to S3 in order to be accessed by SageMaker during training. \n4. Create s3_inputs that our training function can use as a pointer to the files in S3.\n5. Specify the ECR container location for SageMaker's implementation of XGBoost.\n\nWe will shuffle and split the dataset into **Training (70%)**, **Validation (20%)**, and **Test (10%)**. We will use the Training and Validation splits during the training phase, while the 'holdout' Test split will be used to evaluate the model performance after it is deployed to production. \n\nAmazon SageMaker's XGBoost algorithm expects data in the **libSVM** or **CSV** formats. For the CSV format, the following specifications should be met:\n- The first column must be the target variable.\n- No headers should be included.",
"_____no_output_____"
]
],
[
[
"# shuffle and splitting dataset\ntrain_data, validation_data, test_data = np.split(\n df_model_data.sample(frac=1, random_state=1729), \n [int(0.7 * len(df_model_data)), int(0.9*len(df_model_data))],\n) \n\n# create CSV files for Train / Validation / Test\n# XGBoost expects a CSV file with no headers, with the 1st row being the ground truth\n# We are preparing such a CSV file in the following lines\npd.concat([train_data[\"y_yes\"], train_data.drop([\"y_no\", \"y_yes\"], axis=1)], axis=1).to_csv(\"data/train.csv\", index=False, header=False)\npd.concat([validation_data[\"y_yes\"], validation_data.drop([\"y_no\", \"y_yes\"], axis=1)], axis=1).to_csv(\"data/validation.csv\", index=False, header=False)\npd.concat([test_data[\"y_yes\"], test_data.drop([\"y_no\", \"y_yes\"], axis=1)], axis=1).to_csv(\"data/test.csv\", index=False, header=False)\n\n# copy CSV files to S3 for SageMaker training (training files should reside in S3)\nboto3.Session().resource(\"s3\").Bucket(sgmk_bucket).Object(os.path.join(sgmk_prefix, \"train.csv\")).upload_file(\"data/train.csv\")\nboto3.Session().resource(\"s3\").Bucket(sgmk_bucket).Object(os.path.join(sgmk_prefix, \"validation.csv\")).upload_file(\"data/validation.csv\")\n\n# create s3_inputs channels (objects pointing to the S3 locations)\ns3_input_train = sagemaker.s3_input(s3_data=\"s3://{}/{}/train\".format(sgmk_bucket, sgmk_prefix), content_type=\"csv\")\ns3_input_validation = sagemaker.s3_input(s3_data=\"s3://{}/{}/validation\".format(sgmk_bucket, sgmk_prefix), content_type=\"csv\")\n",
"_____no_output_____"
]
],
[
[
"### Specify algorithm container image",
"_____no_output_____"
]
],
[
[
"# specify object of the xgboost container image\nfrom sagemaker.amazon.amazon_estimator import get_image_uri\nxgb_container_image = get_image_uri(sgmk_region, \"xgboost\", repo_version=\"latest\")\n",
"_____no_output_____"
]
],
[
[
"### A small competition: try to predict the best values for 4 hyper-parameters!\nSageMaker's XGBoost includes 38 parameters. You can find more information about them [here](https://docs.aws.amazon.com/sagemaker/latest/dg/xgboost_hyperparameters.html).\nFor simplicity, we choose to experiment only with 6 of them.\n\n**Please select values for the 4 hyperparameters (by replacing the \"?\") based on the provided ranges.** Later we will see which model performed best and compare it with the one from the Hyperparameter Optimization step.",
"_____no_output_____"
]
],
[
[
"sess = sagemaker.Session() # initiate a SageMaker session\n\n# instantiate an XGBoost estimator object\nxgb_estimator = sagemaker.estimator.Estimator(\n image_name=xgb_container_image, # XGBoost algorithm container\n role=sgmk_role, # IAM role to be used\n train_instance_type=\"ml.m4.xlarge\", # type of training instance\n train_instance_count=1, # number of instances to be used\n output_path=\"s3://{}/{}/output\".format(sgmk_bucket, sgmk_prefix),\n sagemaker_session=sess,\n train_use_spot_instances=True, # Use spot instances to reduce cost\n train_max_run=20*60, # Maximum allowed active runtime\n train_max_wait=30*60, # Maximum clock time (including spot delays)\n)\n\n# scale_pos_weight is a paramater that controls the relative weights of the classes.\n# Because the data set is so highly skewed, we set this parameter according to the ratio (y_no/y_yes)\nscale_pos_weight = np.count_nonzero(train_data[\"y_yes\"].values==0) / np.count_nonzero(train_data[\"y_yes\"].values)\n\n# define its hyperparameters\nxgb_estimator.set_hyperparameters(\n num_round=?, # int: [1,300]\n max_depth=?, # int: [1,10]\n alpha=?, # float: [0,5]\n eta=?, # float: [0,1]\n silent=0,\n objective=\"binary:logistic\",\n scale_pos_weight=scale_pos_weight,\n)\n\nxgb_estimator.fit({\"train\": s3_input_train, \"validation\": s3_input_validation}, wait=True) # start a training (fitting) job\n",
"_____no_output_____"
]
],
[
[
"---\n\n## Deploying and evaluating model\n\n### Deployment\nNow that we've trained the xgboost algorithm on our data, deploying the model (hosting it behind a real-time endpoint) is just one line of code!\n\n*Attention! This may take up to 10 minutes, depending on the AWS instance you select*.",
"_____no_output_____"
]
],
[
[
"xgb_predictor = xgb_estimator.deploy(initial_instance_count=1, instance_type=\"ml.m5.large\")\n",
"_____no_output_____"
]
],
[
[
"### Evaluation\n\nFirst we'll need to determine how we pass data into and receive data from our endpoint. Our data is currently stored as NumPy a array in memory of our notebook instance. To send it in an HTTP POST request, we will serialize it as a CSV string and then decode the resulting CSV. \nNote: For inference with CSV format, SageMaker XGBoost requires that the data **does NOT include the target variable.**",
"_____no_output_____"
]
],
[
[
"# Converting strings for HTTP POST requests on inference\nfrom sagemaker.predictor import csv_serializer\n\ndef predict_prob(predictor, data):\n # predictor settings\n predictor.content_type = \"text/csv\"\n predictor.serializer = csv_serializer\n return np.fromstring(predictor.predict(data).decode(\"utf-8\"), sep=\",\") # convert back to numpy \n\n\n# getting the predicted probabilities \npredictions = predict_prob(xgb_predictor, test_data.drop([\"y_no\", \"y_yes\"], axis=1).values)\n\nprint(predictions)\n",
"_____no_output_____"
]
],
[
[
"These numbers are the **predicted probabilities** (in the interval [0,1]) of a potential customer enrolling for a term deposit. \n- 0: the person WILL NOT enroll.\n- 1: the person WILL enroll (which makes him/her good candidate for direct marketing).\n\nNow we will generate a **comprehensive model report**, using the following functions. ",
"_____no_output_____"
]
],
[
[
"generate_classification_report(\n y_actual=test_data[\"y_yes\"].values, \n y_predict_proba=predictions, \n decision_threshold=0.5,\n class_names_list=[\"Did not enroll\",\"Enrolled\"],\n model_info=\"XGBoost SageMaker inbuilt\"\n)\n",
"_____no_output_____"
]
],
[
[
"---\n\n## Hyperparameter Optimization (HPO)\n*Note, with the default setting below, the hyperparameter tuning job can take up to 30 minutes to complete.*\n\nWe will use SageMaker HyperParameter Optimization (HPO) to automate the searching process effectively. Specifically, we **specify a range**, or a list of possible values in the case of categorical hyperparameters, for each of the hyperparameter that we plan to tune. \n\nWe will tune 4 hyperparameters in this example:\n* **eta**: Step size shrinkage used in updates to prevent overfitting. After each boosting step, you can directly get the weights of new features. The eta parameter actually shrinks the feature weights to make the boosting process more conservative. \n* **alpha**: L1 regularization term on weights. Increasing this value makes models more conservative. \n* **min_child_weight**: Minimum sum of instance weight (hessian) needed in a child. If the tree partition step results in a leaf node with the sum of instance weight less than min_child_weight, the building process gives up further partitioning. In linear regression models, this simply corresponds to a minimum number of instances needed in each node. The larger the algorithm, the more conservative it is. \n* **max_depth**: Maximum depth of a tree. Increasing this value makes the model more complex and likely to be overfitted. \n\nSageMaker hyperparameter tuning will automatically launch **multiple training jobs** with different hyperparameter settings, evaluate results of those training jobs based on a predefined \"objective metric\", and select the hyperparameter settings for future attempts based on previous results. For each hyperparameter tuning job, we will specify the maximum number of HPO tries (`max_jobs`) and how many of these can happen in parallel (`max_parallel_jobs`).\n\nTip: `max_parallel_jobs` creates a **trade-off between parformance and speed** (better hyperparameter values vs how long it takes to find these values). If `max_parallel_jobs` is large, then HPO is faster, but the discovered values may not be optimal. Smaller `max_parallel_jobs` will increase the chance of finding optimal values, but HPO will take more time to finish.\n\nNext we'll specify the objective metric that we'd like to tune and its definition, which includes the regular expression (Regex) needed to extract that metric from the CloudWatch logs of the training job. Since we are using built-in XGBoost algorithm here, it emits two predefined metrics: **validation:auc** and **train:auc**, and we elected to monitor *validation:auc* as you can see below. In this case, we only need to specify the metric name and do not need to provide regex. \n\nIf you bring your own algorithm, your algorithm emits metrics by itself. In that case, you'll need to add a MetricDefinition object here to define the format of those metrics through regex, so that SageMaker knows how to extract those metrics from your CloudWatch logs.\n\nFor more information on the documentation of the Sagemaker HPO please refer [here](https://sagemaker.readthedocs.io/en/stable/tuner.html).",
"_____no_output_____"
]
],
[
[
"# import required HPO objects\nfrom sagemaker.tuner import IntegerParameter, CategoricalParameter, ContinuousParameter, HyperparameterTuner\n\n# set up hyperparameter ranges\nranges = {\n \"num_round\": IntegerParameter(1, 300),\n \"max_depth\": IntegerParameter(1, 10),\n \"alpha\": ContinuousParameter(0, 5),\n \"eta\": ContinuousParameter(0, 1)\n}\n\n# set up the objective metric\nobjective = \"validation:auc\"\n\n# instantiate a HPO object\ntuner = HyperparameterTuner(\n estimator=xgb_estimator, # the SageMaker estimator object\n objective_metric_name=objective, # the objective metric to be used for HPO\n hyperparameter_ranges=ranges, # the range of hyperparameters\n max_jobs=20, # total number of HPO jobs\n max_parallel_jobs=4, # how many HPO jobs can run in parallel\n strategy=\"Bayesian\", # the internal optimization strategy of HPO\n objective_type=\"Maximize\" # maximize or minimize the objective metric\n) \n",
"_____no_output_____"
]
],
[
[
"### Launch HPO\nNow we can launch a hyperparameter tuning job by calling *fit()* function. After the hyperparameter tuning job is created, we can go to SageMaker console to track the progress of the hyperparameter tuning job until it is completed.",
"_____no_output_____"
]
],
[
[
"# start HPO\ntuner.fit({\"train\": s3_input_train, \"validation\": s3_input_validation}, include_cls_metadata=False)\n",
"_____no_output_____"
]
],
[
[
"**Important notice**: HPO jobs are expected to take quite long to finsih and as such, **they do not wait by default** (the cell will look as 'done' while the job will still be running on the cloud). As such, all subsequent cells relying on the HPO output cannot run unless the job is finished. In order to check whether the HPO has finished (so we can proceed with executing the subsequent code) we can run the following polling script:",
"_____no_output_____"
]
],
[
[
"# wait, until HPO is finished\nhpo_state = \"InProgress\"\n\nwhile hpo_state == \"InProgress\":\n hpo_state = sgmk_client.describe_hyper_parameter_tuning_job(\n HyperParameterTuningJobName=tuner.latest_tuning_job.job_name)[\"HyperParameterTuningJobStatus\"]\n print(\"-\", end=\"\")\n time.sleep(60) # poll once every 1 min\n\nprint(\"\\nHPO state:\", hpo_state)\n",
"_____no_output_____"
]
],
[
[
"### Deploy and test optimized model\nDeploying the best model is simply one line of code:",
"_____no_output_____"
]
],
[
[
"# deploy the best model from HPO\nbest_model_predictor = tuner.deploy(initial_instance_count=1, instance_type=\"ml.m5.large\")\n",
"_____no_output_____"
]
],
[
[
"Once deployed, we can now evaluate the performance of the best model.",
"_____no_output_____"
]
],
[
[
"# getting the predicted probabilities of the best model\npredictions = predict_prob(best_model_predictor, test_data.drop([\"y_no\", \"y_yes\"], axis=1).values)\nprint(predictions)\n\n# generate report for the best model\ngenerate_classification_report(\n y_actual=test_data[\"y_yes\"].values, \n y_predict_proba=predictions, \n decision_threshold=0.5,\n class_names_list=[\"Did not enroll\",\"Enrolled\"],\n model_info=\"XGBoost SageMaker inbuilt + HPO\"\n)",
"_____no_output_____"
]
],
[
[
"---\n\n## Conclusions\n\nThe optimized HPO model exhibits approximately AUC=0.773.\nDepending on the number of tries, HPO can give a better performing model, compared to simply trying different hyperparameters (by trial and error). \nYou can learn more in-depth details about HPO [here](https://docs.aws.amazon.com/sagemaker/latest/dg/automatic-model-tuning-how-it-works.html).",
"_____no_output_____"
],
[
"---\n\n## Releasing cloud resources\n\nIt is generally a good practice to deactivate all endpoints which are not in use. \nPlease uncomment the following lines and run the cell in order to deactive the 2 endpoints that were created before. ",
"_____no_output_____"
]
],
[
[
"# xgb_predictor.delete_endpoint(delete_endpoint_config=True)\n# best_model_predictor.delete_endpoint(delete_endpoint_config=True)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e7f6f3ac67dec2f18914b82e248c646e0ca22f73 | 3,965 | ipynb | Jupyter Notebook | generated/literated/literatejl.ipynb | rmsrosa/booksjl-franklin-template | 2f70ebe7334b4d27c180b4a84d576caadcd8e5ba | [
"CC0-1.0"
] | 2 | 2022-02-12T12:30:50.000Z | 2022-02-12T13:21:38.000Z | generated/literated/literatejl.ipynb | rmsrosa/booksjl-franklin-template | 2f70ebe7334b4d27c180b4a84d576caadcd8e5ba | [
"CC0-1.0"
] | 8 | 2022-02-08T18:13:48.000Z | 2022-03-15T09:19:28.000Z | generated/literated/literatejl.ipynb | rmsrosa/booksjl-franklin-template | 2f70ebe7334b4d27c180b4a84d576caadcd8e5ba | [
"CC0-1.0"
] | 1 | 2022-02-12T12:30:53.000Z | 2022-02-12T12:30:53.000Z | 20.025253 | 221 | 0.517024 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e7f703395d9829a40d1fe14785340562fb53569c | 64,146 | ipynb | Jupyter Notebook | machineLearning/scikitLearnAndPolyRegression.ipynb | naokishami/Classwork | ac59d640f15e88294804fdb518b6c84b10e0d2bd | [
"MIT"
] | null | null | null | machineLearning/scikitLearnAndPolyRegression.ipynb | naokishami/Classwork | ac59d640f15e88294804fdb518b6c84b10e0d2bd | [
"MIT"
] | null | null | null | machineLearning/scikitLearnAndPolyRegression.ipynb | naokishami/Classwork | ac59d640f15e88294804fdb518b6c84b10e0d2bd | [
"MIT"
] | null | null | null | 64.793939 | 16,548 | 0.829904 | [
[
[
"Naoki Atkins\n\nProject 3",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.metrics import mean_squared_error\nfrom sklearn.preprocessing import PolynomialFeatures\nnp.set_printoptions(suppress=True)",
"_____no_output_____"
]
],
[
[
"***Question 1***",
"_____no_output_____"
]
],
[
[
"data = np.load('./boston.npz')",
"_____no_output_____"
]
],
[
[
"***Question 2***",
"_____no_output_____"
]
],
[
[
"features = data['features']\ntarget = data['target']\n\nX = features\ny = target[:,None]",
"_____no_output_____"
],
[
"X = np.concatenate((np.ones((len(X),1)),X),axis=1)",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=(2021-3-11))",
"_____no_output_____"
]
],
[
[
"***Question 3***",
"_____no_output_____"
]
],
[
[
"plt.plot(X_train[:,13], y_train, 'ro')",
"_____no_output_____"
]
],
[
[
"The relationship seems to follow more of a negative quadratic than a linear line.",
"_____no_output_____"
],
[
"***Question 4***",
"_____no_output_____"
]
],
[
[
"LSTAT = X_train[:,13][:,None]\nMEDV = y_train",
"_____no_output_____"
],
[
"reg = LinearRegression().fit(LSTAT, MEDV)",
"_____no_output_____"
],
[
"reg.coef_",
"_____no_output_____"
],
[
"reg.intercept_",
"_____no_output_____"
]
],
[
[
"MEDV = 34.991133021969475 + (-0.98093888)(LSTAT)",
"_____no_output_____"
],
[
"***Question 5***",
"_____no_output_____"
]
],
[
[
"abline = np.array([reg.intercept_, reg.coef_], dtype=object)",
"_____no_output_____"
],
[
"testx = np.linspace(0,40,100)[:,None]\ntestX = np.hstack((np.ones_like(testx),testx))\ntestt = np.dot(testX,abline)",
"_____no_output_____"
],
[
"plt.figure()\nplt.plot(LSTAT,MEDV,'ro')\nplt.plot(testx,testt,'b')",
"_____no_output_____"
]
],
[
[
"The model fits decently well along the center of the mass of data. Around the extremes, the line is a little bit off.",
"_____no_output_____"
],
[
"***Question 6***",
"_____no_output_____"
]
],
[
[
"pred = reg.predict(LSTAT)",
"_____no_output_____"
],
[
"mean_squared_error(y_train, pred)",
"_____no_output_____"
]
],
[
[
"Average Loss = 38.47893344802523",
"_____no_output_____"
],
[
"***Question 7***",
"_____no_output_____"
]
],
[
[
"pred_test = reg.predict(X_test[:,13][:,None])",
"_____no_output_____"
],
[
"mean_squared_error(y_test, pred_test)",
"_____no_output_____"
]
],
[
[
"Test MSE is slightly higher, which means that there is a slight overfit",
"_____no_output_____"
],
[
"***Question 8***",
"_____no_output_____"
]
],
[
[
"LSTAT_sqr = np.hstack((np.ones_like(LSTAT), LSTAT, LSTAT**2))",
"_____no_output_____"
],
[
"reg = LinearRegression().fit(LSTAT_sqr, MEDV)",
"_____no_output_____"
],
[
"pred_train_LSTAT_sqr = reg.predict(LSTAT_sqr)",
"_____no_output_____"
],
[
"MSE_train_sqr = mean_squared_error(y_train, pred_train_LSTAT_sqr)",
"_____no_output_____"
],
[
"MSE_train_sqr",
"_____no_output_____"
],
[
"LSTAT_sqr_test = np.hstack((np.ones_like(X_test[:,13][:,None]), X_test[:,13][:,None], X_test[:,13][:,None]**2))",
"_____no_output_____"
],
[
"pred_test_LSTAT_sqr = reg.predict(LSTAT_sqr_test)",
"_____no_output_____"
],
[
"MSE_test_sqr = mean_squared_error(y_test, pred_test_LSTAT_sqr)",
"_____no_output_____"
],
[
"MSE_test_sqr",
"_____no_output_____"
]
],
[
[
"The test set has a lower MSE compared to the training set which means the model is fitting well.",
"_____no_output_____"
],
[
"***Question 9***",
"_____no_output_____"
]
],
[
[
"reg.coef_",
"_____no_output_____"
],
[
"reg.intercept_",
"_____no_output_____"
],
[
"squared_line = [reg.intercept_, reg.coef_[0][1], reg.coef_[0][2]]",
"_____no_output_____"
],
[
"testx = np.linspace(0,40,100)[:,None]\ntestX = np.hstack((np.ones_like(testx),testx, testx**2))\ntestt = np.dot(testX,squared_line)",
"_____no_output_____"
],
[
"plt.figure()\nplt.plot(LSTAT,MEDV,'ro')\nplt.plot(testx,testt,'b')",
"_____no_output_____"
]
],
[
[
"Model fits pretty well. Better than the line.",
"_____no_output_____"
],
[
"***Question 10***",
"_____no_output_____"
]
],
[
[
"reg = LinearRegression().fit(X_train, y_train)",
"_____no_output_____"
],
[
"reg.coef_",
"_____no_output_____"
],
[
"reg.intercept_",
"_____no_output_____"
],
[
"pred = reg.predict(X_train)",
"_____no_output_____"
],
[
"mean_squared_error(y_train, pred)",
"_____no_output_____"
]
],
[
[
"The above mean square error is for the training set",
"_____no_output_____"
]
],
[
[
"pred_test = reg.predict(X_test)",
"_____no_output_____"
],
[
"mean_squared_error(y_test, pred_test)",
"_____no_output_____"
]
],
[
[
"This model with polynomial features fits better as compared to the linear model with just a single feature. Making the model more complex allows it to fit the data more flexibly. This causes the MSE to go lower. ",
"_____no_output_____"
],
[
"***Question 11***",
"_____no_output_____"
]
],
[
[
"train_square_matrix = np.hstack((X_train, X_train**2))",
"_____no_output_____"
],
[
"model = LinearRegression().fit(train_square_matrix, MEDV)",
"_____no_output_____"
],
[
"pred_train_sqr = model.predict(train_square_matrix)",
"_____no_output_____"
],
[
"MSE_train_sqr = mean_squared_error(y_train, pred_train_sqr)\nMSE_train_sqr",
"_____no_output_____"
],
[
"test_square_matrix = np.hstack((X_test, X_test**2))",
"_____no_output_____"
],
[
"pred = model.predict(test_square_matrix)",
"_____no_output_____"
],
[
"mean_squared_error(y_test, pred)",
"_____no_output_____"
]
],
[
[
"The MSE's for the matrix of the squares of all the 13 input features performs better than the just the matrix of the features themselves. However, the testing set shows that the model is overfitting a little",
"_____no_output_____"
],
[
"***Question 12***",
"_____no_output_____"
]
],
[
[
"poly = PolynomialFeatures(degree = 2)\nX_train_poly = poly.fit_transform(X_train)\nX_test_poly = poly.fit_transform(X_test)\n\nmodel = LinearRegression().fit(X_train_poly, y_train)",
"_____no_output_____"
],
[
"pred = model.predict(X_train_poly)\nmean_squared_error(y_train, pred)",
"_____no_output_____"
],
[
"pred = model.predict(X_test_poly)\nmean_squared_error(y_test, pred)",
"_____no_output_____"
]
],
[
[
"The model is now overfitting the data after fitting a polynomial matrix with interaction terms added.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
e7f70b85c058d55a489ea35b64538f8a2c7e1830 | 1,988 | ipynb | Jupyter Notebook | code/05-bids-derivatives.ipynb | josephmje/SDC-BIDS-IntroMRI | cd29c90afe317f6a9a214872f15bd32859b48e93 | [
"CC-BY-4.0"
] | null | null | null | code/05-bids-derivatives.ipynb | josephmje/SDC-BIDS-IntroMRI | cd29c90afe317f6a9a214872f15bd32859b48e93 | [
"CC-BY-4.0"
] | null | null | null | code/05-bids-derivatives.ipynb | josephmje/SDC-BIDS-IntroMRI | cd29c90afe317f6a9a214872f15bd32859b48e93 | [
"CC-BY-4.0"
] | 1 | 2019-11-08T17:55:34.000Z | 2019-11-08T17:55:34.000Z | 24.54321 | 123 | 0.544266 | [
[
[
"## BIDS Apps\n\n[BIDS Apps](https://bids-apps.neuroimaging.io/) are containerized applications that run on BIDS data structures. \n\nSome examples include:\n- mriqc\n- fmriprep\n- freesurfer\n- ciftify\n- SPM\n- MRtrix3_connectome\n\nThey rely on 2 technologies for container computing:\n- **Docker**\n - for building, hosting, and running containers on local hardware (Windows, Mac OS, Linux) or in the cloud\n- **Singularity**\n - for running containers on high performance compute clusters\n \n<img src=\"../fig/bids_app.png\" alt=\"Drawing\" align=\"middle\" width=\"500px\"/>",
"_____no_output_____"
],
[
"All BIDS Apps use the same command line format to run them:\n\n`<app_name> /data /output participant [options]`",
"_____no_output_____"
],
[
"Building a singularity container is as easy as:\n \n `singularity build mriqc-0.16.1.simg docker://poldracklab/mriqc:0.16.1`\n \nTo run the container:\n\n```\nsingularity run --cleanenv \\\n -B bids_folder:/data \\\n mriqc-0.16.1.simg \\\n /data /data/derivatives participant\n```",
"_____no_output_____"
]
]
] | [
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
]
] |
e7f7136c5ae6677be020aaa9291af3bfcbb5485a | 96,680 | ipynb | Jupyter Notebook | 3. Landmark Detection and Tracking.ipynb | takam5f2/CVN_SLAM | 3c0d3a22cede7a0ae1dbd2a69cef3adcebb04ab3 | [
"MIT"
] | null | null | null | 3. Landmark Detection and Tracking.ipynb | takam5f2/CVN_SLAM | 3c0d3a22cede7a0ae1dbd2a69cef3adcebb04ab3 | [
"MIT"
] | null | null | null | 3. Landmark Detection and Tracking.ipynb | takam5f2/CVN_SLAM | 3c0d3a22cede7a0ae1dbd2a69cef3adcebb04ab3 | [
"MIT"
] | null | null | null | 108.873874 | 31,128 | 0.811409 | [
[
[
"# Project 3: Implement SLAM \n\n---\n\n## Project Overview\n\nIn this project, you'll implement SLAM for robot that moves and senses in a 2 dimensional, grid world!\n\nSLAM gives us a way to both localize a robot and build up a map of its environment as a robot moves and senses in real-time. This is an active area of research in the fields of robotics and autonomous systems. Since this localization and map-building relies on the visual sensing of landmarks, this is a computer vision problem. \n\nUsing what you've learned about robot motion, representations of uncertainty in motion and sensing, and localization techniques, you will be tasked with defining a function, `slam`, which takes in six parameters as input and returns the vector `mu`. \n> `mu` contains the (x,y) coordinate locations of the robot as it moves, and the positions of landmarks that it senses in the world\n\nYou can implement helper functions as you see fit, but your function must return `mu`. The vector, `mu`, should have (x, y) coordinates interlaced, for example, if there were 2 poses and 2 landmarks, `mu` will look like the following, where `P` is the robot position and `L` the landmark position:\n```\nmu = matrix([[Px0],\n [Py0],\n [Px1],\n [Py1],\n [Lx0],\n [Ly0],\n [Lx1],\n [Ly1]])\n```\n\nYou can see that `mu` holds the poses first `(x0, y0), (x1, y1), ...,` then the landmark locations at the end of the matrix; we consider a `nx1` matrix to be a vector.\n\n## Generating an environment\n\nIn a real SLAM problem, you may be given a map that contains information about landmark locations, and in this example, we will make our own data using the `make_data` function, which generates a world grid with landmarks in it and then generates data by placing a robot in that world and moving and sensing over some numer of time steps. The `make_data` function relies on a correct implementation of robot move/sense functions, which, at this point, should be complete and in the `robot_class.py` file. The data is collected as an instantiated robot moves and senses in a world. Your SLAM function will take in this data as input. So, let's first create this data and explore how it represents the movement and sensor measurements that our robot takes.\n\n---",
"_____no_output_____"
],
[
"## Create the world\n\nUse the code below to generate a world of a specified size with randomly generated landmark locations. You can change these parameters and see how your implementation of SLAM responds! \n\n`data` holds the sensors measurements and motion of your robot over time. It stores the measurements as `data[i][0]` and the motion as `data[i][1]`.\n\n#### Helper functions\n\nYou will be working with the `robot` class that may look familiar from the first notebook, \n\nIn fact, in the `helpers.py` file, you can read the details of how data is made with the `make_data` function. It should look very similar to the robot move/sense cycle you've seen in the first notebook.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom helpers import make_data\n\n# your implementation of slam should work with the following inputs\n# feel free to change these input values and see how it responds!\n\n# world parameters\nnum_landmarks = 5 # number of landmarks\nN = 20 # time steps\nworld_size = 100.0 # size of world (square)\n\n# robot parameters\nmeasurement_range = 50.0 # range at which we can sense landmarks\nmotion_noise = 2.0 # noise in robot motion\nmeasurement_noise = 2.0 # noise in the measurements\ndistance = 20.0 # distance by which robot (intends to) move each iteratation \n\n\n# make_data instantiates a robot, AND generates random landmarks for a given world size and number of landmarks\ndata = make_data(N, num_landmarks, world_size, measurement_range, motion_noise, measurement_noise, distance)",
" \nLandmarks: [[21, 13], [16, 21], [70, 38], [38, 75], [87, 50]]\nRobot: [x=21.10550 y=65.76182]\n"
]
],
[
[
"### A note on `make_data`\n\nThe function above, `make_data`, takes in so many world and robot motion/sensor parameters because it is responsible for:\n1. Instantiating a robot (using the robot class)\n2. Creating a grid world with landmarks in it\n\n**This function also prints out the true location of landmarks and the *final* robot location, which you should refer back to when you test your implementation of SLAM.**\n\nThe `data` this returns is an array that holds information about **robot sensor measurements** and **robot motion** `(dx, dy)` that is collected over a number of time steps, `N`. You will have to use *only* these readings about motion and measurements to track a robot over time and find the determine the location of the landmarks using SLAM. We only print out the true landmark locations for comparison, later.\n\n\nIn `data` the measurement and motion data can be accessed from the first and second index in the columns of the data array. See the following code for an example, where `i` is the time step:\n```\nmeasurement = data[i][0]\nmotion = data[i][1]\n```\n",
"_____no_output_____"
]
],
[
[
"# print out some stats about the data\ntime_step = 0\n\nprint('Example measurements: \\n', data[time_step][0])\nprint('\\n')\nprint('Example motion: \\n', data[time_step][1])",
"Example measurements: \n [[0, -27.336665046464944, -35.10866490452625], [1, -33.31752573050853, -30.61726091048749], [2, 19.910918480109437, -10.91254402509894], [3, -10.810346809363109, 24.042261189064593], [4, 35.51407538801196, -0.736122885336894]]\n\n\nExample motion: \n [-19.11495180327814, 5.883758795052183]\n"
]
],
[
[
"Try changing the value of `time_step`, you should see that the list of measurements varies based on what in the world the robot sees after it moves. As you know from the first notebook, the robot can only sense so far and with a certain amount of accuracy in the measure of distance between its location and the location of landmarks. The motion of the robot always is a vector with two values: one for x and one for y displacement. This structure will be useful to keep in mind as you traverse this data in your implementation of slam.",
"_____no_output_____"
],
[
"## Initialize Constraints\n\nOne of the most challenging tasks here will be to create and modify the constraint matrix and vector: omega and xi. In the second notebook, you saw an example of how omega and xi could hold all the values the define the relationships between robot poses `xi` and landmark positions `Li` in a 1D world, as seen below, where omega is the blue matrix and xi is the pink vector.\n\n<img src='images/motion_constraint.png' width=50% height=50% />\n\n\nIn *this* project, you are tasked with implementing constraints for a 2D world. We are referring to robot poses as `Px, Py` and landmark positions as `Lx, Ly`, and one way to approach this challenge is to add *both* x and y locations in the constraint matrices.\n\n<img src='images/constraints2D.png' width=50% height=50% />\n\nYou may also choose to create two of each omega and xi (one for x and one for y positions).",
"_____no_output_____"
],
[
"### TODO: Write a function that initializes omega and xi\n\nComplete the function `initialize_constraints` so that it returns `omega` and `xi` constraints for the starting position of the robot. Any values that we do not yet know should be initialized with the value `0`. You may assume that our robot starts out in exactly the middle of the world with 100% confidence (no motion or measurement noise at this point). The inputs `N` time steps, `num_landmarks`, and `world_size` should give you all the information you need to construct intial constraints of the correct size and starting values.\n\n*Depending on your approach you may choose to return one omega and one xi that hold all (x,y) positions *or* two of each (one for x values and one for y); choose whichever makes most sense to you!*",
"_____no_output_____"
]
],
[
[
"def initialize_constraints(N, num_landmarks, world_size):\n ''' This function takes in a number of time steps N, number of landmarks, and a world_size,\n and returns initialized constraint matrices, omega and xi.'''\n \n ## Recommended: Define and store the size (rows/cols) of the constraint matrix in a variable\n mat_size = 2 * (N + num_landmarks) # multiply 2 because of 2-dimension expression\n ## TODO: Define the constraint matrix, Omega, with two initial \"strength\" values\n ## for the initial x, y location of our robot\n omega = np.zeros((mat_size, mat_size))\n omega[0][0] = 1 # x in initial position\n omega[1][1] = 1 # y in initial position\n \n ## TODO: Define the constraint *vector*, xi\n ## you can assume that the robot starts out in the middle of the world with 100% confidence\n xi = np.zeros((mat_size, 1))\n xi[0] = world_size/2\n xi[1] = world_size/2\n \n return omega, xi\n\n",
"_____no_output_____"
]
],
[
[
"### Test as you go\n\nIt's good practice to test out your code, as you go. Since `slam` relies on creating and updating constraint matrices, `omega` and `xi` to account for robot sensor measurements and motion, let's check that they initialize as expected for any given parameters.\n\nBelow, you'll find some test code that allows you to visualize the results of your function `initialize_constraints`. We are using the [seaborn](https://seaborn.pydata.org/) library for visualization.\n\n**Please change the test values of N, landmarks, and world_size and see the results**. Be careful not to use these values as input into your final smal function.\n\nThis code assumes that you have created one of each constraint: `omega` and `xi`, but you can change and add to this code, accordingly. The constraints should vary in size with the number of time steps and landmarks as these values affect the number of poses a robot will take `(Px0,Py0,...Pxn,Pyn)` and landmark locations `(Lx0,Ly0,...Lxn,Lyn)` whose relationships should be tracked in the constraint matrices. Recall that `omega` holds the weights of each variable and `xi` holds the value of the sum of these variables, as seen in Notebook 2. You'll need the `world_size` to determine the starting pose of the robot in the world and fill in the initial values for `xi`.",
"_____no_output_____"
]
],
[
[
"# import data viz resources\nimport matplotlib.pyplot as plt\nfrom pandas import DataFrame\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
],
[
"# define a small N and world_size (small for ease of visualization)\nN_test = 5\nnum_landmarks_test = 2\nsmall_world = 10\n\n# initialize the constraints\ninitial_omega, initial_xi = initialize_constraints(N_test, num_landmarks_test, small_world)",
"_____no_output_____"
],
[
"# define figure size\nplt.rcParams[\"figure.figsize\"] = (10,7)\n\n# display omega\nsns.heatmap(DataFrame(initial_omega), cmap='Blues', annot=True, linewidths=.5)",
"_____no_output_____"
],
[
"# define figure size\nplt.rcParams[\"figure.figsize\"] = (1,7)\n\n# display xi\nsns.heatmap(DataFrame(initial_xi), cmap='Oranges', annot=True, linewidths=.5)",
"_____no_output_____"
]
],
[
[
"---\n## SLAM inputs \n\nIn addition to `data`, your slam function will also take in:\n* N - The number of time steps that a robot will be moving and sensing\n* num_landmarks - The number of landmarks in the world\n* world_size - The size (w/h) of your world\n* motion_noise - The noise associated with motion; the update confidence for motion should be `1.0/motion_noise`\n* measurement_noise - The noise associated with measurement/sensing; the update weight for measurement should be `1.0/measurement_noise`\n\n#### A note on noise\n\nRecall that `omega` holds the relative \"strengths\" or weights for each position variable, and you can update these weights by accessing the correct index in omega `omega[row][col]` and *adding/subtracting* `1.0/noise` where `noise` is measurement or motion noise. `Xi` holds actual position values, and so to update `xi` you'll do a similar addition process only using the actual value of a motion or measurement. So for a vector index `xi[row][0]` you will end up adding/subtracting one measurement or motion divided by their respective `noise`.\n\n### TODO: Implement Graph SLAM\n\nFollow the TODO's below to help you complete this slam implementation (these TODO's are in the recommended order), then test out your implementation! \n\n#### Updating with motion and measurements\n\nWith a 2D omega and xi structure as shown above (in earlier cells), you'll have to be mindful about how you update the values in these constraint matrices to account for motion and measurement constraints in the x and y directions. Recall that the solution to these matrices (which holds all values for robot poses `P` and landmark locations `L`) is the vector, `mu`, which can be computed at the end of the construction of omega and xi as the inverse of omega times xi: $\\mu = \\Omega^{-1}\\xi$\n\n**You may also choose to return the values of `omega` and `xi` if you want to visualize their final state!**",
"_____no_output_____"
]
],
[
[
"## TODO: Complete the code to implement SLAM\n\n## slam takes in 6 arguments and returns mu, \n## mu is the entire path traversed by a robot (all x,y poses) *and* all landmarks locations\ndef slam(data, N, num_landmarks, world_size, motion_noise, measurement_noise):\n \n ## TODO: Use your initilization to create constraint matrices, omega and xi\n omega, xi = initialize_constraints(N, num_landmarks, world_size)\n \n ## TODO: Iterate through each time step in the data\n ## get all the motion and measurement data as you iterate\n for i in range(0, N-1): # loop for steps\n pos_index = 2 * i # index for 2-dimension\n measurement = data[i][0]\n ## TODO: update the constraint matrix/vector to account for all *measurements*\n ## this should be a series of additions that take into account the measurement noise\n for j in range(len(measurement)): # observed landmarks loop\n land_index = 2 * ( N + measurement[j][0] )\n for k in range(2): # 2-dimension loop\n # update omega matrix\n assert pos_index < 2*N, \"pos_index: {}\".format(pos_index)\n assert land_index < 2*(N+num_landmarks), \"land_index: {}\".format(land_index)\n omega[pos_index+k][pos_index+k] += (1 / measurement_noise)\n omega[pos_index+k][land_index+k] -= (1 / measurement_noise)\n omega[land_index+k][pos_index+k] -= (1 / measurement_noise)\n omega[land_index+k][land_index+k] += (1 / measurement_noise)\n # update xi vector\n xi[pos_index+k] -= (measurement[j][k+1] / measurement_noise)\n xi[land_index+k] += (measurement[j][k+1] / measurement_noise)\n \n \n ## TODO: update the constraint matrix/vector to account for all *motion* and motion noise\n for i in range(0, N-1): # loop for steps\n cpos_index = 2 * i # index for 2-dimension current position.\n npos_index = 2 * (i + 1) # index for 2-dimension next position.\n motion = data[i][1]\n for j in range(2): # loop for 2-dimension\n omega[cpos_index+j][cpos_index+j] += (1 / motion_noise)\n omega[cpos_index+j][npos_index+j] -= (1 / motion_noise)\n omega[npos_index+j][cpos_index+j] -= (1 / motion_noise)\n omega[npos_index+j][npos_index+j] += (1 / motion_noise)\n xi[cpos_index+j] -= (motion[j] / motion_noise)\n xi[npos_index+j] += (motion[j] / motion_noise)\n \n ## TODO: After iterating through all the data\n ## Compute the best estimate of poses and landmark positions\n ## using the formula, omega_inverse * Xi\n mu = np.linalg.inv(np.matrix(omega)) * xi\n \n return mu # return `mu`\n",
"_____no_output_____"
]
],
[
[
"## Helper functions\n\nTo check that your implementation of SLAM works for various inputs, we have provided two helper functions that will help display the estimated pose and landmark locations that your function has produced. First, given a result `mu` and number of time steps, `N`, we define a function that extracts the poses and landmarks locations and returns those as their own, separate lists. \n\nThen, we define a function that nicely print out these lists; both of these we will call, in the next step.\n",
"_____no_output_____"
]
],
[
[
"# a helper function that creates a list of poses and of landmarks for ease of printing\n# this only works for the suggested constraint architecture of interlaced x,y poses\ndef get_poses_landmarks(mu, N):\n # create a list of poses\n poses = []\n for i in range(N):\n poses.append((mu[2*i].item(), mu[2*i+1].item()))\n\n # create a list of landmarks\n landmarks = []\n for i in range(num_landmarks):\n landmarks.append((mu[2*(N+i)].item(), mu[2*(N+i)+1].item()))\n\n # return completed lists\n return poses, landmarks\n",
"_____no_output_____"
],
[
"def print_all(poses, landmarks):\n print('\\n')\n print('Estimated Poses:')\n for i in range(len(poses)):\n print('['+', '.join('%.3f'%p for p in poses[i])+']')\n print('\\n')\n print('Estimated Landmarks:')\n for i in range(len(landmarks)):\n print('['+', '.join('%.3f'%l for l in landmarks[i])+']')\n",
"_____no_output_____"
]
],
[
[
"## Run SLAM\n\nOnce you've completed your implementation of `slam`, see what `mu` it returns for different world sizes and different landmarks!\n\n### What to Expect\n\nThe `data` that is generated is random, but you did specify the number, `N`, or time steps that the robot was expected to move and the `num_landmarks` in the world (which your implementation of `slam` should see and estimate a position for. Your robot should also start with an estimated pose in the very center of your square world, whose size is defined by `world_size`.\n\nWith these values in mind, you should expect to see a result that displays two lists:\n1. **Estimated poses**, a list of (x, y) pairs that is exactly `N` in length since this is how many motions your robot has taken. The very first pose should be the center of your world, i.e. `[50.000, 50.000]` for a world that is 100.0 in square size.\n2. **Estimated landmarks**, a list of landmark positions (x, y) that is exactly `num_landmarks` in length. \n\n#### Landmark Locations\n\nIf you refer back to the printout of *exact* landmark locations when this data was created, you should see values that are very similar to those coordinates, but not quite (since `slam` must account for noise in motion and measurement).",
"_____no_output_____"
]
],
[
[
"# call your implementation of slam, passing in the necessary parameters\nmu = slam(data, N, num_landmarks, world_size, motion_noise, measurement_noise)\n\n# print out the resulting landmarks and poses\nif(mu is not None):\n # get the lists of poses and landmarks\n # and print them out\n poses, landmarks = get_poses_landmarks(mu, N)\n print_all(poses, landmarks)",
"\n\nEstimated Poses:\n[50.000, 50.000]\n[30.932, 55.524]\n[10.581, 60.210]\n[1.931, 44.539]\n[13.970, 30.030]\n[26.721, 15.510]\n[38.852, 1.865]\n[23.640, 16.597]\n[7.988, 30.490]\n[16.829, 49.508]\n[26.632, 68.416]\n[34.588, 85.257]\n[15.954, 87.795]\n[14.410, 68.543]\n[11.222, 48.268]\n[7.365, 29.064]\n[4.241, 8.590]\n[10.207, 28.434]\n[14.889, 47.138]\n[20.159, 66.431]\n\n\nEstimated Landmarks:\n[21.992, 13.723]\n[16.718, 21.137]\n[69.826, 37.990]\n[38.311, 75.271]\n[87.067, 48.906]\n"
]
],
[
[
"## Visualize the constructed world\n\nFinally, using the `display_world` code from the `helpers.py` file (which was also used in the first notebook), we can actually visualize what you have coded with `slam`: the final position of the robot and the positon of landmarks, created from only motion and measurement data!\n\n**Note that these should be very similar to the printed *true* landmark locations and final pose from our call to `make_data` early in this notebook.**",
"_____no_output_____"
]
],
[
[
"# import the helper function\nfrom helpers import display_world\n\n# Display the final world!\n\n# define figure size\nplt.rcParams[\"figure.figsize\"] = (20,20)\n\n# check if poses has been created\nif 'poses' in locals():\n # print out the last pose\n print('Last pose: ', poses[-1])\n # display the last position of the robot *and* the landmark positions\n display_world(int(world_size), poses[-1], landmarks)",
"Last pose: (20.15873442355911, 66.43079160194176)\n"
]
],
[
[
"### Question: How far away is your final pose (as estimated by `slam`) compared to the *true* final pose? Why do you think these poses are different?\n\nYou can find the true value of the final pose in one of the first cells where `make_data` was called. You may also want to look at the true landmark locations and compare them to those that were estimated by `slam`. Ask yourself: what do you think would happen if we moved and sensed more (increased N)? Or if we had lower/higher noise parameters.",
"_____no_output_____"
],
[
"**Answer**: \n\nGround truth position was displayed as the following.\n\n`Landmarks: [[21, 13], [16, 21], [70, 38], [38, 75], [87, 50]]\nRobot: [x=21.10550 y=65.76182]`\n\n\nEstimated Landmarks was obtained as below.\n\n`Estimated Poses:\n[50.000, 50.000]\n... (after 20 iterations)\n[20.159, 66.431]\n\n\nEstimated Landmarks:\n[21.992, 13.723]\n[16.718, 21.137]\n[69.826, 37.990]\n[38.311, 75.271]\n[87.067, 48.906]`\n\n\nEstimated poses and ground truth poses are similar to each other.\nThere are also much similarity between estimated landmarks and that of ground truth.\nRMSE between ground truth and estimation is 0.6315.\nThis is calculated in the following cell of this notebook.\nRMSE is very small number and SLAM estimated robot position and landmarks.\n\n\nThough only robot moved at only 20 steps in this case, more motion steps will contribute more accurate estimation.\nA lot of sample of measurements will decrease the influence of measurment noise and motion noise.\nWith more measurement sample, SLAM can ignore noise deviation.\n\n\nWhen noise deviation is less, estimation accuracy of SLAM is higher.\nNoise deviation prevents robot from estimating its position and landmarks' position with high accuracy.\n",
"_____no_output_____"
]
],
[
[
"# calculate RMSE\nimport math\ndef getRMSE(ground_truth, estimation):\n sum_rmse = 0\n for i, element_est in enumerate(estimation):\n diff = ground_truth[i] - element_est\n diff_square = diff * diff\n sum_rmse += diff_square\n rmse = math.sqrt(sum_rmse / len(ground_truth))\n return rmse\nflatten = lambda x: [z for y in x for z in (flatten(y) if hasattr(y, '__iter__') else (y,))]\nground_truth = [[21.10550, 65.76182], [21, 13], [16, 21], [70, 38], [38, 75], [87, 50]]\nestimation = [[20.159, 66.431], [21.992, 13.723], [16.718, 21.137], [69.826, 37.990], [38.311, 75.271], [87.067, 48.906]]\nground_truth = flatten(ground_truth)\nestimation = flatten(estimation)\nrmse = getRMSE(ground_truth, estimation)\nprint(rmse)",
"0.6315729782587813\n"
]
],
[
[
"## Testing\n\nTo confirm that your slam code works before submitting your project, it is suggested that you run it on some test data and cases. A few such cases have been provided for you, in the cells below. When you are ready, uncomment the test cases in the next cells (there are two test cases, total); your output should be **close-to or exactly** identical to the given results. If there are minor discrepancies it could be a matter of floating point accuracy or in the calculation of the inverse matrix.\n\n### Submit your project\n\nIf you pass these tests, it is a good indication that your project will pass all the specifications in the project rubric. Follow the submission instructions to officially submit!",
"_____no_output_____"
]
],
[
[
"# Here is the data and estimated outputs for test case 1\n\ntest_data1 = [[[[1, 19.457599255548065, 23.8387362100849], [2, -13.195807561967236, 11.708840328458608], [3, -30.0954905279171, 15.387879242505843]], [-12.2607279422326, -15.801093326936487]], [[[2, -0.4659930049620491, 28.088559771215664], [4, -17.866382374890936, -16.384904503932]], [-12.2607279422326, -15.801093326936487]], [[[4, -6.202512900833806, -1.823403210274639]], [-12.2607279422326, -15.801093326936487]], [[[4, 7.412136480918645, 15.388585962142429]], [14.008259661173426, 14.274756084260822]], [[[4, -7.526138813444998, -0.4563942429717849]], [14.008259661173426, 14.274756084260822]], [[[2, -6.299793150150058, 29.047830407717623], [4, -21.93551130411791, -13.21956810989039]], [14.008259661173426, 14.274756084260822]], [[[1, 15.796300959032276, 30.65769689694247], [2, -18.64370821983482, 17.380022987031367]], [14.008259661173426, 14.274756084260822]], [[[1, 0.40311325410337906, 14.169429532679855], [2, -35.069349468466235, 2.4945558982439957]], [14.008259661173426, 14.274756084260822]], [[[1, -16.71340983241936, -2.777000269543834]], [-11.006096015782283, 16.699276945166858]], [[[1, -3.611096830835776, -17.954019226763958]], [-19.693482634035977, 3.488085684573048]], [[[1, 18.398273354362416, -22.705102332550947]], [-19.693482634035977, 3.488085684573048]], [[[2, 2.789312482883833, -39.73720193121324]], [12.849049222879723, -15.326510824972983]], [[[1, 21.26897046581808, -10.121029799040915], [2, -11.917698965880655, -23.17711662602097], [3, -31.81167947898398, -16.7985673023331]], [12.849049222879723, -15.326510824972983]], [[[1, 10.48157743234859, 5.692957082575485], [2, -22.31488473554935, -5.389184118551409], [3, -40.81803984305378, -2.4703329790238118]], [12.849049222879723, -15.326510824972983]], [[[0, 10.591050242096598, -39.2051798967113], [1, -3.5675572049297553, 22.849456408289125], [2, -38.39251065320351, 7.288990306029511]], [12.849049222879723, -15.326510824972983]], [[[0, -3.6225556479370766, -25.58006865235512]], [-7.8874682868419965, -18.379005523261092]], [[[0, 1.9784503557879374, -6.5025974151499]], [-7.8874682868419965, -18.379005523261092]], [[[0, 10.050665232782423, 11.026385307998742]], [-17.82919359778298, 9.062000642947142]], [[[0, 26.526838150174818, -0.22563393232425621], [4, -33.70303936886652, 2.880339841013677]], [-17.82919359778298, 9.062000642947142]]]\n\n## Test Case 1\n##\n# Estimated Pose(s):\n# [50.000, 50.000]\n# [37.858, 33.921]\n# [25.905, 18.268]\n# [13.524, 2.224]\n# [27.912, 16.886]\n# [42.250, 30.994]\n# [55.992, 44.886]\n# [70.749, 59.867]\n# [85.371, 75.230]\n# [73.831, 92.354]\n# [53.406, 96.465]\n# [34.370, 100.134]\n# [48.346, 83.952]\n# [60.494, 68.338]\n# [73.648, 53.082]\n# [86.733, 38.197]\n# [79.983, 20.324]\n# [72.515, 2.837]\n# [54.993, 13.221]\n# [37.164, 22.283]\n\n\n# Estimated Landmarks:\n# [82.679, 13.435]\n# [70.417, 74.203]\n# [36.688, 61.431]\n# [18.705, 66.136]\n# [20.437, 16.983]\n\n\n### Uncomment the following three lines for test case 1 and compare the output to the values above ###\n\nmu_1 = slam(test_data1, 20, 5, 100.0, 2.0, 2.0)\nposes, landmarks = get_poses_landmarks(mu_1, 20)\nprint_all(poses, landmarks)",
"\n\nEstimated Poses:\n[50.000, 50.000]\n[37.973, 33.652]\n[26.185, 18.155]\n[13.745, 2.116]\n[28.097, 16.783]\n[42.384, 30.902]\n[55.831, 44.497]\n[70.857, 59.699]\n[85.697, 75.543]\n[74.011, 92.434]\n[53.544, 96.454]\n[34.525, 100.080]\n[48.623, 83.953]\n[60.197, 68.107]\n[73.778, 52.935]\n[87.132, 38.538]\n[80.303, 20.508]\n[72.798, 2.945]\n[55.245, 13.255]\n[37.416, 22.317]\n\n\nEstimated Landmarks:\n[82.956, 13.539]\n[70.495, 74.141]\n[36.740, 61.281]\n[18.698, 66.060]\n[20.635, 16.875]\n"
],
[
"# Here is the data and estimated outputs for test case 2\n\ntest_data2 = [[[[0, 26.543274387283322, -6.262538160312672], [3, 9.937396825799755, -9.128540360867689]], [18.92765331253674, -6.460955043986683]], [[[0, 7.706544739722961, -3.758467215445748], [1, 17.03954411948937, 31.705489938553438], [3, -11.61731288777497, -6.64964096716416]], [18.92765331253674, -6.460955043986683]], [[[0, -12.35130507136378, 2.585119104239249], [1, -2.563534536165313, 38.22159657838369], [3, -26.961236804740935, -0.4802312626141525]], [-11.167066095509824, 16.592065417497455]], [[[0, 1.4138633151721272, -13.912454837810632], [1, 8.087721200818589, 20.51845934354381], [3, -17.091723454402302, -16.521500551709707], [4, -7.414211721400232, 38.09191602674439]], [-11.167066095509824, 16.592065417497455]], [[[0, 12.886743222179561, -28.703968411636318], [1, 21.660953298391387, 3.4912891084614914], [3, -6.401401414569506, -32.321583037341625], [4, 5.034079343639034, 23.102207946092893]], [-11.167066095509824, 16.592065417497455]], [[[1, 31.126317672358578, -10.036784369535214], [2, -38.70878528420893, 7.4987265861424595], [4, 17.977218575473767, 6.150889254289742]], [-6.595520680493778, -18.88118393939265]], [[[1, 41.82460922922086, 7.847527392202475], [3, 15.711709540417502, -30.34633659912818]], [-6.595520680493778, -18.88118393939265]], [[[0, 40.18454208294434, -6.710999804403755], [3, 23.019508919299156, -10.12110867290604]], [-6.595520680493778, -18.88118393939265]], [[[3, 27.18579315312821, 8.067219022708391]], [-6.595520680493778, -18.88118393939265]], [[], [11.492663265706092, 16.36822198838621]], [[[3, 24.57154567653098, 13.461499960708197]], [11.492663265706092, 16.36822198838621]], [[[0, 31.61945290413707, 0.4272295085799329], [3, 16.97392299158991, -5.274596836133088]], [11.492663265706092, 16.36822198838621]], [[[0, 22.407381798735177, -18.03500068379259], [1, 29.642444125196995, 17.3794951934614], [3, 4.7969752441371645, -21.07505361639969], [4, 14.726069092569372, 32.75999422300078]], [11.492663265706092, 16.36822198838621]], [[[0, 10.705527984670137, -34.589764174299596], [1, 18.58772336795603, -0.20109708164787765], [3, -4.839806195049413, -39.92208742305105], [4, 4.18824810165454, 14.146847823548889]], [11.492663265706092, 16.36822198838621]], [[[1, 5.878492140223764, -19.955352450942357], [4, -7.059505455306587, -0.9740849280550585]], [19.628527845173146, 3.83678180657467]], [[[1, -11.150789592446378, -22.736641053247872], [4, -28.832815721158255, -3.9462962046291388]], [-19.841703647091965, 2.5113335861604362]], [[[1, 8.64427397916182, -20.286336970889053], [4, -5.036917727942285, -6.311739993868336]], [-5.946642674882207, -19.09548221169787]], [[[0, 7.151866679283043, -39.56103232616369], [1, 16.01535401373368, -3.780995345194027], [4, -3.04801331832137, 13.697362774960865]], [-5.946642674882207, -19.09548221169787]], [[[0, 12.872879480504395, -19.707592098123207], [1, 22.236710716903136, 16.331770792606406], [3, -4.841206109583004, -21.24604435851242], [4, 4.27111163223552, 32.25309748614184]], [-5.946642674882207, -19.09548221169787]]] \n\n\n## Test Case 2\n##\n# Estimated Pose(s):\n# [50.000, 50.000]\n# [69.035, 45.061]\n# [87.655, 38.971]\n# [76.084, 55.541]\n# [64.283, 71.684]\n# [52.396, 87.887]\n# [44.674, 68.948]\n# [37.532, 49.680]\n# [31.392, 30.893]\n# [24.796, 12.012]\n# [33.641, 26.440]\n# [43.858, 43.560]\n# [54.735, 60.659]\n# [65.884, 77.791]\n# [77.413, 94.554]\n# [96.740, 98.020]\n# [76.149, 99.586]\n# [70.211, 80.580]\n# [64.130, 61.270]\n# [58.183, 42.175]\n\n\n# Estimated Landmarks:\n# [76.777, 42.415]\n# [85.109, 76.850]\n# [13.687, 95.386]\n# [59.488, 39.149]\n# [69.283, 93.654]\n\n\n### Uncomment the following three lines for test case 2 and compare to the values above ###\n\nmu_2 = slam(test_data2, 20, 5, 100.0, 2.0, 2.0)\nposes, landmarks = get_poses_landmarks(mu_2, 20)\nprint_all(poses, landmarks)\n",
"\n\nEstimated Poses:\n[50.000, 50.000]\n[69.181, 45.665]\n[87.743, 39.703]\n[76.270, 56.311]\n[64.317, 72.176]\n[52.257, 88.154]\n[44.059, 69.401]\n[37.002, 49.918]\n[30.924, 30.955]\n[23.508, 11.419]\n[34.180, 27.133]\n[44.155, 43.846]\n[54.806, 60.920]\n[65.698, 78.546]\n[77.468, 95.626]\n[96.802, 98.821]\n[75.957, 99.971]\n[70.200, 81.181]\n[64.054, 61.723]\n[58.107, 42.628]\n\n\nEstimated Landmarks:\n[76.779, 42.887]\n[85.065, 77.438]\n[13.548, 95.652]\n[59.449, 39.595]\n[69.263, 94.240]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e7f7159e32ce8acf95211161dfcd84b1de2549ac | 25,649 | ipynb | Jupyter Notebook | notebooks/develop/Cluster_Heatmaps.ipynb | michaelfedell/instacart | dc7a49d1247e0a894cc1b1efa5fa876df6bd5683 | [
"MIT"
] | null | null | null | notebooks/develop/Cluster_Heatmaps.ipynb | michaelfedell/instacart | dc7a49d1247e0a894cc1b1efa5fa876df6bd5683 | [
"MIT"
] | 1 | 2019-06-13T01:25:54.000Z | 2019-06-13T01:25:54.000Z | notebooks/develop/Cluster_Heatmaps.ipynb | michaelfedell/instacart | dc7a49d1247e0a894cc1b1efa5fa876df6bd5683 | [
"MIT"
] | null | null | null | 75.660767 | 15,968 | 0.747982 | [
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport matplotlib as mpl\nimport seaborn as sns\nfrom scipy.stats import mode\nfrom sklearn.preprocessing import scale\n\n%matplotlib inline\nmpl.rcParams['figure.figsize'] = [12, 8]",
"_____no_output_____"
],
[
"order_types = pd.read_csv('../../data/features/order_types.csv', index_col='label')",
"_____no_output_____"
],
[
"order_types.head()",
"_____no_output_____"
],
[
"sub = order_types.columns[:11]\nplot_data = pd.DataFrame(scale(order_types[sub]), columns=sub).transpose()\nax = sns.heatmap(plot_data, center=0, xticklabels=list(order_types.index))\nplt.xlabel('cluster label')\n# plt.show()",
"_____no_output_____"
],
[
"plt.savefig('test.png')",
"_____no_output_____"
],
[
"import os\nos.getcwd()",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f725d53b4750965b3bcd49b9756a27d0220797 | 170,849 | ipynb | Jupyter Notebook | ml/Random Forests/RandomForests.ipynb | Siddhant-K-code/AlgoBook | a37f1fbcb11ae27801bc47c01357ed90035a4e82 | [
"MIT"
] | 191 | 2020-09-28T10:00:20.000Z | 2022-03-06T14:36:55.000Z | ml/Random Forests/RandomForests.ipynb | Siddhant-K-code/AlgoBook | a37f1fbcb11ae27801bc47c01357ed90035a4e82 | [
"MIT"
] | 210 | 2020-09-28T10:06:36.000Z | 2022-03-05T03:44:24.000Z | ml/Random Forests/RandomForests.ipynb | Siddhant-K-code/AlgoBook | a37f1fbcb11ae27801bc47c01357ed90035a4e82 | [
"MIT"
] | 320 | 2020-09-28T09:56:14.000Z | 2022-02-12T16:45:57.000Z | 349.384458 | 159,728 | 0.933029 | [
[
[
"# Random Forests",
"_____no_output_____"
],
[
"### Import Libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn import datasets,metrics\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.model_selection import train_test_split\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Load the [iris_data](https://archive.ics.uci.edu/ml/datasets/iris)",
"_____no_output_____"
]
],
[
[
"iris_data = datasets.load_iris()\n\nprint(iris_data.target_names)\nprint(iris_data.feature_names)",
"['setosa' 'versicolor' 'virginica']\n['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']\n"
]
],
[
[
"### Preprocess the data",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n{\n 'sepal_length':iris_data.data[:,0],\n 'sepal_width':iris_data.data[:,1],\n 'petal_length':iris_data.data[:,2],\n 'petal_width':iris_data.data[:,3],\n 'species':iris_data.target\n})\ndf.head()",
"_____no_output_____"
],
[
"#Number of instances per class\ndf.groupby('species').size()",
"_____no_output_____"
],
[
"# species -> target column\nfeatures = df.iloc[:,:4].values\ntargets = df['species']",
"_____no_output_____"
]
],
[
[
"### Visualization",
"_____no_output_____"
]
],
[
[
"#pair_plot\n#To explore the relationship between the features\nplt.figure()\nsns.pairplot(df,hue = \"species\", height=3, markers=[\"o\", \"s\", \"D\"])\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Fitting the model",
"_____no_output_____"
]
],
[
[
"X_train, X_test, Y_train, Y_test = train_test_split(features,targets,test_size = 0.3,random_state = 1)\nmodel_1 = RandomForestClassifier(n_estimators = 100,random_state = 1)\nmodel_1.fit(X_train, Y_train)",
"_____no_output_____"
],
[
"Y_pred = model_1.predict(X_test)",
"_____no_output_____"
],
[
"metrics.accuracy_score(Y_test,Y_pred)",
"_____no_output_____"
]
],
[
[
"#### Accuracy is around 95.6%",
"_____no_output_____"
],
[
"### Improving the model",
"_____no_output_____"
],
[
"#### Hyperparameter selection",
"_____no_output_____"
]
],
[
[
"#using Exhaustive Grid Search\nn_estimators = [2, 10, 100,500]\nmax_depth = [2, 10, 15,20]\nmin_samples_split = [1,2, 5, 10]\nmin_samples_leaf = [1, 2, 10,20]\n\nhyper_param = dict(n_estimators = n_estimators, max_depth = max_depth, \n min_samples_split = min_samples_split, \n min_samples_leaf = min_samples_leaf)",
"_____no_output_____"
],
[
"gridF = GridSearchCV(RandomForestClassifier(random_state = 1), hyper_param, cv = 3, verbose = 1, \n n_jobs = -1)\nbestF = gridF.fit(X_train, Y_train)",
"Fitting 3 folds for each of 256 candidates, totalling 768 fits\n"
],
[
"grid.best_params_",
"_____no_output_____"
],
[
"#using these parameters\nmodel_2 = RandomForestClassifier(n_estimators = 2,max_depth = 15, min_samples_leaf = 2, min_samples_split = 2)\nmodel_2.fit(X_train,Y_train)",
"_____no_output_____"
],
[
"Y_pred_2 = model_2.predict(X_test)",
"_____no_output_____"
],
[
"metrics.accuracy_score(Y_test,Y_pred_2)",
"_____no_output_____"
],
[
"#Other such Hyperparameter tuning methods can also be used. ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f725f9209f0c668db7300fef8a6a2c677b606d | 13,396 | ipynb | Jupyter Notebook | Jupyter/cpp/Thread.ipynb | tedi21/SisypheReview | f7c05bad1ccc036f45870535149d9685e1120c2c | [
"Unlicense"
] | null | null | null | Jupyter/cpp/Thread.ipynb | tedi21/SisypheReview | f7c05bad1ccc036f45870535149d9685e1120c2c | [
"Unlicense"
] | null | null | null | Jupyter/cpp/Thread.ipynb | tedi21/SisypheReview | f7c05bad1ccc036f45870535149d9685e1120c2c | [
"Unlicense"
] | null | null | null | 27.73499 | 322 | 0.475142 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e7f73123470ec41c0d04a6bdbb5db2c86779c4fc | 181 | ipynb | Jupyter Notebook | lessons/02_spacetime/Untitled0.ipynb | Yurlungur/numerical-mooc | f40dce7b545c8dc33fc9d88b9a2442b091175be4 | [
"CC-BY-3.0"
] | null | null | null | lessons/02_spacetime/Untitled0.ipynb | Yurlungur/numerical-mooc | f40dce7b545c8dc33fc9d88b9a2442b091175be4 | [
"CC-BY-3.0"
] | null | null | null | lessons/02_spacetime/Untitled0.ipynb | Yurlungur/numerical-mooc | f40dce7b545c8dc33fc9d88b9a2442b091175be4 | [
"CC-BY-3.0"
] | null | null | null | 20.111111 | 88 | 0.690608 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e7f75c66604c1e5951489eddeb4805fcf2506725 | 4,411 | ipynb | Jupyter Notebook | Flask_Apps/Climate_Analysis/data_engineering.ipynb | bnonni/Python | 9ebd18caa4e2d805028b557e8b77ea65a9ee1a3d | [
"Apache-2.0"
] | 4 | 2019-10-05T03:41:20.000Z | 2020-11-04T00:39:13.000Z | Flask_Apps/Climate_Analysis/data_engineering.ipynb | bnonni/Python | 9ebd18caa4e2d805028b557e8b77ea65a9ee1a3d | [
"Apache-2.0"
] | null | null | null | Flask_Apps/Climate_Analysis/data_engineering.ipynb | bnonni/Python | 9ebd18caa4e2d805028b557e8b77ea65a9ee1a3d | [
"Apache-2.0"
] | 2 | 2019-10-02T14:08:51.000Z | 2019-10-03T20:49:09.000Z | 22.973958 | 90 | 0.530719 | [
[
[
"### BEGIN SOLUTION\nimport os\nimport pandas as pd\nimport numpy as np\n### END SOLUTION",
"_____no_output_____"
],
[
"# Grab a reference to the current directory\n### BEGIN SOLUTION\nrootdir = os.getcwd()\n### END SOLUTION",
"_____no_output_____"
],
[
"# Use `os.scandir` to get a list of all files in the current directory\n### BEGIN SOLUTION\ncsvs = os.scandir(rootdir)\n### END SOLUTION",
"_____no_output_____"
],
[
"# Iterate through the list and clean/process any CSV file using Pandas\n### BEGIN SOLUTION\nfor csv in csvs:\n # Only open CSV file extensions\n if csv.name.endswith('.csv'):\n # Read the CSV file\n df = pd.read_csv(csv.path, dtype=object)\n\n # Drop the location column since lat, lon, and elev already exist\n if 'location' in df.columns:\n df = df.drop(['location'], axis=1).reset_index(drop=True)\n \n # Use the mean to fill in any NaNs\n df.fillna(df.mean(), inplace=True)\n\n # Save the cleaned files with a `clean_` prefix\n df.to_csv(os.path.join(rootdir, f\"clean_{csv.name}\"), index=False)\n### END SOLUTION",
"_____no_output_____"
],
[
"# Verify that the cleaned files were created\n!ls",
"clean_hawaii_measurements.csv hawaii.sqlite\nclean_hawaii_stations.csv \u001b[31mhawaii_measurements.csv\u001b[m\u001b[m\nclimate_analysis.ipynb \u001b[31mhawaii_stations.csv\u001b[m\u001b[m\ndata_engineering.ipynb stats.ipynb\ndb_engineering.ipynb\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7f75daeee51eb5e7acc9f493775e0d8f15fd190 | 13,877 | ipynb | Jupyter Notebook | how-to-use-azureml/deployment/onnx/onnx-modelzoo-aml-deploy-resnet50.ipynb | MustAl-Du/MachineLearningNotebooks | a85cf47a5b923463bdc5a14bfd7a3ec0d46dd35d | [
"MIT"
] | null | null | null | how-to-use-azureml/deployment/onnx/onnx-modelzoo-aml-deploy-resnet50.ipynb | MustAl-Du/MachineLearningNotebooks | a85cf47a5b923463bdc5a14bfd7a3ec0d46dd35d | [
"MIT"
] | null | null | null | how-to-use-azureml/deployment/onnx/onnx-modelzoo-aml-deploy-resnet50.ipynb | MustAl-Du/MachineLearningNotebooks | a85cf47a5b923463bdc5a14bfd7a3ec0d46dd35d | [
"MIT"
] | 1 | 2021-06-02T06:31:15.000Z | 2021-06-02T06:31:15.000Z | 33.438554 | 462 | 0.520574 | [
[
[
"Copyright (c) Microsoft Corporation. All rights reserved. \n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# ResNet50 Image Classification using ONNX and AzureML\n\nThis example shows how to deploy the ResNet50 ONNX model as a web service using Azure Machine Learning services and the ONNX Runtime.\n\n## What is ONNX\nONNX is an open format for representing machine learning and deep learning models. ONNX enables open and interoperable AI by enabling data scientists and developers to use the tools of their choice without worrying about lock-in and flexibility to deploy to a variety of platforms. ONNX is developed and supported by a community of partners including Microsoft, Facebook, and Amazon. For more information, explore the [ONNX website](http://onnx.ai).\n\n## ResNet50 Details\nResNet classifies the major object in an input image into a set of 1000 pre-defined classes. For more information about the ResNet50 model and how it was created can be found on the [ONNX Model Zoo github](https://github.com/onnx/models/tree/master/vision/classification/resnet). ",
"_____no_output_____"
],
[
"## Prerequisites\n\nTo make the best use of your time, make sure you have done the following:\n\n* Understand the [architecture and terms](https://docs.microsoft.com/azure/machine-learning/service/concept-azure-machine-learning-architecture) introduced by Azure Machine Learning\n* If you are using an Azure Machine Learning Notebook VM, you are all set. Otherwise, go through the [configuration notebook](../../../configuration.ipynb) to:\n * install the AML SDK\n * create a workspace and its configuration file (config.json)",
"_____no_output_____"
]
],
[
[
"# Check core SDK version number\nimport azureml.core\n\nprint(\"SDK version:\", azureml.core.VERSION)",
"_____no_output_____"
]
],
[
[
"#### Download pre-trained ONNX model from ONNX Model Zoo.\n\nDownload the [ResNet50v2 model and test data](https://s3.amazonaws.com/onnx-model-zoo/resnet/resnet50v2/resnet50v2.tar.gz) and extract it in the same folder as this tutorial notebook.\n",
"_____no_output_____"
]
],
[
[
"import urllib.request\n\nonnx_model_url = \"https://s3.amazonaws.com/onnx-model-zoo/resnet/resnet50v2/resnet50v2.tar.gz\"\nurllib.request.urlretrieve(onnx_model_url, filename=\"resnet50v2.tar.gz\")\n\n!tar xvzf resnet50v2.tar.gz",
"_____no_output_____"
]
],
[
[
"## Deploying as a web service with Azure ML",
"_____no_output_____"
],
[
"### Load your Azure ML workspace\n\nWe begin by instantiating a workspace object from the existing workspace created earlier in the configuration notebook.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Workspace\n\nws = Workspace.from_config()\nprint(ws.name, ws.location, ws.resource_group, sep = '\\n')",
"_____no_output_____"
]
],
[
[
"### Register your model with Azure ML\n\nNow we upload the model and register it in the workspace.",
"_____no_output_____"
]
],
[
[
"from azureml.core.model import Model\n\nmodel = Model.register(model_path = \"resnet50v2/resnet50v2.onnx\",\n model_name = \"resnet50v2\",\n tags = {\"onnx\": \"demo\"},\n description = \"ResNet50v2 from ONNX Model Zoo\",\n workspace = ws)",
"_____no_output_____"
]
],
[
[
"#### Displaying your registered models\n\nYou can optionally list out all the models that you have registered in this workspace.",
"_____no_output_____"
]
],
[
[
"models = ws.models\nfor name, m in models.items():\n print(\"Name:\", name,\"\\tVersion:\", m.version, \"\\tDescription:\", m.description, m.tags)",
"_____no_output_____"
]
],
[
[
"### Write scoring file\n\nWe are now going to deploy our ONNX model on Azure ML using the ONNX Runtime. We begin by writing a score.py file that will be invoked by the web service call. The `init()` function is called once when the container is started so we load the model using the ONNX Runtime into a global session object.",
"_____no_output_____"
]
],
[
[
"%%writefile score.py\nimport json\nimport time\nimport sys\nimport os\nimport numpy as np # we're going to use numpy to process input and output data\nimport onnxruntime # to inference ONNX models, we use the ONNX Runtime\n\ndef softmax(x):\n x = x.reshape(-1)\n e_x = np.exp(x - np.max(x))\n return e_x / e_x.sum(axis=0)\n\ndef init():\n global session\n # AZUREML_MODEL_DIR is an environment variable created during deployment.\n # It is the path to the model folder (./azureml-models/$MODEL_NAME/$VERSION)\n # For multiple models, it points to the folder containing all deployed models (./azureml-models)\n model = os.path.join(os.getenv('AZUREML_MODEL_DIR'), 'resnet50v2.onnx')\n session = onnxruntime.InferenceSession(model, None)\n\ndef preprocess(input_data_json):\n # convert the JSON data into the tensor input\n img_data = np.array(json.loads(input_data_json)['data']).astype('float32')\n \n #normalize\n mean_vec = np.array([0.485, 0.456, 0.406])\n stddev_vec = np.array([0.229, 0.224, 0.225])\n norm_img_data = np.zeros(img_data.shape).astype('float32')\n for i in range(img_data.shape[0]):\n norm_img_data[i,:,:] = (img_data[i,:,:]/255 - mean_vec[i]) / stddev_vec[i]\n\n return norm_img_data\n\ndef postprocess(result):\n return softmax(np.array(result)).tolist()\n\ndef run(input_data_json):\n try:\n start = time.time()\n # load in our data which is expected as NCHW 224x224 image\n input_data = preprocess(input_data_json)\n input_name = session.get_inputs()[0].name # get the id of the first input of the model \n result = session.run([], {input_name: input_data})\n end = time.time() # stop timer\n return {\"result\": postprocess(result),\n \"time\": end - start}\n except Exception as e:\n result = str(e)\n return {\"error\": result}",
"_____no_output_____"
]
],
[
[
"### Create inference configuration",
"_____no_output_____"
],
[
"First we create a YAML file that specifies which dependencies we would like to see in our container.",
"_____no_output_____"
]
],
[
[
"from azureml.core.conda_dependencies import CondaDependencies \n\nmyenv = CondaDependencies.create(pip_packages=[\"numpy\",\"onnxruntime\",\"azureml-core\"])\n\nwith open(\"myenv.yml\",\"w\") as f:\n f.write(myenv.serialize_to_string())",
"_____no_output_____"
]
],
[
[
"Create the inference configuration object",
"_____no_output_____"
]
],
[
[
"from azureml.core.model import InferenceConfig\n\ninference_config = InferenceConfig(runtime= \"python\", \n entry_script=\"score.py\",\n conda_file=\"myenv.yml\",\n extra_docker_file_steps = \"Dockerfile\")",
"_____no_output_____"
]
],
[
[
"### Deploy the model",
"_____no_output_____"
]
],
[
[
"from azureml.core.webservice import AciWebservice\n\naciconfig = AciWebservice.deploy_configuration(cpu_cores = 1, \n memory_gb = 1, \n tags = {'demo': 'onnx'}, \n description = 'web service for ResNet50 ONNX model')",
"_____no_output_____"
]
],
[
[
"The following cell will likely take a few minutes to run as well.",
"_____no_output_____"
]
],
[
[
"from random import randint\n\naci_service_name = 'onnx-demo-resnet50'+str(randint(0,100))\nprint(\"Service\", aci_service_name)\naci_service = Model.deploy(ws, aci_service_name, [model], inference_config, aciconfig)\naci_service.wait_for_deployment(True)\nprint(aci_service.state)",
"_____no_output_____"
]
],
[
[
"In case the deployment fails, you can check the logs. Make sure to delete your aci_service before trying again.",
"_____no_output_____"
]
],
[
[
"if aci_service.state != 'Healthy':\n # run this command for debugging.\n print(aci_service.get_logs())\n aci_service.delete()",
"_____no_output_____"
]
],
[
[
"## Success!\n\nIf you've made it this far, you've deployed a working web service that does image classification using an ONNX model. You can get the URL for the webservice with the code below.",
"_____no_output_____"
]
],
[
[
"print(aci_service.scoring_uri)",
"_____no_output_____"
]
],
[
[
"When you are eventually done using the web service, remember to delete it.",
"_____no_output_____"
]
],
[
[
"#aci_service.delete()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f76e297653fc2e5193b0b0eda51d5a295d75e5 | 9,071 | ipynb | Jupyter Notebook | my_colabs/stbl_team/saving_loading_a2c.ipynb | guyk1971/stable-baselines | ac7a1f3c32851577d5a4fc76e2c42760b9379634 | [
"MIT"
] | null | null | null | my_colabs/stbl_team/saving_loading_a2c.ipynb | guyk1971/stable-baselines | ac7a1f3c32851577d5a4fc76e2c42760b9379634 | [
"MIT"
] | null | null | null | my_colabs/stbl_team/saving_loading_a2c.ipynb | guyk1971/stable-baselines | ac7a1f3c32851577d5a4fc76e2c42760b9379634 | [
"MIT"
] | null | null | null | 29.643791 | 265 | 0.498953 | [
[
[
"# Stable Baselines, a Fork of OpenAI Baselines - Training, Saving and Loading\n\nGithub Repo: [https://github.com/hill-a/stable-baselines](https://github.com/hill-a/stable-baselines)\n\nMedium article: [https://medium.com/@araffin/stable-baselines-a-fork-of-openai-baselines-df87c4b2fc82](https://medium.com/@araffin/stable-baselines-a-fork-of-openai-baselines-df87c4b2fc82)\n\n## Install Dependencies and Stable Baselines Using Pip\n\nList of full dependencies can be found in the [README](https://github.com/hill-a/stable-baselines).\n\n```\n\nsudo apt-get update && sudo apt-get install cmake libopenmpi-dev zlib1g-dev\n```\n\n\n```\n\npip install stable-baselines\n```",
"_____no_output_____"
]
],
[
[
"!apt install swig cmake libopenmpi-dev zlib1g-dev\n!pip install stable-baselines==2.5.1 box2d box2d-kengz",
"_____no_output_____"
]
],
[
[
"## Import policy, RL agent, ...",
"_____no_output_____"
]
],
[
[
"import gym\nimport numpy as np\n\nfrom stable_baselines.common.policies import MlpPolicy\nfrom stable_baselines.common.vec_env import DummyVecEnv\nfrom stable_baselines import A2C",
"_____no_output_____"
]
],
[
[
"## Create the Gym env and instantiate the agent\n\nFor this example, we will use Lunar Lander environment.\n\n\"Landing outside landing pad is possible. Fuel is infinite, so an agent can learn to fly and then land on its first attempt. Four discrete actions available: do nothing, fire left orientation engine, fire main engine, fire right orientation engine. \"\n\nLunar Lander environment: [https://gym.openai.com/envs/LunarLander-v2/](https://gym.openai.com/envs/LunarLander-v2/)\n\n\n\nNote: vectorized environments allow to easily multiprocess training. In this example, we are using only one process, hence the DummyVecEnv.\n\nWe chose the MlpPolicy because input of CartPole is a feature vector, not images.\n\nThe type of action to use (discrete/continuous) will be automatically deduced from the environment action space\n\n",
"_____no_output_____"
]
],
[
[
"env = gym.make('LunarLander-v2')\n# vectorized environments allow to easily multiprocess training\n# we demonstrate its usefulness in the next examples\nenv = DummyVecEnv([lambda: env]) # The algorithms require a vectorized environment to run\n\nmodel = A2C(MlpPolicy, env, ent_coef=0.1, verbose=0)\n",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n"
]
],
[
[
"We create a helper function to evaluate the agent:",
"_____no_output_____"
]
],
[
[
"def evaluate(model, num_steps=1000):\n \"\"\"\n Evaluate a RL agent\n :param model: (BaseRLModel object) the RL Agent\n :param num_steps: (int) number of timesteps to evaluate it\n :return: (float) Mean reward for the last 100 episodes\n \"\"\"\n episode_rewards = [0.0]\n obs = env.reset()\n for i in range(num_steps):\n # _states are only useful when using LSTM policies\n action, _states = model.predict(obs)\n # here, action, rewards and dones are arrays\n # because we are using vectorized env\n obs, rewards, dones, info = env.step(action)\n \n # Stats\n episode_rewards[-1] += rewards[0]\n if dones[0]:\n obs = env.reset()\n episode_rewards.append(0.0)\n # Compute mean reward for the last 100 episodes\n mean_100ep_reward = round(np.mean(episode_rewards[-100:]), 1)\n print(\"Mean reward:\", mean_100ep_reward, \"Num episodes:\", len(episode_rewards))\n \n return mean_100ep_reward",
"_____no_output_____"
]
],
[
[
"Let's evaluate the un-trained agent, this should be a random agent.",
"_____no_output_____"
]
],
[
[
"# Random Agent, before training\nmean_reward_before_train = evaluate(model, num_steps=10000)",
"Mean reward: -210.3 Num episodes: 107\n"
]
],
[
[
"## Train the agent and save it\n\nWarning: this may take a while",
"_____no_output_____"
]
],
[
[
"# Train the agent\nmodel.learn(total_timesteps=10000)\n# Save the agent\nmodel.save(\"a2c_lunar\")\ndel model # delete trained model to demonstrate loading",
"_____no_output_____"
]
],
[
[
"## Load the trained agent",
"_____no_output_____"
]
],
[
[
"model = A2C.load(\"a2c_lunar\")",
"_____no_output_____"
],
[
"# Evaluate the trained agent\nmean_reward = evaluate(model, num_steps=10000)",
"Mean reward: -310.2 Num episodes: 68\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e7f7733919b7f68feb50a8a734bf3746ac7ad34b | 540,303 | ipynb | Jupyter Notebook | notebooks/analyses_reports/2019-03-15_to_03-19_ab3_node2vec_i_loved.ipynb | alphagov/govuk_ab_analysis | fec954d9c90be09e1a74ced64551c2eb68b05d56 | [
"MIT"
] | 9 | 2019-02-04T08:45:50.000Z | 2021-04-22T04:08:49.000Z | notebooks/analyses_reports/2019-03-15_to_03-19_ab3_node2vec_i_loved.ipynb | ukgovdatascience/govuk_ab_analysis | 26e24f38b2811eb0f25d9cd97dbd1732823dbc4c | [
"MIT"
] | 18 | 2019-02-04T14:32:33.000Z | 2019-06-12T10:08:35.000Z | notebooks/analyses_reports/2019-03-15_to_03-19_ab3_node2vec_i_loved.ipynb | alphagov/govuk_ab_analysis | fec954d9c90be09e1a74ced64551c2eb68b05d56 | [
"MIT"
] | 1 | 2021-04-11T08:56:05.000Z | 2021-04-11T08:56:05.000Z | 252.006996 | 102,700 | 0.917278 | [
[
[
"# A/B test 3 - loved journeys, control vs node2vec\n\nThis related links B/C test (ab3) was conducted from 15-20th 2019.\n\nThe data used in this report are 15-19th Mar 2019 because the test was ended on 20th mar.\n\nThe test compared the existing related links (where available) to links generated using node2vec algorithm ",
"_____no_output_____"
],
[
"## Import",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n\nimport os \nimport pandas as pd\nimport numpy as np\nimport ast\nimport re\n\n# z test\nfrom statsmodels.stats.proportion import proportions_ztest\n\n# bayesian bootstrap and vis\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport bayesian_bootstrap.bootstrap as bb\nfrom astropy.utils import NumpyRNGContext\n\n# progress bar\nfrom tqdm import tqdm, tqdm_notebook\n\nfrom scipy import stats\nfrom collections import Counter\n\nimport sys\nsys.path.insert(0, '../../src' )\nimport analysis as analysis",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"# set up the style for our plots\nsns.set(style='white', palette='colorblind', font_scale=1.3,\n rc={'figure.figsize':(12,9), \n \"axes.facecolor\": (0, 0, 0, 0)})\n\n# instantiate progress bar goodness\ntqdm.pandas(tqdm_notebook)\n\npd.set_option('max_colwidth',500)\n\n# the number of bootstrap means used to generate a distribution\nboot_reps = 10000\n\n# alpha - false positive rate\nalpha = 0.05\n# number of tests\nm = 4\n# Correct alpha for multiple comparisons\nalpha = alpha / m\n\n# The Bonferroni correction can be used to adjust confidence intervals also. \n# If one establishes m confidence intervals, and wishes to have an overall confidence level of 1-alpha,\n# each individual confidence interval can be adjusted to the level of 1-(alpha/m).\n\n# reproducible\nseed = 1337",
"_____no_output_____"
]
],
[
[
"## File/dir locations\n### Processed journey data",
"_____no_output_____"
]
],
[
[
"DATA_DIR = os.getenv(\"DATA_DIR\")\nfilename = \"full_sample_loved_947858.csv.gz\"\nfilepath = os.path.join(\n DATA_DIR, \"sampled_journey\", \"20190315_20190319\",\n filename)\nfilepath",
"_____no_output_____"
],
[
"VARIANT_DICT = {\n 'CONTROL_GROUP':'B',\n 'INTERVENTION_GROUP':'C'\n}",
"_____no_output_____"
],
[
"# read in processed sampled journey with just the cols we need for related links\ndf = pd.read_csv(filepath, sep =\"\\t\", compression=\"gzip\")\n# convert from str to list\ndf['Event_cat_act_agg']= df['Event_cat_act_agg'].progress_apply(ast.literal_eval)\ndf['Page_Event_List'] = df['Page_Event_List'].progress_apply(ast.literal_eval)\ndf['Page_List'] = df['Page_List'].progress_apply(ast.literal_eval)",
"100%|██████████| 740885/740885 [00:40<00:00, 18428.22it/s]\n100%|██████████| 740885/740885 [01:09<00:00, 10736.62it/s]\n100%|██████████| 740885/740885 [00:15<00:00, 47142.59it/s]\n"
],
[
"# drop dodgy rows, where page variant is not A or B.\ndf = df.query('ABVariant in [@CONTROL_GROUP, @INTERVENTION_GROUP]')",
"_____no_output_____"
],
[
"df[['Occurrences', 'ABVariant']].groupby('ABVariant').sum()",
"_____no_output_____"
],
[
"df['Page_List_Length'] = df['Page_List'].progress_apply(len)\n",
"100%|██████████| 740885/740885 [00:00<00:00, 766377.92it/s]\n"
]
],
[
[
"### Nav type of page lookup - is it a finding page? if not it's a thing page",
"_____no_output_____"
]
],
[
[
"filename = \"document_types.csv.gz\"\n\n# created a metadata dir in the DATA_DIR to hold this data\nfilepath = os.path.join(\n DATA_DIR, \"metadata\",\n filename)\nprint(filepath)\n\ndf_finding_thing = pd.read_csv(filepath, sep=\"\\t\", compression=\"gzip\")\n\ndf_finding_thing.head()",
"/Users/ellieking/Documents/govuk_ab_analysis/data/metadata/document_types.csv.gz\n"
],
[
"thing_page_paths = df_finding_thing[\n df_finding_thing['is_finding']==0]['pagePath'].tolist()\n\n\nfinding_page_paths = df_finding_thing[\n df_finding_thing['is_finding']==1]['pagePath'].tolist()",
"_____no_output_____"
]
],
[
[
"## Outliers\nSome rows should be removed before analysis. For example rows with journey lengths of 500 or very high related link click rates. This process might have to happen once features have been created.",
"_____no_output_____"
],
[
"# Derive variables",
"_____no_output_____"
],
[
"## journey_click_rate\nThere is no difference in the proportion of journeys using at least one related link (journey_click_rate) between page variant A and page variant B.\n\n",
"_____no_output_____"
],
[
"\\begin{equation*}\n\\frac{\\text{total number of journeys including at least one click on a related link}}{\\text{total number of journeys}}\n\\end{equation*}",
"_____no_output_____"
]
],
[
[
"# get the number of related links clicks per Sequence\ndf['Related Links Clicks per seq'] = df['Event_cat_act_agg'].map(analysis.sum_related_click_events)",
"_____no_output_____"
],
[
"# map across the Sequence variable, which includes pages and Events\n# we want to pass all the list elements to a function one-by-one and then collect the output.\ndf[\"Has_Related\"] = df[\"Related Links Clicks per seq\"].map(analysis.is_related)\n\ndf['Related Links Clicks row total'] = df['Related Links Clicks per seq'] * df['Occurrences']\n",
"_____no_output_____"
],
[
"df.head(3)",
"_____no_output_____"
]
],
[
[
"## count of clicks on navigation elements\n\nThere is no statistically significant difference in the count of clicks on navigation elements per journey between page variant A and page variant B.\n\n\\begin{equation*}\n{\\text{total number of navigation element click events from content pages}}\n\\end{equation*}",
"_____no_output_____"
],
[
"### Related link counts",
"_____no_output_____"
]
],
[
[
"# get the total number of related links clicks for that row (clicks per sequence multiplied by occurrences)\ndf['Related Links Clicks row total'] = df['Related Links Clicks per seq'] * df['Occurrences']",
"_____no_output_____"
]
],
[
[
"### Navigation events",
"_____no_output_____"
]
],
[
[
"def count_nav_events(page_event_list):\n \"\"\"Counts the number of nav events from a content page in a Page Event List.\"\"\"\n content_page_nav_events = 0\n for pair in page_event_list:\n if analysis.is_nav_event(pair[1]):\n if pair[0] in thing_page_paths:\n content_page_nav_events += 1\n return content_page_nav_events",
"_____no_output_____"
],
[
"# needs finding_thing_df read in from document_types.csv.gz\ndf['Content_Page_Nav_Event_Count'] = df['Page_Event_List'].progress_map(count_nav_events)",
"100%|██████████| 740885/740885 [15:57<00:00, 773.42it/s]\n"
],
[
"def count_search_from_content(page_list):\n search_from_content = 0\n for i, page in enumerate(page_list):\n if i > 0:\n if '/search?q=' in page:\n if page_list[i-1] in thing_page_paths:\n search_from_content += 1\n return search_from_content",
"_____no_output_____"
],
[
"df['Content_Search_Event_Count'] = df['Page_List'].progress_map(count_search_from_content)",
"100%|██████████| 740885/740885 [37:56<00:00, 325.50it/s]\n"
],
[
"# count of nav or search clicks\ndf['Content_Nav_or_Search_Count'] = df['Content_Page_Nav_Event_Count'] + df['Content_Search_Event_Count']\n# occurrences is accounted for by the group by bit in our bayesian boot analysis function\ndf['Content_Nav_Search_Event_Sum_row_total'] = df['Content_Nav_or_Search_Count'] * df['Occurrences']\n# required for journeys with no nav later\ndf['Has_No_Nav_Or_Search'] = df['Content_Nav_Search_Event_Sum_row_total'] == 0",
"_____no_output_____"
]
],
[
[
"## Temporary df file in case of crash\n### Save",
"_____no_output_____"
]
],
[
[
"df.to_csv(os.path.join(\n DATA_DIR, \n \"ab3_loved_temp.csv.gz\"), sep=\"\\t\", compression=\"gzip\", index=False)",
"_____no_output_____"
],
[
"df = pd.read_csv(os.path.join(\n DATA_DIR, \n \"ab3_loved_temp.csv.gz\"), sep=\"\\t\", compression=\"gzip\")",
"_____no_output_____"
]
],
[
[
"### Frequentist statistics",
"_____no_output_____"
],
[
"#### Statistical significance",
"_____no_output_____"
]
],
[
[
"# help(proportions_ztest)",
"_____no_output_____"
],
[
"has_rel = analysis.z_prop(df, 'Has_Related', VARIANT_DICT)\nhas_rel",
"_____no_output_____"
],
[
"has_rel['p-value'] < alpha",
"_____no_output_____"
]
],
[
[
"#### Practical significance - uplift",
"_____no_output_____"
]
],
[
[
"# Due to multiple testing we used the Bonferroni correction for alpha\nci_low,ci_upp = analysis.zconf_interval_two_samples(has_rel['x_a'], has_rel['n_a'],\n has_rel['x_b'], has_rel['n_b'], alpha = alpha)\nprint(' difference in proportions = {0:.2f}%'.format(100*(has_rel['p_b']-has_rel['p_a'])))\nprint(' % relative change in proportions = {0:.2f}%'.format(100*((has_rel['p_b']-has_rel['p_a'])/has_rel['p_a'])))\nprint(' 95% Confidence Interval = ( {0:.2f}% , {1:.2f}% )'\n .format(100*ci_low, 100*ci_upp))",
" difference in proportions = 1.53%\n % relative change in proportions = 44.16%\n 95% Confidence Interval = ( 1.46% , 1.61% )\n"
]
],
[
[
"### Bayesian statistics ",
"_____no_output_____"
],
[
"Based on [this](https://medium.com/@thibalbo/coding-bayesian-ab-tests-in-python-e89356b3f4bd) blog",
"_____no_output_____"
],
[
"To be developed, a Bayesian approach can provide a simpler interpretation.",
"_____no_output_____"
],
[
"### Bayesian bootstrap",
"_____no_output_____"
]
],
[
[
"analysis.compare_total_searches(df, VARIANT_DICT)",
"total searches in control group = 55473\ntotal searches in intervention group = 52052\nintervention has 3421 fewer navigation or searches than control;\na 3.18% overall difference\nThe relative change was -6.17% from control to intervention\n"
],
[
"fig, ax = plt.subplots()\nplot_df_B = df[df.ABVariant == VARIANT_DICT['INTERVENTION_GROUP']].groupby(\n 'Content_Nav_or_Search_Count').sum().iloc[:, 0]\nplot_df_A = df[df.ABVariant == VARIANT_DICT['CONTROL_GROUP']].groupby(\n 'Content_Nav_or_Search_Count').sum().iloc[:, 0]\n\nax.set_yscale('log')\nwidth =0.4\nax = plot_df_B.plot.bar(label='B', position=1, width=width)\nax = plot_df_A.plot.bar(label='A', color='salmon', position=0, width=width)\nplt.title(\"loved journeys\")\nplt.ylabel(\"Log(number of journeys)\")\nplt.xlabel(\"Number of uses of search/nav elements in journey\")\n\nlegend = plt.legend(frameon=True)\nframe = legend.get_frame()\nframe.set_facecolor('white')\nplt.savefig('nav_counts_loved_bar.png', dpi = 900, bbox_inches = 'tight')",
"_____no_output_____"
],
[
"a_bootstrap, b_bootstrap = analysis.bayesian_bootstrap_analysis(df, col_name='Content_Nav_or_Search_Count', boot_reps=boot_reps, seed = seed, variant_dict=VARIANT_DICT)",
"_____no_output_____"
],
[
"np.array(a_bootstrap).mean()",
"_____no_output_____"
],
[
"np.array(a_bootstrap).mean() - (0.05 * np.array(a_bootstrap).mean())",
"_____no_output_____"
],
[
"np.array(b_bootstrap).mean()",
"_____no_output_____"
],
[
"print(\"A relative change of {0:.2f}% from control to intervention\".format((np.array(b_bootstrap).mean()-np.array(a_bootstrap).mean())/np.array(a_bootstrap).mean()*100))",
"A relative change of -6.17% from control to intervention\n"
],
[
"# ratio is vestigial but we keep it here for convenience\n# it's actually a count but considers occurrences\nratio_stats = analysis.bb_hdi(a_bootstrap, b_bootstrap, alpha=alpha)\nratio_stats",
"_____no_output_____"
],
[
"ax = sns.distplot(b_bootstrap, label='B')\nax.errorbar(x=[ratio_stats['b_ci_low'], ratio_stats['b_ci_hi']], y=[2, 2], linewidth=5, c='teal', marker='o', \n label='95% HDI B')\n\nax = sns.distplot(a_bootstrap, label='A', ax=ax, color='salmon')\nax.errorbar(x=[ratio_stats['a_ci_low'], ratio_stats['a_ci_hi']], y=[5, 5], linewidth=5, c='salmon', marker='o', \n label='95% HDI A')\n\nax.set(xlabel='mean search/nav count per journey', ylabel='Density')\nsns.despine()\nlegend = plt.legend(frameon=True, bbox_to_anchor=(0.75, 1), loc='best')\nframe = legend.get_frame()\nframe.set_facecolor('white')\nplt.title(\"loved journeys\")\n\nplt.savefig('nav_counts_loved.png', dpi = 900, bbox_inches = 'tight')",
"_____no_output_____"
],
[
"# calculate the posterior for the difference between A's and B's ratio\n# ypa prefix is vestigial from blog post\nypa_diff = np.array(b_bootstrap) - np.array(a_bootstrap)\n# get the hdi\nypa_diff_ci_low, ypa_diff_ci_hi = bb.highest_density_interval(ypa_diff)\n\n# the mean of the posterior\nprint('mean:', ypa_diff.mean())\n\nprint('low ci:', ypa_diff_ci_low, '\\nhigh ci:', ypa_diff_ci_hi)",
"mean: -0.0036118066005200915\nlow ci: -0.004526914146546164 \nhigh ci: -0.002678271951587055\n"
],
[
"ax = sns.distplot(ypa_diff)\nax.plot([ypa_diff_ci_low, ypa_diff_ci_hi], [0, 0], linewidth=10, c='k', marker='o', \n label='95% HDI')\nax.set(xlabel='Content_Nav_or_Search_Count', ylabel='Density', \n title='The difference between B\\'s and A\\'s mean counts times occurrences')\nsns.despine()\nlegend = plt.legend(frameon=True)\nframe = legend.get_frame()\nframe.set_facecolor('white')\nplt.show();",
"_____no_output_____"
],
[
"# We count the number of values greater than 0 and divide by the total number\n# of observations\n# which returns us the the proportion of values in the distribution that are\n# greater than 0, could act a bit like a p-value\n(ypa_diff > 0).sum() / ypa_diff.shape[0]",
"_____no_output_____"
],
[
"# We count the number of values less than 0 and divide by the total number\n# of observations\n# which returns us the the proportion of values in the distribution that are\n# less than 0, could act a bit like a p-value\n(ypa_diff < 0).sum() / ypa_diff.shape[0]",
"_____no_output_____"
],
[
"(ypa_diff>0).sum()",
"_____no_output_____"
],
[
"(ypa_diff<0).sum()",
"_____no_output_____"
]
],
[
[
"## proportion of journeys with a page sequence including content and related links only\n\nThere is no statistically significant difference in the proportion of journeys with a page sequence including content and related links only (including loops) between page variant A and page variant B",
"_____no_output_____"
],
[
"\\begin{equation*}\n\\frac{\\text{total number of journeys that only contain content pages and related links (i.e. no nav pages)}}{\\text{total number of journeys}}\n\\end{equation*}",
"_____no_output_____"
],
[
"### Overall",
"_____no_output_____"
]
],
[
[
"# if (Content_Nav_Search_Event_Sum == 0) that's our success\n# Has_No_Nav_Or_Search == 1 is a success\n# the problem is symmetrical so doesn't matter too much\nsum(df.Has_No_Nav_Or_Search * df.Occurrences) / df.Occurrences.sum()",
"_____no_output_____"
],
[
"sns.distplot(df.Content_Nav_or_Search_Count.values);",
"_____no_output_____"
]
],
[
[
"### Frequentist statistics\n#### Statistical significance",
"_____no_output_____"
]
],
[
[
"nav = analysis.z_prop(df, 'Has_No_Nav_Or_Search', VARIANT_DICT)\nnav",
"_____no_output_____"
]
],
[
[
"#### Practical significance - uplift",
"_____no_output_____"
]
],
[
[
"# Due to multiple testing we used the Bonferroni correction for alpha\nci_low,ci_upp = analysis.zconf_interval_two_samples(nav['x_a'], nav['n_a'],\n nav['x_b'], nav['n_b'], alpha = alpha)\ndiff = 100*(nav['x_b']/nav['n_b']-nav['x_a']/nav['n_a'])\nprint(' difference in proportions = {0:.2f}%'.format(diff))\nprint(' 95% Confidence Interval = ( {0:.2f}% , {1:.2f}% )'\n .format(100*ci_low, 100*ci_upp))",
" difference in proportions = 0.17%\n 95% Confidence Interval = ( 0.10% , 0.24% )\n"
],
[
"print(\"There was a {0: .2f}% relative change in the proportion of journeys not using search/nav elements\".format(100 * ((nav['p_b']-nav['p_a'])/nav['p_a'])))",
"There was a 0.18% relative change in the proportion of journeys not using search/nav elements\n"
]
],
[
[
"## Average Journey Length (number of page views)\nThere is no statistically significant difference in the average page list length of journeys (including loops) between page variant A and page variant B.",
"_____no_output_____"
]
],
[
[
"length_B = df[df.ABVariant == VARIANT_DICT['INTERVENTION_GROUP']].groupby(\n 'Page_List_Length').sum().iloc[:, 0]\nlengthB_2 = length_B.reindex(np.arange(1, 501, 1), fill_value=0)\n\nlength_A = df[df.ABVariant == VARIANT_DICT['CONTROL_GROUP']].groupby(\n 'Page_List_Length').sum().iloc[:, 0]\nlengthA_2 = length_A.reindex(np.arange(1, 501, 1), fill_value=0)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(100, 30))\n\nax.set_yscale('log')\nwidth = 0.4\nax = lengthB_2.plot.bar(label='B', position=1, width=width)\nax = lengthA_2.plot.bar(label='A', color='salmon', position=0, width=width)\nplt.xlabel('length', fontsize=1)\nlegend = plt.legend(frameon=True)\nframe = legend.get_frame()\nframe.set_facecolor('white')\nplt.show();",
"_____no_output_____"
]
],
[
[
"### Bayesian bootstrap for non-parametric hypotheses",
"_____no_output_____"
]
],
[
[
"# http://savvastjortjoglou.com/nfl-bayesian-bootstrap.html",
"_____no_output_____"
],
[
"# let's use mean journey length (could probably model parametrically but we use it for demonstration here)\n# some journeys have length 500 and should probably be removed as they are liekely bots or other weirdness",
"_____no_output_____"
],
[
"#exclude journeys of longer than 500 as these could be automated traffic",
"_____no_output_____"
],
[
"df_short = df[df['Page_List_Length'] < 500]",
"_____no_output_____"
],
[
"print(\"The mean number of pages in an loved journey is {0:.3f}\".format(sum(df.Page_List_Length*df.Occurrences)/df.Occurrences.sum()))",
"The mean number of pages in an loved journey is 2.925\n"
],
[
"# for reproducibility, set the seed within this context\na_bootstrap, b_bootstrap = analysis.bayesian_bootstrap_analysis(df, col_name='Page_List_Length', boot_reps=boot_reps, seed = seed, variant_dict=VARIANT_DICT)\na_bootstrap_short, b_bootstrap_short = analysis.bayesian_bootstrap_analysis(df_short, col_name='Page_List_Length', boot_reps=boot_reps, seed = seed, variant_dict=VARIANT_DICT)",
"_____no_output_____"
],
[
"np.array(a_bootstrap).mean()",
"_____no_output_____"
],
[
"np.array(b_bootstrap).mean()",
"_____no_output_____"
],
[
"print(\"There's a relative change in page length of {0:.2f}% from A to B\".format((np.array(b_bootstrap).mean()-np.array(a_bootstrap).mean())/np.array(a_bootstrap).mean()*100))",
"There's a relative change in page length of 0.66% from A to B\n"
],
[
"print(np.array(a_bootstrap_short).mean())\nprint(np.array(b_bootstrap_short).mean())",
"2.915517748374824\n2.934754208109922\n"
],
[
"# Calculate a 95% HDI\na_ci_low, a_ci_hi = bb.highest_density_interval(a_bootstrap)\nprint('low ci:', a_ci_low, '\\nhigh ci:', a_ci_hi)",
"low ci: 2.9067864815468747 \nhigh ci: 2.9243293080699546\n"
],
[
"ax = sns.distplot(a_bootstrap, color='salmon')\nax.plot([a_ci_low, a_ci_hi], [0, 0], linewidth=10, c='k', marker='o', \n label='95% HDI')\nax.set(xlabel='Journey Length', ylabel='Density', title='Page Variant A Mean Journey Length')\nsns.despine()\nplt.legend();",
"_____no_output_____"
],
[
"# Calculate a 95% HDI\nb_ci_low, b_ci_hi = bb.highest_density_interval(b_bootstrap)\nprint('low ci:', b_ci_low, '\\nhigh ci:', b_ci_hi)",
"low ci: 2.925637198049479 \nhigh ci: 2.9435603551617002\n"
],
[
"ax = sns.distplot(b_bootstrap)\nax.plot([b_ci_low, b_ci_hi], [0, 0], linewidth=10, c='k', marker='o', \n label='95% HDI')\nax.set(xlabel='Journey Length', ylabel='Density', title='Page Variant B Mean Journey Length')\nsns.despine()\nlegend = plt.legend(frameon=True)\nframe = legend.get_frame()\nframe.set_facecolor('white')\nplt.show();",
"_____no_output_____"
],
[
"ax = sns.distplot(b_bootstrap, label='B')\nax = sns.distplot(a_bootstrap, label='A', ax=ax, color='salmon')\nax.set(xlabel='Journey Length', ylabel='Density')\nsns.despine()\nlegend = plt.legend(frameon=True)\nframe = legend.get_frame()\nframe.set_facecolor('white')\nplt.title(\"loved journeys\")\n\nplt.savefig('journey_length_loved.png', dpi = 900, bbox_inches = 'tight')",
"_____no_output_____"
],
[
"ax = sns.distplot(b_bootstrap_short, label='B')\nax = sns.distplot(a_bootstrap_short, label='A', ax=ax, color='salmon')\nax.set(xlabel='Journey Length', ylabel='Density')\nsns.despine()\nlegend = plt.legend(frameon=True)\nframe = legend.get_frame()\nframe.set_facecolor('white')\nplt.show();",
"_____no_output_____"
]
],
[
[
"We can also measure the uncertainty in the difference between the Page Variants's Journey Length by subtracting their posteriors.\n\n",
"_____no_output_____"
]
],
[
[
"# calculate the posterior for the difference between A's and B's YPA\nypa_diff = np.array(b_bootstrap) - np.array(a_bootstrap)\n# get the hdi\nypa_diff_ci_low, ypa_diff_ci_hi = bb.highest_density_interval(ypa_diff)",
"_____no_output_____"
],
[
"# the mean of the posterior\nypa_diff.mean()",
"_____no_output_____"
],
[
"print('low ci:', ypa_diff_ci_low, '\\nhigh ci:', ypa_diff_ci_hi)\n",
"low ci: 0.0068061407105859395 \nhigh ci: 0.031673290308020796\n"
],
[
"ax = sns.distplot(ypa_diff)\nax.plot([ypa_diff_ci_low, ypa_diff_ci_hi], [0, 0], linewidth=10, c='k', marker='o', \n label='95% HDI')\nax.set(xlabel='Journey Length', ylabel='Density', \n title='The difference between B\\'s and A\\'s mean Journey Length')\nsns.despine()\nlegend = plt.legend(frameon=True)\nframe = legend.get_frame()\nframe.set_facecolor('white')\nplt.show();",
"_____no_output_____"
]
],
[
[
"We can actually calculate the probability that B's mean Journey Length was greater than A's mean Journey Length by measuring the proportion of values greater than 0 in the above distribution.",
"_____no_output_____"
]
],
[
[
"# We count the number of values greater than 0 and divide by the total number\n# of observations\n# which returns us the the proportion of values in the distribution that are\n# greater than 0, could act a bit like a p-value\n(ypa_diff > 0).sum() / ypa_diff.shape[0]",
"_____no_output_____"
],
[
"# We count the number of values greater than 0 and divide by the total number\n# of observations\n# which returns us the the proportion of values in the distribution that are\n# greater than 0, could act a bit like a p-value\n(ypa_diff < 0).sum() / ypa_diff.shape[0]",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e7f7758d3a176124ed1478b82abf86737b5f6e8d | 133,312 | ipynb | Jupyter Notebook | DesignYourNeuralNet.ipynb | Ursinus-CS477-F2021/Week11_Convexity_NNIntro | f9f1c5974b38052d68864e8caf82327b1917967b | [
"Apache-2.0"
] | 1 | 2021-11-11T05:31:18.000Z | 2021-11-11T05:31:18.000Z | DesignYourNeuralNet.ipynb | Ursinus-CS477-F2021/Week11_Convexity_NNIntro | f9f1c5974b38052d68864e8caf82327b1917967b | [
"Apache-2.0"
] | null | null | null | DesignYourNeuralNet.ipynb | Ursinus-CS477-F2021/Week11_Convexity_NNIntro | f9f1c5974b38052d68864e8caf82327b1917967b | [
"Apache-2.0"
] | null | null | null | 574.62069 | 41,844 | 0.944341 | [
[
[
"# Design Your Own Neural Net",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\nlogistic = lambda u: 1/(1+np.exp(-u))",
"_____no_output_____"
],
[
"def get_challenge1():\n np.random.seed(0)\n X = np.random.randn(100, 2)\n d = np.sqrt(np.sum(X**2, axis=1))\n y = np.array(d < 1, dtype=float)\n return X, y\n\ndef get_challenge2():\n X, y = get_challenge1()\n X = np.concatenate((X+np.array([[-2, 0]]), X+np.array([[2, 0]])), axis=0)\n y = np.concatenate((y, y))\n return X, y\n\n\nX, y = get_challenge1()\nplt.scatter(X[y==0, 0], X[y==0, 1])\nplt.scatter(X[y==1, 0], X[y==1, 1])\nplt.axis(\"equal\")",
"_____no_output_____"
],
[
"def plot_net(X, y, mynet, res=100):\n rg = [np.min(X[:, 0]), np.max(X[:, 0])]\n dr = rg[1] - rg[0]\n pixx = np.linspace(rg[0], rg[1], res)\n rg = [np.min(X[:, 1]), np.max(X[:, 1])]\n dr = rg[1]- rg[0]\n pixy = np.linspace(rg[0], rg[1], res)\n xx, yy = np.meshgrid(pixx, pixy)\n I = mynet(xx, yy)\n \n plt.figure(figsize=(12, 6))\n plt.subplot(121)\n plt.imshow(I, cmap='gray', extent=(pixx[0], pixx[-1], pixy[-1], pixy[0]))\n plt.colorbar()\n plt.scatter(X[y == 0, 0], X[y == 0, 1], c='C0')\n plt.scatter(X[y == 1, 0], X[y == 1, 1], c='C1')\n plt.gca().invert_yaxis()\n plt.subplot(122)\n plt.imshow(I > 0.5, cmap='gray', extent=(pixx[0], pixx[-1], pixy[-1], pixy[0]))\n plt.colorbar()\n \n pred = mynet(X[:, 0], X[:, 1]) > 0.5\n plt.scatter(X[(y == 0)*(pred == 0), 0], X[(y == 0)*(pred == 0), 1], c='C0')\n plt.scatter(X[(y == 0)*(pred == 1), 0], X[(y == 0)*(pred == 1), 1], c='C0', marker='x')\n plt.scatter(X[(y == 1)*(pred == 0), 0], X[(y == 1)*(pred == 0), 1], c='C1', marker='x')\n plt.scatter(X[(y == 1)*(pred == 1), 0], X[(y == 1)*(pred == 1), 1], c='C1')\n plt.gca().invert_yaxis()\n num_correct = np.sum((y==0)*(pred==0)) + np.sum((y==1)*(pred==1))\n perc = 100*num_correct/X.shape[0]\n plt.title(\"{} Correct ({}%)\".format(num_correct, perc))",
"_____no_output_____"
],
[
"def fn1(x, y):\n return logistic(-2*x+2)\n\ndef fn2(x, y):\n return logistic(2*x+2)\n\ndef myfn(x, y):\n return logistic(fn1(x, y) + fn2(x, y) - 1.5)\n\nplt.figure()\nplot_net(X, y, fn1)\nplt.figure()\nplot_net(X, y, fn2)\nplt.figure()\nplot_net(X, y, myfn)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7f7763d938c79344078637d013437e2ae50934c | 10,171 | ipynb | Jupyter Notebook | ml/visualizations/Gaussian Process Regression.ipynb | kingreatwill/penter | 2d027fd2ae639ac45149659a410042fe76b9dab0 | [
"MIT"
] | 110 | 2018-01-26T10:31:10.000Z | 2022-03-20T21:30:23.000Z | ml/visualizations/Gaussian Process Regression.ipynb | kingreatwill/penter | 2d027fd2ae639ac45149659a410042fe76b9dab0 | [
"MIT"
] | 6 | 2022-01-24T10:04:51.000Z | 2022-01-31T13:02:41.000Z | ml/visualizations/Gaussian Process Regression.ipynb | kingreatwill/penter | 2d027fd2ae639ac45149659a410042fe76b9dab0 | [
"MIT"
] | 35 | 2018-03-04T10:04:49.000Z | 2022-02-12T09:19:40.000Z | 41.012097 | 181 | 0.556877 | [
[
[
"## Gaussian Process Regression\n\n### Gaussian Process\n* Random process where any point $\\large 𝑥∈\\mathbb{R}^𝑑$ is assigned random variable $\\large \\mathbb{f}(𝑥)$\n* Joint distribution of such finite number of variables is given by:\n\n$$\\large 𝑝(\\mathbb{f}│𝑋)=𝒩(\\mathbb{f}|𝜇,𝐾)$$ where\n$$ \\mathbb{f} = (\\mathbb{f}(𝑥_1 ), …, \\mathbb{f}(𝑥_𝑁 )) $$\n$$ \\mu = (𝑚(𝑥_1 ),…, 𝑚(𝑥_𝑁 )) $$\n$$ 𝐾_{𝑖𝑗} = \\kappa(𝑥_𝑖, 𝑥_𝑗) $$ where $\\kappa$ is a PSD kernel function\n\n### Gaussian Process Regression\n* Joint distribution of observed values $\\large \\mathbb{f} $ and predictions $\\large \\mathbb{f}_∗ $ is Gaussian with\n$$\n\\begin{pmatrix} \\large \\mathbb{f} \\\\ \\large \\mathbb{f}_* \\end{pmatrix} \\sim N\\Bigg( \\large 0, \\begin{pmatrix} K & K_* \\\\ K_*^T & K_{**} \\end{pmatrix} \\Bigg)\n$$\nwhere $𝐾 = \\kappa(𝑋, 𝑋)$, $𝐾_∗ = \\kappa(𝑋, 𝑋_∗)$ and $𝐾_{∗*}=\\kappa(𝑋_∗, 𝑋_∗)$\n\n* Posterior/predictive distribution for $\\large 𝑦=f+\\epsilon$ with $\\large \\epsilon \\sim N(0, \\sigma_𝑦^2 \\mathbb{I})$ is given by\n\n$$ \\large 𝑝(\\mathbb{𝕗}_∗│𝑋_∗, 𝑋, 𝑦) = N(\\mu_∗, \\Sigma_∗ )$$\nwhere \n$$\\large \\mu_∗=𝐾_∗(𝐾+\\sigma_𝑛^2 𝐼)^{−1} 𝑦$$\n$$\\large \\Sigma_∗=𝐾_{∗∗}−(𝐾_∗ (𝐾+\\sigma_𝑛^2 \\mathbb{I})^{−1} 𝐾_∗^𝑇$$\n\n* Regression line is the mean of the posterior distribution $\\large\\mu_*$\n* Diagonal entries of the covariance matrix $\\large \\Sigma_*$ can be used for confidence intervals surrounding the regression line\n\n## Gaussian Process Regression Dashboard\nThe dashboard below helps us better understand GP regression\n* Ground truth (or the function GPR is trying to learn) is shown as a white dotted line\n* The regression line in magenta is the zero line (mean of the prior distribution) to start with\n* `Display 5 Priors?` checkbox shows/hides 5 realizations from prior distribution\n* Training samples can be added by clicking anywhere on the figure or can be updated by dragging the existing points\n* `Display 5 Posteriors?` checkbox shows/hides 5 realizations from the posterior distribution\n* `Display Std Bands?` checkbox shows/hides 2 std bands from the posterior mean (aka regression line)\n* $\\sigma_{noise}$ slider controls noise around the training samples\n* Add a few points close to the white line at different places to see the regression line and the confidence intervals update in real time!\n* Impact of RBF kernel hyper-params ($\\sigma$ and $l$) can be seen by updating their values below the figure",
"_____no_output_____"
]
],
[
[
"import inspect\nimport numpy as np\n\nimport ipywidgets as w\nimport bqplot.pyplot as plt\nimport bqplot as bq",
"_____no_output_____"
],
[
"# kernels\ndef rbf(x1, x2, sigma=1., l=1.):\n z = (x1 - x2[:, np.newaxis]) / l\n return sigma**2 * np.exp(-.5 * z ** 2)",
"_____no_output_____"
],
[
"def gp_regression(X_train, y_train, X_test,\n kernel=rbf,\n sigma_noise=.1,\n kernel_params=dict(sigma=1., l=1.)):\n # compute the kernel matrices for train, train_test, test combinations\n K = kernel(X_train, X_train, **kernel_params)\n K_s = kernel(X_train, X_test, **kernel_params)\n K_ss = kernel(X_test, X_test, **kernel_params)\n \n n, p = len(X_train), len(X_test)\n \n # compute the posterior mean and cov\n mu_s = np.dot(K_s, np.linalg.solve(K + sigma_noise**2 * np.eye(n), y_train))\n cov_s = K_ss - np.dot(K_s, np.linalg.solve(K + sigma_noise**2 * np.eye(n), K_s.T))\n \n # prior and posterior moments\n mu_prior, cov_prior = np.zeros(p), K_ss\n mu_post, cov_post = mu_s, cov_s + sigma_noise**2\n \n return dict(prior=(mu_prior, cov_prior), \n posterior=(mu_post, cov_post))",
"_____no_output_____"
],
[
"xmin, xmax = -1, 2\nkernel = rbf\nparams = dict(sigma=1., l=1.)\n\nX_test = np.arange(xmin, xmax, .05)\np = len(X_test)\nK_ss = kernel(X_test, X_test, **params)\nmu_prior, cov_prior = np.zeros(p), K_ss\n\nN = 5\nf_priors = np.random.multivariate_normal(mu_prior, cov_prior, N)",
"_____no_output_____"
],
[
"# kernel controls\nkernel_label = w.HTML(description='RBF Kernel')\nequation_label = w.Label(\"$\\kappa(x_1, x_2) = \\sigma^2 exp(-\\\\frac{(x_1 - x_2)^2}{2l^2})$\")\nsigma_slider = w.FloatText(description=\"$\\sigma$\", min=0, value=1, step=1)\nl_slider = w.FloatText(description=\"$l$\", min=0, value=1, step=1)\nkernel_controls = w.HBox([kernel_label, equation_label, sigma_slider, l_slider])\n\nfig_margin=dict(top=60, bottom=40, left=50, right=0)\nfig = plt.figure(title='Gaussian Process Regression', \n layout=w.Layout(width='1200px', height='700px'),\n animation_duration=750,\n fig_margin=fig_margin)\n\nplt.scales(scales={'x': bq.LinearScale(min=xmin, max=xmax),\n 'y': bq.LinearScale(min=-2, max=2)})\n\n# ground truth line\ny = -np.sin(3 * X_test) - X_test ** 2 + .3 * X_test + .5\nf_line = plt.plot(X_test, y, colors=['white'], line_style='dash_dotted')\nstd_bands = plt.plot(X_test, [],\n fill='between',\n fill_colors=['yellow'],\n apply_clip=False,\n fill_opacities=[.2], stroke_width=0)\n\ntrain_scat = plt.scatter([], [], colors=['magenta'], \n enable_move=True,\n interactions={'click': 'add'},\n marker_size=1, marker='square')\n\nprior_lines = plt.plot(X_test, f_priors, stroke_width=1, \n colors=['#ccc'], apply_clip=False)\nposterior_lines = plt.plot(X_test, [], stroke_width=1, apply_clip=False)\n\nmean_line = plt.plot(X_test, [], 'm')\n\nplt.xlabel('X')\nplt.ylabel('Y')\n\n# reset btn\nreset_button = w.Button(description='Reset Points', button_style='success')\nreset_button.layout.margin = '20px 0px 0px 70px'\n\ndata_noise_slider = w.FloatSlider(description='$\\sigma_{noise}$', value=0, step=.01, max=1)\n\n# controls for the plot\nf_priors_cb = w.Checkbox(description='Display 5 Priors?')\nf_posteriors_cb = w.Checkbox(description='Display 5 Posteriors?')\nstd_bands_cb = w.Checkbox(description='Display Std Bands?')\ncheck_boxes = [f_priors_cb, f_posteriors_cb, std_bands_cb]\n\nlabel = w.Label('*Click on the figure to add training samples')\ncontrols = w.VBox(check_boxes + [reset_button, label, data_noise_slider])\n\n# link widgets\n_ = w.jslink((f_priors_cb, 'value'), (prior_lines, 'visible'))\n_ = w.jslink((f_posteriors_cb, 'value'), (posterior_lines, 'visible'))\n_ = w.jslink((std_bands_cb, 'value'), (std_bands, 'visible'))\n\ndef update_plot(change): \n X_train = train_scat.x\n y_train = train_scat.y\n \n gp_res = gp_regression(X_train, y_train, X_test,\n sigma_noise=data_noise_slider.value,\n kernel=rbf,\n kernel_params=dict(sigma=sigma_slider.value, l=l_slider.value))\n mu_post, cov_post = gp_res['posterior']\n \n # simulate N samples from the posterior distribution\n posterior_lines.y = np.random.multivariate_normal(mu_post, cov_post, N)\n sig_post = np.sqrt(np.diag(cov_post))\n\n # update the regression line to the mean of the posterior distribution\n mean_line.y = mu_post\n \n # update the std bands to +/- 2 sigmas from the posterior mean\n std_bands.y = [mu_post - 2 * sig_post, mu_post + 2 * sig_post]\n\ntrain_scat.observe(update_plot, names=['x', 'y'])\n\n# redraw plot whenever controls are updated\nfor widget in [sigma_slider, l_slider, data_noise_slider]:\n widget.observe(update_plot)\n\ndef reset_points(*args):\n with train_scat.hold_trait_notifications():\n train_scat.x = []\n train_scat.y = []\nreset_button.on_click(lambda btn: reset_points())\n\nfig.on_displayed(update_plot)\nw.HBox([w.VBox([fig, kernel_controls]), controls])",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7f776ed3ae112938dc21274dc9b020f0ed6cf61 | 85,715 | ipynb | Jupyter Notebook | data_processing/lidar_data_processing.ipynb | abhitoronto/KITTI_ROAD_SEGMENTATION | b9387e706ee49a3b03360176a42eca3201d95d8b | [
"MIT"
] | 1 | 2021-07-12T11:00:42.000Z | 2021-07-12T11:00:42.000Z | data_processing/lidar_data_processing.ipynb | abhitoronto/KITTI_ROAD_SEGMENTATION | b9387e706ee49a3b03360176a42eca3201d95d8b | [
"MIT"
] | null | null | null | data_processing/lidar_data_processing.ipynb | abhitoronto/KITTI_ROAD_SEGMENTATION | b9387e706ee49a3b03360176a42eca3201d95d8b | [
"MIT"
] | null | null | null | 33.812623 | 2,077 | 0.557907 | [
[
[
"import json\nimport pprint\n\nimport numpy as np\nimport numpy.linalg as la\n\n# File IO\nimport os\nfrom os.path import join\nimport glob\nimport pickle\nfrom pathlib import Path\n\nimport cv2\n%matplotlib inline\nimport matplotlib.pylab as pt\n\nimport math\nimport time\n\n\nDEBUG = False\n",
"_____no_output_____"
]
],
[
[
"# Tranformation helper Functions",
"_____no_output_____"
],
[
"### Getter function for axes and origin of a sensors coordinate system\n\n`Note: view is a field extracted from the config of sensors.`\n\nFor example, `view = config['cameras']['front_left']['view']`",
"_____no_output_____"
]
],
[
[
"def get_axes_of_a_view(view):\n \"\"\"\n Extract the normalized axes of a sensor in the vehicle coordinate system\n \n view: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n \"\"\"\n x_axis = view['x-axis']\n y_axis = view['y-axis']\n \n x_axis_norm = la.norm(x_axis)\n y_axis_norm = la.norm(y_axis)\n \n if (x_axis_norm < EPSILON or y_axis_norm < EPSILON):\n raise ValueError(\"Norm of input vector(s) too small.\")\n \n # normalize the axes\n x_axis = x_axis / x_axis_norm\n y_axis = y_axis / y_axis_norm\n \n # make a new y-axis which lies in the original x-y plane, but is orthogonal to x-axis\n y_axis = y_axis - x_axis * np.dot(y_axis, x_axis)\n \n # create orthogonal z-axis\n z_axis = np.cross(x_axis, y_axis)\n \n # calculate and check y-axis and z-axis norms\n y_axis_norm = la.norm(y_axis)\n z_axis_norm = la.norm(z_axis)\n \n if (y_axis_norm < EPSILON) or (z_axis_norm < EPSILON):\n raise ValueError(\"Norm of view axis vector(s) too small.\")\n \n # make x/y/z-axes orthonormal\n y_axis = y_axis / y_axis_norm\n z_axis = z_axis / z_axis_norm\n \n return x_axis, y_axis, z_axis\n\ndef get_origin_of_a_view(view):\n \"\"\"\n Extract the origin of a sensor configuration in the vehicle coordinate system\n \n view: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n \"\"\"\n return view['origin']",
"_____no_output_____"
]
],
[
[
"### Getter functions for Coordinate tranformation matrix: \n$$\n\\begin{bmatrix}\n R & T \\\\ 0 & 1\n\\end{bmatrix}\n$$",
"_____no_output_____"
]
],
[
[
"def get_transform_to_global(view):\n \"\"\"\n Get the Tranformation matrix to convert sensor coordinates to global coordinates\n from the view object of a sensor\n \n view: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n \"\"\"\n # get axes\n x_axis, y_axis, z_axis = get_axes_of_a_view(view)\n \n # get origin \n origin = get_origin_of_a_view(view)\n transform_to_global = np.eye(4)\n \n # rotation\n transform_to_global[0:3, 0] = x_axis\n transform_to_global[0:3, 1] = y_axis\n transform_to_global[0:3, 2] = z_axis\n \n # origin\n transform_to_global[0:3, 3] = origin\n \n return transform_to_global",
"_____no_output_____"
],
[
"def get_transform_from_global(view):\n \"\"\"\n Get the Tranformation matrix to convert global coordinates to sensor coordinates \n from the view object of a sensor\n \n view: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n \"\"\"\n # get transform to global\n transform_to_global = get_transform_to_global(view)\n trans = np.eye(4)\n rot = np.transpose(transform_to_global[0:3, 0:3])\n trans[0:3, 0:3] = rot\n trans[0:3, 3] = np.dot(rot, -transform_to_global[0:3, 3])\n\n return trans",
"_____no_output_____"
],
[
"def transform_from_to(src, target):\n \"\"\"\n Get the Tranformation matrix to convert from source sensor view to target sensor view\n \n src: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n target: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n \"\"\"\n transform = np.dot(get_transform_from_global(target), \\\n get_transform_to_global(src))\n \n return transform",
"_____no_output_____"
]
],
[
[
"### Getter Functions for Rotation Matrix \n$$R_{3x3}$$",
"_____no_output_____"
]
],
[
[
"def get_rot_from_global(view):\n \"\"\"\n Get the only the Rotation matrix to rotate sensor coordinates to global coordinates\n from the view object of a sensor\n \n view: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n \"\"\"\n # get transform to global\n transform_to_global = get_transform_to_global(view)\n # get rotation\n rot = np.transpose(transform_to_global[0:3, 0:3])\n \n return rot\n\ndef get_rot_to_global(view):\n \"\"\"\n Get only the Rotation matrix to rotate global coordinates to sensor coordinates \n from the view object of a sensor\n \n view: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n \"\"\"\n # get transform to global\n transform_to_global = get_transform_to_global(view)\n # get rotation\n rot = transform_to_global[0:3, 0:3]\n \n return rot\n\ndef rot_from_to(src, target):\n \"\"\"\n Get only the rotation matrix to rotate from source sensor view to target sensor view\n \n src: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n target: 'view object'\n is a dictionary of the x-axis, y-axis and origin of a sensor\n \"\"\"\n rot = np.dot(get_rot_from_global(target), get_rot_to_global(src))\n \n return rot",
"_____no_output_____"
]
],
[
[
"# Helper Functions for (image/Lidar/label) file names",
"_____no_output_____"
]
],
[
[
"def extract_sensor_file_name(file_name, root_path, sensor_name, ext):\n file_name_split = file_name.split('/')\n \n seq_name = file_name_split[-4]\n data_viewpoint = file_name_split[-2]\n \n file_name_sensor = file_name_split[-1].split('.')[0]\n file_name_sensor = file_name_sensor.split('_')\n file_name_sensor = file_name_sensor[0] + '_' + \\\n sensor_name + '_' + \\\n file_name_sensor[2] + '_' + \\\n file_name_sensor[3] + '.' + ext\n file_path_sensor = join(root_path, seq_name, sensor_name, data_viewpoint, file_name_sensor)\n\n return file_path_sensor",
"_____no_output_____"
],
[
"def extract_image_file_name_from_any_file_name(file_name, root_path):\n return extract_sensor_file_name(file_name, root_path, 'camera', 'png')",
"_____no_output_____"
],
[
"def extract_semantic_file_name_from_any_file_name(file_name, root_path):\n return extract_sensor_file_name(file_name, root_path, 'label', 'png')",
"_____no_output_____"
],
[
"def get_prev_directory(file_name):\n file_name_split = file_name.split('/')\n it = -1\n if not file_name_split[it]:\n it = it - 1\n return file_name.replace(file_name_split[it], '')",
"_____no_output_____"
],
[
"def create_unique_dir(dir_name):\n if dir_name[-1] == '/':\n try:\n os.mkdir(dir_name)\n if DEBUG:\n print(f'New directory created: {dir_name}')\n except FileExistsError :\n if DEBUG:\n print(f'{dir_name} Already Exists. Directory creation skipped')\n else:\n if DEBUG:\n print(f'ERROR: {dir_name} is not a Valid Directory')\n ",
"_____no_output_____"
],
[
"def get_cam_name_from_file_name(file_name):\n file_name_array = file_name.split('/')\n view_point = file_name_array[-2]\n view_point_array = view_point.split('_')\n cam_name = view_point_array[-2] + '_' + view_point_array[-1]\n \n return cam_name",
"_____no_output_____"
]
],
[
[
"# Helper Functions for Images",
"_____no_output_____"
]
],
[
[
"def get_cv2_image(file_name_image, color_transform):\n # Create Image object and correct image color\n image = cv2.imread(file_name_image)\n image = cv2.cvtColor(image, color_transform)\n \n return image",
"_____no_output_____"
],
[
"def get_undistorted_cv2_image(file_name_image, config, color_transform):\n \n # Create Image object and correct image color\n image = get_cv2_image(file_name_image, color_transform)\n \n # Extract cam_name\n cam_name = get_cam_name_from_file_name(file_name_image)\n \n if cam_name in ['front_left', 'front_center', \\\n 'front_right', 'side_left', \\\n 'side_right', 'rear_center']:\n # get parameters from config file\n intr_mat_undist = \\\n np.asarray(config['cameras'][cam_name]['CamMatrix'])\n intr_mat_dist = \\\n np.asarray(config['cameras'][cam_name]['CamMatrixOriginal'])\n dist_parms = \\\n np.asarray(config['cameras'][cam_name]['Distortion'])\n lens = config['cameras'][cam_name]['Lens']\n \n if (lens == 'Fisheye'):\n return cv2.fisheye.undistortImage(image, intr_mat_dist,\\\n D=dist_parms, Knew=intr_mat_undist)\n elif (lens == 'Telecam'):\n return cv2.undistort(image, intr_mat_dist, \\\n distCoeffs=dist_parms, newCameraMatrix=intr_mat_undist)\n else:\n return image\n else:\n print(\"Invalid camera name. Returning original image\")\n return image",
"_____no_output_____"
],
[
"def hsv_to_rgb(h, s, v):\n \"\"\"\n Colour format conversion from Hue Saturation Value to RGB.\n \"\"\"\n if s == 0.0:\n return v, v, v\n \n i = int(h * 6.0)\n f = (h * 6.0) - i\n p = v * (1.0 - s)\n q = v * (1.0 - s * f)\n t = v * (1.0 - s * (1.0 - f))\n i = i % 6\n \n if i == 0:\n return v, t, p\n if i == 1:\n return q, v, p\n if i == 2:\n return p, v, t\n if i == 3:\n return p, q, v\n if i == 4:\n return t, p, v\n if i == 5:\n return v, p, q",
"_____no_output_____"
],
[
"def normalize_vector(vector, lb, ub):\n minimum = np.min(vector)\n maximum = np.max(vector)\n \n return lb + (ub - lb)*(vector - minimum)/(maximum-minimum)",
"_____no_output_____"
]
],
[
[
"# LIDAR Helper Function",
"_____no_output_____"
],
[
"## Using LIDAR data\n\n- LiDAR data is provided in a camera reference frame.\n- `np.load(file_name_lidar)` loads the LIDAR points dictionary\n- LIDAR info\n - azimuth: \n - row: y axis image location of the lidar point\n - lidar_id: id of the LIDAR that the point belongs to\n - depth: Point Depth\n - reflectance: \n - col: x axis image location of the lidar point\n - points: \n - timestamp: \n - distance: ",
"_____no_output_____"
],
[
"LIDAR dictionary loading Example: \n```\nroot_path = './camera_lidar_semantic/'\n\n# get the list of files in lidar directory\nfile_names = sorted(glob.glob(join(root_path, '*/lidar/*/*.npz')))\n\n# read the lidar data\nlidar_front_center = np.load(file_names[0])\n```",
"_____no_output_____"
]
],
[
[
"def get_lidar_on_image(file_name_lidar, config, root_path, pixel_size=3, pixel_opacity=1):\n file_name_image = extract_image_file_name_from_any_file_name(file_name_lidar, root_path)\n image = get_undistorted_cv2_image(file_name_image, config, cv2.COLOR_BGR2RGB)\n \n lidar = np.load(file_name_lidar)\n \n # get rows and cols\n rows = (lidar['row'] + 0.5).astype(np.int)\n cols = (lidar['col'] + 0.5).astype(np.int)\n \n # lowest distance values to be accounted for in colour code\n MIN_DISTANCE = np.min(lidar['distance'])\n # largest distance values to be accounted for in colour code\n MAX_DISTANCE = np.max(lidar['distance'])\n\n # get distances\n distances = lidar['distance'] \n # determine point colours from distance\n colours = (distances - MIN_DISTANCE) / (MAX_DISTANCE - MIN_DISTANCE)\n colours = np.asarray([np.asarray(hsv_to_rgb(0.75 * c, \\\n np.sqrt(pixel_opacity), 1.0)) for c in colours])\n pixel_rowoffs = np.indices([pixel_size, pixel_size])[0] - pixel_size // 2\n pixel_coloffs = np.indices([pixel_size, pixel_size])[1] - pixel_size // 2\n canvas_rows = image.shape[0]\n canvas_cols = image.shape[1]\n for i in range(len(rows)):\n pixel_rows = np.clip(rows[i] + pixel_rowoffs, 0, canvas_rows - 1)\n pixel_cols = np.clip(cols[i] + pixel_coloffs, 0, canvas_cols - 1)\n image[pixel_rows, pixel_cols, :] = \\\n (1. - pixel_opacity) * \\\n np.multiply(image[pixel_rows, pixel_cols, :], \\\n colours[i]) + pixel_opacity * 255 * colours[i]\n return image.astype(np.uint8), lidar",
"_____no_output_____"
]
],
[
[
"# MAIN",
"_____no_output_____"
]
],
[
[
"# # Pick a random LIDAR file from the custom data set\n# np.random.seed()\n# idx = np.random.randint(0, len(custom_lidar_files)-1)\n# file_name_lidar = custom_lidar_files[idx]\n\n# # Visualize LIDAR on image\n# lidar_on_image, lidar = get_lidar_on_image(file_name_lidar, config, root_path)\n# pt.fig = pt.figure(figsize=(15, 15))\n# pt.title('number of points are '+ str(len(lidar['row'])) )\n# pt.imshow(lidar_on_image)\n# pt.axis('off')",
"_____no_output_____"
],
[
"# # Visualize Semantic Image\n# label_image = get_undistorted_cv2_image(extract_semantic_file_name_from_any_file_name(file_name_lidar, root_path) ,\\\n# config, cv2.COLOR_BGR2RGB)\n\n# pt.fig = pt.figure(figsize=(15, 15))\n# pt.imshow(label_image)\n# pt.axis('off')",
"_____no_output_____"
]
],
[
[
"### LIDAR data loading",
"_____no_output_____"
]
],
[
[
"# Open Config File\nwith open ('cams_lidars.json', 'r') as f:\n config = json.load(f)\n \n# pprint.pprint(config)",
"_____no_output_____"
],
[
"# Create Root Path\nroot_path = '/hdd/a2d2-data/camera_lidar_semantic/'",
"_____no_output_____"
],
[
"# Count Number of LIDAR points in each file\ndef get_num_lidar_pts_list(file_names_lidar):\n num_lidar_points = []\n start = time.time()\n for file_lidar in file_names_lidar:\n n_points = len(np.load(file_lidar)['points'])\n num_lidar_points.append(n_points)\n end = time.time() - start\n return num_lidar_points\n",
"_____no_output_____"
],
[
"# Create a histogram\ndef create_hist_pts(points, xlabel='number of points', ylabel='freq', title='Histogram of points'):\n fig = pt.hist(points, 1000)\n pt.xlabel(xlabel)\n pt.ylabel(ylabel)\n pt.title(title)\n pt.show()\n return fig",
"_____no_output_____"
],
[
"# Save this list in a file\ndef save_list_to_pfile(list_, file_name='file.pkl'):\n with open(file_name, 'wb') as filehandle:\n pickle.dump(list_, filehandle)",
"_____no_output_____"
],
[
"# Load Lidar data\nN = 10000\nlidar_file_list = root_path + f'../dataset/lidar_files_{N}.pkl'\n\nif Path(lidar_file_list).is_file():\n with open(lidar_file_list, 'rb') as handle:\n file_names_lidar = pickle.load(handle)\nelse:\n # Get the list of files in lidar directory\n lidar_dirs = '*/lidar/*/*.npz' # ALL LIDAR\n # lidar_dirs = '20180925_124435/lidar/*/*.npz' # 1 - Front and Sides\n # lidar_dirs = '*/lidar/cam_front_center/*.npz' # ALL front center\n file_names_lidar = sorted(glob.glob(join(root_path, lidar_dirs)))\n \n # Extract Lidar files with minimum N points\n num_lidar_points_list = get_num_lidar_pts_list(file_names_lidar)\n\n # Create Histogram\n create_hist_pts(num_lidar_points_list, title='Histogram of Lidar data-points')\n file_names_lidar = [file_names_lidar[_] for _ in range(len(num_lidar_points_list))\\\n if num_lidar_points_list[_] >= N ]\n print(f'There are {len(file_names_lidar)} files greater than {N} points')\n \n # Save list to file\n save_list_to_pfile(file_names_lidar, lidar_file_list)",
"_____no_output_____"
]
],
[
[
"### LIDAR DATA PROCESSING",
"_____no_output_____"
]
],
[
[
"def get_image_files(lidar_file, method_type):\n # Create Lidar_x Lidar_y Lidar_z directory\n lx_file = extract_sensor_file_name(lidar_file, root_path, f'lidar-x-{method_type}', 'png')\n ly_file = extract_sensor_file_name(lidar_file, root_path, f'lidar-y-{method_type}', 'png')\n lz_file = extract_sensor_file_name(lidar_file, root_path, f'lidar-z-{method_type}', 'png')\n l_color_file = extract_sensor_file_name(lidar_file, root_path, 'lidar-image', 'png')\n img_file = extract_image_file_name_from_any_file_name(lidar_file, root_path)\n \n return img_file, lx_file, ly_file, lz_file, l_color_file",
"_____no_output_____"
],
[
"# Create Upsampled LIDAR image\n\n# Iterate over Lidar Files\n# for lidar_file in file_names_lidar:\ndef create_dense_lidar_images_upsample(lidar_file, project_lidar=False):\n if project_lidar:\n lidar_on_image, lidar_data = get_lidar_on_image(lidar_file, config, root_path)\n else:\n lidar_data = np.load(lidar_file)\n \n ## CONSTANTS\n NEIGHBOUR_RADIUS = 40 #Pixels\n INVERSE_COFF = 0.5\n DEPTH_COFF = 0\n CUTOFF_THRESH = 0.4\n PIXEL_THRESH = 1/(1+INVERSE_COFF*NEIGHBOUR_RADIUS)\n \n lidar_on_image, lidar_data = get_lidar_on_image(lidar_file, config, root_path)\n \n # Create Lidar_x Lidar_y Lidar_z directory\n img_file, lx_file, ly_file, lz_file, l_color_file = get_image_files(lidar_file, 'upsample')\n \n # TODO: Check if files already exist\n\n lx_cam_dir = get_prev_directory(lx_file)\n ly_cam_dir = get_prev_directory(ly_file)\n lz_cam_dir = get_prev_directory(lz_file)\n l_color_cam_dir = get_prev_directory(l_color_file)\n \n lx_dir = get_prev_directory(lx_cam_dir)\n ly_dir = get_prev_directory(ly_cam_dir)\n lz_dir = get_prev_directory(lz_cam_dir)\n l_color_dir = get_prev_directory(l_color_cam_dir)\n\n create_unique_dir(lx_dir)\n create_unique_dir(ly_dir)\n create_unique_dir(lz_dir)\n create_unique_dir(l_color_dir)\n \n create_unique_dir(lx_cam_dir)\n create_unique_dir(ly_cam_dir)\n create_unique_dir(lz_cam_dir)\n create_unique_dir(l_color_cam_dir)\n \n # Load Lidar Data and find max distance\n rows = (lidar_data['row'] + 0.5).astype(np.int)\n cols = (lidar_data['col'] + 0.5).astype(np.int)\n rows_float = np.array(lidar_data['row'])\n cols_float = np.array(lidar_data['col'])\n lidar_points = np.array(lidar_data['points'])\n lidar_depth = np.array(lidar_data['distance'])\n max_distance = np.max(lidar_depth)\n if DEBUG:\n print(f'max distance: {max_distance}')\n \n if DEBUG:\n print(f'Processing {lx_file}')\n \n # create X,Y,Z images\n img_file = extract_image_file_name_from_any_file_name(lidar_file, root_path)\n img_x = get_cv2_image(img_file ,cv2.COLOR_BGR2GRAY) # Grayscale image only has one channel\n img_dim = np.shape(img_x)\n\n img_x_num = np.zeros(img_dim)\n img_y_num = img_x_num.copy()\n img_z_num = img_x_num.copy()\n img_den = np.zeros(img_dim)\n \n x_or = np.zeros(img_dim)\n \n # Iterate Over LIDAR points\n if DEBUG:\n print(f'total Lidar Points: {len(rows)}')\n for lid_idx in range(len(rows)):\n idx_a = np.arange(np.maximum(rows[lid_idx] - NEIGHBOUR_RADIUS, 0),\\\n np.minimum(rows[lid_idx] + NEIGHBOUR_RADIUS + 1, img_dim[0]))\n idx_b = np.arange(np.maximum(cols[lid_idx] - NEIGHBOUR_RADIUS, 0),\\\n np.minimum(cols[lid_idx] + NEIGHBOUR_RADIUS + 1, img_dim[1]))\n \n dist_row = (rows_float[lid_idx] - idx_a)\n dist_col = (cols_float[lid_idx] - idx_b)\n \n if len(idx_a) != len(dist_row) or len(idx_b) != len(dist_col):\n print(str(rows_float[lid_idx]) + \", \" + str(cols_float[lid_idx]))\n print(f'{len(idx_a)}, {len(idx_b)}')\n print(f'{len(dist_row)}, {len(dist_col)}')\n break\n \n dist_row_mat = np.array([dist_row]).T * np.ones(len(dist_col))\n dist_col_mat = np.ones((len(dist_row), 1)) * np.array([dist_col])\n\n temp_mat = ( 1 - DEPTH_COFF*lidar_depth[lid_idx]/max_distance)/\\\n ( 1 + INVERSE_COFF*np.sqrt( np.square(dist_row_mat) + np.square(dist_col_mat)))\n \n # Cap the lowest value of denominator \n temp_mat[temp_mat < PIXEL_THRESH ] = 0.0\n \n img_den[np.ix_(idx_a,idx_b)] += temp_mat\n \n# img_den[np.ix_(idx_a,idx_b)] += ( 1 - DEPTH_COFF*lidar_data['distance'][lid_idx]/max_distance)/\\\n# ( 1 + INVERSE_COFF*np.sqrt( np.square(dist_row_mat) + np.square(dist_col_mat)))\n \n img_x_num[np.ix_(idx_a,idx_b)] += img_den[idx_a][:,idx_b] * lidar_points[lid_idx,0]\n img_y_num[np.ix_(idx_a,idx_b)] += img_den[idx_a][:,idx_b] * lidar_points[lid_idx,1]\n img_z_num[np.ix_(idx_a,idx_b)] += img_den[idx_a][:,idx_b] * lidar_points[lid_idx,2]\n \n print(f'Creating Image: {lx_file}\\n')\n \n # Cap the lowest value of denominator \n img_den[img_den < CUTOFF_THRESH] = 0.0\n \n img_x_num = np.divide(img_x_num, img_den, out=np.zeros_like(img_x_num), where=img_den!=0) # Divide by 0 is a 0\n img_y_num = np.divide(img_y_num, img_den, out=np.zeros_like(img_y_num), where=img_den!=0) # Divide by 0 is a 0\n img_z_num = np.divide(img_z_num, img_den, out=np.zeros_like(img_z_num), where=img_den!=0) # Divide by 0 is a 0\n \n img_x_num = normalize_vector(img_x_num, 0.0, 2**16).astype(np.uint16)\n img_y_num = normalize_vector(img_y_num, 0.0, 2**16).astype(np.uint16)\n img_z_num = normalize_vector(img_z_num, 0.0, 2**16).astype(np.uint16)\n \n# img_x_num[np.argwhere(img_x_num == 0.0)] = 255.0\n \n cv2.imwrite(lx_file, img_x_num)\n cv2.imwrite(ly_file, img_y_num)\n cv2.imwrite(lz_file, img_z_num)\n if project_lidar:\n cv2.imwrite(l_color_file, lidar_on_image)\n \n if DEBUG:\n print(f'Saving {lx_file}')\n print(f'Saving {ly_file}')\n print(f'Saving {lz_file}')\n \n return img_file, lx_file, ly_file, lz_file, l_color_file",
"_____no_output_____"
],
[
"# Using Waslander code here\nfrom ip_basic import depth_map_utils\n\ndef create_dense_lidar_images_ip_basic(lidar_file, project_lidar=False):\n if project_lidar:\n lidar_on_image, lidar_data = get_lidar_on_image(lidar_file, config, root_path)\n else:\n lidar_data = np.load(lidar_file)\n \n # Create Lidar_x Lidar_y Lidar_z directory\n img_file, lx_file, ly_file, lz_file, l_color_file = get_image_files(lidar_file, 'ip')\n \n if False and Path(lx_file).is_file() and Path(ly_file).is_file() and Path(lz_file).is_file():\n return img_file, lx_file, ly_file, lz_file, l_color_file\n \n # TODO: Check if files already exist\n\n lx_cam_dir = get_prev_directory(lx_file)\n ly_cam_dir = get_prev_directory(ly_file)\n lz_cam_dir = get_prev_directory(lz_file)\n l_color_cam_dir = get_prev_directory(l_color_file)\n \n lx_dir = get_prev_directory(lx_cam_dir)\n ly_dir = get_prev_directory(ly_cam_dir)\n lz_dir = get_prev_directory(lz_cam_dir)\n l_color_dir = get_prev_directory(l_color_cam_dir)\n\n create_unique_dir(lx_dir)\n create_unique_dir(ly_dir)\n create_unique_dir(lz_dir)\n create_unique_dir(l_color_dir)\n \n create_unique_dir(lx_cam_dir)\n create_unique_dir(ly_cam_dir)\n create_unique_dir(lz_cam_dir)\n create_unique_dir(l_color_cam_dir)\n \n # Load Lidar Data and find max distance\n rows = (lidar_data['row'] + 0.5).astype(np.int)\n cols = (lidar_data['col'] + 0.5).astype(np.int)\n lidar_points = np.array(lidar_data['points'])\n lidar_points_x = normalize_vector(lidar_points[:,0], 2**8 * 0.1 + 1, 2**16 - 1)\n lidar_points_y = normalize_vector(lidar_points[:,1], 2**8 * 0.1 + 1, 2**16 - 1)\n lidar_points_z = normalize_vector(lidar_points[:,2], 2**8 * 0.1 + 1, 2**16 - 1)\n \n # create X,Y,Z images\n img_x = get_cv2_image(img_file ,cv2.COLOR_BGR2GRAY) # Grayscale image only has one channel\n img_dim = np.shape(img_x)\n\n img_x_num = np.zeros(img_dim, dtype=np.uint16)\n img_y_num = img_x_num.copy()\n img_z_num = img_x_num.copy()\n \n # Iterate Over LIDAR points\n if DEBUG:\n print(f'total Lidar Points: {len(rows)}')\n \n if DEBUG:\n print(f'Processing {lx_file}')\n \n for lid_idx in range(len(rows)):\n idx_a = np.clip(rows[lid_idx], 0, img_dim[0]-1)\n idx_b = np.clip(cols[lid_idx], 0, img_dim[1]-1)\n \n img_x_num[idx_a,idx_b] = lidar_points_x[lid_idx]\n img_y_num[idx_a,idx_b] = lidar_points_y[lid_idx]\n img_z_num[idx_a,idx_b] = lidar_points_z[lid_idx]\n \n projected_x = np.float32(img_x_num/256.0)\n projected_y = np.float32(img_y_num/256.0)\n projected_z = np.float32(img_z_num/256.0)\n\n projected_x = depth_map_utils.fill_in_fast( projected_x, max_depth=2**8 + 1)\n projected_y = depth_map_utils.fill_in_fast( projected_y, max_depth=2**8 + 1)\n projected_z = depth_map_utils.fill_in_fast( projected_z, max_depth=2**8 + 1)\n\n img_x_num = (projected_x * 256.0).astype(np.uint16)\n img_y_num = (projected_y * 256.0).astype(np.uint16)\n img_z_num = (projected_z * 256.0).astype(np.uint16)\n\n print(f'Creating Image: {lx_file}\\n')\n\n cv2.imwrite(lx_file, img_x_num)\n cv2.imwrite(ly_file, img_y_num)\n cv2.imwrite(lz_file, img_z_num)\n if project_lidar:\n cv2.imwrite(l_color_file, lidar_on_image)\n \n return img_file, lx_file, ly_file, lz_file, l_color_file",
"_____no_output_____"
],
[
"# from multiprocessing import Pool\nfrom multiprocessing import Pool\n\nNUM_WORKERS = 6\n\ndef create_dense_lidar_images(file_names_lidar, image_range = (0, 100), use_mp=True,n_worker = NUM_WORKERS): \n process_files = file_names_lidar[image_range[0]:image_range[1]]\n ip_files = []\n upsample_files = []\n \n if use_mp:\n pool1 = Pool(n_worker)\n start = time.time()\n pool1.map(create_dense_lidar_images_ip_basic, process_files)\n compute_time_ip = (time.time() - start)/num_images\n start = time.time()\n pool1.map(create_dense_lidar_images_upsample, process_files)\n compute_time_upsample = (time.time() - start)/num_images\n\n else:\n start = time.time()\n for lidar_file in process_files:\n out_ip = create_dense_lidar_images_ip_basic(lidar_file)\n ip_files.append(list(out_ip)[:-1])\n compute_time_ip = (time.time() - start)/num_images\n\n start = time.time()\n for lidar_file in process_files:\n out_upsample = create_dense_lidar_images_upsample(lidar_file)\n upsample_files.append(out_upsample)\n compute_time_upsample = (time.time() - start)/num_images\n\n print(f'Processing time per image (ip_basic): {compute_time_ip} seconds')\n print(f'Processing time per image (upsampling): {compute_time_upsample} seconds')\n \n return ip_files, upsample_files",
"_____no_output_____"
],
[
"# LIDAR data Processing\nnum_images = 20\n\nip_files = []\nupsample_files = []\nprocessed_lidar_files = file_names_lidar[0:num_images]\nip_files, upsample_files = \\\n create_dense_lidar_images(file_names_lidar, image_range=(0,num_images), use_mp=True)",
"Creating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009806.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009489.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000006176.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009789.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000006128.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009786.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009813.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009820.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009861.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009899.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009944.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000009912.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000010313.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000010193.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000012121.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000014481.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000014548.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000014772.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000014962.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-ip/cam_front_center/20180807145028_lidar-x-ip_frontcenter_000014943.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000006128.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009789.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009786.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009489.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000006176.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009806.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009820.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009813.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009861.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009912.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009899.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000009944.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000010313.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000010193.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000012121.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000014548.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000014481.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000014772.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000014943.png\n\nCreating Image: /hdd/a2d2-data/camera_lidar_semantic/20180807_145028/lidar-x-upsample/cam_front_center/20180807145028_lidar-x-upsample_frontcenter_000014962.png\n\nProcessing time per image (ip_basic): 0.30456315279006957 seconds\nProcessing time per image (upsampling): 3.206155979633331 seconds\n"
]
],
[
[
"### Dataset saving",
"_____no_output_____"
]
],
[
[
"# Create a list of all input images\nif not len(ip_files) or not len(upsample_files):\n for lidar_file in processed_lidar_files:\n if 'front' in lidar_file:\n out_ip = get_image_files(lidar_file, 'ip')\n out_upsample = get_image_files(lidar_file, 'upsample')\n ip_files.append(list(out_ip)[:-1])\n upsample_files.append(list(out_upsample)[:-1])\n\nsave_list_to_pfile(ip_files, root_path + '../dataset/ip_inputs.pkl')\nsave_list_to_pfile(upsample_files, root_path + '../dataset/upsample_inputs.pkl')",
"_____no_output_____"
]
],
[
[
"### Ground Truth Conditioning",
"_____no_output_____"
]
],
[
[
"def create_binary_gt(lidar_files, color=(255,0,255)):\n gt_files = []\n for lidar_file in lidar_files:\n if 'front' in lidar_file:\n assert Path(lidar_file).is_file(), f'{lidar_file} is not a file'\n # Get Label file\n label_file = extract_semantic_file_name_from_any_file_name(lidar_file, root_path)\n gt_file = extract_sensor_file_name(lidar_file, root_path, 'image-gt', 'png')\n\n # Skip if file exists\n if Path(gt_file).is_file():\n gt_files.append(gt_file)\n continue\n\n # Mask for color\n label_img = cv2.imread(str(label_file), 1)\n B = label_img[:,:,0] == color[0]\n G = label_img[:,:,1] == color[1]\n R = label_img[:,:,2] == color[2]\n road_area = B & G & R\n gt_img = road_area.astype(dtype=np.uint8) * 255\n\n # Create GT folder and save file\n if not Path(gt_file).is_file():\n gt_cam_dir = get_prev_directory(gt_file)\n gt_dir = get_prev_directory(gt_cam_dir)\n create_unique_dir(gt_dir)\n create_unique_dir(gt_cam_dir)\n cv2.imwrite(gt_file, gt_img)\n\n # Append file to list\n gt_files.append(gt_file)\n \n return gt_files",
"_____no_output_____"
],
[
"gt_files = create_binary_gt(processed_lidar_files)\nsave_list_to_pfile(gt_files, root_path + '../dataset/outputs.pkl')",
"_____no_output_____"
],
[
"len(gt_files)",
"_____no_output_____"
],
[
"means_img = np.array([0.0, 0.0, 0.0])\nmeans_lidar = np.array([0.0, 0.0, 0.0])\n\nmeanss_img = np.array([0.0, 0.0, 0.0])\nmeanss_lidar = np.array([0.0, 0.0, 0.0])\n\nidx = 0\n\nfor ip in ip_files:\n if Path(ip[0]).is_file():\n idx = idx+1\n img = cv2.imread(str(ip[0]), cv2.COLOR_BGR2RGB)\n img_x = cv2.imread(str(ip[1]), 1)\n img_y = cv2.imread(str(ip[2]), 1)\n img_z = cv2.imread(str(ip[3]), 1)\n img_lidar = cv2.merge((img_x, img_y, img_z))\n \n means_img += np.array([(img[0]).mean(),(img[1]).mean(), (img[2]).mean()])\n means_lidar += np.array([(img_lidar[0]).mean(),(img_lidar[1]).mean(), (img_lidar[2]).mean()])\n \n img_s = img.astype(np.uint32)**2\n img_lidar_s = img_lidar.astype(np.uint32)**2\n meanss_img += np.array([(img_s[0]).mean(),(img_s[1]).mean(), (img_s[2]).mean()])\n meanss_lidar += np.array([(img_lidar_s[0]).mean(),(img_lidar_s[1]).mean(), (img_lidar_s[2]).mean()])\n \n print(f'Done with img{idx}')\n ",
"Done with img1\nDone with img2\nDone with img3\nDone with img4\nDone with img5\nDone with img6\nDone with img7\nDone with img8\nDone with img9\nDone with img10\nDone with img11\nDone with img12\nDone with img13\nDone with img14\nDone with img15\nDone with img16\nDone with img17\nDone with img18\nDone with img19\nDone with img20\nDone with img21\nDone with img22\nDone with img23\nDone with img24\nDone with img25\nDone with img26\nDone with img27\nDone with img28\nDone with img29\nDone with img30\nDone with img31\nDone with img32\nDone with img33\nDone with img34\nDone with img35\nDone with img36\nDone with img37\nDone with img38\nDone with img39\nDone with img40\nDone with img41\nDone with img42\nDone with img43\nDone with img44\nDone with img45\nDone with img46\nDone with img47\nDone with img48\nDone with img49\nDone with img50\nDone with img51\nDone with img52\nDone with img53\nDone with img54\nDone with img55\nDone with img56\nDone with img57\nDone with img58\nDone with img59\nDone with img60\nDone with img61\nDone with img62\nDone with img63\nDone with img64\nDone with img65\nDone with img66\nDone with img67\nDone with img68\nDone with img69\nDone with img70\nDone with img71\nDone with img72\nDone with img73\nDone with img74\nDone with img75\nDone with img76\nDone with img77\nDone with img78\nDone with img79\nDone with img80\nDone with img81\nDone with img82\nDone with img83\nDone with img84\nDone with img85\nDone with img86\nDone with img87\nDone with img88\nDone with img89\nDone with img90\nDone with img91\nDone with img92\nDone with img93\nDone with img94\nDone with img95\nDone with img96\nDone with img97\nDone with img98\nDone with img99\nDone with img100\nDone with img101\nDone with img102\nDone with img103\nDone with img104\nDone with img105\nDone with img106\nDone with img107\nDone with img108\nDone with img109\nDone with img110\nDone with img111\nDone with img112\nDone with img113\nDone with img114\nDone with img115\nDone with img116\nDone with img117\nDone with img118\nDone with img119\nDone with img120\nDone with img121\nDone with img122\nDone with img123\nDone with img124\nDone with img125\nDone with img126\nDone with img127\nDone with img128\nDone with img129\nDone with img130\nDone with img131\nDone with img132\nDone with img133\nDone with img134\nDone with img135\nDone with img136\nDone with img137\nDone with img138\nDone with img139\nDone with img140\nDone with img141\nDone with img142\nDone with img143\nDone with img144\nDone with img145\nDone with img146\nDone with img147\nDone with img148\nDone with img149\nDone with img150\nDone with img151\nDone with img152\nDone with img153\nDone with img154\nDone with img155\nDone with img156\nDone with img157\nDone with img158\nDone with img159\nDone with img160\nDone with img161\nDone with img162\nDone with img163\nDone with img164\nDone with img165\nDone with img166\nDone with img167\nDone with img168\nDone with img169\nDone with img170\nDone with img171\nDone with img172\nDone with img173\nDone with img174\nDone with img175\nDone with img176\nDone with img177\nDone with img178\nDone with img179\nDone with img180\nDone with img181\nDone with img182\nDone with img183\nDone with img184\nDone with img185\nDone with img186\nDone with img187\nDone with img188\nDone with img189\nDone with img190\nDone with img191\nDone with img192\nDone with img193\nDone with img194\nDone with img195\nDone with img196\nDone with img197\nDone with img198\nDone with img199\nDone with img200\nDone with img201\nDone with img202\nDone with img203\nDone with img204\nDone with img205\nDone with img206\nDone with img207\nDone with img208\nDone with img209\nDone with img210\nDone with img211\nDone with img212\nDone with img213\nDone with img214\nDone with img215\nDone with img216\nDone with img217\nDone with img218\nDone with img219\nDone with img220\nDone with img221\nDone with img222\nDone with img223\nDone with img224\nDone with img225\nDone with img226\nDone with img227\nDone with img228\nDone with img229\nDone with img230\nDone with img231\nDone with img232\nDone with img233\nDone with img234\nDone with img235\nDone with img236\nDone with img237\nDone with img238\nDone with img239\nDone with img240\nDone with img241\nDone with img242\nDone with img243\nDone with img244\nDone with img245\nDone with img246\nDone with img247\nDone with img248\nDone with img249\nDone with img250\nDone with img251\nDone with img252\nDone with img253\nDone with img254\nDone with img255\nDone with img256\nDone with img257\nDone with img258\nDone with img259\nDone with img260\nDone with img261\nDone with img262\nDone with img263\nDone with img264\nDone with img265\nDone with img266\nDone with img267\nDone with img268\nDone with img269\nDone with img270\nDone with img271\nDone with img272\nDone with img273\nDone with img274\nDone with img275\nDone with img276\nDone with img277\nDone with img278\nDone with img279\nDone with img280\nDone with img281\nDone with img282\nDone with img283\nDone with img284\nDone with img285\nDone with img286\nDone with img287\nDone with img288\nDone with img289\nDone with img290\nDone with img291\nDone with img292\nDone with img293\nDone with img294\nDone with img295\nDone with img296\nDone with img297\nDone with img298\nDone with img299\nDone with img300\nDone with img301\nDone with img302\nDone with img303\nDone with img304\nDone with img305\nDone with img306\nDone with img307\nDone with img308\nDone with img309\nDone with img310\nDone with img311\nDone with img312\nDone with img313\nDone with img314\nDone with img315\nDone with img316\nDone with img317\nDone with img318\nDone with img319\nDone with img320\nDone with img321\nDone with img322\nDone with img323\nDone with img324\nDone with img325\nDone with img326\nDone with img327\nDone with img328\nDone with img329\nDone with img330\nDone with img331\nDone with img332\nDone with img333\nDone with img334\nDone with img335\nDone with img336\nDone with img337\nDone with img338\nDone with img339\nDone with img340\nDone with img341\nDone with img342\nDone with img343\nDone with img344\nDone with img345\nDone with img346\nDone with img347\nDone with img348\nDone with img349\nDone with img350\nDone with img351\nDone with img352\nDone with img353\nDone with img354\nDone with img355\nDone with img356\nDone with img357\nDone with img358\nDone with img359\nDone with img360\nDone with img361\nDone with img362\nDone with img363\nDone with img364\nDone with img365\nDone with img366\nDone with img367\nDone with img368\nDone with img369\nDone with img370\nDone with img371\nDone with img372\nDone with img373\nDone with img374\nDone with img375\nDone with img376\nDone with img377\nDone with img378\nDone with img379\nDone with img380\nDone with img381\nDone with img382\nDone with img383\nDone with img384\nDone with img385\nDone with img386\nDone with img387\nDone with img388\nDone with img389\nDone with img390\nDone with img391\nDone with img392\nDone with img393\nDone with img394\nDone with img395\nDone with img396\nDone with img397\nDone with img398\nDone with img399\nDone with img400\nDone with img401\nDone with img402\nDone with img403\nDone with img404\nDone with img405\nDone with img406\nDone with img407\nDone with img408\nDone with img409\nDone with img410\nDone with img411\nDone with img412\nDone with img413\nDone with img414\nDone with img415\nDone with img416\nDone with img417\nDone with img418\nDone with img419\nDone with img420\nDone with img421\nDone with img422\nDone with img423\nDone with img424\nDone with img425\nDone with img426\nDone with img427\nDone with img428\nDone with img429\nDone with img430\nDone with img431\nDone with img432\nDone with img433\nDone with img434\nDone with img435\nDone with img436\nDone with img437\nDone with img438\nDone with img439\nDone with img440\nDone with img441\nDone with img442\nDone with img443\nDone with img444\nDone with img445\nDone with img446\nDone with img447\nDone with img448\nDone with img449\nDone with img450\nDone with img451\nDone with img452\nDone with img453\nDone with img454\nDone with img455\nDone with img456\nDone with img457\nDone with img458\nDone with img459\nDone with img460\nDone with img461\nDone with img462\nDone with img463\nDone with img464\nDone with img465\nDone with img466\nDone with img467\nDone with img468\nDone with img469\nDone with img470\nDone with img471\nDone with img472\nDone with img473\nDone with img474\nDone with img475\nDone with img476\nDone with img477\nDone with img478\nDone with img479\nDone with img480\nDone with img481\nDone with img482\nDone with img483\nDone with img484\nDone with img485\nDone with img486\nDone with img487\nDone with img488\nDone with img489\n"
],
[
"std_img = np.sqrt(meanss_img/idx - (means_img/idx)**2)\nstd_lidar = np.sqrt(meanss_lidar/idx - (means_lidar/idx)**2)\nmean_img = means_img/idx\nmean_lidar = means_lidar/idx",
"_____no_output_____"
],
[
"mean_img/255, std_img/255",
"_____no_output_____"
],
[
"mean_lidar/255, std_lidar/255",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f77f9e05b47b39204b053399664bcb540cbc09 | 873,867 | ipynb | Jupyter Notebook | docs/tutorials/beyond-sir.ipynb | collectif-codata/pyepidemics | 36ca85a58e1c991b8ada419f8b07eb9fcddedf59 | [
"MIT"
] | 1 | 2020-06-08T14:35:32.000Z | 2020-06-08T14:35:32.000Z | docs/tutorials/beyond-sir.ipynb | collectif-codata/pyepidemics | 36ca85a58e1c991b8ada419f8b07eb9fcddedf59 | [
"MIT"
] | null | null | null | docs/tutorials/beyond-sir.ipynb | collectif-codata/pyepidemics | 36ca85a58e1c991b8ada419f8b07eb9fcddedf59 | [
"MIT"
] | 1 | 2020-06-09T08:31:08.000Z | 2020-06-09T08:31:08.000Z | 683.242377 | 46,388 | 0.950748 | [
[
[
"# Beyond SIR modeling",
"_____no_output_____"
],
[
"[](https://colab.research.google.com/github/collectif-codata/pyepidemics/blob/master/docs/tutorials/beyond-sir.ipynb)",
"_____no_output_____"
],
[
"<div class=\"admonition note\">\n<p class=\"admonition-title\">Note</p>\n<p>\nIn this tutorial we will see how we can build differential equations models and go from simple SIR modeling to add more states and model public policies such as lockdown\n</p>\n</div>",
"_____no_output_____"
],
[
"##### Developer import",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n\n# Developer import\nimport sys\nsys.path.append(\"../../\")",
"_____no_output_____"
]
],
[
[
"##### On Google Colab\nUncomment the following line to install the library locally",
"_____no_output_____"
]
],
[
[
"# !pip install pyepidemics",
"_____no_output_____"
]
],
[
[
"##### Verify the library is correctly installed",
"_____no_output_____"
]
],
[
[
"import pyepidemics\nfrom pyepidemics.models import SIR,SEIR,SEIDR,SEIHDR",
"_____no_output_____"
]
],
[
[
"# Introduction ",
"_____no_output_____"
],
[
"<div class=\"admonition tip\">\n<p class=\"admonition-title\">Tip</p>\n<p>\nThis tutorial is largely inspired from this great article [Infectious Disease Modelling: Beyond the Basic SIR Model](https://towardsdatascience.com/infectious-disease-modelling-beyond-the-basic-sir-model-216369c584c4) by Henri Froese, from which actually a huge part of the code from this library is inspired. \n</div>",
"_____no_output_____"
],
[
"# Simple models by complexity",
"_____no_output_____"
],
[
"## SIR model",
"_____no_output_____"
],
[
"Differential equations models represents transitions between population states. <br>\nSIR is one the most simple model used for many epidemics, in which you suppose three population states : \n \n- ``S`` - Susceptible state, all people that can still be infected\n- ``I`` - Infected state, contaminated people that will recover\n- ``R`` - Removed state, people that are removed from the models, ie that cannot be infected again which is either you recover and you are immune, or unfortunately you are deceased\n\nBetween each state you consider three information : \n- The **population** considered\n- The temporal **rate** (ie 1/duration) representing the number of persons transitioning per day\n- The **probability** to go to the next state\n\nYou can also notice the **epidemiological parameters** such as $\\beta$ or $\\gamma$",
"_____no_output_____"
],
[
"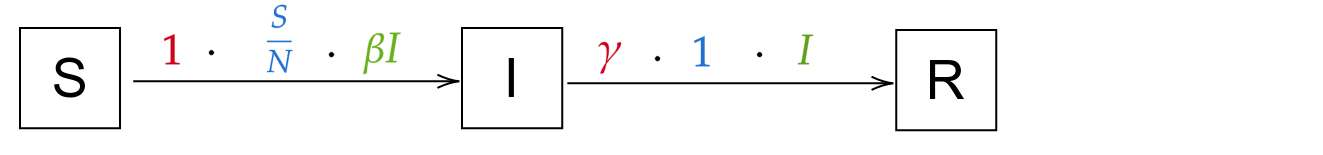",
"_____no_output_____"
]
],
[
[
"N = 1000\nbeta = 1\ngamma = 1/4\n\n# Define model\nsir = SIR(N,beta,gamma)\n\n# Solve the equations\nstates = sir.solve(init_state = 1)\nstates.show(plotly = False)",
"_____no_output_____"
]
],
[
[
"You can visualize the transitions by compartments, with the command ``.network.show()`` (which is not super useful for SIR models, but can be interesting to check more complex models)",
"_____no_output_____"
]
],
[
[
"sir.network.show()",
"[INFO] Displaying only the largest graph component, graphs may be repeated for each category\n"
]
],
[
[
"## SEIR model",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"# Population \nN = 1e6\nbeta = 1\ndelta = 1/3\ngamma = 1/4\n\n# Define the model\nseir = SEIR(N,beta,delta,gamma)\n\n# Solve the equations\nstates = seir.solve(init_state = 1)\nstates.show(plotly = False)",
"_____no_output_____"
],
[
"seir.network.show()",
"[INFO] Displaying only the largest graph component, graphs may be repeated for each category\n"
]
],
[
[
"## SEIDR model",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"# Population \nN = 1e6\ngamma = 1/4\nbeta = 3/4\ndelta = 1/3\nalpha = 0.2 # probability to die\nrho = 1/9 # 9 ndays before death \n\n# Define the model\nseidr = SEIDR(N,beta,delta,gamma,rho,alpha)\n\n# Solve the equations\nstates = seidr.solve(init_state = 1)\nstates.show(plotly = False)",
"_____no_output_____"
],
[
"seidr.network.show()",
"[INFO] Displaying only the largest graph component, graphs may be repeated for each category\n"
]
],
[
[
"## SEIHDR model",
"_____no_output_____"
]
],
[
[
"# Population \nN = 1e6\nbeta = 1/4 * 5 # R0 = 2.5\ndelta = 1/5\ngamma = 1/4\ntheta = 1/5 # ndays before complication\nkappa = 1/10 # ndays before symptoms disappear\nphi = 0.5 # probability of complications\nalpha = 0.2 # probability to die\nrho = 1/9 # 9 ndays before death \n\n# Define the model\nseihdr = SEIHDR(N,beta,delta,gamma,rho,alpha,theta,phi,kappa)\n\n# Solve the equations\nstates = seihdr.solve(init_state = 1,n_days = 100)\nstates.show(plotly = False)",
"_____no_output_____"
],
[
"seihdr.network.show()",
"[INFO] Displaying only the largest graph component, graphs may be repeated for each category\n"
]
],
[
[
"## Towards COVID19 modeling",
"_____no_output_____"
],
[
"To model COVID19 epidemics, we can use a more complex compartmental model to account for different levels of symptoms and patients going to ICU. You can read more about it in this [tutorial](https://collectif-codata.github.io/pyepidemics/tutorials/covid/)",
"_____no_output_____"
],
[
"# Modeling policies",
"_____no_output_____"
],
[
"## Simulating parameters change over time",
"_____no_output_____"
],
[
"To model any policy with macro-epidemiological models we can play with the parameters or the equations. One simple way to model the implementation of a public policy is to make one parameter vary over time when it's implemented. For example to model a lockdown (or any equivalent policy such as social distancing, masks, ...) we can make the parameter ``beta`` vary.",
"_____no_output_____"
],
[
"### Piecewise evolution",
"_____no_output_____"
],
[
"One option is to take a piecewise function that can be as simple as shown here",
"_____no_output_____"
]
],
[
[
"date_lockdown = 53\n\ndef beta(t):\n if t < date_lockdown:\n return 3.3/4\n else:\n return 1/4",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\nx = np.linspace(0,100)\ny = np.vectorize(beta)(x)\n\nplt.figure(figsize = (15,4))\nplt.plot(x,y);",
"_____no_output_____"
]
],
[
[
"For convenience we can use the helper function defined in pyepidemics",
"_____no_output_____"
]
],
[
[
"from pyepidemics.policies.utils import make_dynamic_fn\n\npolicies = [\n 3.3/4,\n (1/4,53),\n]\n\nfn = make_dynamic_fn(policies,sigmoid = False)\n\n# Visualize policies\nx = np.linspace(0,100)\ny = np.vectorize(fn)(x)\nplt.figure(figsize = (15,4))\nplt.plot(x,y);",
"_____no_output_____"
]
],
[
[
"The result is the same, but we can use this function for more complex policies",
"_____no_output_____"
]
],
[
[
"from pyepidemics.policies.utils import make_dynamic_fn\n\npolicies = [\n 3.3/4,\n (1/4,53),\n (2/4,80),\n]\n\nfn = make_dynamic_fn(policies,sigmoid = False)\n\n# Visualize policies\nx = np.linspace(0,100)\ny = np.vectorize(fn)(x)\nplt.figure(figsize = (15,4))\nplt.plot(x,y);",
"_____no_output_____"
]
],
[
[
"### Gradual transitions with sigmoid\nBehaviors don't change over a day, to model this phenomenon we could prefer gradual transitions from one value to the next using sigmoid functions. We can use the previous function for that : ",
"_____no_output_____"
]
],
[
[
"from pyepidemics.policies.utils import make_dynamic_fn\n\npolicies = [\n 3.3/4,\n (1/4,53),\n (2/4,80),\n]\n\nfn = make_dynamic_fn(policies,sigmoid = True)\n\n# Visualize policies\nx = np.linspace(0,100)\ny = np.vectorize(fn)(x)\nplt.figure(figsize = (15,4))\nplt.plot(x,y);",
"_____no_output_____"
]
],
[
[
"We can even specify the transitions durations as followed",
"_____no_output_____"
]
],
[
[
"from pyepidemics.policies.utils import make_dynamic_fn\n\npolicies = [\n 3.3/4,\n (1/4,53),\n (2/4,80),\n]\n\nfn = make_dynamic_fn(policies,sigmoid = True,transition = 8)\n\n# Visualize policies\nx = np.linspace(0,100)\ny = np.vectorize(fn)(x)\nplt.figure(figsize = (15,4))\nplt.plot(x,y);",
"_____no_output_____"
]
],
[
[
"Or even for each transition",
"_____no_output_____"
]
],
[
[
"from pyepidemics.policies.utils import make_dynamic_fn\n\npolicies = [\n 3.3/4,\n (1/4,53,15),\n (2/4,80,5),\n]\n\nfn = make_dynamic_fn(policies,sigmoid = True)\n\n# Visualize policies\nx = np.linspace(0,100)\ny = np.vectorize(fn)(x)\nplt.figure(figsize = (15,4))\nplt.plot(x,y);",
"_____no_output_____"
]
],
[
[
"## Lockdown",
"_____no_output_____"
],
[
"Instead of passing a constant as beta in the previous SEIHDR model, we can pass any function depending over time",
"_____no_output_____"
]
],
[
[
"lockdown_date = 53\n\npolicies = [\n 3.3/4,\n (1/4,lockdown_date),\n]\n\nfn = make_dynamic_fn(policies,sigmoid = True)\n\nbeta = lambda y,t : fn(t)",
"_____no_output_____"
],
[
"# Population \nN = 1e6\ndelta = 1/5\ngamma = 1/4\ntheta = 1/5 # ndays before complication\nkappa = 1/10 # ndays before symptoms disappear\nphi = 0.5 # probability of complications\nalpha = 0.2 # probability to die\nrho = 1/9 # 9 ndays before death \n\n# Define the model\nseihdr = SEIHDR(N,beta,delta,gamma,rho,alpha,theta,phi,kappa)\n\n# Solve the equations\nstates = seihdr.solve(init_state = 1,n_days = 100)\n\n# Visualize the epidemic curves\nstates.show(plotly = False,show = False)\nplt.axvline(lockdown_date,c = \"black\")\nplt.show()",
"_____no_output_____"
],
[
"for Rlockdown in [0.1,0.5,1,2,3.3]:\n \n lockdown_date = 53\n policies = [\n 3.3/4,\n (Rlockdown/4,lockdown_date),\n ]\n\n fn = make_dynamic_fn(policies,sigmoid = True)\n beta = lambda y,t : fn(t)\n \n \n # Define the model\n seihdr = SEIHDR(N,beta,delta,gamma,rho,alpha,theta,phi,kappa)\n states = seihdr.solve(init_state = 1,n_days = 100)\n\n # Visualize the epidemic curves\n states.show(plotly = False,show = False)\n plt.axvline(lockdown_date,c = \"black\")\n plt.title(f\"Lockdown with R={Rlockdown}\")\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Lockdown exit",
"_____no_output_____"
],
[
"Now that you've understood how to change a parameter over time, it's easy to simulate a lockdown exit by adding a new parameter. ",
"_____no_output_____"
]
],
[
[
"for R_post_lockdown in [0.1,0.5,1,2,3.3]:\n \n lockdown_date = 53\n duration_lockdown = 60\n \n policies = [\n 3.3/4,\n (0.6/4,lockdown_date),\n (R_post_lockdown/4,lockdown_date+duration_lockdown),\n\n ]\n\n fn = make_dynamic_fn(policies,sigmoid = True)\n beta = lambda y,t : fn(t)\n \n \n # Define the model\n seihdr = SEIHDR(N,beta,delta,gamma,rho,alpha,theta,phi,kappa)\n states = seihdr.solve(init_state = 1,n_days = 200)\n\n # Visualize the epidemic curves\n states.show(plotly = False,show = False)\n plt.axvline(lockdown_date,c = \"black\")\n plt.axvline(lockdown_date+duration_lockdown,c = \"black\")\n plt.title(f\"Lockdown of {duration_lockdown} days with R_post_lockdown={R_post_lockdown}\")\n plt.show()",
"_____no_output_____"
],
[
"for duration_lockdown in [20,40,60,90]:\n \n lockdown_date = 53\n R_post_lockdown = 2\n \n policies = [\n 3.3/4,\n (0.6/4,lockdown_date),\n (R_post_lockdown/4,lockdown_date+duration_lockdown),\n\n ]\n\n fn = make_dynamic_fn(policies,sigmoid = True)\n beta = lambda y,t : fn(t)\n \n \n # Define the model\n seihdr = SEIHDR(N,beta,delta,gamma,rho,alpha,theta,phi,kappa)\n states = seihdr.solve(init_state = 1,n_days = 200)\n\n # Visualize the epidemic curves\n states.show(plotly = False,show = False)\n plt.axvline(lockdown_date,c = \"black\")\n plt.axvline(lockdown_date+duration_lockdown,c = \"black\")\n plt.title(f\"Lockdown of {duration_lockdown} days with R_post_lockdown={R_post_lockdown}\")\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Note on epidemiological parameters",
"_____no_output_____"
],
[
"Parameters such as beta, R0, or probabilities between states are the main component in differential equations models. You have a few methods to evaluate them: \n\n- Either you estimate the parameters on real-data, this is called **calibration**, you can read more in this [tutorial](https://collectif-codata.github.io/pyepidemics/tutorials/calibration/)\n- Some of them are already pretty well estimated by researchers, like $R0$",
"_____no_output_____"
],
[
"Here in this example the lockdown is simulated by varying the beta, but actually beta encompass individual behavior change, social distancing, self isolation, masks, etc... To better estimate scenarios other options are available from using contact matrices to switching to individual-center models (agent based modeling). ",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e7f7884b957df437c6537bd08089318ed52365e3 | 5,844 | ipynb | Jupyter Notebook | Amuse community codes.ipynb | rieder/exeter-amuse-tutorial | d7db083eedd2cd5b446aba45c4c4d4801a2b39f0 | [
"MIT"
] | null | null | null | Amuse community codes.ipynb | rieder/exeter-amuse-tutorial | d7db083eedd2cd5b446aba45c4c4d4801a2b39f0 | [
"MIT"
] | null | null | null | Amuse community codes.ipynb | rieder/exeter-amuse-tutorial | d7db083eedd2cd5b446aba45c4c4d4801a2b39f0 | [
"MIT"
] | null | null | null | 33.976744 | 259 | 0.627823 | [
[
[
"# AMUSE: Community codes",
"_____no_output_____"
]
],
[
[
"import numpy\nnumpy.random.seed(11)\nfrom amuse.lab import *\nfrom amuse.support.console import set_printing_strategy\nset_printing_strategy(\n \"custom\",\n preferred_units=[units.MSun, units.parsec, units.Myr, units.kms],\n precision=6, prefix=\"\", separator=\" [\", suffix=\"]\",\n)\nconverter = nbody_system.nbody_to_si(1 | units.parsec, 1000 | units.MSun)",
"_____no_output_____"
]
],
[
[
"Amuse contains many community codes, which can be found in amuse.community.\nThese are often codes that have been in use as standalone codes for a long time (e.g. Gadget2), but some are unique to AMUSE (e.g. ph4, a 4th order parallel Hermite N-body integrator with GPU support).\n\nEach community code must be instantiated to start it, after which parameters can be set and particles added.\n\nThe code can then be instructed to evolve the particles to a specific time. Once it reaches this time, the code can be called again, or it can be stopped, removing it from memory and stopping the running process(es).",
"_____no_output_____"
]
],
[
[
"test_sphere = new_plummer_model(1000, converter)\ntest_sphere.mass = new_salpeter_mass_distribution(1000, mass_min=0.3 | units.MSun)\ndef new_gravity(particles):\n gravity = ph4(converter, number_of_workers=1)\n gravity.parameters.epsilon_squared = (0.01 | units.parsec)**2\n gravity.particles.add_particles(particles)\n gravity_to_model = gravity.particles.new_channel_to(particles)\n return gravity, gravity_to_model\ngravity, gravity_to_model = new_gravity(test_sphere)\n\nprint(test_sphere.center_of_mass())\nprint(gravity.particles.center_of_mass())\ngravity.evolve_model(0.1 | units.Myr)\nprint(gravity.particles.center_of_mass())\nprint(test_sphere.center_of_mass())\n\ngravity.stop()",
"_____no_output_____"
]
],
[
[
"Note that the original particles (`test_sphere`) were not modified, while those maintained by the code were (for performance reasons). Also, small numerical errors can arise at this point, the magnitude of which depends on the chosen converter units.\n\nTo synchronise the particle sets, AMUSE uses \"channels\". These can copy the required data when needed, e.g. when synchronising changes in particle properties to other codes.",
"_____no_output_____"
]
],
[
[
"gravity, gravity_to_model = new_gravity(test_sphere)\n\nprint(gravity.particles.center_of_mass())\ngravity.evolve_model(0.1 | units.Myr)\ngravity_to_model.copy()\nprint(gravity.particles.center_of_mass())\nprint(test_sphere.center_of_mass())\n\ngravity.stop()",
"_____no_output_____"
]
],
[
[
"## Combining codes: gravity and stellar evolution",
"_____no_output_____"
],
[
"In a simulation of a star cluster, we may want to combine several codes to address different parts of the problem:\n- an N-body code for gravity,\n- a stellar evolution code\n\nIn the simplest case, these interact only via the stellar mass, which is changed over time by the stellar evolution code and then updated in the gravity code.",
"_____no_output_____"
]
],
[
[
"def new_evolution(particles):\n evolution = SSE()\n evolution.parameters.metallicity = 0.01\n evolution.particles.add_particles(particles)\n evolution_to_model = evolution.particles.new_channel_to(particles)\n return evolution, evolution_to_model\n\nevolution, evolution_to_model = new_evolution(test_sphere)\ngravity, gravity_to_model = new_gravity(test_sphere)\nmodel_to_gravity = test_sphere.new_channel_to(gravity.particles)\n\ntime = gravity.model_time\nend_time = 1 | units.Myr\nwhile time < end_time:\n timestep = evolution.particles.time_step.min()\n gravity.evolve_model(time+timestep/2)\n evolution.evolve_model(time+timestep)\n evolution_to_model.copy()\n model_to_gravity.copy()\n gravity.evolve_model(time+timestep)\n time += timestep\n print(\"Now at time %s.\" % gravity.model_time, end=\" \")\n print(\"The most massive star is now %s\" % test_sphere.mass.max())\nevolution.stop()\ngravity.stop()",
"_____no_output_____"
]
],
[
[
"Note that the timestep is now set by the stellar evolution code, and is based on the evolution timescale of the stellar mass.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e7f7894134f2b81cd731de81e42b4f41870e6315 | 16,671 | ipynb | Jupyter Notebook | tutorials/SingleQubitGates/Workbook_SingleQubitGates.ipynb | bzu13/QuantumKatas | 1db369f3c0d7ee66c6482da307086aa27e4af6f9 | [
"MIT"
] | 1 | 2020-12-29T19:39:50.000Z | 2020-12-29T19:39:50.000Z | tutorials/SingleQubitGates/Workbook_SingleQubitGates.ipynb | bzu13/QuantumKatas | 1db369f3c0d7ee66c6482da307086aa27e4af6f9 | [
"MIT"
] | null | null | null | tutorials/SingleQubitGates/Workbook_SingleQubitGates.ipynb | bzu13/QuantumKatas | 1db369f3c0d7ee66c6482da307086aa27e4af6f9 | [
"MIT"
] | null | null | null | 41.781955 | 427 | 0.57597 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e7f78958ee6bf6d4579c20b496235f1ff540e2ae | 4,884 | ipynb | Jupyter Notebook | Data Science Academy/Cap04/Notebooks/DSA-Python-Cap04-10-Enumerate.ipynb | srgbastos/Artificial-Intelligence | 546935bdb1c57bffaf696fe0256052031dea5981 | [
"MIT"
] | null | null | null | Data Science Academy/Cap04/Notebooks/DSA-Python-Cap04-10-Enumerate.ipynb | srgbastos/Artificial-Intelligence | 546935bdb1c57bffaf696fe0256052031dea5981 | [
"MIT"
] | null | null | null | Data Science Academy/Cap04/Notebooks/DSA-Python-Cap04-10-Enumerate.ipynb | srgbastos/Artificial-Intelligence | 546935bdb1c57bffaf696fe0256052031dea5981 | [
"MIT"
] | null | null | null | 18.292135 | 116 | 0.457207 | [
[
[
"# <font color='blue'>Data Science Academy - Python Fundamentos - Capítulo 4</font>\n\n## Download: http://github.com/dsacademybr",
"_____no_output_____"
]
],
[
[
"# Versão da Linguagem Python\nfrom platform import python_version\nprint('Versão da Linguagem Python Usada Neste Jupyter Notebook:', python_version())",
"Versão da Linguagem Python Usada Neste Jupyter Notebook: 3.7.6\n"
]
],
[
[
"## Enumerate",
"_____no_output_____"
]
],
[
[
"# Criando uma lista\nseq = ['a','b','c']",
"_____no_output_____"
],
[
"enumerate(seq)",
"_____no_output_____"
],
[
"list(enumerate(seq))",
"_____no_output_____"
],
[
"# Imprimindo os valores de uma lista com a função enumerate() e seus respectivos índices\nfor indice, valor in enumerate(seq):\n print (indice, valor)",
"0 a\n1 b\n2 c\n"
],
[
"for indice, valor in enumerate(seq):\n if indice >= 2:\n break\n else:\n print (valor)",
"a\nb\n"
],
[
"lista = ['Marketing', 'Tecnologia', 'Business']",
"_____no_output_____"
],
[
"for i, item in enumerate(lista):\n print(i, item)",
"0 Marketing\n1 Tecnologia\n2 Business\n"
],
[
"for i, item in enumerate('Isso é uma string'):\n print(i, item)",
"0 I\n1 s\n2 s\n3 o\n4 \n5 é\n6 \n7 u\n8 m\n9 a\n10 \n11 s\n12 t\n13 r\n14 i\n15 n\n16 g\n"
],
[
"for i, item in enumerate(range(10)):\n print(i, item)",
"0 0\n1 1\n2 2\n3 3\n4 4\n5 5\n6 6\n7 7\n8 8\n9 9\n"
]
],
[
[
"# FIM",
"_____no_output_____"
],
[
"Interessado(a) em conhecer os cursos e formações da DSA? Confira aqui nosso catálogo de cursos:\n \nhttps://www.datascienceacademy.com.br/pages/todos-os-cursos-dsa ",
"_____no_output_____"
],
[
"### Obrigado - Data Science Academy - <a href=\"http://facebook.com/dsacademybr\">facebook.com/dsacademybr</a>",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
e7f79c7266b46b486f669ddc08b9a70f96987ba1 | 30,238 | ipynb | Jupyter Notebook | 18_convolutional_neural_nets/05_engineer_cnn_features.ipynb | driscolljt/Machine-Learning-for-Algorithmic-Trading-Second-Edition_Original | 2a39f17a6112618bb0fe5455328edd3b2881e4a6 | [
"MIT"
] | 336 | 2020-09-24T01:35:33.000Z | 2022-03-29T18:35:31.000Z | 18_convolutional_neural_nets/05_engineer_cnn_features.ipynb | ikamanu/Machine-Learning-for-Algorithmic-Trading-Second-Edition_Original | ca5817ad00890fa6d6321a27277ee9a1a4f2fcf4 | [
"MIT"
] | 10 | 2020-12-18T02:45:32.000Z | 2021-12-17T19:21:09.000Z | 18_convolutional_neural_nets/05_engineer_cnn_features.ipynb | ikamanu/Machine-Learning-for-Algorithmic-Trading-Second-Edition_Original | ca5817ad00890fa6d6321a27277ee9a1a4f2fcf4 | [
"MIT"
] | 143 | 2020-09-25T08:35:04.000Z | 2022-03-31T01:39:34.000Z | 24.209768 | 142 | 0.504398 | [
[
[
"# Engineer features and convert time series data to images",
"_____no_output_____"
],
[
"## Imports & Settings",
"_____no_output_____"
],
[
"To install `talib` with Python 3.7 follow [these](https://medium.com/@joelzhang/install-ta-lib-in-python-3-7-51219acacafb) instructions.",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"from talib import (RSI, BBANDS, MACD,\n NATR, WILLR, WMA,\n EMA, SMA, CCI, CMO,\n MACD, PPO, ROC,\n ADOSC, ADX, MOM)\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nfrom statsmodels.regression.rolling import RollingOLS\nimport statsmodels.api as sm\nimport pandas_datareader.data as web\nimport pandas as pd\nimport numpy as np\nfrom pathlib import Path\n%matplotlib inline",
"_____no_output_____"
],
[
"DATA_STORE = '../data/assets.h5'",
"_____no_output_____"
],
[
"MONTH = 21\nYEAR = 12 * MONTH",
"_____no_output_____"
],
[
"START = '2000-01-01'\nEND = '2017-12-31'",
"_____no_output_____"
],
[
"sns.set_style('whitegrid')\nidx = pd.IndexSlice",
"_____no_output_____"
],
[
"T = [1, 5, 10, 21, 42, 63]",
"_____no_output_____"
],
[
"results_path = Path('results', 'cnn_for_trading')\nif not results_path.exists():\n results_path.mkdir(parents=True)",
"_____no_output_____"
]
],
[
[
"## Loading Quandl Wiki Stock Prices & Meta Data",
"_____no_output_____"
]
],
[
[
"adj_ohlcv = ['adj_open', 'adj_close', 'adj_low', 'adj_high', 'adj_volume']",
"_____no_output_____"
],
[
"with pd.HDFStore(DATA_STORE) as store:\n prices = (store['quandl/wiki/prices']\n .loc[idx[START:END, :], adj_ohlcv]\n .rename(columns=lambda x: x.replace('adj_', ''))\n .swaplevel()\n .sort_index()\n .dropna())\n metadata = (store['us_equities/stocks'].loc[:, ['marketcap', 'sector']])\nohlcv = prices.columns.tolist()",
"_____no_output_____"
],
[
"prices.volume /= 1e3\nprices.index.names = ['symbol', 'date']\nmetadata.index.name = 'symbol'",
"_____no_output_____"
]
],
[
[
"## Rolling universe: pick 500 most-traded stocks",
"_____no_output_____"
]
],
[
[
"dollar_vol = prices.close.mul(prices.volume).unstack('symbol').sort_index()",
"_____no_output_____"
],
[
"years = sorted(np.unique([d.year for d in prices.index.get_level_values('date').unique()]))",
"_____no_output_____"
],
[
"train_window = 5 # years\nuniverse_size = 500",
"_____no_output_____"
],
[
"universe = []\nfor i, year in enumerate(years[5:], 5):\n start = str(years[i-5])\n end = str(years[i])\n most_traded = dollar_vol.loc[start:end, :].dropna(thresh=1000, axis=1).median().nlargest(universe_size).index\n universe.append(prices.loc[idx[most_traded, start:end], :])\nuniverse = pd.concat(universe)",
"_____no_output_____"
],
[
"universe = universe.loc[~universe.index.duplicated()]",
"_____no_output_____"
],
[
"universe.info(null_counts=True)",
"<class 'pandas.core.frame.DataFrame'>\nMultiIndex: 2530228 entries, ('A', Timestamp('2000-01-03 00:00:00')) to ('ZTS', Timestamp('2017-12-29 00:00:00'))\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 open 2530228 non-null float64\n 1 close 2530228 non-null float64\n 2 low 2530228 non-null float64\n 3 high 2530228 non-null float64\n 4 volume 2530228 non-null float64\ndtypes: float64(5)\nmemory usage: 106.2+ MB\n"
],
[
"universe.groupby('symbol').size().describe()",
"_____no_output_____"
],
[
"universe.to_hdf('data.h5', 'universe')",
"_____no_output_____"
]
],
[
[
"## Generate Technical Indicators Factors",
"_____no_output_____"
]
],
[
[
"T = list(range(6, 21))",
"_____no_output_____"
]
],
[
[
"### Relative Strength Index",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_RSI'] = universe.groupby(level='symbol').close.apply(RSI, timeperiod=t)",
"_____no_output_____"
]
],
[
[
"### Williams %R",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_WILLR'] = (universe.groupby(level='symbol', group_keys=False)\n .apply(lambda x: WILLR(x.high, x.low, x.close, timeperiod=t)))",
"_____no_output_____"
]
],
[
[
"### Compute Bollinger Bands",
"_____no_output_____"
]
],
[
[
"def compute_bb(close, timeperiod):\n high, mid, low = BBANDS(close, timeperiod=timeperiod)\n return pd.DataFrame({f'{timeperiod:02}_BBH': high, f'{timeperiod:02}_BBL': low}, index=close.index)",
"_____no_output_____"
],
[
"for t in T:\n bbh, bbl = f'{t:02}_BBH', f'{t:02}_BBL'\n universe = (universe.join(\n universe.groupby(level='symbol').close.apply(compute_bb,\n timeperiod=t)))\n universe[bbh] = universe[bbh].sub(universe.close).div(universe[bbh]).apply(np.log1p)\n universe[bbl] = universe.close.sub(universe[bbl]).div(universe.close).apply(np.log1p)",
"_____no_output_____"
]
],
[
[
"### Normalized Average True Range",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_NATR'] = universe.groupby(level='symbol', \n group_keys=False).apply(lambda x: \n NATR(x.high, x.low, x.close, timeperiod=t))",
"_____no_output_____"
]
],
[
[
"### Percentage Price Oscillator",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_PPO'] = universe.groupby(level='symbol').close.apply(PPO, fastperiod=t, matype=1)",
"_____no_output_____"
]
],
[
[
"### Moving Average Convergence/Divergence",
"_____no_output_____"
]
],
[
[
"def compute_macd(close, signalperiod):\n macd = MACD(close, signalperiod=signalperiod)[0]\n return (macd - np.mean(macd))/np.std(macd)",
"_____no_output_____"
],
[
"for t in T:\n universe[f'{t:02}_MACD'] = (universe\n .groupby('symbol', group_keys=False)\n .close\n .apply(compute_macd, signalperiod=t))",
"_____no_output_____"
]
],
[
[
"### Momentum",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_MOM'] = universe.groupby(level='symbol').close.apply(MOM, timeperiod=t)",
"_____no_output_____"
]
],
[
[
"### Weighted Moving Average",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_WMA'] = universe.groupby(level='symbol').close.apply(WMA, timeperiod=t)",
"_____no_output_____"
]
],
[
[
"### Exponential Moving Average",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_EMA'] = universe.groupby(level='symbol').close.apply(EMA, timeperiod=t)",
"_____no_output_____"
]
],
[
[
"### Commodity Channel Index",
"_____no_output_____"
]
],
[
[
"for t in T: \n universe[f'{t:02}_CCI'] = (universe.groupby(level='symbol', group_keys=False)\n .apply(lambda x: CCI(x.high, x.low, x.close, timeperiod=t)))",
"_____no_output_____"
]
],
[
[
"### Chande Momentum Oscillator",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_CMO'] = universe.groupby(level='symbol').close.apply(CMO, timeperiod=t)",
"_____no_output_____"
]
],
[
[
"### Rate of Change",
"_____no_output_____"
],
[
"Rate of change is a technical indicator that illustrates the speed of price change over a period of time.",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_ROC'] = universe.groupby(level='symbol').close.apply(ROC, timeperiod=t)",
"_____no_output_____"
]
],
[
[
"### Chaikin A/D Oscillator",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_ADOSC'] = (universe.groupby(level='symbol', group_keys=False)\n .apply(lambda x: ADOSC(x.high, x.low, x.close, x.volume, fastperiod=t-3, slowperiod=4+t)))",
"_____no_output_____"
]
],
[
[
"### Average Directional Movement Index",
"_____no_output_____"
]
],
[
[
"for t in T:\n universe[f'{t:02}_ADX'] = universe.groupby(level='symbol', \n group_keys=False).apply(lambda x: \n ADX(x.high, x.low, x.close, timeperiod=t))",
"_____no_output_____"
],
[
"universe.drop(ohlcv, axis=1).to_hdf('data.h5', 'features')",
"_____no_output_____"
]
],
[
[
"## Compute Historical Returns",
"_____no_output_____"
],
[
"### Historical Returns",
"_____no_output_____"
]
],
[
[
"by_sym = universe.groupby(level='symbol').close\nfor t in [1,5]:\n universe[f'r{t:02}'] = by_sym.pct_change(t)",
"_____no_output_____"
]
],
[
[
"### Remove outliers",
"_____no_output_____"
]
],
[
[
"universe[[f'r{t:02}' for t in [1, 5]]].describe()",
"_____no_output_____"
],
[
"outliers = universe[universe.r01>1].index.get_level_values('symbol').unique()\nlen(outliers)",
"_____no_output_____"
],
[
"universe = universe.drop(outliers, level='symbol')",
"_____no_output_____"
]
],
[
[
"### Historical return quantiles",
"_____no_output_____"
]
],
[
[
"for t in [1, 5]:\n universe[f'r{t:02}dec'] = (universe[f'r{t:02}'].groupby(level='date')\n .apply(lambda x: pd.qcut(x, q=10, labels=False, duplicates='drop')))",
"_____no_output_____"
]
],
[
[
"## Rolling Factor Betas",
"_____no_output_____"
]
],
[
[
"factor_data = (web.DataReader('F-F_Research_Data_5_Factors_2x3_daily', 'famafrench', \n start=START)[0].rename(columns={'Mkt-RF': 'Market'}))\nfactor_data.index.names = ['date']",
"_____no_output_____"
],
[
"factor_data.info()",
"<class 'pandas.core.frame.DataFrame'>\nDatetimeIndex: 5114 entries, 2000-01-03 to 2020-04-30\nData columns (total 6 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Market 5114 non-null float64\n 1 SMB 5114 non-null float64\n 2 HML 5114 non-null float64\n 3 RMW 5114 non-null float64\n 4 CMA 5114 non-null float64\n 5 RF 5114 non-null float64\ndtypes: float64(6)\nmemory usage: 279.7 KB\n"
],
[
"windows = list(range(15, 90, 5))\nlen(windows)",
"_____no_output_____"
],
[
"t = 1\nret = f'r{t:02}'\nfactors = ['Market', 'SMB', 'HML', 'RMW', 'CMA']\nwindows = list(range(15, 90, 5))\nfor window in windows:\n print(window)\n betas = []\n for symbol, data in universe.groupby(level='symbol'):\n model_data = data[[ret]].merge(factor_data, on='date').dropna()\n model_data[ret] -= model_data.RF\n\n rolling_ols = RollingOLS(endog=model_data[ret], \n exog=sm.add_constant(model_data[factors]), window=window)\n factor_model = rolling_ols.fit(params_only=True).params.drop('const', axis=1)\n result = factor_model.assign(symbol=symbol).set_index('symbol', append=True)\n betas.append(result)\n betas = pd.concat(betas).rename(columns=lambda x: f'{window:02}_{x}')\n universe = universe.join(betas)",
"_____no_output_____"
]
],
[
[
"## Compute Forward Returns",
"_____no_output_____"
]
],
[
[
"for t in [1, 5]:\n universe[f'r{t:02}_fwd'] = universe.groupby(level='symbol')[f'r{t:02}'].shift(-t)\n universe[f'r{t:02}dec_fwd'] = universe.groupby(level='symbol')[f'r{t:02}dec'].shift(-t)",
"_____no_output_____"
]
],
[
[
"## Store Model Data",
"_____no_output_____"
]
],
[
[
"universe = universe.drop(ohlcv, axis=1)",
"_____no_output_____"
],
[
"universe.info(null_counts=True)",
"<class 'pandas.core.frame.DataFrame'>\nMultiIndex: 2499265 entries, ('A', Timestamp('2000-01-03 00:00:00')) to ('ZTS', Timestamp('2017-12-29 00:00:00'))\nColumns: 308 entries, 06_RSI to r05dec_fwd\ndtypes: float64(308)\nmemory usage: 5.7+ GB\n"
],
[
"drop_cols = ['r01', 'r01dec', 'r05', 'r05dec']",
"_____no_output_____"
],
[
"outcomes = universe.filter(like='_fwd').columns",
"_____no_output_____"
],
[
"universe = universe.sort_index()\nwith pd.HDFStore('data.h5') as store:\n store.put('features', universe.drop(drop_cols, axis=1).drop(outcomes, axis=1).loc[idx[:, '2001':], :])\n store.put('targets', universe.loc[idx[:, '2001':], outcomes])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7f7adb248de25fc275a164356dd4005b9933d64 | 962 | ipynb | Jupyter Notebook | Unordered-tensorflow-examples/aymericdamien-Examples/examples/a10-multigpu_basics-notebook.ipynb | hpssjellis/deeplearnjs-javascript-examples | 1888c1313d977e18837fa6fea3be272bfc79830e | [
"MIT"
] | 10 | 2017-12-07T15:53:45.000Z | 2021-02-19T10:08:51.000Z | Unordered-tensorflow-examples/aymericdamien-Examples/examples/a10-multigpu_basics-notebook.ipynb | hpssjellis/deeplearnjs-javascript-examples | 1888c1313d977e18837fa6fea3be272bfc79830e | [
"MIT"
] | null | null | null | Unordered-tensorflow-examples/aymericdamien-Examples/examples/a10-multigpu_basics-notebook.ipynb | hpssjellis/deeplearnjs-javascript-examples | 1888c1313d977e18837fa6fea3be272bfc79830e | [
"MIT"
] | 2 | 2017-11-18T04:00:06.000Z | 2019-10-24T09:37:54.000Z | 18.862745 | 84 | 0.550936 | [
[
[
"%autosave 0\n#Enter a bash command below and click run to activate\n!python multigpu_basics.py",
"_____no_output_____"
],
[
"#Enter a python file to load and edit. Changes will be automatically saved!\n%load multigpu_basics.py",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
e7f7c25223ad6aec2095b46800a7e9137bce7767 | 106,345 | ipynb | Jupyter Notebook | Project/SageMaker Project.ipynb | simonmijares/Sagemaker | 0aad43a2eada8e4851a476d70f5434574acd7efd | [
"MIT"
] | null | null | null | Project/SageMaker Project.ipynb | simonmijares/Sagemaker | 0aad43a2eada8e4851a476d70f5434574acd7efd | [
"MIT"
] | null | null | null | Project/SageMaker Project.ipynb | simonmijares/Sagemaker | 0aad43a2eada8e4851a476d70f5434574acd7efd | [
"MIT"
] | null | null | null | 54.930269 | 2,613 | 0.627552 | [
[
[
"# Creating a Sentiment Analysis Web App\n## Using PyTorch and SageMaker\n\n_Deep Learning Nanodegree Program | Deployment_\n\n---\n\nNow that we have a basic understanding of how SageMaker works we will try to use it to construct a complete project from end to end. Our goal will be to have a simple web page which a user can use to enter a movie review. The web page will then send the review off to our deployed model which will predict the sentiment of the entered review.\n\n## Instructions\n\nSome template code has already been provided for you, and you will need to implement additional functionality to successfully complete this notebook. You will not need to modify the included code beyond what is requested. Sections that begin with '**TODO**' in the header indicate that you need to complete or implement some portion within them. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a `# TODO: ...` comment. Please be sure to read the instructions carefully!\n\nIn addition to implementing code, there will be questions for you to answer which relate to the task and your implementation. Each section where you will answer a question is preceded by a '**Question:**' header. Carefully read each question and provide your answer below the '**Answer:**' header by editing the Markdown cell.\n\n> **Note**: Code and Markdown cells can be executed using the **Shift+Enter** keyboard shortcut. In addition, a cell can be edited by typically clicking it (double-click for Markdown cells) or by pressing **Enter** while it is highlighted.\n\n## General Outline\n\nRecall the general outline for SageMaker projects using a notebook instance.\n\n1. Download or otherwise retrieve the data.\n2. Process / Prepare the data.\n3. Upload the processed data to S3.\n4. Train a chosen model.\n5. Test the trained model (typically using a batch transform job).\n6. Deploy the trained model.\n7. Use the deployed model.\n\nFor this project, you will be following the steps in the general outline with some modifications. \n\nFirst, you will not be testing the model in its own step. You will still be testing the model, however, you will do it by deploying your model and then using the deployed model by sending the test data to it. One of the reasons for doing this is so that you can make sure that your deployed model is working correctly before moving forward.\n\nIn addition, you will deploy and use your trained model a second time. In the second iteration you will customize the way that your trained model is deployed by including some of your own code. In addition, your newly deployed model will be used in the sentiment analysis web app.",
"_____no_output_____"
]
],
[
[
"# Make sure that we use SageMaker 1.x\n!pip install sagemaker==1.72.0",
"Requirement already satisfied: sagemaker==1.72.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (1.72.0)\nRequirement already satisfied: protobuf>=3.1 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (3.11.4)\nRequirement already satisfied: boto3>=1.14.12 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (1.17.16)\nRequirement already satisfied: scipy>=0.19.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (1.4.1)\nRequirement already satisfied: protobuf3-to-dict>=0.1.5 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (0.1.5)\nRequirement already satisfied: smdebug-rulesconfig==0.1.4 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (0.1.4)\nRequirement already satisfied: importlib-metadata>=1.4.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (3.4.0)\nRequirement already satisfied: packaging>=20.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (20.1)\nRequirement already satisfied: numpy>=1.9.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from sagemaker==1.72.0) (1.18.1)\nRequirement already satisfied: botocore<1.21.0,>=1.20.16 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from boto3>=1.14.12->sagemaker==1.72.0) (1.20.16)\nRequirement already satisfied: s3transfer<0.4.0,>=0.3.0 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from boto3>=1.14.12->sagemaker==1.72.0) (0.3.4)\nRequirement already satisfied: jmespath<1.0.0,>=0.7.1 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from boto3>=1.14.12->sagemaker==1.72.0) (0.10.0)\nRequirement already satisfied: python-dateutil<3.0.0,>=2.1 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from botocore<1.21.0,>=1.20.16->boto3>=1.14.12->sagemaker==1.72.0) (2.8.1)\nRequirement already satisfied: urllib3<1.27,>=1.25.4 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from botocore<1.21.0,>=1.20.16->boto3>=1.14.12->sagemaker==1.72.0) (1.25.10)\nRequirement already satisfied: zipp>=0.5 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from importlib-metadata>=1.4.0->sagemaker==1.72.0) (2.2.0)\nRequirement already satisfied: typing-extensions>=3.6.4 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from importlib-metadata>=1.4.0->sagemaker==1.72.0) (3.7.4.3)\nRequirement already satisfied: pyparsing>=2.0.2 in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from packaging>=20.0->sagemaker==1.72.0) (2.4.6)\nRequirement already satisfied: six in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from packaging>=20.0->sagemaker==1.72.0) (1.14.0)\nRequirement already satisfied: setuptools in /home/ec2-user/anaconda3/envs/pytorch_p36/lib/python3.6/site-packages (from protobuf>=3.1->sagemaker==1.72.0) (45.2.0.post20200210)\n"
]
],
[
[
"## Step 1: Downloading the data\n\nAs in the XGBoost in SageMaker notebook, we will be using the [IMDb dataset](http://ai.stanford.edu/~amaas/data/sentiment/)\n\n> Maas, Andrew L., et al. [Learning Word Vectors for Sentiment Analysis](http://ai.stanford.edu/~amaas/data/sentiment/). In _Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies_. Association for Computational Linguistics, 2011.",
"_____no_output_____"
]
],
[
[
"%mkdir ../data\n!wget -O ../data/aclImdb_v1.tar.gz http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz\n!tar -zxf ../data/aclImdb_v1.tar.gz -C ../data",
"mkdir: cannot create directory ‘../data’: File exists\n--2021-03-07 19:37:15-- http://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz\nResolving ai.stanford.edu (ai.stanford.edu)... 171.64.68.10\nConnecting to ai.stanford.edu (ai.stanford.edu)|171.64.68.10|:80... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 84125825 (80M) [application/x-gzip]\nSaving to: ‘../data/aclImdb_v1.tar.gz’\n\n../data/aclImdb_v1. 100%[===================>] 80.23M 23.8MB/s in 4.6s \n\n2021-03-07 19:37:20 (17.5 MB/s) - ‘../data/aclImdb_v1.tar.gz’ saved [84125825/84125825]\n\n"
]
],
[
[
"## Step 2: Preparing and Processing the data\n\nAlso, as in the XGBoost notebook, we will be doing some initial data processing. The first few steps are the same as in the XGBoost example. To begin with, we will read in each of the reviews and combine them into a single input structure. Then, we will split the dataset into a training set and a testing set.",
"_____no_output_____"
]
],
[
[
"import os\nimport glob\n\ndef read_imdb_data(data_dir='../data/aclImdb'):\n data = {}\n labels = {}\n \n for data_type in ['train', 'test']:\n data[data_type] = {}\n labels[data_type] = {}\n \n for sentiment in ['pos', 'neg']:\n data[data_type][sentiment] = []\n labels[data_type][sentiment] = []\n \n path = os.path.join(data_dir, data_type, sentiment, '*.txt')\n files = glob.glob(path)\n \n for f in files:\n with open(f) as review:\n data[data_type][sentiment].append(review.read())\n # Here we represent a positive review by '1' and a negative review by '0'\n labels[data_type][sentiment].append(1 if sentiment == 'pos' else 0)\n \n assert len(data[data_type][sentiment]) == len(labels[data_type][sentiment]), \\\n \"{}/{} data size does not match labels size\".format(data_type, sentiment)\n \n return data, labels",
"_____no_output_____"
],
[
"data, labels = read_imdb_data()\nprint(\"IMDB reviews: train = {} pos / {} neg, test = {} pos / {} neg\".format(\n len(data['train']['pos']), len(data['train']['neg']),\n len(data['test']['pos']), len(data['test']['neg'])))",
"IMDB reviews: train = 12500 pos / 12500 neg, test = 12500 pos / 12500 neg\n"
]
],
[
[
"Now that we've read the raw training and testing data from the downloaded dataset, we will combine the positive and negative reviews and shuffle the resulting records.",
"_____no_output_____"
]
],
[
[
"from sklearn.utils import shuffle\n\ndef prepare_imdb_data(data, labels):\n \"\"\"Prepare training and test sets from IMDb movie reviews.\"\"\"\n \n #Combine positive and negative reviews and labels\n data_train = data['train']['pos'] + data['train']['neg']\n data_test = data['test']['pos'] + data['test']['neg']\n labels_train = labels['train']['pos'] + labels['train']['neg']\n labels_test = labels['test']['pos'] + labels['test']['neg']\n \n #Shuffle reviews and corresponding labels within training and test sets\n data_train, labels_train = shuffle(data_train, labels_train)\n data_test, labels_test = shuffle(data_test, labels_test)\n \n # Return a unified training data, test data, training labels, test labets\n return data_train, data_test, labels_train, labels_test",
"_____no_output_____"
],
[
"train_X, test_X, train_y, test_y = prepare_imdb_data(data, labels)\nprint(\"IMDb reviews (combined): train = {}, test = {}\".format(len(train_X), len(test_X)))",
"IMDb reviews (combined): train = 25000, test = 25000\n"
]
],
[
[
"Now that we have our training and testing sets unified and prepared, we should do a quick check and see an example of the data our model will be trained on. This is generally a good idea as it allows you to see how each of the further processing steps affects the reviews and it also ensures that the data has been loaded correctly.",
"_____no_output_____"
]
],
[
[
"print(train_X[100])\nprint(train_y[100])",
"Think of this pilot as \"Hawaii Five-O Lite\". It's set in Hawaii, it's an action/adventure crime drama, lots of scenes feature boats and palm trees and polyester fabrics and garish shirts...it even stars the character actor \"Zulu\" in a supporting role. Oh, there are some minor differences - Roy Thinnes is supposed to be some front-line undercover agent, and the supporting cast is much smaller (and less interesting), but basically the atmosphere is still the same. Problem is, \"Hawaii Five-O\" (another QM product) already existed at the time and had run for years. It filled the market demand for Hawaii-based crime dramas quite adequately. Code Name: Diamond Head may have been intended as the hier to H50 as the older series eventually dwindled away...but it comes across as a superfluous, 2nd rate copy. It doesn't suck, but it's completely derivative and doesn't do anything as well as the original.<br /><br />There is some decent acting talent involved here. Thinnes is an old pro, and he gives the role his best shot, and he isn't bad. But Thinnes is only as good as his material and his director. Ian McShane is in here as an evil spy master named \"Tree\", and McShane tends to be the most interesting actor in any scene he appears in. But he's phoning his part in here. Frances Ngyuen is reasonably exotic looking, but her astounding skinniness, opaque features, thick accent and wooden delivery aren't the stuff of which dreams are made. Relying on her to supply the 'romantic interest' for Thinnes was probably the series' biggest mistake. At least for for a series aimed at white audiences brought up with Marsha Brady and Peggy Lee as our love goddesses. Give her another 30 lbs and a year with a dialog/voice coach, and she might cut it. Zulu is, well, his usual self - enjoyable in bit parts, but he isn't a person who can carry a feature by himself. <br /><br />In addition, the plot and dialog are strictly by-the-numbers, with nothing to distinguish them from any other Quinn Martin production. And by this point, the American TV audience had seen a whoooole lot of QM productions....I think \"CN: DH\" was one too many, and it sank without a trace. It wasn't the really the actors' fault, and I hope they walked away from this with a decent paycheck and one more entry on their C.V.s. <br /><br />MST3000 revived this for their treatment in their sixth season, and they had a lot of good natured fun with it. Worth seeking out in that version if you enjoy the MST approach to movie japery and lampoon, but I can't imagine anyone caring about this pilot for any other reason.\n0\n"
]
],
[
[
"The first step in processing the reviews is to make sure that any html tags that appear should be removed. In addition we wish to tokenize our input, that way words such as *entertained* and *entertaining* are considered the same with regard to sentiment analysis.",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk.corpus import stopwords\nfrom nltk.stem.porter import *\n\nimport re\nfrom bs4 import BeautifulSoup\n\ndef review_to_words(review):\n nltk.download(\"stopwords\", quiet=True)\n stemmer = PorterStemmer()\n \n text = BeautifulSoup(review, \"html.parser\").get_text() # Remove HTML tags\n text = re.sub(r\"[^a-zA-Z0-9]\", \" \", text.lower()) # Convert to lower case\n words = text.split() # Split string into words\n words = [w for w in words if w not in stopwords.words(\"english\")] # Remove stopwords\n words = [PorterStemmer().stem(w) for w in words] # stem\n \n return words",
"_____no_output_____"
]
],
[
[
"The `review_to_words` method defined above uses `BeautifulSoup` to remove any html tags that appear and uses the `nltk` package to tokenize the reviews. As a check to ensure we know how everything is working, try applying `review_to_words` to one of the reviews in the training set.",
"_____no_output_____"
]
],
[
[
"# TODO: Apply review_to_words to a review (train_X[100] or any other review)\nprint('Original review:')\nprint(train_X[100])\nprint('Tokenized review:')\nprint(review_to_words(train_X[100]))",
"Original review:\nThink of this pilot as \"Hawaii Five-O Lite\". It's set in Hawaii, it's an action/adventure crime drama, lots of scenes feature boats and palm trees and polyester fabrics and garish shirts...it even stars the character actor \"Zulu\" in a supporting role. Oh, there are some minor differences - Roy Thinnes is supposed to be some front-line undercover agent, and the supporting cast is much smaller (and less interesting), but basically the atmosphere is still the same. Problem is, \"Hawaii Five-O\" (another QM product) already existed at the time and had run for years. It filled the market demand for Hawaii-based crime dramas quite adequately. Code Name: Diamond Head may have been intended as the hier to H50 as the older series eventually dwindled away...but it comes across as a superfluous, 2nd rate copy. It doesn't suck, but it's completely derivative and doesn't do anything as well as the original.<br /><br />There is some decent acting talent involved here. Thinnes is an old pro, and he gives the role his best shot, and he isn't bad. But Thinnes is only as good as his material and his director. Ian McShane is in here as an evil spy master named \"Tree\", and McShane tends to be the most interesting actor in any scene he appears in. But he's phoning his part in here. Frances Ngyuen is reasonably exotic looking, but her astounding skinniness, opaque features, thick accent and wooden delivery aren't the stuff of which dreams are made. Relying on her to supply the 'romantic interest' for Thinnes was probably the series' biggest mistake. At least for for a series aimed at white audiences brought up with Marsha Brady and Peggy Lee as our love goddesses. Give her another 30 lbs and a year with a dialog/voice coach, and she might cut it. Zulu is, well, his usual self - enjoyable in bit parts, but he isn't a person who can carry a feature by himself. <br /><br />In addition, the plot and dialog are strictly by-the-numbers, with nothing to distinguish them from any other Quinn Martin production. And by this point, the American TV audience had seen a whoooole lot of QM productions....I think \"CN: DH\" was one too many, and it sank without a trace. It wasn't the really the actors' fault, and I hope they walked away from this with a decent paycheck and one more entry on their C.V.s. <br /><br />MST3000 revived this for their treatment in their sixth season, and they had a lot of good natured fun with it. Worth seeking out in that version if you enjoy the MST approach to movie japery and lampoon, but I can't imagine anyone caring about this pilot for any other reason.\nTokenized review:\n['think', 'pilot', 'hawaii', 'five', 'lite', 'set', 'hawaii', 'action', 'adventur', 'crime', 'drama', 'lot', 'scene', 'featur', 'boat', 'palm', 'tree', 'polyest', 'fabric', 'garish', 'shirt', 'even', 'star', 'charact', 'actor', 'zulu', 'support', 'role', 'oh', 'minor', 'differ', 'roy', 'thinn', 'suppos', 'front', 'line', 'undercov', 'agent', 'support', 'cast', 'much', 'smaller', 'less', 'interest', 'basic', 'atmospher', 'still', 'problem', 'hawaii', 'five', 'anoth', 'qm', 'product', 'alreadi', 'exist', 'time', 'run', 'year', 'fill', 'market', 'demand', 'hawaii', 'base', 'crime', 'drama', 'quit', 'adequ', 'code', 'name', 'diamond', 'head', 'may', 'intend', 'hier', 'h50', 'older', 'seri', 'eventu', 'dwindl', 'away', 'come', 'across', 'superflu', '2nd', 'rate', 'copi', 'suck', 'complet', 'deriv', 'anyth', 'well', 'origin', 'decent', 'act', 'talent', 'involv', 'thinn', 'old', 'pro', 'give', 'role', 'best', 'shot', 'bad', 'thinn', 'good', 'materi', 'director', 'ian', 'mcshane', 'evil', 'spi', 'master', 'name', 'tree', 'mcshane', 'tend', 'interest', 'actor', 'scene', 'appear', 'phone', 'part', 'franc', 'ngyuen', 'reason', 'exot', 'look', 'astound', 'skinni', 'opaqu', 'featur', 'thick', 'accent', 'wooden', 'deliveri', 'stuff', 'dream', 'made', 'reli', 'suppli', 'romant', 'interest', 'thinn', 'probabl', 'seri', 'biggest', 'mistak', 'least', 'seri', 'aim', 'white', 'audienc', 'brought', 'marsha', 'bradi', 'peggi', 'lee', 'love', 'goddess', 'give', 'anoth', '30', 'lb', 'year', 'dialog', 'voic', 'coach', 'might', 'cut', 'zulu', 'well', 'usual', 'self', 'enjoy', 'bit', 'part', 'person', 'carri', 'featur', 'addit', 'plot', 'dialog', 'strictli', 'number', 'noth', 'distinguish', 'quinn', 'martin', 'product', 'point', 'american', 'tv', 'audienc', 'seen', 'whooool', 'lot', 'qm', 'product', 'think', 'cn', 'dh', 'one', 'mani', 'sank', 'without', 'trace', 'realli', 'actor', 'fault', 'hope', 'walk', 'away', 'decent', 'paycheck', 'one', 'entri', 'c', 'v', 'mst3000', 'reviv', 'treatment', 'sixth', 'season', 'lot', 'good', 'natur', 'fun', 'worth', 'seek', 'version', 'enjoy', 'mst', 'approach', 'movi', 'japeri', 'lampoon', 'imagin', 'anyon', 'care', 'pilot', 'reason']\n"
]
],
[
[
"**Question:** Above we mentioned that `review_to_words` method removes html formatting and allows us to tokenize the words found in a review, for example, converting *entertained* and *entertaining* into *entertain* so that they are treated as though they are the same word. What else, if anything, does this method do to the input?",
"_____no_output_____"
],
[
"**Answer:** The mentioned function also remove articles, connectives, common vebs like \"to be\", possesives an othe grammatical tools not relevant to detect sentiment in the sentence. Additionaly vectorized it.",
"_____no_output_____"
],
[
"The method below applies the `review_to_words` method to each of the reviews in the training and testing datasets. In addition it caches the results. This is because performing this processing step can take a long time. This way if you are unable to complete the notebook in the current session, you can come back without needing to process the data a second time.",
"_____no_output_____"
]
],
[
[
"import pickle\n\ncache_dir = os.path.join(\"../cache\", \"sentiment_analysis\") # where to store cache files\nos.makedirs(cache_dir, exist_ok=True) # ensure cache directory exists\n\ndef preprocess_data(data_train, data_test, labels_train, labels_test,\n cache_dir=cache_dir, cache_file=\"preprocessed_data.pkl\"):\n \"\"\"Convert each review to words; read from cache if available.\"\"\"\n\n # If cache_file is not None, try to read from it first\n cache_data = None\n if cache_file is not None:\n try:\n with open(os.path.join(cache_dir, cache_file), \"rb\") as f:\n cache_data = pickle.load(f)\n print(\"Read preprocessed data from cache file:\", cache_file)\n except:\n pass # unable to read from cache, but that's okay\n \n # If cache is missing, then do the heavy lifting\n if cache_data is None:\n # Preprocess training and test data to obtain words for each review\n #words_train = list(map(review_to_words, data_train))\n #words_test = list(map(review_to_words, data_test))\n words_train = [review_to_words(review) for review in data_train]\n words_test = [review_to_words(review) for review in data_test]\n \n # Write to cache file for future runs\n if cache_file is not None:\n cache_data = dict(words_train=words_train, words_test=words_test,\n labels_train=labels_train, labels_test=labels_test)\n with open(os.path.join(cache_dir, cache_file), \"wb\") as f:\n pickle.dump(cache_data, f)\n print(\"Wrote preprocessed data to cache file:\", cache_file)\n else:\n # Unpack data loaded from cache file\n words_train, words_test, labels_train, labels_test = (cache_data['words_train'],\n cache_data['words_test'], cache_data['labels_train'], cache_data['labels_test'])\n \n return words_train, words_test, labels_train, labels_test",
"_____no_output_____"
],
[
"# Preprocess data\ntrain_X, test_X, train_y, test_y = preprocess_data(train_X, test_X, train_y, test_y)",
"Read preprocessed data from cache file: preprocessed_data.pkl\n"
]
],
[
[
"## Transform the data\n\nIn the XGBoost notebook we transformed the data from its word representation to a bag-of-words feature representation. For the model we are going to construct in this notebook we will construct a feature representation which is very similar. To start, we will represent each word as an integer. Of course, some of the words that appear in the reviews occur very infrequently and so likely don't contain much information for the purposes of sentiment analysis. The way we will deal with this problem is that we will fix the size of our working vocabulary and we will only include the words that appear most frequently. We will then combine all of the infrequent words into a single category and, in our case, we will label it as `1`.\n\nSince we will be using a recurrent neural network, it will be convenient if the length of each review is the same. To do this, we will fix a size for our reviews and then pad short reviews with the category 'no word' (which we will label `0`) and truncate long reviews.",
"_____no_output_____"
],
[
"### (TODO) Create a word dictionary\n\nTo begin with, we need to construct a way to map words that appear in the reviews to integers. Here we fix the size of our vocabulary (including the 'no word' and 'infrequent' categories) to be `5000` but you may wish to change this to see how it affects the model.\n\n> **TODO:** Complete the implementation for the `build_dict()` method below. Note that even though the vocab_size is set to `5000`, we only want to construct a mapping for the most frequently appearing `4998` words. This is because we want to reserve the special labels `0` for 'no word' and `1` for 'infrequent word'.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef build_dict(data, vocab_size = 5000):\n \"\"\"Construct and return a dictionary mapping each of the most frequently appearing words to a unique integer.\"\"\"\n \n # TODO: Determine how often each word appears in `data`. Note that `data` is a list of sentences and that a\n # sentence is a list of words.\n \n word_count = {} # A dict storing the words that appear in the reviews along with how often they occur\n \n # Solution:\n for sentence in data:\n for word in sentence:\n word_count[word]=word_count.get(word,0)+1\n\n # TODO: Sort the words found in `data` so that sorted_words[0] is the most frequently appearing word and\n # sorted_words[-1] is the least frequently appearing word.\n \n sorted_words = None\n \n # Solution:\n sorted_words = sorted(word_count, key=word_count.get, reverse=True)\n \n word_dict = {} # This is what we are building, a dictionary that translates words into integers\n for idx, word in enumerate(sorted_words[:vocab_size - 2]): # The -2 is so that we save room for the 'no word'\n word_dict[word] = idx + 2 # 'infrequent' labels\n \n return word_dict",
"_____no_output_____"
],
[
"word_dict = build_dict(train_X)",
"_____no_output_____"
]
],
[
[
"**Question:** What are the five most frequently appearing (tokenized) words in the training set? Does it makes sense that these words appear frequently in the training set?",
"_____no_output_____"
],
[
"**Answer:**\nThe most common tokenized words apearing in the training set are 'movi', 'film', 'one', 'like' and 'time'. The first two words are quite obvious, _movies_ and _films_ are the topics of the reviews. The rest three are frequent in english: _one_ could be use to avoid repeating the movie name, _like_ could be use in a positive and negative review and _time_ might be just common.",
"_____no_output_____"
]
],
[
[
"# TODO: Use this space to determine the five most frequently appearing words in the training set.\nlist(word_dict)[0:5]",
"_____no_output_____"
]
],
[
[
"### Save `word_dict`\n\nLater on when we construct an endpoint which processes a submitted review we will need to make use of the `word_dict` which we have created. As such, we will save it to a file now for future use.",
"_____no_output_____"
]
],
[
[
"data_dir = '../data/pytorch' # The folder we will use for storing data\nif not os.path.exists(data_dir): # Make sure that the folder exists\n os.makedirs(data_dir)",
"_____no_output_____"
],
[
"with open(os.path.join(data_dir, 'word_dict.pkl'), \"wb\") as f:\n pickle.dump(word_dict, f)",
"_____no_output_____"
]
],
[
[
"### Transform the reviews\n\nNow that we have our word dictionary which allows us to transform the words appearing in the reviews into integers, it is time to make use of it and convert our reviews to their integer sequence representation, making sure to pad or truncate to a fixed length, which in our case is `500`.",
"_____no_output_____"
]
],
[
[
"def convert_and_pad(word_dict, sentence, pad=500):\n NOWORD = 0 # We will use 0 to represent the 'no word' category\n INFREQ = 1 # and we use 1 to represent the infrequent words, i.e., words not appearing in word_dict\n \n working_sentence = [NOWORD] * pad\n \n for word_index, word in enumerate(sentence[:pad]):\n if word in word_dict:\n working_sentence[word_index] = word_dict[word]\n else:\n working_sentence[word_index] = INFREQ\n \n return working_sentence, min(len(sentence), pad)\n\ndef convert_and_pad_data(word_dict, data, pad=500):\n result = []\n lengths = []\n \n for sentence in data:\n converted, leng = convert_and_pad(word_dict, sentence, pad)\n result.append(converted)\n lengths.append(leng)\n \n return np.array(result), np.array(lengths)",
"_____no_output_____"
],
[
"train_X, train_X_len = convert_and_pad_data(word_dict, train_X)\ntest_X, test_X_len = convert_and_pad_data(word_dict, test_X)",
"_____no_output_____"
]
],
[
[
"As a quick check to make sure that things are working as intended, check to see what one of the reviews in the training set looks like after having been processeed. Does this look reasonable? What is the length of a review in the training set?",
"_____no_output_____"
]
],
[
[
"# Use this cell to examine one of the processed reviews to make sure everything is working as intended.\nn_sample=15\nprint(train_X[n_sample])\nprint(len(train_X[n_sample]))",
"[ 641 4 174 2 56 47 8 175 2663 168 2 19 5 1\n 632 341 154 4 1 1 349 977 82 1108 134 60 3756 1\n 189 111 1408 17 320 13 672 2529 501 1 551 1 1 85\n 318 52 1632 1 1438 1 3416 85 3441 258 718 296 1 130\n 31 82 7 25 892 496 212 214 91 51 56 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0]\n500\n"
]
],
[
[
"**Question:** In the cells above we use the `preprocess_data` and `convert_and_pad_data` methods to process both the training and testing set. Why or why not might this be a problem?",
"_____no_output_____"
],
[
"**Answer:** It's important to use the same function to both proccesses in order to assure there will be no missalignment in the codification.",
"_____no_output_____"
],
[
"## Step 3: Upload the data to S3\n\nAs in the XGBoost notebook, we will need to upload the training dataset to S3 in order for our training code to access it. For now we will save it locally and we will upload to S3 later on.\n\n### Save the processed training dataset locally\n\nIt is important to note the format of the data that we are saving as we will need to know it when we write the training code. In our case, each row of the dataset has the form `label`, `length`, `review[500]` where `review[500]` is a sequence of `500` integers representing the words in the review.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n \npd.concat([pd.DataFrame(train_y), pd.DataFrame(train_X_len), pd.DataFrame(train_X)], axis=1) \\\n .to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)",
"_____no_output_____"
]
],
[
[
"### Uploading the training data\n\n\nNext, we need to upload the training data to the SageMaker default S3 bucket so that we can provide access to it while training our model.",
"_____no_output_____"
]
],
[
[
"import sagemaker\n\nsagemaker_session = sagemaker.Session()\n\nbucket = sagemaker_session.default_bucket()\nprefix = 'sagemaker/sentiment_rnn'\n\nrole = sagemaker.get_execution_role()",
"_____no_output_____"
],
[
"input_data = sagemaker_session.upload_data(path=data_dir, bucket=bucket, key_prefix=prefix)",
"_____no_output_____"
]
],
[
[
"**NOTE:** The cell above uploads the entire contents of our data directory. This includes the `word_dict.pkl` file. This is fortunate as we will need this later on when we create an endpoint that accepts an arbitrary review. For now, we will just take note of the fact that it resides in the data directory (and so also in the S3 training bucket) and that we will need to make sure it gets saved in the model directory.",
"_____no_output_____"
],
[
"## Step 4: Build and Train the PyTorch Model\n\nIn the XGBoost notebook we discussed what a model is in the SageMaker framework. In particular, a model comprises three objects\n\n - Model Artifacts,\n - Training Code, and\n - Inference Code,\n \neach of which interact with one another. In the XGBoost example we used training and inference code that was provided by Amazon. Here we will still be using containers provided by Amazon with the added benefit of being able to include our own custom code.\n\nWe will start by implementing our own neural network in PyTorch along with a training script. For the purposes of this project we have provided the necessary model object in the `model.py` file, inside of the `train` folder. You can see the provided implementation by running the cell below.",
"_____no_output_____"
]
],
[
[
"!pygmentize train/model.py",
"\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch\u001b[39;49;00m\u001b[04m\u001b[36m.\u001b[39;49;00m\u001b[04m\u001b[36mnn\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mnn\u001b[39;49;00m\r\n\r\n\u001b[34mclass\u001b[39;49;00m \u001b[04m\u001b[32mLSTMClassifier\u001b[39;49;00m(nn.Module):\r\n \u001b[33m\"\"\"\u001b[39;49;00m\r\n\u001b[33m This is the simple RNN model we will be using to perform Sentiment Analysis.\u001b[39;49;00m\r\n\u001b[33m \"\"\"\u001b[39;49;00m\r\n\r\n \u001b[34mdef\u001b[39;49;00m \u001b[32m__init__\u001b[39;49;00m(\u001b[36mself\u001b[39;49;00m, embedding_dim, hidden_dim, vocab_size):\r\n \u001b[33m\"\"\"\u001b[39;49;00m\r\n\u001b[33m Initialize the model by settingg up the various layers.\u001b[39;49;00m\r\n\u001b[33m \"\"\"\u001b[39;49;00m\r\n \u001b[36msuper\u001b[39;49;00m(LSTMClassifier, \u001b[36mself\u001b[39;49;00m).\u001b[32m__init__\u001b[39;49;00m()\r\n\r\n \u001b[36mself\u001b[39;49;00m.embedding = nn.Embedding(vocab_size, embedding_dim, padding_idx=\u001b[34m0\u001b[39;49;00m)\r\n \u001b[36mself\u001b[39;49;00m.lstm = nn.LSTM(embedding_dim, hidden_dim)\r\n \u001b[36mself\u001b[39;49;00m.dense = nn.Linear(in_features=hidden_dim, out_features=\u001b[34m1\u001b[39;49;00m)\r\n \u001b[36mself\u001b[39;49;00m.sig = nn.Sigmoid()\r\n \r\n \u001b[36mself\u001b[39;49;00m.word_dict = \u001b[34mNone\u001b[39;49;00m\r\n\r\n \u001b[34mdef\u001b[39;49;00m \u001b[32mforward\u001b[39;49;00m(\u001b[36mself\u001b[39;49;00m, x):\r\n \u001b[33m\"\"\"\u001b[39;49;00m\r\n\u001b[33m Perform a forward pass of our model on some input.\u001b[39;49;00m\r\n\u001b[33m \"\"\"\u001b[39;49;00m\r\n x = x.t()\r\n lengths = x[\u001b[34m0\u001b[39;49;00m,:]\r\n reviews = x[\u001b[34m1\u001b[39;49;00m:,:]\r\n embeds = \u001b[36mself\u001b[39;49;00m.embedding(reviews)\r\n lstm_out, _ = \u001b[36mself\u001b[39;49;00m.lstm(embeds)\r\n out = \u001b[36mself\u001b[39;49;00m.dense(lstm_out)\r\n out = out[lengths - \u001b[34m1\u001b[39;49;00m, \u001b[36mrange\u001b[39;49;00m(\u001b[36mlen\u001b[39;49;00m(lengths))]\r\n \u001b[34mreturn\u001b[39;49;00m \u001b[36mself\u001b[39;49;00m.sig(out.squeeze())\r\n"
]
],
[
[
"The important takeaway from the implementation provided is that there are three parameters that we may wish to tweak to improve the performance of our model. These are the embedding dimension, the hidden dimension and the size of the vocabulary. We will likely want to make these parameters configurable in the training script so that if we wish to modify them we do not need to modify the script itself. We will see how to do this later on. To start we will write some of the training code in the notebook so that we can more easily diagnose any issues that arise.\n\nFirst we will load a small portion of the training data set to use as a sample. It would be very time consuming to try and train the model completely in the notebook as we do not have access to a gpu and the compute instance that we are using is not particularly powerful. However, we can work on a small bit of the data to get a feel for how our training script is behaving.",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.utils.data\n\n# Read in only the first 250 rows\ntrain_sample = pd.read_csv(os.path.join(data_dir, 'train.csv'), header=None, names=None, nrows=250)\n\n# Turn the input pandas dataframe into tensors\ntrain_sample_y = torch.from_numpy(train_sample[[0]].values).float().squeeze()\ntrain_sample_X = torch.from_numpy(train_sample.drop([0], axis=1).values).long()\n\n# Build the dataset\ntrain_sample_ds = torch.utils.data.TensorDataset(train_sample_X, train_sample_y)\n# Build the dataloader\ntrain_sample_dl = torch.utils.data.DataLoader(train_sample_ds, batch_size=50)",
"_____no_output_____"
]
],
[
[
"### (TODO) Writing the training method\n\nNext we need to write the training code itself. This should be very similar to training methods that you have written before to train PyTorch models. We will leave any difficult aspects such as model saving / loading and parameter loading until a little later.",
"_____no_output_____"
]
],
[
[
"def train(model, train_loader, epochs, optimizer, loss_fn, device):\n for epoch in range(1, epochs + 1):\n model.train()\n total_loss = 0\n for batch in train_loader: \n batch_X, batch_y = batch\n \n batch_X = batch_X.to(device)\n batch_y = batch_y.to(device)\n \n # TODO: Complete this train method to train the model provided.\n # Reference https://towardsdatascience.com/lstm-text-classification-using-pytorch-2c6c657f8fc0\n # Solution:\n optimizer.zero_grad()\n output = model(batch_X)\n loss=loss_fn(output, batch_y)\n\n loss.backward()\n optimizer.step()\n total_loss += loss.data.item()\n print(\"Epoch: {}, BCELoss: {}\".format(epoch, total_loss / len(train_loader)))",
"_____no_output_____"
]
],
[
[
"Supposing we have the training method above, we will test that it is working by writing a bit of code in the notebook that executes our training method on the small sample training set that we loaded earlier. The reason for doing this in the notebook is so that we have an opportunity to fix any errors that arise early when they are easier to diagnose.",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim\nfrom train.model import LSTMClassifier\n\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\nmodel = LSTMClassifier(32, 100, 5000).to(device)\noptimizer = optim.Adam(model.parameters())\nloss_fn = torch.nn.BCELoss()\n\ntrain(model, train_sample_dl, 5, optimizer, loss_fn, device)",
"Epoch: 1, BCELoss: 0.6889122724533081\nEpoch: 2, BCELoss: 0.6780008792877197\nEpoch: 3, BCELoss: 0.6685242891311646\nEpoch: 4, BCELoss: 0.6583548784255981\nEpoch: 5, BCELoss: 0.6465497970581054\n"
]
],
[
[
"In order to construct a PyTorch model using SageMaker we must provide SageMaker with a training script. We may optionally include a directory which will be copied to the container and from which our training code will be run. When the training container is executed it will check the uploaded directory (if there is one) for a `requirements.txt` file and install any required Python libraries, after which the training script will be run.",
"_____no_output_____"
],
[
"### (TODO) Training the model\n\nWhen a PyTorch model is constructed in SageMaker, an entry point must be specified. This is the Python file which will be executed when the model is trained. Inside of the `train` directory is a file called `train.py` which has been provided and which contains most of the necessary code to train our model. The only thing that is missing is the implementation of the `train()` method which you wrote earlier in this notebook.\n\n**TODO**: Copy the `train()` method written above and paste it into the `train/train.py` file where required.\n\nThe way that SageMaker passes hyperparameters to the training script is by way of arguments. These arguments can then be parsed and used in the training script. To see how this is done take a look at the provided `train/train.py` file.",
"_____no_output_____"
]
],
[
[
"from sagemaker.pytorch import PyTorch\n\nestimator = PyTorch(entry_point=\"train.py\",\n source_dir=\"train\",\n role=role,\n framework_version='0.4.0',\n py_version=\"py3\",\n train_instance_count=1,\n train_instance_type='ml.p2.xlarge',\n hyperparameters={\n 'epochs': 10,\n 'hidden_dim': 200,\n })",
"_____no_output_____"
],
[
"estimator.fit({'training': input_data})",
"'create_image_uri' will be deprecated in favor of 'ImageURIProvider' class in SageMaker Python SDK v2.\n's3_input' class will be renamed to 'TrainingInput' in SageMaker Python SDK v2.\n'create_image_uri' will be deprecated in favor of 'ImageURIProvider' class in SageMaker Python SDK v2.\n"
]
],
[
[
"## Step 5: Testing the model\n\nAs mentioned at the top of this notebook, we will be testing this model by first deploying it and then sending the testing data to the deployed endpoint. We will do this so that we can make sure that the deployed model is working correctly.\n\n## Step 6: Deploy the model for testing\n\nNow that we have trained our model, we would like to test it to see how it performs. Currently our model takes input of the form `review_length, review[500]` where `review[500]` is a sequence of `500` integers which describe the words present in the review, encoded using `word_dict`. Fortunately for us, SageMaker provides built-in inference code for models with simple inputs such as this.\n\nThere is one thing that we need to provide, however, and that is a function which loads the saved model. This function must be called `model_fn()` and takes as its only parameter a path to the directory where the model artifacts are stored. This function must also be present in the python file which we specified as the entry point. In our case the model loading function has been provided and so no changes need to be made.\n\n**NOTE**: When the built-in inference code is run it must import the `model_fn()` method from the `train.py` file. This is why the training code is wrapped in a main guard ( ie, `if __name__ == '__main__':` )\n\nSince we don't need to change anything in the code that was uploaded during training, we can simply deploy the current model as-is.\n\n**NOTE:** When deploying a model you are asking SageMaker to launch an compute instance that will wait for data to be sent to it. As a result, this compute instance will continue to run until *you* shut it down. This is important to know since the cost of a deployed endpoint depends on how long it has been running for.\n\nIn other words **If you are no longer using a deployed endpoint, shut it down!**\n\n**TODO:** Deploy the trained model.",
"_____no_output_____"
]
],
[
[
"# TODO: Deploy the trained model\n\n# Solution:\n# Deploy my estimator to a SageMaker Endpoint and get a Predictor\npredictor = estimator.deploy(instance_type='ml.m4.xlarge',\n initial_instance_count=1)\n",
"Parameter image will be renamed to image_uri in SageMaker Python SDK v2.\n'create_image_uri' will be deprecated in favor of 'ImageURIProvider' class in SageMaker Python SDK v2.\n"
]
],
[
[
"## Step 7 - Use the model for testing\n\nOnce deployed, we can read in the test data and send it off to our deployed model to get some results. Once we collect all of the results we can determine how accurate our model is.",
"_____no_output_____"
]
],
[
[
"test_X = pd.concat([pd.DataFrame(test_X_len), pd.DataFrame(test_X)], axis=1)",
"_____no_output_____"
],
[
"# We split the data into chunks and send each chunk seperately, accumulating the results.\n\ndef predict(data, rows=512):\n split_array = np.array_split(data, int(data.shape[0] / float(rows) + 1))\n predictions = np.array([])\n for array in split_array:\n predictions = np.append(predictions, predictor.predict(array))\n \n return predictions",
"_____no_output_____"
],
[
"predictions = predict(test_X.values)\npredictions = [round(num) for num in predictions]",
"_____no_output_____"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(test_y, predictions)",
"_____no_output_____"
]
],
[
[
"**Question:** How does this model compare to the XGBoost model you created earlier? Why might these two models perform differently on this dataset? Which do *you* think is better for sentiment analysis?",
"_____no_output_____"
],
[
"**Answer:** It was quite good results for the pytorch model in comparison with the XGBoost. The advantage of the pytorch model is the possibility of model specifically to the application. The advantage of the XGBoost is that is ready to work.\nSince the pytorch model is tailored to the application, should perform better.",
"_____no_output_____"
],
[
"### (TODO) More testing\n\nWe now have a trained model which has been deployed and which we can send processed reviews to and which returns the predicted sentiment. However, ultimately we would like to be able to send our model an unprocessed review. That is, we would like to send the review itself as a string. For example, suppose we wish to send the following review to our model.",
"_____no_output_____"
]
],
[
[
"test_review = 'The simplest pleasures in life are the best, and this film is one of them. Combining a rather basic storyline of love and adventure this movie transcends the usual weekend fair with wit and unmitigated charm.'",
"_____no_output_____"
]
],
[
[
"The question we now need to answer is, how do we send this review to our model?\n\nRecall in the first section of this notebook we did a bunch of data processing to the IMDb dataset. In particular, we did two specific things to the provided reviews.\n - Removed any html tags and stemmed the input\n - Encoded the review as a sequence of integers using `word_dict`\n \nIn order process the review we will need to repeat these two steps.\n\n**TODO**: Using the `review_to_words` and `convert_and_pad` methods from section one, convert `test_review` into a numpy array `test_data` suitable to send to our model. Remember that our model expects input of the form `review_length, review[500]`.",
"_____no_output_____"
]
],
[
[
"# TODO: Convert test_review into a form usable by the model and save the results in test_data\ntest_data=[]\ntest_data, test_data_len = convert_and_pad_data(word_dict, [review_to_words(test_review)])\ntest_data_full = pd.concat([pd.DataFrame(test_data_len), pd.DataFrame(test_data)], axis=1)\nprint(test_data_full)\nlen(test_data_full)",
" 0 0 1 2 3 4 5 6 7 8 ... 490 491 492 493 \\\n0 20 1 1376 49 53 3 4 878 173 392 ... 0 0 0 0 \n\n 494 495 496 497 498 499 \n0 0 0 0 0 0 0 \n\n[1 rows x 501 columns]\n"
]
],
[
[
"Now that we have processed the review, we can send the resulting array to our model to predict the sentiment of the review.",
"_____no_output_____"
]
],
[
[
"predict(test_data_full.values)",
"_____no_output_____"
]
],
[
[
"Since the return value of our model is close to `1`, we can be certain that the review we submitted is positive.",
"_____no_output_____"
],
[
"### Delete the endpoint\n\nOf course, just like in the XGBoost notebook, once we've deployed an endpoint it continues to run until we tell it to shut down. Since we are done using our endpoint for now, we can delete it.",
"_____no_output_____"
]
],
[
[
"estimator.delete_endpoint()",
"estimator.delete_endpoint() will be deprecated in SageMaker Python SDK v2. Please use the delete_endpoint() function on your predictor instead.\n"
]
],
[
[
"## Step 6 (again) - Deploy the model for the web app\n\nNow that we know that our model is working, it's time to create some custom inference code so that we can send the model a review which has not been processed and have it determine the sentiment of the review.\n\nAs we saw above, by default the estimator which we created, when deployed, will use the entry script and directory which we provided when creating the model. However, since we now wish to accept a string as input and our model expects a processed review, we need to write some custom inference code.\n\nWe will store the code that we write in the `serve` directory. Provided in this directory is the `model.py` file that we used to construct our model, a `utils.py` file which contains the `review_to_words` and `convert_and_pad` pre-processing functions which we used during the initial data processing, and `predict.py`, the file which will contain our custom inference code. Note also that `requirements.txt` is present which will tell SageMaker what Python libraries are required by our custom inference code.\n\nWhen deploying a PyTorch model in SageMaker, you are expected to provide four functions which the SageMaker inference container will use.\n - `model_fn`: This function is the same function that we used in the training script and it tells SageMaker how to load our model.\n - `input_fn`: This function receives the raw serialized input that has been sent to the model's endpoint and its job is to de-serialize and make the input available for the inference code.\n - `output_fn`: This function takes the output of the inference code and its job is to serialize this output and return it to the caller of the model's endpoint.\n - `predict_fn`: The heart of the inference script, this is where the actual prediction is done and is the function which you will need to complete.\n\nFor the simple website that we are constructing during this project, the `input_fn` and `output_fn` methods are relatively straightforward. We only require being able to accept a string as input and we expect to return a single value as output. You might imagine though that in a more complex application the input or output may be image data or some other binary data which would require some effort to serialize.\n\n### (TODO) Writing inference code\n\nBefore writing our custom inference code, we will begin by taking a look at the code which has been provided.",
"_____no_output_____"
]
],
[
[
"!pygmentize serve/predict.py",
"\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36margparse\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mjson\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mos\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mpickle\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36msys\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36msagemaker_containers\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mpandas\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mpd\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mnumpy\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mnp\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch\u001b[39;49;00m\u001b[04m\u001b[36m.\u001b[39;49;00m\u001b[04m\u001b[36mnn\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36mnn\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch\u001b[39;49;00m\u001b[04m\u001b[36m.\u001b[39;49;00m\u001b[04m\u001b[36moptim\u001b[39;49;00m \u001b[34mas\u001b[39;49;00m \u001b[04m\u001b[36moptim\u001b[39;49;00m\r\n\u001b[34mimport\u001b[39;49;00m \u001b[04m\u001b[36mtorch\u001b[39;49;00m\u001b[04m\u001b[36m.\u001b[39;49;00m\u001b[04m\u001b[36mutils\u001b[39;49;00m\u001b[04m\u001b[36m.\u001b[39;49;00m\u001b[04m\u001b[36mdata\u001b[39;49;00m\r\n\r\n\u001b[34mfrom\u001b[39;49;00m \u001b[04m\u001b[36mmodel\u001b[39;49;00m \u001b[34mimport\u001b[39;49;00m LSTMClassifier\r\n\r\n\u001b[34mfrom\u001b[39;49;00m \u001b[04m\u001b[36mutils\u001b[39;49;00m \u001b[34mimport\u001b[39;49;00m review_to_words, convert_and_pad\r\n\r\n\u001b[34mdef\u001b[39;49;00m \u001b[32mmodel_fn\u001b[39;49;00m(model_dir):\r\n \u001b[33m\"\"\"Load the PyTorch model from the `model_dir` directory.\"\"\"\u001b[39;49;00m\r\n \u001b[36mprint\u001b[39;49;00m(\u001b[33m\"\u001b[39;49;00m\u001b[33mLoading model.\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\r\n\r\n \u001b[37m# First, load the parameters used to create the model.\u001b[39;49;00m\r\n model_info = {}\r\n model_info_path = os.path.join(model_dir, \u001b[33m'\u001b[39;49;00m\u001b[33mmodel_info.pth\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mwith\u001b[39;49;00m \u001b[36mopen\u001b[39;49;00m(model_info_path, \u001b[33m'\u001b[39;49;00m\u001b[33mrb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m) \u001b[34mas\u001b[39;49;00m f:\r\n model_info = torch.load(f)\r\n\r\n \u001b[36mprint\u001b[39;49;00m(\u001b[33m\"\u001b[39;49;00m\u001b[33mmodel_info: \u001b[39;49;00m\u001b[33m{}\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m.format(model_info))\r\n\r\n \u001b[37m# Determine the device and construct the model.\u001b[39;49;00m\r\n device = torch.device(\u001b[33m\"\u001b[39;49;00m\u001b[33mcuda\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m \u001b[34mif\u001b[39;49;00m torch.cuda.is_available() \u001b[34melse\u001b[39;49;00m \u001b[33m\"\u001b[39;49;00m\u001b[33mcpu\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\r\n model = LSTMClassifier(model_info[\u001b[33m'\u001b[39;49;00m\u001b[33membedding_dim\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m], model_info[\u001b[33m'\u001b[39;49;00m\u001b[33mhidden_dim\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m], model_info[\u001b[33m'\u001b[39;49;00m\u001b[33mvocab_size\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m])\r\n\r\n \u001b[37m# Load the store model parameters.\u001b[39;49;00m\r\n model_path = os.path.join(model_dir, \u001b[33m'\u001b[39;49;00m\u001b[33mmodel.pth\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mwith\u001b[39;49;00m \u001b[36mopen\u001b[39;49;00m(model_path, \u001b[33m'\u001b[39;49;00m\u001b[33mrb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m) \u001b[34mas\u001b[39;49;00m f:\r\n model.load_state_dict(torch.load(f))\r\n\r\n \u001b[37m# Load the saved word_dict.\u001b[39;49;00m\r\n word_dict_path = os.path.join(model_dir, \u001b[33m'\u001b[39;49;00m\u001b[33mword_dict.pkl\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mwith\u001b[39;49;00m \u001b[36mopen\u001b[39;49;00m(word_dict_path, \u001b[33m'\u001b[39;49;00m\u001b[33mrb\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m) \u001b[34mas\u001b[39;49;00m f:\r\n model.word_dict = pickle.load(f)\r\n\r\n model.to(device).eval()\r\n\r\n \u001b[36mprint\u001b[39;49;00m(\u001b[33m\"\u001b[39;49;00m\u001b[33mDone loading model.\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\r\n \u001b[34mreturn\u001b[39;49;00m model\r\n\r\n\u001b[34mdef\u001b[39;49;00m \u001b[32minput_fn\u001b[39;49;00m(serialized_input_data, content_type):\r\n \u001b[36mprint\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mDeserializing the input data.\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mif\u001b[39;49;00m content_type == \u001b[33m'\u001b[39;49;00m\u001b[33mtext/plain\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m:\r\n data = serialized_input_data.decode(\u001b[33m'\u001b[39;49;00m\u001b[33mutf-8\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mreturn\u001b[39;49;00m data\r\n \u001b[34mraise\u001b[39;49;00m \u001b[36mException\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mRequested unsupported ContentType in content_type: \u001b[39;49;00m\u001b[33m'\u001b[39;49;00m + content_type)\r\n\r\n\u001b[34mdef\u001b[39;49;00m \u001b[32moutput_fn\u001b[39;49;00m(prediction_output, accept):\r\n \u001b[36mprint\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mSerializing the generated output:\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \u001b[34mreturn\u001b[39;49;00m \u001b[36mstr\u001b[39;49;00m(prediction_output)\r\n\r\n\u001b[34mdef\u001b[39;49;00m \u001b[32mpredict_fn\u001b[39;49;00m(input_data, model):\r\n \u001b[36mprint\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mInferring sentiment of input data.\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n\r\n device = torch.device(\u001b[33m\"\u001b[39;49;00m\u001b[33mcuda\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m \u001b[34mif\u001b[39;49;00m torch.cuda.is_available() \u001b[34melse\u001b[39;49;00m \u001b[33m\"\u001b[39;49;00m\u001b[33mcpu\u001b[39;49;00m\u001b[33m\"\u001b[39;49;00m)\r\n \r\n \u001b[34mif\u001b[39;49;00m model.word_dict \u001b[35mis\u001b[39;49;00m \u001b[34mNone\u001b[39;49;00m:\r\n \u001b[34mraise\u001b[39;49;00m \u001b[36mException\u001b[39;49;00m(\u001b[33m'\u001b[39;49;00m\u001b[33mModel has not been loaded properly, no word_dict.\u001b[39;49;00m\u001b[33m'\u001b[39;49;00m)\r\n \r\n \u001b[37m# TODO: Process input_data so that it is ready to be sent to our model.\u001b[39;49;00m\r\n \u001b[37m# You should produce two variables:\u001b[39;49;00m\r\n \u001b[37m# data_X - A sequence of length 500 which represents the converted review\u001b[39;49;00m\r\n \u001b[37m# data_len - The length of the review\u001b[39;49;00m\r\n \r\n data_X = \u001b[34mNone\u001b[39;49;00m\r\n data_len = \u001b[34mNone\u001b[39;49;00m\r\n \r\n \u001b[37m# SOLUTION:\u001b[39;49;00m\r\n data_X, data_len = convert_and_pad(model.word_dict, review_to_words(input_data))\r\n\r\n \u001b[37m# Using data_X and data_len we construct an appropriate input tensor. Remember\u001b[39;49;00m\r\n \u001b[37m# that our model expects input data of the form 'len, review[500]'.\u001b[39;49;00m\r\n data_pack = np.hstack((data_len, data_X))\r\n data_pack = data_pack.reshape(\u001b[34m1\u001b[39;49;00m, -\u001b[34m1\u001b[39;49;00m)\r\n \r\n data = torch.from_numpy(data_pack)\r\n data = data.to(device)\r\n\r\n \u001b[37m# Make sure to put the model into evaluation mode\u001b[39;49;00m\r\n model.eval()\r\n\r\n \u001b[37m# TODO: Compute the result of applying the model to the input data. The variable `result` should\u001b[39;49;00m\r\n \u001b[37m# be a numpy array which contains a single integer which is either 1 or 0\u001b[39;49;00m\r\n\r\n result = \u001b[34mNone\u001b[39;49;00m\r\n \r\n \u001b[37m# Solution:\u001b[39;49;00m\r\n \r\n result = \u001b[36mround\u001b[39;49;00m(model(data).item())\r\n \r\n \u001b[34mreturn\u001b[39;49;00m result\r\n"
]
],
[
[
"As mentioned earlier, the `model_fn` method is the same as the one provided in the training code and the `input_fn` and `output_fn` methods are very simple and your task will be to complete the `predict_fn` method. Make sure that you save the completed file as `predict.py` in the `serve` directory.\n\n**TODO**: Complete the `predict_fn()` method in the `serve/predict.py` file.",
"_____no_output_____"
],
[
"### Deploying the model\n\nNow that the custom inference code has been written, we will create and deploy our model. To begin with, we need to construct a new PyTorchModel object which points to the model artifacts created during training and also points to the inference code that we wish to use. Then we can call the deploy method to launch the deployment container.\n\n**NOTE**: The default behaviour for a deployed PyTorch model is to assume that any input passed to the predictor is a `numpy` array. In our case we want to send a string so we need to construct a simple wrapper around the `RealTimePredictor` class to accomodate simple strings. In a more complicated situation you may want to provide a serialization object, for example if you wanted to sent image data.",
"_____no_output_____"
]
],
[
[
"from sagemaker.predictor import RealTimePredictor\nfrom sagemaker.pytorch import PyTorchModel\n\nclass StringPredictor(RealTimePredictor):\n def __init__(self, endpoint_name, sagemaker_session):\n super(StringPredictor, self).__init__(endpoint_name, sagemaker_session, content_type='text/plain')\n\nmodel = PyTorchModel(model_data=estimator.model_data,\n role = role,\n framework_version='0.4.0',\n entry_point='predict.py',\n source_dir='serve',\n predictor_cls=StringPredictor)\npredictor = model.deploy(initial_instance_count=1, instance_type='ml.m4.xlarge')",
"Parameter image will be renamed to image_uri in SageMaker Python SDK v2.\n'create_image_uri' will be deprecated in favor of 'ImageURIProvider' class in SageMaker Python SDK v2.\n"
]
],
[
[
"### Testing the model\n\nNow that we have deployed our model with the custom inference code, we should test to see if everything is working. Here we test our model by loading the first `250` positive and negative reviews and send them to the endpoint, then collect the results. The reason for only sending some of the data is that the amount of time it takes for our model to process the input and then perform inference is quite long and so testing the entire data set would be prohibitive.",
"_____no_output_____"
]
],
[
[
"import glob\n\ndef test_reviews(data_dir='../data/aclImdb', stop=250):\n \n results = []\n ground = []\n \n # We make sure to test both positive and negative reviews \n for sentiment in ['pos', 'neg']:\n \n path = os.path.join(data_dir, 'test', sentiment, '*.txt')\n files = glob.glob(path)\n \n files_read = 0\n \n print('Starting ', sentiment, ' files')\n \n # Iterate through the files and send them to the predictor\n for f in files:\n with open(f) as review:\n # First, we store the ground truth (was the review positive or negative)\n if sentiment == 'pos':\n ground.append(1)\n else:\n ground.append(0)\n # Read in the review and convert to 'utf-8' for transmission via HTTP\n review_input = review.read().encode('utf-8')\n # Send the review to the predictor and store the results\n results.append(float(predictor.predict(review_input)))\n \n # Sending reviews to our endpoint one at a time takes a while so we\n # only send a small number of reviews\n files_read += 1\n if files_read == stop:\n break\n \n return ground, results",
"_____no_output_____"
],
[
"ground, results = test_reviews()",
"Starting pos files\nStarting neg files\n"
],
[
"from sklearn.metrics import accuracy_score\naccuracy_score(ground, results)",
"_____no_output_____"
]
],
[
[
"As an additional test, we can try sending the `test_review` that we looked at earlier.",
"_____no_output_____"
]
],
[
[
"predictor.predict(test_review)",
"_____no_output_____"
]
],
[
[
"Now that we know our endpoint is working as expected, we can set up the web page that will interact with it. If you don't have time to finish the project now, make sure to skip down to the end of this notebook and shut down your endpoint. You can deploy it again when you come back.",
"_____no_output_____"
],
[
"## Step 7 (again): Use the model for the web app\n\n> **TODO:** This entire section and the next contain tasks for you to complete, mostly using the AWS console.\n\nSo far we have been accessing our model endpoint by constructing a predictor object which uses the endpoint and then just using the predictor object to perform inference. What if we wanted to create a web app which accessed our model? The way things are set up currently makes that not possible since in order to access a SageMaker endpoint the app would first have to authenticate with AWS using an IAM role which included access to SageMaker endpoints. However, there is an easier way! We just need to use some additional AWS services.\n\n<img src=\"Web App Diagram.svg\">\n\nThe diagram above gives an overview of how the various services will work together. On the far right is the model which we trained above and which is deployed using SageMaker. On the far left is our web app that collects a user's movie review, sends it off and expects a positive or negative sentiment in return.\n\nIn the middle is where some of the magic happens. We will construct a Lambda function, which you can think of as a straightforward Python function that can be executed whenever a specified event occurs. We will give this function permission to send and recieve data from a SageMaker endpoint.\n\nLastly, the method we will use to execute the Lambda function is a new endpoint that we will create using API Gateway. This endpoint will be a url that listens for data to be sent to it. Once it gets some data it will pass that data on to the Lambda function and then return whatever the Lambda function returns. Essentially it will act as an interface that lets our web app communicate with the Lambda function.\n\n### Setting up a Lambda function\n\nThe first thing we are going to do is set up a Lambda function. This Lambda function will be executed whenever our public API has data sent to it. When it is executed it will receive the data, perform any sort of processing that is required, send the data (the review) to the SageMaker endpoint we've created and then return the result.\n\n#### Part A: Create an IAM Role for the Lambda function\n\nSince we want the Lambda function to call a SageMaker endpoint, we need to make sure that it has permission to do so. To do this, we will construct a role that we can later give the Lambda function.\n\nUsing the AWS Console, navigate to the **IAM** page and click on **Roles**. Then, click on **Create role**. Make sure that the **AWS service** is the type of trusted entity selected and choose **Lambda** as the service that will use this role, then click **Next: Permissions**.\n\nIn the search box type `sagemaker` and select the check box next to the **AmazonSageMakerFullAccess** policy. Then, click on **Next: Review**.\n\nLastly, give this role a name. Make sure you use a name that you will remember later on, for example `LambdaSageMakerRole`. Then, click on **Create role**.\n\n#### Part B: Create a Lambda function\n\nNow it is time to actually create the Lambda function.\n\nUsing the AWS Console, navigate to the AWS Lambda page and click on **Create a function**. When you get to the next page, make sure that **Author from scratch** is selected. Now, name your Lambda function, using a name that you will remember later on, for example `sentiment_analysis_func`. Make sure that the **Python 3.6** runtime is selected and then choose the role that you created in the previous part. Then, click on **Create Function**.\n\nOn the next page you will see some information about the Lambda function you've just created. If you scroll down you should see an editor in which you can write the code that will be executed when your Lambda function is triggered. In our example, we will use the code below. \n\n```python\n# We need to use the low-level library to interact with SageMaker since the SageMaker API\n# is not available natively through Lambda.\nimport boto3\n\ndef lambda_handler(event, context):\n\n # The SageMaker runtime is what allows us to invoke the endpoint that we've created.\n runtime = boto3.Session().client('sagemaker-runtime')\n\n # Now we use the SageMaker runtime to invoke our endpoint, sending the review we were given\n response = runtime.invoke_endpoint(EndpointName = '**ENDPOINT NAME HERE**', # The name of the endpoint we created\n ContentType = 'text/plain', # The data format that is expected\n Body = event['body']) # The actual review\n\n # The response is an HTTP response whose body contains the result of our inference\n result = response['Body'].read().decode('utf-8')\n\n return {\n 'statusCode' : 200,\n 'headers' : { 'Content-Type' : 'text/plain', 'Access-Control-Allow-Origin' : '*' },\n 'body' : result\n }\n```\n\nOnce you have copy and pasted the code above into the Lambda code editor, replace the `**ENDPOINT NAME HERE**` portion with the name of the endpoint that we deployed earlier. You can determine the name of the endpoint using the code cell below.",
"_____no_output_____"
]
],
[
[
"predictor.endpoint",
"_____no_output_____"
]
],
[
[
"Once you have added the endpoint name to the Lambda function, click on **Save**. Your Lambda function is now up and running. Next we need to create a way for our web app to execute the Lambda function.\n\n### Setting up API Gateway\n\nNow that our Lambda function is set up, it is time to create a new API using API Gateway that will trigger the Lambda function we have just created.\n\nUsing AWS Console, navigate to **Amazon API Gateway** and then click on **Get started**.\n\nOn the next page, make sure that **New API** is selected and give the new api a name, for example, `sentiment_analysis_api`. Then, click on **Create API**.\n\nNow we have created an API, however it doesn't currently do anything. What we want it to do is to trigger the Lambda function that we created earlier.\n\nSelect the **Actions** dropdown menu and click **Create Method**. A new blank method will be created, select its dropdown menu and select **POST**, then click on the check mark beside it.\n\nFor the integration point, make sure that **Lambda Function** is selected and click on the **Use Lambda Proxy integration**. This option makes sure that the data that is sent to the API is then sent directly to the Lambda function with no processing. It also means that the return value must be a proper response object as it will also not be processed by API Gateway.\n\nType the name of the Lambda function you created earlier into the **Lambda Function** text entry box and then click on **Save**. Click on **OK** in the pop-up box that then appears, giving permission to API Gateway to invoke the Lambda function you created.\n\nThe last step in creating the API Gateway is to select the **Actions** dropdown and click on **Deploy API**. You will need to create a new Deployment stage and name it anything you like, for example `prod`.\n\nYou have now successfully set up a public API to access your SageMaker model. Make sure to copy or write down the URL provided to invoke your newly created public API as this will be needed in the next step. This URL can be found at the top of the page, highlighted in blue next to the text **Invoke URL**.",
"_____no_output_____"
],
[
"## Step 4: Deploying our web app\n\nNow that we have a publicly available API, we can start using it in a web app. For our purposes, we have provided a simple static html file which can make use of the public api you created earlier.\n\nIn the `website` folder there should be a file called `index.html`. Download the file to your computer and open that file up in a text editor of your choice. There should be a line which contains **\\*\\*REPLACE WITH PUBLIC API URL\\*\\***. Replace this string with the url that you wrote down in the last step and then save the file.\n\nNow, if you open `index.html` on your local computer, your browser will behave as a local web server and you can use the provided site to interact with your SageMaker model.\n\nIf you'd like to go further, you can host this html file anywhere you'd like, for example using github or hosting a static site on Amazon's S3. Once you have done this you can share the link with anyone you'd like and have them play with it too!\n\n> **Important Note** In order for the web app to communicate with the SageMaker endpoint, the endpoint has to actually be deployed and running. This means that you are paying for it. Make sure that the endpoint is running when you want to use the web app but that you shut it down when you don't need it, otherwise you will end up with a surprisingly large AWS bill.\n\n**TODO:** Make sure that you include the edited `index.html` file in your project submission.",
"_____no_output_____"
],
[
"Now that your web app is working, trying playing around with it and see how well it works.\n\n**Question**: Give an example of a review that you entered into your web app. What was the predicted sentiment of your example review?",
"_____no_output_____"
],
[
"**Answer:**\nReview: The special effects are magnificents. The ships looks so real and credible. Would see it thousand times.\nResult: Your review was POSITIVE!",
"_____no_output_____"
],
[
"### Delete the endpoint\n\nRemember to always shut down your endpoint if you are no longer using it. You are charged for the length of time that the endpoint is running so if you forget and leave it on you could end up with an unexpectedly large bill.",
"_____no_output_____"
]
],
[
[
"predictor.delete_endpoint()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
e7f7d0b510106d2fd29c6a651ac2d53762ff7311 | 7,510 | ipynb | Jupyter Notebook | poets_scraper.ipynb | Vihanga97/poets-engine | 7ce04ebed1b258046d46fb81929acdef93b15a68 | [
"Apache-2.0"
] | null | null | null | poets_scraper.ipynb | Vihanga97/poets-engine | 7ce04ebed1b258046d46fb81929acdef93b15a68 | [
"Apache-2.0"
] | null | null | null | poets_scraper.ipynb | Vihanga97/poets-engine | 7ce04ebed1b258046d46fb81929acdef93b15a68 | [
"Apache-2.0"
] | null | null | null | 33.67713 | 263 | 0.494407 | [
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"_____no_output_____"
],
[
"base_path = '/content/drive/My Drive/DM Challenge'\nsave_path = base_path + '/poets_with_imgs.csv'",
"_____no_output_____"
],
[
"import requests\nsource = 'http://famouspoetsandpoems.com'\npage = requests.get(source+'/poets.html')\npage",
"_____no_output_____"
],
[
"from bs4 import BeautifulSoup\nmain_soup = BeautifulSoup(page.content, 'html.parser')\npoets = main_soup.find('table', cellspacing='0').find('table').find_all('td', valign='top')\nprint (poets)",
"_____no_output_____"
],
[
"!pip install deep-translator\nfrom deep_translator import GoogleTranslator",
"_____no_output_____"
],
[
"'''fields:\n1. name\n2. birth_year\n3. death_year\n4. bio\n5. poem\n6. quote\n7. category (school)\n8. similar poets\n9. image\n'''",
"_____no_output_____"
],
[
"from bs4 import NavigableString, Tag\nimport pandas as pd\nimport re\npoets_df = pd.DataFrame(columns=['name', 'birth_year', 'death_year', 'categories', 'bio', 'poem', 'quote', 'similar_poets', 'image'])\n\nfor i in range(1000):\n try:\n td = poets[i]\n\n url = td.find('a')['href']\n text = td.text.split('(')\n\n name = text[0].strip()\n name_sin = GoogleTranslator(source='en', target='si').translate(name)\n birth_year = text[2].split('-')[0].strip()\n if not birth_year.isnumeric():\n birth_year = GoogleTranslator(source='en', target='si').translate(birth_year)\n death_year = text[2].split('-')[1].strip(')').strip()\n if not death_year.isnumeric():\n death_year = GoogleTranslator(source='en', target='si').translate(death_year)\n \n bio_page = requests.get('https://poets.org/poet/'+'-'.join(name.split()))\n bio_soup = BeautifulSoup(bio_page.content, 'html.parser')\n bio = bio_soup.find('div', class_='poet__body-content').text\n bio_sin = GoogleTranslator(source='en', target='si').translate(bio)\n\n try:\n img_page = requests.get(source+url+'/photo')\n img_soup = BeautifulSoup(img_page.content, 'html.parser')\n img = source+img_soup.find('img', alt=name)['src']\n except:\n img = 'http://famouspoetsandpoems.com/images/_no_photo.gif'\n\n categories = [c.text.strip() for c in bio_soup.find_all('div', class_='school')]\n categories_sin = ', '.join([GoogleTranslator(source='en', target='si').translate(c) for c in categories])\n \n similar_poets = [p.text.strip() for p in bio_soup.find_all('div', class_='poet__sidebar-related-poets-poet')]\n similar_poets_sin = ', '.join([GoogleTranslator(source='en', target='si').translate(p) for p in similar_poets])\n\n poems_page = requests.get(source+url+'/poems')\n poems_soup = BeautifulSoup(poems_page.content, 'html.parser')\n poem_url = poems_soup.find_all('table', width='436')[1].find_all('a')[0]['href']\n poem_page = requests.get(source+poem_url)\n poem_soup = BeautifulSoup(poem_page.content, 'html.parser')\n poem = (str(poem_soup.find('div', style='padding-left:14px;padding-top:20px;font-family:Arial;font-size:13px;')).strip('<div style=\"padding-left:14px;padding-top:20px;font-family:Arial;font-size:13px;\">').strip('</div>').strip().split('<br/>'))\n poem_sin = '\\n'.join([GoogleTranslator(source='en', target='si').translate(i) for i in poem if i!=''])\n \n quotes_page = requests.get(source+url+'/quotes')\n quotes_soup = BeautifulSoup(quotes_page.content, 'html.parser')\n quotes = quotes_soup.find_all('div', style='padding-right:15px;padding-left:16px;padding-bottom:20px;')\n if len(quotes)>0:\n quote = quotes[0].text\n quote_sin = GoogleTranslator(source='en', target='si').translate(quote)\n else:\n quote = None\n quote_sin = None\n \n if re.search('[a-zA-Z]', name_sin):\n print (name_sin)\n continue\n \n poets_df.loc[i] = [name_sin, birth_year, death_year, categories_sin, bio_sin, poem_sin, quote_sin, similar_poets_sin, img]\n print (i, name_sin)\n\n except:\n continue\n ",
"_____no_output_____"
],
[
"poets_df",
"_____no_output_____"
],
[
"poets_df.to_csv(save_path)",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f7d2fb07a99ffb5e58d9bdab361a715b711666 | 45,743 | ipynb | Jupyter Notebook | Informatics/Deep Learning/TensorFlow - deeplearning.ai/2. CNN/utf-8''Exercise_4_Multi_class_classifier_Question-FINAL.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
] | null | null | null | Informatics/Deep Learning/TensorFlow - deeplearning.ai/2. CNN/utf-8''Exercise_4_Multi_class_classifier_Question-FINAL.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
] | null | null | null | Informatics/Deep Learning/TensorFlow - deeplearning.ai/2. CNN/utf-8''Exercise_4_Multi_class_classifier_Question-FINAL.ipynb | MarcosSalib/Cocktail_MOOC | 46279c2ec642554537c639702ed8e540ea49afdf | [
"MIT"
] | null | null | null | 135.33432 | 18,120 | 0.866122 | [
[
[
"# ATTENTION: Please do not alter any of the provided code in the exercise. Only add your own code where indicated\n# ATTENTION: Please do not add or remove any cells in the exercise. The grader will check specific cells based on the cell position.\n# ATTENTION: Please use the provided epoch values when training.\n\nimport csv\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\nfrom os import getcwd",
"_____no_output_____"
],
[
"def get_data(filename):\n # You will need to write code that will read the file passed\n # into this function. The first line contains the column headers\n # so you should ignore it\n # Each successive line contians 785 comma separated values between 0 and 255\n # The first value is the label\n # The rest are the pixel values for that picture\n # The function will return 2 np.array types. One with all the labels\n # One with all the images\n #\n # Tips: \n # If you read a full line (as 'row') then row[0] has the label\n # and row[1:785] has the 784 pixel values\n # Take a look at np.array_split to turn the 784 pixels into 28x28\n # You are reading in strings, but need the values to be floats\n # Check out np.array().astype for a conversion\n with open(filename) as training_file:\n L_lst, I_lst = [], []\n for line in training_file.readlines()[1:]:\n line = line.split(',')\n I_lst.append(np.array_split(np.array(line[1:]), 28))\n L_lst.append(line[0])\n \n labels = np.array(L_lst).astype('float')\n images = np.array(I_lst).astype('float')\n return images, labels\n\npath_sign_mnist_train = f\"{getcwd()}/../tmp2/sign_mnist_train.csv\"\npath_sign_mnist_test = f\"{getcwd()}/../tmp2/sign_mnist_test.csv\"\ntraining_images, training_labels = get_data(path_sign_mnist_train)\ntesting_images, testing_labels = get_data(path_sign_mnist_test)\n\n# Keep these\nprint(training_images.shape)\nprint(training_labels.shape)\nprint(testing_images.shape)\nprint(testing_labels.shape)\n\n# Their output should be:\n# (27455, 28, 28)\n# (27455,)\n# (7172, 28, 28)\n# (7172,)",
"(27455, 28, 28)\n(27455,)\n(7172, 28, 28)\n(7172,)\n"
],
[
"# In this section you will have to add another dimension to the data\n# So, for example, if your array is (10000, 28, 28)\n# You will need to make it (10000, 28, 28, 1)\n# Hint: np.expand_dims\n\ntraining_images = np.expand_dims(training_images, axis=3)\ntesting_images = np.expand_dims(testing_images, axis=3)\n\n# Create an ImageDataGenerator and do Image Augmentation\ntrain_datagen = ImageDataGenerator(rescale=1/255,\n rotation_range = 40,\n width_shift_range = 0.2,\n height_shift_range = 0.2,\n shear_range = 0.2,\n zoom_range = 0.2,\n horizontal_flip = True\n )\n\nvalidation_datagen = ImageDataGenerator(rescale=1/255)\n \n# Keep These\nprint(training_images.shape)\nprint(testing_images.shape)\n \n# Their output should be:\n# (27455, 28, 28, 1)\n# (7172, 28, 28, 1)",
"(27455, 28, 28, 1)\n(7172, 28, 28, 1)\n"
],
[
"# Define the model\n# Use no more than 2 Conv2D and 2 MaxPooling2D\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Conv2D(64, (3,3), activation='relu', input_shape=(28,28,1)),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Conv2D(64, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(512, activation='relu'),\n tf.keras.layers.Dense(26, activation='softmax')\n ])\n\n# generators\ntrain_generator = train_datagen.flow(training_images,\n training_labels,\n batch_size=32\n )\n\nvalidation_generator = validation_datagen.flow(testing_images,\n testing_labels,\n batch_size=32\n )\n\n# Compile Model. \nmodel.compile(optimizer='rmsprop', loss='sparse_categorical_crossentropy', metrics=['accuracy'])\n\n# Train the Model\nhistory = model.fit_generator(train_generator, validation_data=validation_generator, epochs=2)\n\nmodel.evaluate(testing_images, testing_labels, verbose=0)",
"Epoch 1/2\n858/858 [==============================] - 81s 94ms/step - loss: 2.6034 - accuracy: 0.2069 - val_loss: 1.6327 - val_accuracy: 0.4520\nEpoch 2/2\n858/858 [==============================] - 79s 92ms/step - loss: 1.7157 - accuracy: 0.4555 - val_loss: 0.8937 - val_accuracy: 0.6817\n"
],
[
"# Plot the chart for accuracy and loss on both training and validation\n%matplotlib inline\nimport matplotlib.pyplot as plt\nacc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(len(acc))\n\nplt.plot(epochs, acc, 'r', label='Training accuracy')\nplt.plot(epochs, val_acc, 'b', label='Validation accuracy')\nplt.title('Training and validation accuracy')\nplt.legend()\nplt.figure()\n\nplt.plot(epochs, loss, 'r', label='Training Loss')\nplt.plot(epochs, val_loss, 'b', label='Validation Loss')\nplt.title('Training and validation loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Submission Instructions",
"_____no_output_____"
]
],
[
[
"# Now click the 'Submit Assignment' button above.",
"_____no_output_____"
]
],
[
[
"# When you're done or would like to take a break, please run the two cells below to save your work and close the Notebook. This will free up resources for your fellow learners. ",
"_____no_output_____"
]
],
[
[
"%%javascript\n<!-- Save the notebook -->\nIPython.notebook.save_checkpoint();",
"_____no_output_____"
],
[
"%%javascript\nIPython.notebook.session.delete();\nwindow.onbeforeunload = null\nsetTimeout(function() { window.close(); }, 1000);",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e7f7dfc030a676157ceec0aa71e4e2225194e11e | 153,475 | ipynb | Jupyter Notebook | tests/Demo_Microstripline.ipynb | sarang-IITKgp/scikit-microwave-design | a8567c2d40eebde93af5989c43d6e3008167e137 | [
"BSD-3-Clause"
] | null | null | null | tests/Demo_Microstripline.ipynb | sarang-IITKgp/scikit-microwave-design | a8567c2d40eebde93af5989c43d6e3008167e137 | [
"BSD-3-Clause"
] | null | null | null | tests/Demo_Microstripline.ipynb | sarang-IITKgp/scikit-microwave-design | a8567c2d40eebde93af5989c43d6e3008167e137 | [
"BSD-3-Clause"
] | null | null | null | 275.5386 | 71,540 | 0.922209 | [
[
[
"# `Microstripline` object in `structure` module. ",
"_____no_output_____"
],
[
"## Analytical modeling of Microstripline in Scikit-microwave-design.\n\nIn this file, we show how `scikit-microwave-design` library can be used to implement and analyze basic microstrip line structures. ",
"_____no_output_____"
],
[
"### Defining a microstrip line in `skmd`\n\nThere are two ways in which we can define a microstrip line (msl). \n1. We define the msl width. And then compute its characteristic impedance in the analytical formulation.\n2. We define the characteristic impedance of the msl, and then compute the physical dimension that gives the desired characteristic impedance. \n\nIn both the methods, the effective dielectric constant becomes a function of the msl width - in addition to the substrate thickness and substrate dielectric constant. And since the effective dielectric constant is one of the determining factors in the propagation constant, the propagation constant is also dependent on the characteristics impedance indirectly. \n\n\nIn `skmd` you can define a microstrip line by both the methods. ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport skmd as md\n\nimport matplotlib.pyplot as plt\n\n### Define frequency \npts_freq = 1000\nfreq = np.linspace(1e9,3e9,pts_freq)\nomega = 2*np.pi*freq\n\n\n\n#### define substrate\nepsilon_r = 10.8 # dielectric constant or the effective dielectric constant\nh_subs = 1.27*md.MILLI # meters. \n\n",
"_____no_output_____"
]
],
[
[
"### 1. Defining msl with characteristic impedance. ",
"_____no_output_____"
]
],
[
[
"msl1 = md.structure.Microstripline(er=epsilon_r,h=h_subs,Z0=93,text_tag='Line-abc')\n",
" ============ \n Defining Line-abc\nLine-abc defined with Z0\n==============\n"
]
],
[
[
"`Microstripline` object is defined in the `structure` module of the `skmd` librayr. With the above command, we have defined a _msl_ by giving the characteristic impedance $Z_0$ with a text identifier 'Line-abc'. The library will compute the required line width to achieve the desired characteristic impedance for the given values of substrate thickness and dielectric constant. Therefore this code can also be used to get the design parameters for desired specifications. \n\nThe computed width of the microstrip line is stored in the attribute `w` of the `Microstripline` object, and can be displayed by print(msl1.w). The units are in meters. You can also print all the specifications by `msl1.print_specs()`.",
"_____no_output_____"
]
],
[
[
"msl1.print_specs()",
"--------- Line-abc Specifications---------\n-----Substrate-----\nEpsilon_r 10.8\nsubstrate thickness 0.00127\n-------------------\nline width W= 0.00019296747453793648\nCharacteristics impedance= 93\nLength of the line = 1\nEffective dielectric constant er_eff = 6.555924417931664\nFrequency defined ?: False\n-------------------\n"
]
],
[
[
"### 2. Defining the msl by width. \nWe can also define the msl by giving the width at the time of definition. The characteristic impedance will be computed by the code in this case. ",
"_____no_output_____"
]
],
[
[
"msl2 = md.structure.Microstripline(er=epsilon_r,h=h_subs,w = 1.1*md.MILLI,text_tag='Line-xyz')\nmsl2.print_specs()",
" ============ \n Defining Line-xyz\nLine-xyz defined with width.\n==============\n--------- Line-xyz Specifications---------\n-----Substrate-----\nEpsilon_r 10.8\nsubstrate thickness 0.00127\n-------------------\nline width W= 0.0011\nCharacteristics impedance= 50.466917262179905\nLength of the line = 1\nEffective dielectric constant er_eff = 7.12610312997174\nFrequency defined ?: False\n-------------------\n"
]
],
[
[
"At least either width or characteristic impedance must be defined, else an error will be generated. \nIf both characteristic impedance and width are given, than width is used in the definitiona and characertistic impedance is computed.",
"_____no_output_____"
],
[
"### Defining frequency range and network parameters for the microstrip line. \n\nWe can also give the frequency values at which we want to perform the analysis. When frequency values are given, the corresponding two-port microwave `network` object also gets defined for the microstrip transmission line. If the length of the transmission line is not defined, a default length of 1 meter is considered. \n\nThe frequency can be defined at the time of `Microstripline` definition, or can be added later using the object function `fun_add_frequency(omega)`. However, it is recommended to be defined during the initial object definition itself. ",
"_____no_output_____"
]
],
[
[
"msl3 = md.structure.Microstripline(er=epsilon_r,h=h_subs,w = 1.1*md.MILLI,omega = omega,text_tag='Line-with-frequency')\nmsl3.print_specs()\n# msl.",
" ============ \n Defining Line-with-frequency\nLine-with-frequency defined with width.\nFrequency given. Network defined.\n==============\n--------- Line-with-frequency Specifications---------\n-----Substrate-----\nEpsilon_r 10.8\nsubstrate thickness 0.00127\n-------------------\nline width W= 0.0011\nCharacteristics impedance= 50.466917262179905\nLength of the line = 1\nEffective dielectric constant er_eff = 7.12610312997174\nFrequency defined ?: True\n-------------------\n"
],
[
"msl2.print_specs()\nmsl2.fun_add_frequency(omega)\n",
"--------- Line-xyz Specifications---------\n-----Substrate-----\nEpsilon_r 10.8\nsubstrate thickness 0.00127\n-------------------\nline width W= 0.0011\nCharacteristics impedance= 50.466917262179905\nLength of the line = 1\nEffective dielectric constant er_eff = 7.12610312997174\nFrequency defined ?: False\n-------------------\nFrequency added (override old values). Network defined.\n"
]
],
[
[
"# Microstrip-line filters. \n\nDesigning microstrip line filters and their analytical computation becomes very simple in `scikit-microwave-design` library. Since a microwave network object is created for a microstrip-line section, it becomes a matter of few lines of coding to implement and test filters. In addition excellent plotting features available in the `plot` module of the `skmd` library make visualization of the filer response very easy. ",
"_____no_output_____"
],
[
"## Open quarter-stub filter. \n\nLet us design a T-shaped open stub filter, which acts as notch filter at quarter wavelength. \n\nIf the resonant frequency is $f_0$, then the length of the open-stub corresponding to the resonant frequency will be given by,\n\n\n$l_{stub} = \\frac{\\lambda_0}{4}$\n\nwhere, \n$\\lambda_0 = \\frac{c}{f_0\\sqrt{\\epsilon_{eff}}}$\n\nHere, note that $\\epsilon_{eff}$ is the effective dielectric constant of the substrate for a given width. If the characteristic impedance - and the corresponding width - changes then the effective dielectric constant, and therefore the effective electrical length of the stub will also change. Using this library it is very easy to take care of these issues. \n\n\nFor example, the library can easily compute for us the required stub-length, for a desired combination of characteristic impedance and resonant frequency. \n\nThe following codes shows the implementation of a simple quarter-stub filter with for different values of characteristic impedances, the corresponding stub widths, and the required stub lengths to keep the resonant frequency fixed. \n\n\n\n",
"_____no_output_____"
]
],
[
[
"f0 = 1.5*md.GIGA\n\nomega0 = md.f2omega(f0)\n\nmsl_Tx1 = md.structure.Microstripline(er=epsilon_r,h=h_subs,w=1.1*md.MILLI,l=5*md.MILLI,text_tag='Left-line',omega=omega)\nmsl_Tx2 = md.structure.Microstripline(er=epsilon_r,h=h_subs,w=1.1*md.MILLI,l=5*md.MILLI,text_tag='Right-line',omega=omega)\n\nmsl_Tx1.print_specs()\n\nw_stub = 1*md.MILLI\n\nlambda_g_stub = md.structure.Microstripline(er=epsilon_r,h=h_subs,w=w_stub,text_tag='stub-resonant-length',omega=omega0).lambda_g\n\n\n\nw_stub = 1*md.MILLI\n\nL_stub = lambda_g_stub/4\n\nmsl_stub = md.structure.Microstripline(er=epsilon_r,h=h_subs,w=w_stub,l=L_stub,text_tag='stub',omega=omega)\n\ndef define_NW_for_stub(msl_stub,ZL_stub):\n\tY_stub = msl_stub.NW.input_admittance(1/ZL_stub)\n\tNW_stub = md.network.from_shunt_Y(Y_stub)\n\treturn NW_stub\n\t\n\nZL_stub = md.OPEN\nNW_stub = md.network.from_shunt_Y(msl_stub.NW.input_admittance(1/ZL_stub))\n\n\n\n\nNW_filter = msl_Tx1.NW*NW_stub*msl_Tx2.NW",
" ============ \n Defining Left-line\nLeft-line defined with width.\nFrequency given. Network defined.\n==============\n ============ \n Defining Right-line\nRight-line defined with width.\nFrequency given. Network defined.\n==============\n--------- Left-line Specifications---------\n-----Substrate-----\nEpsilon_r 10.8\nsubstrate thickness 0.00127\n-------------------\nline width W= 0.0011\nCharacteristics impedance= 50.466917262179905\nLength of the line = 0.005\nEffective dielectric constant er_eff = 7.12610312997174\nFrequency defined ?: True\n-------------------\n ============ \n Defining stub-resonant-length\nstub-resonant-length defined with width.\nFrequency given. Network defined.\n==============\n ============ \n Defining stub\nstub defined with width.\nFrequency given. Network defined.\n==============\n"
],
[
"## Plot commands\n\nfig1 = plt.figure('LPF')\nax1_f1 = fig1.add_subplot(111)\nax1_f1.plot(omega/(2*np.pi*md.GIGA),np.abs(NW_filter.S11),linewidth='3',label='$|S_{11}|$')\nax1_f1.plot(omega/(2*np.pi*md.GIGA),np.abs(NW_filter.S21),linewidth='3',label='$|S_{21}|$')\nax1_f1.grid(1)\n\nax1_f1.legend()",
"_____no_output_____"
],
[
"fig2 = plt.figure('LPF-mag-phase')\nax1_f2 = fig2.add_subplot(311)\nax1_cmap_f2 = fig2.add_axes([0.92, 0.1, 0.02, 0.7])\nax2_f2 = fig2.add_subplot(313)\n\nmd.plot.plot_colored_line(md.omega2f(omega)/md.GIGA,np.abs(NW_filter.S11),np.angle(NW_filter.S11)*180/np.pi,ax=ax1_f2,color_axis = ax1_cmap_f2)\nmd.plot.plot_colored_line(md.omega2f(omega)/md.GIGA,np.abs(NW_filter.S21),np.angle(NW_filter.S21)*180/np.pi,ax=ax2_f2)\n\nax1_f2.grid(1)\nax2_f2.grid(1)",
"_____no_output_____"
],
[
"fig3 = plt.figure('LPF-mag-phase-dB')\nax1_f3 = fig3.add_subplot(311)\nax1_cmap_f3 = fig3.add_axes([0.92, 0.5, 0.02, 0.3])\nax2_f3 = fig3.add_subplot(313)\nax2_cmap_f3 = fig3.add_axes([0.92, 0.1, 0.02, 0.3])\n\nmd.plot.plot_colored_line(md.omega2f(omega)/md.GIGA,md.dB_mag(NW_filter.S21),np.angle(NW_filter.S21)*180/np.pi,ax=ax1_f3,color_axis = ax1_cmap_f3)\nmd.plot.plot_colored_line(md.omega2f(omega)/md.GIGA,np.rad2deg(np.angle(NW_filter.S21)),md.dB_mag(NW_filter.S21),ax=ax2_f3,color_axis = ax2_cmap_f3)\n\nax1_f3.grid(1)\nax2_f3.grid(1)",
"_____no_output_____"
],
[
"fig4 = plt.figure('Smith-chart')\n\n\nax1_f4 = md.plot.plot_smith_chart(md.omega2f(omega)/md.GIGA,NW_filter.S21,fig4,use_colormap='inferno',linewidth=10)\n# ax1_f4 = md.plot.plot_smith_chart(md.omega2f(omega)/md.GIGA,NW_filter.S11,fig4,use_colormap='inferno',linewidth=10)\n\n# snap_cursor_2 = md.plot.SnaptoCursor_polar(ax1_f4,md.omega2f(omega), NW_filter.S21)\n# fig4.canvas.mpl_connect('motion_notify_event', snap_cursor_2.mouse_move)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7f7f010b01d60501a6692660f9e79955b854ff2 | 211,785 | ipynb | Jupyter Notebook | examples/tutorials/07_sampler.ipynb | abrikoseg/batchflow | f1060f452b9407477ac61cea2a658792deca29a6 | [
"Apache-2.0"
] | 101 | 2017-06-05T07:33:54.000Z | 2018-10-28T04:55:23.000Z | examples/tutorials/07_sampler.ipynb | abrikoseg/batchflow | f1060f452b9407477ac61cea2a658792deca29a6 | [
"Apache-2.0"
] | 243 | 2018-11-29T02:03:55.000Z | 2022-02-21T08:28:29.000Z | examples/tutorials/07_sampler.ipynb | abrikoseg/batchflow | f1060f452b9407477ac61cea2a658792deca29a6 | [
"Apache-2.0"
] | 35 | 2019-01-29T14:26:14.000Z | 2021-12-30T01:39:02.000Z | 298.70945 | 41,400 | 0.931294 | [
[
[
"# `Sampler`",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append('../..')\nimport matplotlib.pyplot as plt\nimport numpy as np\n%matplotlib inline\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"## Intro",
"_____no_output_____"
],
[
"Welcome! In this section you'll learn about `Sampler`-class. Instances of `Sampler` can be used for flexible sampling of multivariate distributions.\n\nTo begin with, `Sampler` gives rise to several building-blocks classes such as\n- `NumpySampler`, or `NS`\n- `ScipySampler` - `SS`\n\nWhat's more, `Sampler` incorporates a set of operations on `Sampler`-instances, among which are\n- \"`|`\" for building a mixture of two samplers: `s = s1 | s2`\n- \"`&`\" for setting a mixture-weight of a sampler: `s = 0.6 & s1 | 0.4 & s2`\n- \" `truncate`\" for truncating the support of underlying sampler's distribution: `s.truncate(high=[1.0, 1.5])`\n- ..all arithmetic operations: `s = s1 + s2` or `s = s1 + 0.5`\n\nThese operations can be used for combining building-blocks samplers into complex multivariate-samplers, just like that:",
"_____no_output_____"
]
],
[
[
"from batchflow import NumpySampler as NS\n\n# truncated normal and uniform\nns1 = NS('n', dim=2).truncate(2.0, 0.8, lambda m: np.sum(np.abs(m), axis=1)) + 4\nns2 = 2 * NS('u', dim=2).truncate(1, expr=lambda m: np.sum(m, axis=1)) - (1, 1)\nns3 = NS('n', dim=2).truncate(1.5, expr=lambda m: np.sum(np.square(m), axis=1)) + (4, 0)\nns4 = ((NS('n', dim=2).truncate(2.5, expr=lambda m: np.sum(np.square(m), axis=1)) * 4)\n .apply(lambda m: m.astype(np.int)) / 4 + (0, 3))\n\n# a mixture of all four\nns = 0.4 & ns1 | 0.2 & ns2 | 0.39 & ns3 | 0.01 & ns4",
"_____no_output_____"
],
[
"# take a look at the heatmap of our sampler:\nh = np.histogramdd(ns.sample(int(1e6)), bins=100, normed=True)\nplt.imshow(h[0])",
"_____no_output_____"
]
],
[
[
"## Building `Samplers`",
"_____no_output_____"
],
[
"### 1. Numpy, Scipy, TensorFlow - `Samplers`",
"_____no_output_____"
],
[
"To build a `NumpySampler`(`NS`) you need to specify a name of distribution from `numpy.random` (or its [alias](https://github.com/analysiscenter/batchflow/blob/master/batchflow/sampler.py#L15)) and the number of independent dimensions:",
"_____no_output_____"
]
],
[
[
"from batchflow import NumpySampler as NS\nns = NS('n', dim=2)",
"_____no_output_____"
]
],
[
[
"take a look at a sample generated by our sampler:",
"_____no_output_____"
]
],
[
[
"smp = ns.sample(size=200)",
"_____no_output_____"
],
[
"plt.scatter(*np.transpose(smp))",
"_____no_output_____"
]
],
[
[
"The same goes for `ScipySampler` based on `scipy.stats`-distributions, or `SS` (\"mvn\" stands for multivariate-normal):",
"_____no_output_____"
]
],
[
[
"from batchflow import ScipySampler as SS\nss = SS('mvn', mean=[0, 0], cov=[[2, 1], [1, 2]]) # note also that you can pass the same params as in\nsmp = ss.sample(2000) # scipy.sample.multivariate_normal, such as `mean` and `cov` \nplt.scatter(*np.transpose(smp))",
"_____no_output_____"
]
],
[
[
"### 2. `HistoSampler` as an estimate of a distribution generating a cloud of points",
"_____no_output_____"
],
[
"`HistoSampler`, or `HS` can be used for building samplers, with underlying distributions given by a histogram. You can either pass a `np.histogram`-output into the initialization of `HS`",
"_____no_output_____"
]
],
[
[
"from batchflow import HistoSampler as HS\nhisto = np.histogramdd(ss.sample(1000000))\nhs = HS(histo)\nplt.scatter(*np.transpose(hs.sample(150)))",
"_____no_output_____"
]
],
[
[
"...or you can specify empty bins and estimate its weights using a method `HS.update` and a cloud of points:",
"_____no_output_____"
]
],
[
[
"hs = HS(edges=2 * [np.linspace(-4, 4)])\nhs.update(ss.sample(1000000))\nplt.imshow(hs.bins, interpolation='bilinear')",
"_____no_output_____"
]
],
[
[
"### 3. Algebra of `Samplers`; operations on `Samplers`",
"_____no_output_____"
],
[
"`Sampler`-instances support artithmetic operations (`+`, `*`, `-`,...). Arithmetics works on either\n* (`Sampler`, `Sampler`) - pair\n* (`Sampler`, `array-like`) - pair",
"_____no_output_____"
]
],
[
[
"# blur using \"+\"\nu = NS('u', dim=2)\nnoise = NS('n', dim=2)\nblurred = u + noise * 0.2 # decrease the magnitude of the noise\nboth = blurred | u + (2, 2)",
"_____no_output_____"
],
[
"plt.imshow(np.histogramdd(both.sample(1000000), bins=100)[0])",
"_____no_output_____"
]
],
[
[
"You may also want to truncate a sampler's distribution so that sampling points belong to a specific region. The common use-case is to sample normal points inside a box.\n\n..or, inside a ring: ",
"_____no_output_____"
]
],
[
[
"n = NS('n', dim=2).truncate(3, 0.3, expr=lambda m: np.sum(m**2, axis=1))\nplt.imshow(np.histogramdd(n.sample(1000000), bins=100)[0])",
"_____no_output_____"
]
],
[
[
"Not infrequently you need to obtain \"normal\" sample in integers. For this you can use `Sampler.apply` method:",
"_____no_output_____"
]
],
[
[
"n = (4 * NS('n', dim=2)).apply(lambda m: m.astype(np.int)).truncate([6, 6], [-6, -6])\nplt.imshow(np.histogramdd(n.sample(1000000), bins=100)[0])",
"_____no_output_____"
]
],
[
[
"Note that `Sampler.apply`-method allows you to add an arbitrary transformation to a sampler. For instance, [Box-Muller](https://en.wikipedia.org/wiki/Box–Muller_transform) transform:",
"_____no_output_____"
]
],
[
[
"bm = lambda vec2: np.sqrt(-2 * np.log(vec2[:, 0:1])) * np.concatenate([np.cos(2 * np.pi * vec2[:, 1:2]),\n np.sin(2 * np.pi * vec2[:, 1:2])], axis=1)\nn = NS('u', dim=2).apply(bm)",
"_____no_output_____"
],
[
"plt.imshow(np.histogramdd(n.sample(1000000), bins=100)[0])",
"_____no_output_____"
]
],
[
[
"Another useful thing is coordinate stacking (\"&\" stands for multiplication of distribution functions):",
"_____no_output_____"
]
],
[
[
"n, u = NS('n'), SS('u') # initialize one-dimensional notrmal and uniform samplers\ns = n & u # stack them together\ns.sample(3)",
"_____no_output_____"
]
],
[
[
"### 4. Alltogether",
"_____no_output_____"
]
],
[
[
"ns1 = NS('n', dim=2).truncate(2.0, 0.8, lambda m: np.sum(np.abs(m), axis=1)) + 4\nns2 = 2 * NS('u', dim=2).truncate(1, expr=lambda m: np.sum(m, axis=1)) - (1, 1)\nns3 = NS('n', dim=2).truncate(1.5, expr=lambda m: np.sum(np.square(m), axis=1)) + (4, 0)\nns4 = ((NS('n', dim=2).truncate(2.5, expr=lambda m: np.sum(np.square(m), axis=1)) * 4)\n .apply(lambda m: m.astype(np.int)) / 4 + (0, 3))\nns = 0.4 & ns1 | 0.2 & ns2 | 0.39 & ns3 | 0.01 & ns4",
"_____no_output_____"
],
[
"plt.imshow(np.histogramdd(ns.sample(int(1e6)), bins=100, normed=True)[0])",
"_____no_output_____"
]
],
[
[
"### 5. Notes",
"_____no_output_____"
],
[
"* parallellism\n\n`Sampler`-objects allow for parallelism with `mutliprocessing`. Just make sure to use explicitly defined functions (not `lambda`s) when running `Sampler.apply` or `Sampler.truncate`:",
"_____no_output_____"
]
],
[
[
"def transform(m):\n return np.sum(np.abs(m), axis=1)\nns = NS('n', dim=2).truncate(2.0, 0.8, expr=transform) + 4",
"_____no_output_____"
],
[
"from multiprocessing import Pool",
"_____no_output_____"
],
[
"def test_func(s):\n return s.sample(2)\n\np = Pool(5)\n\np.map(test_func, [ns, ns, ns])",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
e7f7ffbb57e126d82a535a00bd739043dd75672e | 815 | ipynb | Jupyter Notebook | docs/example.ipynb | AraiYuno/pyspark-delta-utility | 1e9fd792f52fab17cb7059451e242e7675b481ea | [
"MIT"
] | 1 | 2022-03-26T07:06:06.000Z | 2022-03-26T07:06:06.000Z | docs/example.ipynb | AraiYuno/pyspark-delta-utility | 1e9fd792f52fab17cb7059451e242e7675b481ea | [
"MIT"
] | null | null | null | docs/example.ipynb | AraiYuno/pyspark-delta-utility | 1e9fd792f52fab17cb7059451e242e7675b481ea | [
"MIT"
] | null | null | null | 18.111111 | 50 | 0.53865 | [
[
[
"# Example usage\n\nTo use `pyspark_delta_utility` in a project:",
"_____no_output_____"
]
],
[
[
"import pyspark_delta_utility\n\nprint(pyspark_delta_utility.__version__)",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
]
] |
e7f80d952351131a7f6505124718add82487e835 | 149,441 | ipynb | Jupyter Notebook | Length_Study/MLP_01.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
] | null | null | null | Length_Study/MLP_01.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
] | 4 | 2020-03-24T18:05:09.000Z | 2020-12-22T17:42:54.000Z | Length_Study/MLP_01.ipynb | ShepherdCode/ShepherdML | fd8d71c63f7bd788ea0052294d93e43246254a12 | [
"MIT"
] | null | null | null | 113.041604 | 27,440 | 0.722305 | [
[
[
"# MLP train on K=2,3,4\nTrain a generic MLP as binary classifier of protein-coding/non-coding RNA.\nSet aside a 20% test set, stratified shuffle by length.\nOn the non-test, use random shuffle \nto partition train and validation sets.\nTrain on 80% and valuate on 20% validation set.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport tensorflow as tf\nfrom tensorflow import keras\ntf.keras.backend.set_floatx('float64')",
"_____no_output_____"
]
],
[
[
"## K-mer frequency, K=2",
"_____no_output_____"
]
],
[
[
"def read_features(nc_file,pc_file):\n nc = pd.read_csv (nc_file)\n pc = pd.read_csv (pc_file)\n nc['class']=0\n pc['class']=1\n rna_mer=pd.concat((nc,pc),axis=0)\n return rna_mer\nrna_mer = read_features('ncRNA.2mer.features.csv','pcRNA.2mer.features.csv')\nrna_mer",
"_____no_output_____"
],
[
"# Split into train/test stratified by sequence length.\ndef sizebin(df):\n return pd.cut(df[\"seqlen\"],\n bins=[0,1000,2000,4000,8000,16000,np.inf],\n labels=[0,1,2,3,4,5])\ndef make_train_test(data):\n bin_labels= sizebin(data)\n from sklearn.model_selection import StratifiedShuffleSplit\n splitter = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=37863)\n # split(x,y) expects that y is the labels. \n # Trick: Instead of y, give it it the bin labels that we generated.\n for train_index,test_index in splitter.split(data,bin_labels):\n train_set = rna_mer.iloc[train_index]\n test_set = rna_mer.iloc[test_index]\n return (train_set,test_set)\n(train_set,test_set)=make_train_test(rna_mer)\nprint(\"train,test\")\ntrain_set.shape,test_set.shape",
"train,test\n"
],
[
"def prepare_test_set(test_set):\n y_test= test_set[['class']].copy()\n X_test= test_set.div(test_set['seqlen'],axis=0)\n X_test= X_test.drop(columns=['class','seqnum','seqlen'])\n return (X_test,y_test)\n(X_test,y_test)=prepare_test_set(test_set)\n\ndef prepare_train_set(train_set):\n y_train_all= train_set[['class']].copy()\n X_train_all= train_set.div(train_set['seqlen'],axis=0)\n X_train_all= X_train_all.drop(columns=['class','seqnum','seqlen'])\n\n from sklearn.model_selection import ShuffleSplit\n splitter = ShuffleSplit(n_splits=1, test_size=0.2, random_state=37863)\n for train_index,valid_index in splitter.split(X_train_all):\n X_train=X_train_all.iloc[train_index]\n y_train=y_train_all.iloc[train_index]\n X_valid=X_train_all.iloc[valid_index]\n y_valid=y_train_all.iloc[valid_index]\n \n return (X_train,y_train,X_valid,y_valid)\n\n(X_train,y_train,X_valid,y_valid)=prepare_train_set(train_set)\nprint(\"train\")\nprint(X_train.shape,y_train.shape)\nprint(\"validate\")\nprint(X_valid.shape,y_valid.shape)",
"train\n(24232, 16) (24232, 1)\nvalidate\n(6058, 16) (6058, 1)\n"
],
[
"# We tried all these. No difference.\nact=\"relu\" \nact=\"tanh\"\nact=\"sigmoid\"\n\n# Adding non-trained Layer Normalization improved accuracy a tiny bit sometimes.\n# Adding multiple dense layers only hurt.\nmlp2mer = keras.models.Sequential([\n keras.layers.LayerNormalization(trainable=False),\n keras.layers.Dense(32, activation=act,dtype='float32'),\n keras.layers.Dense(32, activation=act,dtype='float32'),\n keras.layers.Dense(1, activation=act,dtype='float32')\n])\n# Error:\n# ValueError: logits and labels must have the same shape ((None, 2) vs (None, 1))\n# This was because the output layer had 2 nodes (0 and 1) not 1 (binary).",
"_____no_output_____"
],
[
"# See page 302 for explanation of these parameters.\n# See also the keras docs e.g. \n# https://www.tensorflow.org/api_docs/python/tf/keras/losses/sparse_categorical_crossentropy\n# Note keras can take parameters for % train vs % validation.\n\n# It seems the BinaryCrossentropy assumes labels are probabilities.\n# Instead of loss=\"binary_crossentropy\",\nbc=tf.keras.losses.BinaryCrossentropy(from_logits=False)\n# Tried optimizers SGD, Adam\nmlp2mer.compile(loss=bc, optimizer=\"Adam\",metrics=[\"accuracy\"])\n\n# With one dense layer and Adam optimizer, accuracy increases slowly.\nhistory2mer = mlp2mer.fit(X_train,y_train,epochs=100,validation_data=(X_valid,y_valid))",
"Epoch 1/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.6446 - accuracy: 0.6193 - val_loss: 0.6149 - val_accuracy: 0.6588\nEpoch 2/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.6022 - accuracy: 0.6728 - val_loss: 0.5992 - val_accuracy: 0.6770\nEpoch 3/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5937 - accuracy: 0.6826 - val_loss: 0.5943 - val_accuracy: 0.6822\nEpoch 4/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5910 - accuracy: 0.6834 - val_loss: 0.5931 - val_accuracy: 0.6806\nEpoch 5/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5896 - accuracy: 0.6857 - val_loss: 0.5948 - val_accuracy: 0.6817\nEpoch 6/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5879 - accuracy: 0.6871 - val_loss: 0.5896 - val_accuracy: 0.6854\nEpoch 7/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5857 - accuracy: 0.6896 - val_loss: 0.5890 - val_accuracy: 0.6842\nEpoch 8/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5837 - accuracy: 0.6903 - val_loss: 0.5859 - val_accuracy: 0.6862\nEpoch 9/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5822 - accuracy: 0.6918 - val_loss: 0.5844 - val_accuracy: 0.6903\nEpoch 10/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5802 - accuracy: 0.6943 - val_loss: 0.5837 - val_accuracy: 0.6895\nEpoch 11/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5785 - accuracy: 0.6944 - val_loss: 0.5811 - val_accuracy: 0.6931\nEpoch 12/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5769 - accuracy: 0.6961 - val_loss: 0.5816 - val_accuracy: 0.6892\nEpoch 13/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5746 - accuracy: 0.6976 - val_loss: 0.5788 - val_accuracy: 0.6976\nEpoch 14/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5722 - accuracy: 0.6969 - val_loss: 0.5750 - val_accuracy: 0.6996\nEpoch 15/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5700 - accuracy: 0.7024 - val_loss: 0.5731 - val_accuracy: 0.6981\nEpoch 16/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5670 - accuracy: 0.7059 - val_loss: 0.5708 - val_accuracy: 0.7001\nEpoch 17/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5644 - accuracy: 0.7072 - val_loss: 0.5683 - val_accuracy: 0.7042\nEpoch 18/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5621 - accuracy: 0.7100 - val_loss: 0.5676 - val_accuracy: 0.7017\nEpoch 19/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5595 - accuracy: 0.7120 - val_loss: 0.5686 - val_accuracy: 0.7014\nEpoch 20/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5576 - accuracy: 0.7146 - val_loss: 0.5616 - val_accuracy: 0.7106\nEpoch 21/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5556 - accuracy: 0.7148 - val_loss: 0.5607 - val_accuracy: 0.7108\nEpoch 22/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5525 - accuracy: 0.7191 - val_loss: 0.5587 - val_accuracy: 0.7146\nEpoch 23/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5496 - accuracy: 0.7209 - val_loss: 0.5568 - val_accuracy: 0.7162\nEpoch 24/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5471 - accuracy: 0.7216 - val_loss: 0.5523 - val_accuracy: 0.7207\nEpoch 25/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5441 - accuracy: 0.7247 - val_loss: 0.5493 - val_accuracy: 0.7243\nEpoch 26/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5413 - accuracy: 0.7276 - val_loss: 0.5465 - val_accuracy: 0.7248\nEpoch 27/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5378 - accuracy: 0.7318 - val_loss: 0.5420 - val_accuracy: 0.7293\nEpoch 28/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5352 - accuracy: 0.7365 - val_loss: 0.5421 - val_accuracy: 0.7291\nEpoch 29/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5319 - accuracy: 0.7374 - val_loss: 0.5367 - val_accuracy: 0.7337\nEpoch 30/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5296 - accuracy: 0.7390 - val_loss: 0.5354 - val_accuracy: 0.7356\nEpoch 31/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5272 - accuracy: 0.7410 - val_loss: 0.5324 - val_accuracy: 0.7346\nEpoch 32/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5248 - accuracy: 0.7422 - val_loss: 0.5323 - val_accuracy: 0.7384\nEpoch 33/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5230 - accuracy: 0.7457 - val_loss: 0.5289 - val_accuracy: 0.7382\nEpoch 34/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5218 - accuracy: 0.7441 - val_loss: 0.5265 - val_accuracy: 0.7398\nEpoch 35/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5189 - accuracy: 0.7457 - val_loss: 0.5245 - val_accuracy: 0.7427\nEpoch 36/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5177 - accuracy: 0.7477 - val_loss: 0.5254 - val_accuracy: 0.7417\nEpoch 37/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5165 - accuracy: 0.7484 - val_loss: 0.5216 - val_accuracy: 0.7430\nEpoch 38/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5145 - accuracy: 0.7489 - val_loss: 0.5210 - val_accuracy: 0.7443accuracy: 0.74\nEpoch 39/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5126 - accuracy: 0.7502 - val_loss: 0.5189 - val_accuracy: 0.7469\nEpoch 40/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5112 - accuracy: 0.7534 - val_loss: 0.5175 - val_accuracy: 0.7474\nEpoch 41/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5102 - accuracy: 0.7543 - val_loss: 0.5180 - val_accuracy: 0.7486\nEpoch 42/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5099 - accuracy: 0.7547 - val_loss: 0.5158 - val_accuracy: 0.7489\nEpoch 43/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5083 - accuracy: 0.7537 - val_loss: 0.5144 - val_accuracy: 0.7512\nEpoch 44/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5075 - accuracy: 0.7550 - val_loss: 0.5197 - val_accuracy: 0.7455\nEpoch 45/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5064 - accuracy: 0.7550 - val_loss: 0.5197 - val_accuracy: 0.7440\nEpoch 46/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5054 - accuracy: 0.7571 - val_loss: 0.5153 - val_accuracy: 0.7483\nEpoch 47/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5045 - accuracy: 0.7566 - val_loss: 0.5124 - val_accuracy: 0.7516\nEpoch 48/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5037 - accuracy: 0.7601 - val_loss: 0.5121 - val_accuracy: 0.7544\nEpoch 49/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5028 - accuracy: 0.7592 - val_loss: 0.5109 - val_accuracy: 0.7540\nEpoch 50/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5019 - accuracy: 0.7598 - val_loss: 0.5100 - val_accuracy: 0.7537\nEpoch 51/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5013 - accuracy: 0.7594 - val_loss: 0.5099 - val_accuracy: 0.7531\nEpoch 52/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5006 - accuracy: 0.7598 - val_loss: 0.5089 - val_accuracy: 0.7552\nEpoch 53/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4998 - accuracy: 0.7600 - val_loss: 0.5082 - val_accuracy: 0.7552\nEpoch 54/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4994 - accuracy: 0.7602 - val_loss: 0.5098 - val_accuracy: 0.7547\nEpoch 55/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4983 - accuracy: 0.7597 - val_loss: 0.5207 - val_accuracy: 0.7435\nEpoch 56/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4972 - accuracy: 0.7621 - val_loss: 0.5084 - val_accuracy: 0.7572\nEpoch 57/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4972 - accuracy: 0.7612 - val_loss: 0.5097 - val_accuracy: 0.7549\n"
],
[
"pd.DataFrame(history2mer.history).plot(figsize=(8,5))\nplt.grid(True)\nplt.gca().set_ylim(0,1)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## K-mer frequency, K=3",
"_____no_output_____"
]
],
[
[
"rna_mer = read_features('ncRNA.3mer.features.csv','pcRNA.3mer.features.csv')\n(train_set,test_set)=make_train_test(rna_mer)\n(X_train,y_train,X_valid,y_valid)=prepare_train_set(train_set)",
"_____no_output_____"
],
[
"act=\"sigmoid\"\nmlp3mer = keras.models.Sequential([\n keras.layers.LayerNormalization(trainable=False),\n keras.layers.Dense(32, activation=act,dtype='float32'),\n keras.layers.Dense(32, activation=act,dtype='float32'),\n keras.layers.Dense(1, activation=act,dtype='float32')\n])\nbc=tf.keras.losses.BinaryCrossentropy(from_logits=False)\nmlp3mer.compile(loss=bc, optimizer=\"Adam\",metrics=[\"accuracy\"])\nhistory3mer = mlp3mer.fit(X_train,y_train,epochs=100,validation_data=(X_valid,y_valid))",
"Epoch 1/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5930 - accuracy: 0.6764 - val_loss: 0.5191 - val_accuracy: 0.7511\nEpoch 2/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.5048 - accuracy: 0.7590 - val_loss: 0.4997 - val_accuracy: 0.7542\nEpoch 3/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4940 - accuracy: 0.7666 - val_loss: 0.4954 - val_accuracy: 0.7565\nEpoch 4/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4904 - accuracy: 0.7675 - val_loss: 0.4929 - val_accuracy: 0.7549\nEpoch 5/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4870 - accuracy: 0.7709 - val_loss: 0.4908 - val_accuracy: 0.7552\nEpoch 6/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4823 - accuracy: 0.7718 - val_loss: 0.4882 - val_accuracy: 0.7567\nEpoch 7/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4769 - accuracy: 0.7739 - val_loss: 0.4815 - val_accuracy: 0.7605\nEpoch 8/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4743 - accuracy: 0.7760 - val_loss: 0.4786 - val_accuracy: 0.7659\nEpoch 9/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4714 - accuracy: 0.7789 - val_loss: 0.4757 - val_accuracy: 0.7674\nEpoch 10/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4690 - accuracy: 0.7790 - val_loss: 0.4767 - val_accuracy: 0.7725\nEpoch 11/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4674 - accuracy: 0.7790 - val_loss: 0.4717 - val_accuracy: 0.7715\nEpoch 12/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4650 - accuracy: 0.7818 - val_loss: 0.4705 - val_accuracy: 0.7715\nEpoch 13/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4637 - accuracy: 0.7829 - val_loss: 0.4690 - val_accuracy: 0.7743\nEpoch 14/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4612 - accuracy: 0.7836 - val_loss: 0.4690 - val_accuracy: 0.7734\nEpoch 15/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4584 - accuracy: 0.7858 - val_loss: 0.4638 - val_accuracy: 0.7763\nEpoch 16/100\n758/758 [==============================] - 3s 3ms/step - loss: 0.4551 - accuracy: 0.7870 - val_loss: 0.4657 - val_accuracy: 0.7772\nEpoch 17/100\n758/758 [==============================] - 3s 3ms/step - loss: 0.4526 - accuracy: 0.7891 - val_loss: 0.4646 - val_accuracy: 0.7773\nEpoch 18/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4484 - accuracy: 0.7921 - val_loss: 0.4574 - val_accuracy: 0.7818\nEpoch 19/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4459 - accuracy: 0.7928 - val_loss: 0.4539 - val_accuracy: 0.7847\nEpoch 20/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4430 - accuracy: 0.7961 - val_loss: 0.4503 - val_accuracy: 0.7861\nEpoch 21/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4390 - accuracy: 0.7989 - val_loss: 0.4482 - val_accuracy: 0.7887\nEpoch 22/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4364 - accuracy: 0.8014 - val_loss: 0.4441 - val_accuracy: 0.7899\nEpoch 23/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4317 - accuracy: 0.8027 - val_loss: 0.4400 - val_accuracy: 0.7943\nEpoch 24/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4293 - accuracy: 0.8047 - val_loss: 0.4365 - val_accuracy: 0.7975\nEpoch 25/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4257 - accuracy: 0.8083 - val_loss: 0.4341 - val_accuracy: 0.7956\nEpoch 26/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4235 - accuracy: 0.8091 - val_loss: 0.4317 - val_accuracy: 0.7993\nEpoch 27/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4201 - accuracy: 0.8114 - val_loss: 0.4286 - val_accuracy: 0.8029\nEpoch 28/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4180 - accuracy: 0.8140 - val_loss: 0.4317 - val_accuracy: 0.8032\nEpoch 29/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4159 - accuracy: 0.8146 - val_loss: 0.4247 - val_accuracy: 0.8059\nEpoch 30/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4131 - accuracy: 0.8165 - val_loss: 0.4300 - val_accuracy: 0.8044\nEpoch 31/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4113 - accuracy: 0.8171 - val_loss: 0.4228 - val_accuracy: 0.8088\nEpoch 32/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4089 - accuracy: 0.8186 - val_loss: 0.4199 - val_accuracy: 0.8080\nEpoch 33/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4084 - accuracy: 0.8193 - val_loss: 0.4254 - val_accuracy: 0.8067\nEpoch 34/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4064 - accuracy: 0.8205 - val_loss: 0.4168 - val_accuracy: 0.8118\nEpoch 35/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4052 - accuracy: 0.8210 - val_loss: 0.4158 - val_accuracy: 0.8113\nEpoch 36/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4034 - accuracy: 0.8225 - val_loss: 0.4135 - val_accuracy: 0.8118\nEpoch 37/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4021 - accuracy: 0.8231 - val_loss: 0.4179 - val_accuracy: 0.8141\nEpoch 38/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4010 - accuracy: 0.8231 - val_loss: 0.4125 - val_accuracy: 0.8143\nEpoch 39/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3990 - accuracy: 0.8234 - val_loss: 0.4166 - val_accuracy: 0.8136\nEpoch 40/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3978 - accuracy: 0.8262 - val_loss: 0.4110 - val_accuracy: 0.8161\nEpoch 41/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3966 - accuracy: 0.8264 - val_loss: 0.4115 - val_accuracy: 0.8176\nEpoch 42/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3955 - accuracy: 0.8263 - val_loss: 0.4107 - val_accuracy: 0.8141\nEpoch 43/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3942 - accuracy: 0.8271 - val_loss: 0.4099 - val_accuracy: 0.8186\nEpoch 44/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3931 - accuracy: 0.8288 - val_loss: 0.4081 - val_accuracy: 0.8176\nEpoch 45/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3919 - accuracy: 0.8270 - val_loss: 0.4067 - val_accuracy: 0.8197\nEpoch 46/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3905 - accuracy: 0.8294 - val_loss: 0.4081 - val_accuracy: 0.8204\nEpoch 47/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3884 - accuracy: 0.8306 - val_loss: 0.4073 - val_accuracy: 0.8197\nEpoch 48/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3879 - accuracy: 0.8315 - val_loss: 0.4067 - val_accuracy: 0.8207\nEpoch 49/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3869 - accuracy: 0.8317 - val_loss: 0.4047 - val_accuracy: 0.8219\nEpoch 50/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3853 - accuracy: 0.8337 - val_loss: 0.4056 - val_accuracy: 0.8217\nEpoch 51/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3848 - accuracy: 0.8326 - val_loss: 0.4054 - val_accuracy: 0.8199\nEpoch 52/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3832 - accuracy: 0.8333 - val_loss: 0.4024 - val_accuracy: 0.8221\nEpoch 53/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3819 - accuracy: 0.8338 - val_loss: 0.4020 - val_accuracy: 0.8240\nEpoch 54/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3815 - accuracy: 0.8336 - val_loss: 0.4026 - val_accuracy: 0.8194\nEpoch 55/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3810 - accuracy: 0.8359 - val_loss: 0.4014 - val_accuracy: 0.8247\nEpoch 56/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3798 - accuracy: 0.8349 - val_loss: 0.4072 - val_accuracy: 0.8216\nEpoch 57/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3786 - accuracy: 0.8365 - val_loss: 0.4028 - val_accuracy: 0.8230\n"
],
[
"pd.DataFrame(history3mer.history).plot(figsize=(8,5))\nplt.grid(True)\nplt.gca().set_ylim(0,1)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## K-mer frequency, K=4",
"_____no_output_____"
]
],
[
[
"rna_mer = read_features('ncRNA.4mer.features.csv','pcRNA.4mer.features.csv')\n(train_set,test_set)=make_train_test(rna_mer)\n(X_train,y_train,X_valid,y_valid)=prepare_train_set(train_set)\nact=\"sigmoid\"\nmlp4mer = keras.models.Sequential([\n keras.layers.LayerNormalization(trainable=False),\n keras.layers.Dense(32, activation=act,dtype='float32'),\n keras.layers.Dense(32, activation=act,dtype='float32'),\n keras.layers.Dense(1, activation=act,dtype='float32')\n])\nbc=tf.keras.losses.BinaryCrossentropy(from_logits=False)\nmlp4mer.compile(loss=bc, optimizer=\"Adam\",metrics=[\"accuracy\"])\nhistory4mer = mlp4mer.fit(X_train,y_train,epochs=100,validation_data=(X_valid,y_valid))",
"Epoch 1/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.5527 - accuracy: 0.7167 - val_loss: 0.4602 - val_accuracy: 0.7862\nEpoch 2/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4407 - accuracy: 0.7984 - val_loss: 0.4426 - val_accuracy: 0.7912\nEpoch 3/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4295 - accuracy: 0.8019 - val_loss: 0.4440 - val_accuracy: 0.7905\nEpoch 4/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4261 - accuracy: 0.8046 - val_loss: 0.4381 - val_accuracy: 0.7983\nEpoch 5/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4246 - accuracy: 0.8063 - val_loss: 0.4361 - val_accuracy: 0.7991\nEpoch 6/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4236 - accuracy: 0.8065 - val_loss: 0.4370 - val_accuracy: 0.7981\nEpoch 7/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4232 - accuracy: 0.8080 - val_loss: 0.4353 - val_accuracy: 0.7986\nEpoch 8/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4221 - accuracy: 0.8076 - val_loss: 0.4343 - val_accuracy: 0.8026\nEpoch 9/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4214 - accuracy: 0.8081 - val_loss: 0.4348 - val_accuracy: 0.7981\nEpoch 10/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4211 - accuracy: 0.8091 - val_loss: 0.4333 - val_accuracy: 0.8031\nEpoch 11/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4209 - accuracy: 0.8092 - val_loss: 0.4337 - val_accuracy: 0.8001\nEpoch 12/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4202 - accuracy: 0.8100 - val_loss: 0.4318 - val_accuracy: 0.8047\nEpoch 13/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4199 - accuracy: 0.8086 - val_loss: 0.4350 - val_accuracy: 0.8004\nEpoch 14/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4188 - accuracy: 0.8101 - val_loss: 0.4299 - val_accuracy: 0.8044\nEpoch 15/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4179 - accuracy: 0.8100 - val_loss: 0.4285 - val_accuracy: 0.8044\nEpoch 16/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4162 - accuracy: 0.8115 - val_loss: 0.4285 - val_accuracy: 0.8034\nEpoch 17/100\n758/758 [==============================] - 3s 3ms/step - loss: 0.4135 - accuracy: 0.8130 - val_loss: 0.4256 - val_accuracy: 0.8065\nEpoch 18/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4102 - accuracy: 0.8144 - val_loss: 0.4203 - val_accuracy: 0.8112\nEpoch 19/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.4063 - accuracy: 0.8169 - val_loss: 0.4174 - val_accuracy: 0.8110\nEpoch 20/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.4018 - accuracy: 0.8197 - val_loss: 0.4143 - val_accuracy: 0.8121\nEpoch 21/100\n758/758 [==============================] - 3s 3ms/step - loss: 0.3971 - accuracy: 0.8225 - val_loss: 0.4095 - val_accuracy: 0.8171\nEpoch 22/100\n758/758 [==============================] - 3s 4ms/step - loss: 0.3937 - accuracy: 0.8251 - val_loss: 0.4076 - val_accuracy: 0.8153\nEpoch 23/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3898 - accuracy: 0.8264 - val_loss: 0.4035 - val_accuracy: 0.8212\nEpoch 24/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3863 - accuracy: 0.8295 - val_loss: 0.4002 - val_accuracy: 0.8224\nEpoch 25/100\n758/758 [==============================] - 3s 4ms/step - loss: 0.3829 - accuracy: 0.8310 - val_loss: 0.3974 - val_accuracy: 0.8227\nEpoch 26/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3801 - accuracy: 0.8334 - val_loss: 0.3954 - val_accuracy: 0.8260\nEpoch 27/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3773 - accuracy: 0.8346 - val_loss: 0.3927 - val_accuracy: 0.8278\nEpoch 28/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3740 - accuracy: 0.8374 - val_loss: 0.3905 - val_accuracy: 0.8288\nEpoch 29/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3720 - accuracy: 0.8358 - val_loss: 0.3867 - val_accuracy: 0.8334\nEpoch 30/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3692 - accuracy: 0.8399 - val_loss: 0.3861 - val_accuracy: 0.8341\nEpoch 31/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3667 - accuracy: 0.8407 - val_loss: 0.3837 - val_accuracy: 0.8328\nEpoch 32/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3646 - accuracy: 0.8419 - val_loss: 0.3818 - val_accuracy: 0.8371\nEpoch 33/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3622 - accuracy: 0.8424 - val_loss: 0.3799 - val_accuracy: 0.8364\nEpoch 34/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3601 - accuracy: 0.8439 - val_loss: 0.3781 - val_accuracy: 0.8377\nEpoch 35/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3588 - accuracy: 0.8464 - val_loss: 0.3780 - val_accuracy: 0.8392\nEpoch 36/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3560 - accuracy: 0.8464 - val_loss: 0.3749 - val_accuracy: 0.8386\nEpoch 37/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3538 - accuracy: 0.8477 - val_loss: 0.3776 - val_accuracy: 0.8407\nEpoch 38/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3520 - accuracy: 0.8491 - val_loss: 0.3728 - val_accuracy: 0.8399\nEpoch 39/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3490 - accuracy: 0.8493 - val_loss: 0.3710 - val_accuracy: 0.8410\nEpoch 40/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3475 - accuracy: 0.8526 - val_loss: 0.3686 - val_accuracy: 0.8435\nEpoch 41/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3450 - accuracy: 0.8537 - val_loss: 0.3683 - val_accuracy: 0.8412\nEpoch 42/100\n758/758 [==============================] - 2s 3ms/step - loss: 0.3427 - accuracy: 0.8549 - val_loss: 0.3665 - val_accuracy: 0.8448\nEpoch 43/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3412 - accuracy: 0.8556 - val_loss: 0.3662 - val_accuracy: 0.8443\nEpoch 44/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3374 - accuracy: 0.8580 - val_loss: 0.3640 - val_accuracy: 0.8424\nEpoch 45/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3352 - accuracy: 0.8589 - val_loss: 0.3668 - val_accuracy: 0.8417\nEpoch 46/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3330 - accuracy: 0.8594 - val_loss: 0.3622 - val_accuracy: 0.8470\nEpoch 47/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3306 - accuracy: 0.8608 - val_loss: 0.3605 - val_accuracy: 0.8458\nEpoch 48/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3291 - accuracy: 0.8632 - val_loss: 0.3623 - val_accuracy: 0.8435\nEpoch 49/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3260 - accuracy: 0.8632 - val_loss: 0.3596 - val_accuracy: 0.8481\nEpoch 50/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3242 - accuracy: 0.8637 - val_loss: 0.3566 - val_accuracy: 0.8486\nEpoch 51/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3224 - accuracy: 0.8653 - val_loss: 0.3608 - val_accuracy: 0.8470\nEpoch 52/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3200 - accuracy: 0.8665 - val_loss: 0.3559 - val_accuracy: 0.8504\nEpoch 53/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3180 - accuracy: 0.8681 - val_loss: 0.3534 - val_accuracy: 0.8521\nEpoch 54/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3157 - accuracy: 0.8697 - val_loss: 0.3540 - val_accuracy: 0.8513\nEpoch 55/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3131 - accuracy: 0.8699 - val_loss: 0.3544 - val_accuracy: 0.8514\nEpoch 56/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3117 - accuracy: 0.8711 - val_loss: 0.3525 - val_accuracy: 0.8518\nEpoch 57/100\n758/758 [==============================] - 2s 2ms/step - loss: 0.3099 - accuracy: 0.8720 - val_loss: 0.3517 - val_accuracy: 0.8537\n"
],
[
"pd.DataFrame(history4mer.history).plot(figsize=(8,5))\nplt.grid(True)\nplt.gca().set_ylim(0,1)\nplt.show()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
e7f81570cab017b877574395bba8fd8627a51867 | 554,125 | ipynb | Jupyter Notebook | App Store Strategy Game Analysis.ipynb | MarceloFischer/App-Store-Dataset-Analysis | 816f272997d4aafa86ebe8b066e6dddef033c0aa | [
"MIT"
] | null | null | null | App Store Strategy Game Analysis.ipynb | MarceloFischer/App-Store-Dataset-Analysis | 816f272997d4aafa86ebe8b066e6dddef033c0aa | [
"MIT"
] | null | null | null | App Store Strategy Game Analysis.ipynb | MarceloFischer/App-Store-Dataset-Analysis | 816f272997d4aafa86ebe8b066e6dddef033c0aa | [
"MIT"
] | null | null | null | 275 | 182,432 | 0.896247 | [
[
[
"# Analysis of Games from the Apple Store\n\n > This dataset was taken from [Kaggle](https://www.kaggle.com/tristan581/17k-apple-app-store-strategy-games) and it was collected on the 3rd of August 2019 using the [iTunes API](https://affiliate.itunes.apple.com/resources/documentation/itunes-store-web-service-search-api/) and the [App Store sitemap](https://apps.apple.com/us/genre/ios-games/id6014).\n \nThe dataset contains 18 columns:\n - **URL**: _URL of the app._\n - **ID**: _ID of the game._\n - **Name**: _Name of the game._\n - **Subtitle**: _Advertisement text of the game._\n - **Icon URL**: _Icon of the game, 512x512 pixels jpg._\n - **Average User Rating**: _Rounded to nearest .5. Requires at least 5 ratings._\n - **User Rating Count**: _Total of user ratings. Null values means it is below 5._\n - **Price**: _Price in USD._\n - **In-app Purchases**: _Prices of available in-app purchases._\n - **Description**: _Game description._\n - **Developer**: _Game developer._\n - **Age Rating**: _Age to play the game. Either 4+, 9+, 12+or 17+._\n - **Languages**: _Languages the game supports in ISO Alpha-2 codes._\n - **Size**: _Size in bytes._\n - **Genre**: _Main genre of the game._\n - **Primary Genre**: _All genre the game fits in._\n - **Original Release Date**: _Date the game was released._\n - **Current Version Release Date**: _Date of last update._\n \nThe questions we are going to answer are:\n\n 1. Does the advance in technology impact the size of the apps?\n 2. Does the advance in technology impact the amount of apps being produced?\n 3. Are most apps free or paid and which category is more popular?\n 4. Is there a better one between free or paid apps?\n 5. How is the distribution of the age restriction?\n 6. Do most games offer more than one language?\n \n#### Below is the sequence I will be following:\n 1. Reading and Understanding the Data\n 2. Exploratory analysis\n -> Missing data\n -> Data types in the dataframe\n -> Sorting by a desired column\n -> Saving a new file after this job is done\n 3. Graphics and insights\n \n## Important note\n > **This notebook is intended exclusively to practicing and learning purposes. Any corrections, comments and suggestions are more than welcome and I would really appreciate it. Feel free to get in touch if you liked it or if you want to colaborate somehow.**",
"_____no_output_____"
],
[
"# 1. Reading and Understanding the Data",
"_____no_output_____"
]
],
[
[
"# Important imports for the analysis of the dataset\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set_style(\"darkgrid\")\n\n# Show the plot in the same window as the notebook\n%matplotlib inline",
"_____no_output_____"
],
[
"# Create the dataframe and check the first 8 rows\napp_df = pd.read_csv(\"appstore_games.csv\")\napp_df.head()",
"_____no_output_____"
],
[
"# Dropping columns that I will not use for this analysis\napp_df_cut = app_df.drop(columns=['URL', 'Subtitle', 'Icon URL'])",
"_____no_output_____"
]
],
[
[
"# 2. Exploratory Analysis",
"_____no_output_____"
]
],
[
[
"app_df_cut.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 17007 entries, 0 to 17006\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 17007 non-null int64 \n 1 Name 17007 non-null object \n 2 Average User Rating 7561 non-null float64\n 3 User Rating Count 7561 non-null float64\n 4 Price 16983 non-null float64\n 5 In-app Purchases 7683 non-null object \n 6 Description 17007 non-null object \n 7 Developer 17007 non-null object \n 8 Age Rating 17007 non-null object \n 9 Languages 16947 non-null object \n 10 Size 17006 non-null float64\n 11 Primary Genre 17007 non-null object \n 12 Genres 17007 non-null object \n 13 Original Release Date 17007 non-null object \n 14 Current Version Release Date 17007 non-null object \ndtypes: float64(4), int64(1), object(10)\nmemory usage: 1.9+ MB\n"
]
],
[
[
"***\n\nFrom the above cell I understand that I should take a closer look into the columns listed below because they have some missing values:\n - Average User Rating\n - User Rating Count\n - Price\n - Languages\n \nAnother important thing to check is if there are any **duplicate ID's** and, if so, remove them. Also, the last two column are not *datetime* type, which they should be.\n\nThe dataframe will be sorted by the \"User Rating Count\" column. This column will be our guide to conclude if a game is successful or not.",
"_____no_output_____"
]
],
[
[
"# Most reviewed app\n#app_df_cut.iloc[app_df_cut[\"User Rating Count\"].idxmax()]\n\n# A better way of seeing the most reviwed apps \napp_df_cut = app_df_cut.sort_values(by=\"User Rating Count\", ascending=False)\napp_df_cut.head(5)",
"_____no_output_____"
]
],
[
[
"### Rating columns\n> I'm going to consider that all the NaN values in the \"User Rating Count\" column means that the game recieved no ratings and therefore is 0. If the app recieved no ratings, then the \"Average User Rating\" will also be zero for these games.",
"_____no_output_____"
]
],
[
[
"# Get the columns \"User Rating Count\" and \"Average User Rating\" where they are both equal to NaN and set the\n# values to 0.\napp_df_cut.loc[(app_df_cut[\"User Rating Count\"].isnull()) | (app_df_cut[\"Average User Rating\"].isnull()),\n [\"Average User Rating\", \"User Rating Count\"]] = 0",
"_____no_output_____"
]
],
[
[
"### In-app Purchases column\n> I'm considering that the null values within the \"In-app Purchases\" column means that there are no in-app purchases available\n\n**Different considerations could have been done, but I will continue with this one for now.**",
"_____no_output_____"
]
],
[
[
"# Get the column \"In-app Purchases\" where the value is NaN and set it to zero\napp_df_cut.loc[app_df_cut[\"In-app Purchases\"].isnull(),\n \"In-app Purchases\"] = 0",
"_____no_output_____"
]
],
[
[
"### ID column\n> Let's check if there are missing or duplicate ID's in the dataset:",
"_____no_output_____"
]
],
[
[
"# Check if there are missing or 0 ID's\napp_df_cut.loc[(app_df_cut[\"ID\"] == 0) | (app_df_cut[\"ID\"].isnull()),\n \"ID\"]",
"_____no_output_____"
],
[
"# Check for duplicates in the ID column\nlen(app_df_cut[\"ID\"]) - len(app_df_cut[\"ID\"].unique())\n\n# The number of unique values is lower than the total amount of ID's, therefore there are duplicates among them. ",
"_____no_output_____"
],
[
"# Drop every duplicate ID row\napp_df_cut.drop_duplicates(subset=\"ID\", inplace=True)\napp_df_cut.shape",
"_____no_output_____"
]
],
[
[
"### Size column\n> I will check if there are any missing or 0 values in the size column. If so, they will be removed from the data since we cannot know it's value.",
"_____no_output_____"
]
],
[
[
"# Check if there are null values in the Size column\napp_df_cut[(app_df_cut[\"Size\"].isnull()) | (app_df_cut['Size'] == 0)]",
"_____no_output_____"
],
[
"# Drop the only row in which the game has no size\napp_df_cut.drop([16782], axis=0, inplace=True)",
"_____no_output_____"
],
[
"# Convert the size to MB\napp_df_cut[\"Size\"] = round(app_df_cut[\"Size\"]/1000000)\napp_df_cut.head(5)",
"_____no_output_____"
]
],
[
[
"### Price column\n > Games with a missing value in the price column will be dropped",
"_____no_output_____"
]
],
[
[
"# Drop the row with NaN values in the \"Price\" column\napp_df_cut = app_df_cut.drop(app_df_cut.loc[app_df_cut[\"Price\"].isnull()].index)",
"_____no_output_____"
]
],
[
[
"### Languages column\n> Games with a missing value in the \"Languages\" column will be dropped",
"_____no_output_____"
]
],
[
[
"# Drop the rows with NaN values in the \"Languages\" column\napp_df_cut = app_df_cut.drop(app_df_cut.loc[app_df_cut[\"Languages\"].isnull()].index)",
"_____no_output_____"
],
[
"app_df_cut.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 16763 entries, 1378 to 17006\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 16763 non-null int64 \n 1 Name 16763 non-null object \n 2 Average User Rating 16763 non-null float64\n 3 User Rating Count 16763 non-null float64\n 4 Price 16763 non-null float64\n 5 In-app Purchases 16763 non-null object \n 6 Description 16763 non-null object \n 7 Developer 16763 non-null object \n 8 Age Rating 16763 non-null object \n 9 Languages 16763 non-null object \n 10 Size 16763 non-null float64\n 11 Primary Genre 16763 non-null object \n 12 Genres 16763 non-null object \n 13 Original Release Date 16763 non-null object \n 14 Current Version Release Date 16763 non-null object \ndtypes: float64(4), int64(1), object(10)\nmemory usage: 2.0+ MB\n"
]
],
[
[
"### Age Rating column\n > I will pad the Age Rating column with a 0 to make it easier to sort the values later",
"_____no_output_____"
]
],
[
[
"# Put a 0 in front of evry value in the 'Age Rating column'\napp_df_cut['Age Rating'] = app_df_cut['Age Rating'].str.pad(width=3, fillchar='0')",
"_____no_output_____"
]
],
[
[
"### Now that the dataset is organized, let's save it into a csv file so that we do not have to redo all the steps above",
"_____no_output_____"
]
],
[
[
"app_df_cut.to_csv(\"app_df_clean.csv\", index=False)",
"_____no_output_____"
],
[
"app_df_clean = pd.read_csv(\"app_df_clean.csv\")\napp_df_clean.head()",
"_____no_output_____"
],
[
"# Transform the string dates into datetime objects\napp_df_clean[\"Original Release Date\"] = pd.to_datetime(app_df_clean[\"Original Release Date\"])\napp_df_clean[\"Current Version Release Date\"] = pd.to_datetime(app_df_clean[\"Current Version Release Date\"])",
"_____no_output_____"
],
[
"app_df_clean.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16763 entries, 0 to 16762\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 ID 16763 non-null int64 \n 1 Name 16763 non-null object \n 2 Average User Rating 16763 non-null float64 \n 3 User Rating Count 16763 non-null float64 \n 4 Price 16763 non-null float64 \n 5 In-app Purchases 16763 non-null object \n 6 Description 16763 non-null object \n 7 Developer 16763 non-null object \n 8 Age Rating 16763 non-null object \n 9 Languages 16763 non-null object \n 10 Size 16763 non-null float64 \n 11 Primary Genre 16763 non-null object \n 12 Genres 16763 non-null object \n 13 Original Release Date 16763 non-null datetime64[ns]\n 14 Current Version Release Date 16763 non-null datetime64[ns]\ndtypes: datetime64[ns](2), float64(4), int64(1), object(8)\nmemory usage: 1.9+ MB\n"
]
],
[
[
"# 3. Graphics and Insights",
"_____no_output_____"
],
[
"### Evolution of the Apps' Size\n> Do the apps get bigger with time?",
"_____no_output_____"
]
],
[
[
"# Make the figure\nplt.figure(figsize=(16,10))\n\n# Variables\nyears = app_df_clean[\"Original Release Date\"].apply(lambda date: date.year)\nsize = app_df_clean[\"Size\"]\n\n# Plot a swarmplot\npalette = sns.color_palette(\"muted\")\nsize = sns.swarmplot(x=years, y=size, palette=palette)\nsize.set_ylabel(\"Size (in MB)\", fontsize=16)\nsize.set_xlabel(\"Original Release Date\", fontsize=16)\nsize.set_title(\"Time Evolution of the Apps' Sizes\", fontsize=20)\n\n# Save the image. Has to be called before plt.show()\n#plt.savefig(\"Time_Evol_App_Size.png\", dpi=300)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"> **With the advance in technology and the internet becoming cheaper and cheaper more people have access to faster networks. As the years go by, it can be seen in the graph above that the games' size gets bigger. Some games that have more than 2GB can be noted, reaching a maximum value of 4GB, but they are not the most common ones. As each game is represented by a different tiny ball in the graph above, the quantity of games seems to grow as well. Let's investigate the number of apps per year to be sure.**",
"_____no_output_____"
],
[
"### How does the Amount of Apps Released Change Over Time?",
"_____no_output_____"
]
],
[
[
"# Make the figure\nplt.figure(figsize=(16,10))\n\n# Plot a countplot\npalette1 = sns.color_palette(\"inferno_r\")\napps_per_year = sns.countplot(x=years, data=app_df_clean, palette=palette1)\napps_per_year.set_xlabel(\"Year of Release\", fontsize=16)\napps_per_year.set_ylabel(\"Amount of Games\", fontsize=16)\napps_per_year.set_title(\"Quantity of Apps per Year\", fontsize=20)\n\n# Write the height of each bar on top of them\nfor p in apps_per_year.patches:\n apps_per_year.annotate(\"{}\".format(p.get_height()),\n (p.get_x() + p.get_width() / 2, p.get_height() + 40),\n va=\"center\", ha=\"center\", fontsize=16)\n\n# Save the figure \n#plt.savefig(\"Quantity_Apps_Per_Year.png\", dpi=300)",
"_____no_output_____"
]
],
[
[
"> **From 2008 to 2016 we can identify a drastic increase in the number of games released each year in which the highest increase occurs between the years 2015 and 2016. After 2016 the amount of games released per year starts to drop down almost linearly for 2 years (2019 cannot be considered yet because the data was collected in August, 4 months of data of the current year is missing).**\n>\n> **Without further analysis, I would argue that after a boom in the production of apps it gets harder to come up with new ideas that are not out there already, making the production and release of new games slow down, but it is important to keep in mind that without further research it cannot be taken as the right explanation.**",
"_____no_output_____"
]
],
[
[
"#Make a list of years from 2014 to 2018\nyears_lst = [year for year in range(2014,2019)]\n\n#For loop to get a picture of the amount of games produced from August to December\nfor year in years_lst:\n from_August = app_df_clean[\"Original Release Date\"].apply(lambda date: (date.year == year) & (date.month >= 8)).sum()\n total = app_df_clean[\"Original Release Date\"].apply(lambda date: date.year == year).sum()\n print(\"In {year}, {percentage}% games were produced from August to December.\"\n .format(year=year,\n percentage=round((from_August/total)*100, 1)))",
"In 2014, 44.1% games were produced from August to December.\nIn 2015, 42.2% games were produced from August to December.\nIn 2016, 39.9% games were produced from August to December.\nIn 2017, 40.8% games were produced from August to December.\nIn 2018, 42.4% games were produced from August to December.\n"
]
],
[
[
"> **Having checked the previous five years we can see that the amount of games released from August to December represents a significant portion of the whole and that it can be considered roughly constant at 42%. Nevertheless, the last two years show a tendency for a linear decrease in the quantity of games released per year and taking into account that we still have 42% of the games of this year to be released, the total amount in the present year (2019) would be 2617. This is bigger than 2018, but this was not an elaborate calculation as we took the average of games being prouced between the months 8-12 to be 42%.**",
"_____no_output_____"
],
[
"### Now, can we observe a trend in the age restriction of games released each year?",
"_____no_output_____"
]
],
[
[
"# Make the figure\nplt.figure(figsize=(16,10))\n\n# Variables. Sort by age to put the legend in order.\ndata = app_df_clean.sort_values(by='Age Rating')\n\n# Plot a countplot\npalette1 = sns.color_palette(\"viridis\")\napps_per_year = sns.countplot(x=years, data=data, palette=palette1, hue='Age Rating')\napps_per_year.set_xlabel(\"Year of Release per Age\", fontsize=16)\napps_per_year.set_ylabel(\"Amount of Games\", fontsize=16)\napps_per_year.set_title(\"Quantity of Apps per Year & Age\", fontsize=20)\n\nplt.legend(title='Age Restrictions', fontsize=13, title_fontsize=14, loc='upper left')\n# Save the figure \n#plt.savefig(\"Quantity_Apps_Per_Year_&_Age.png\", dpi=300)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"> **As shown above, most apps tend to target all ages.**",
"_____no_output_____"
],
[
"### The amount of apps had a considerable increase in the past years indicating that producing an app has been a trend and possibly a lucrative market. That being said, it is important to analyse if there is a preference for free or paid games and the range of prices they are in.",
"_____no_output_____"
]
],
[
[
"# Make the figure\nplt.figure(figsize=(16,10))\n\n# Variables\nprice = app_df_clean[\"Price\"]\n\n# Plot a Countplot\npalette2 = sns.light_palette(\"green\", reverse=True)\nprice_vis = sns.countplot(x=price, palette=palette2)\nprice_vis.set_xlabel(\"Price (in US dollars)\", fontsize=16)\nprice_vis.set_xticklabels(price_vis.get_xticklabels(), fontsize=12, rotation=45)\nprice_vis.set_ylabel(\"Amount of Games\", fontsize=16)\nprice_vis.set_title(\"Quantity of Apps per Price\", fontsize=20)\n\n# Write the height of the bars on top\nfor p in price_vis.patches:\n price_vis.annotate(\"{:.0f}\".format(p.get_height()), # Text that will appear on the screen\n (p.get_x() + p.get_width() / 2 + 0.1, p.get_height()), # (x, y) has to be a tuple\n ha='center', va='center', fontsize=14, color='black', xytext=(0, 10), # Customizations\n textcoords='offset points')\n\n# Save the figure\n#plt.savefig(\"Quantity_Each_App_Per_Price.png\", dpi=300)",
"_____no_output_____"
]
],
[
[
"> **We can see that the majority of the games are free. That leads me to analyse if the free apps have more in-app purchases then the paid ones, meaning that this might be their source of income.**",
"_____no_output_____"
]
],
[
[
"# Make the figure\nplt.figure(figsize=(16,10))\n\n# Variables\nin_app_purchases = app_df_clean[\"In-app Purchases\"].str.split(\",\").apply(lambda lst: len(lst))\n\n# Plot a stripplot\npalette3 = sns.color_palette(\"BuGn_r\", 23)\nin_app_purchases_vis = sns.stripplot(x=price, y=in_app_purchases, palette=palette3)\nin_app_purchases_vis.set_xlabel(\"Game Price (in US dollars)\", fontsize=16)\nin_app_purchases_vis.set_xticklabels(in_app_purchases_vis.get_xticklabels(), fontsize=12, rotation=45)\nin_app_purchases_vis.set_ylabel(\"In-app Purchases Available\", fontsize=16)\nin_app_purchases_vis.set_title(\"Quantity of In-app Purchases per Game Price\", fontsize=20)\n\n# Save the image. Has to be called before plt.show()\n#plt.savefig(\"Quantity_In_App_Purchase.png\", dpi=300)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"> **As expected, free and lower-priced apps provide more items to be purchased than expensive games. Two reasons can be named:**\n>\n>> **1.The developers have to invest money into making the games and updating them, therefore they need a source of income. In the case of free games, this comes with the in-app purchases available.**\n>\n>> **2. People who have spent a lot of money on an app would not be happy or willing to spend more, given that they have already made an initial high investment.**",
"_____no_output_____"
],
[
"### We know that most of the apps are free. Let's see if there are any links between an app being paid and being better than the free ones:",
"_____no_output_____"
]
],
[
[
"# Plot a distribution of the top 200 apps by their price\n\n# Make the figure\nplt.figure(figsize=(16,10))\n\n# Plot a Countplot\npalette4 = sns.color_palette(\"BuPu_r\")\ntop_prices = sns.countplot(app_df_clean.iloc[:200][\"Price\"], palette=palette4)\ntop_prices.set_xlabel(\"Price (in US dollars)\", fontsize=16)\ntop_prices.set_xticklabels(top_prices.get_xticklabels(), fontsize=12)\ntop_prices.set_ylabel(\"Amount of Games\", fontsize=16)\ntop_prices.set_title(\"Quantity of Apps per Price for the Top 200\", fontsize=20)\n\n# Write the height of the bars on top\nfor p in top_prices.patches:\n top_prices.annotate(\"{:.0f}\".format(p.get_height()), \n (p.get_x() + p.get_width() / 2., p.get_height()),\n ha='center', va='center', fontsize=14, color='black', xytext=(0, 8),\n textcoords='offset points')\n \n# Save the image.\n#plt.savefig(\"Quantity_App_Per_Price.png\", dpi=300)",
"_____no_output_____"
]
],
[
[
"> **The graph above shows that among the top 200 games, the vast majority are free. This result makes sense considering you don't have to invest any money to start playing and can spend afterward if you would like to invest in it.**",
"_____no_output_____"
],
[
"### Even though most games are free we should take a look if a type of app (paid or free) is better. Let's do that by checking the average user rating.",
"_____no_output_____"
]
],
[
[
"# Create the DataFrames needed\npaid = app_df_clean[app_df_clean[\"Price\"] > 0]\ntotal_paid = len(paid)\nfree = app_df_clean[app_df_clean[\"Price\"] == 0]\ntotal_free = len(free)\n\n# Make the figure and the axes (1 row, 2 columns)\nfig, axes = plt.subplots(1, 2, figsize=(16,10))\npalette5 = sns.color_palette(\"gist_yarg\", 10)\n\n# Free apps countplot\nfree_vis = sns.countplot(x=\"Average User Rating\", data=free, ax=axes[0], palette=palette5)\nfree_vis.set_xlabel(\"Average User Rating\", fontsize=16)\nfree_vis.set_ylabel(\"Amount of Games\", fontsize=16)\nfree_vis.set_title(\"Free Apps\", fontsize=20)\n\n# Display the percentages on top of the bars\nfor p in free_vis.patches:\n free_vis.annotate(\"{:.1f}%\".format(100 * (p.get_height()/total_free)),\n (p.get_x() + p.get_width() / 2 + 0.1, p.get_height()),\n ha='center', va='center', fontsize=14, color='black', xytext=(0, 8),\n textcoords='offset points')\n \n# Paid apps countplot\npaid_vis = sns.countplot(x=\"Average User Rating\", data=paid, ax=axes[1], palette=palette5)\npaid_vis.set_xlabel(\"Average User Rating\", fontsize=16)\npaid_vis.set_ylabel(\" \", fontsize=16)\npaid_vis.set_title(\"Paid Apps\", fontsize=20)\n\n# Display the percentages on top of the bars\nfor p in paid_vis.patches:\n paid_vis.annotate(\"{:.1f}%\".format(100 * (p.get_height()/total_paid)),\n (p.get_x() + p.get_width() / 2 + 0.1, p.get_height()),\n ha='center', va='center', fontsize=14, color='black', xytext=(0, 8),\n textcoords='offset points')\n \n# Save the image.\n#plt.savefig(\"Free_VS_Paid.png\", dpi=300)",
"_____no_output_____"
]
],
[
[
"> **There are no indications of whether a paid or a free game is better. Actually, the pattern of user ratings is pretty much equal for both types of games. The graph above shows that both categories seem to deliver a good service and mostly satisfy their costumers as most of the ratings are between 4-5 stars. We can also identify that the majority of the users do not rate the games.**",
"_____no_output_____"
],
[
"# Age Rating\n> Is there a preference for permitted age to the games?",
"_____no_output_____"
]
],
[
[
"# Make the figure\nplt.figure(figsize=(16,10))\n\n# Make a countplot\npalette6 = sns.color_palette(\"BuGn_r\")\nage_vis = sns.countplot(x=app_df_clean[\"Age Rating\"], order=[\"04+\", \"09+\", \"12+\", \"17+\"], palette=palette6)\nage_vis.set_xlabel(\"Age Rating\", fontsize=16)\nage_vis.set_ylabel(\"Amount of Games\", fontsize=16)\nage_vis.set_title(\"Amount of Games per Age Restriction\", fontsize=20)\n\n# Write the height of the bars on top\nfor p in age_vis.patches:\n age_vis.annotate(\"{:.0f}\".format(p.get_height()), \n (p.get_x() + p.get_width() / 2., p.get_height()),\n ha='center', va='center', fontsize=14, color='black', xytext=(0, 8),\n textcoords='offset points')\n \n# Save the image.\n#plt.savefig(\"Amount_Games_Per_Age.png\", dpi=300)",
"_____no_output_____"
]
],
[
[
"> **Most of the apps are in the +4 age category, which can be translated as \"everyone can play\". This ensures that the developers are targeting a much broader audience with their games.**",
"_____no_output_____"
],
[
"# Languages\n> Do most games have various choices of languages?",
"_____no_output_____"
]
],
[
[
"# Create a new column that contains the amount of languages that app has available\napp_df_clean[\"numLang\"] = app_df_clean[\"Languages\"].apply(lambda x: len(x.split(\",\")))",
"_____no_output_____"
],
[
"#Make the figure\nplt.figure(figsize=(16,10))\n\n#Variables\nlang = app_df_clean.loc[app_df_clean[\"numLang\"] <= 25, \"numLang\"]\n\n#Plot a countplot\npalette7 = sns.color_palette(\"PuBuGn_r\")\nnumLang_vis = sns.countplot(x=lang, data=app_df_clean, palette=palette7)\nnumLang_vis.set_xlabel(\"Quantity of Languages\", fontsize=16)\nnumLang_vis.set_ylabel(\"Amount of Games\", fontsize=16)\nnumLang_vis.set_title(\"Quantity of Languages Available per Game\", fontsize=20)\n\n# Write the height of the bars on top\nfor p in numLang_vis.patches:\n numLang_vis.annotate(\"{:.0f}\".format(p.get_height()), \n (p.get_x() + p.get_width() / 2. + .1, p.get_height()),\n ha='center', va='center', fontsize=12, color='black', xytext=(0, 12),\n textcoords='offset points')\n \n# Save the image.\n#plt.savefig(\"Quantity_Lang_Per_Game.png\", dpi=300)",
"_____no_output_____"
],
[
"#Amount of games that have only the English language\nlen(app_df_clean[(app_df_clean[\"numLang\"] == 1) & (app_df_clean[\"Languages\"] == \"EN\")])",
"_____no_output_____"
],
[
"#Amount of games that have only one language and is not English\nlen(app_df_clean[(app_df_clean[\"numLang\"] == 1) & (app_df_clean[\"Languages\"] != \"EN\")])",
"_____no_output_____"
]
],
[
[
"> **The vast majority of the games - 12.431 - have only one language available and more than 99% of these use the English language. After that, there is a huge drop and only 1089 games have two languages available. Note that not all the data is shown in the graph above, but games with more than 25 languages were left out and they don't represent a huge number overall. It is interesting to point out that there is a strange increase in the number of games with 16 languages and then another one when we reach 25 languages. The explanation for that is unknown and it will not be investigated in this notebook.**",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e7f8175750cff7faafb7b0a6fccbd06d0d194b43 | 30,380 | ipynb | Jupyter Notebook | Assignments/hw3/HW3_Generalized_Linear_Model_finished/plot_iris_logistic1.ipynb | Leon23N/Leon23N.github.io | bfa1cf19a14da7cb13842fa0567c6c555d4abab4 | [
"CC-BY-3.0"
] | null | null | null | Assignments/hw3/HW3_Generalized_Linear_Model_finished/plot_iris_logistic1.ipynb | Leon23N/Leon23N.github.io | bfa1cf19a14da7cb13842fa0567c6c555d4abab4 | [
"CC-BY-3.0"
] | null | null | null | Assignments/hw3/HW3_Generalized_Linear_Model_finished/plot_iris_logistic1.ipynb | Leon23N/Leon23N.github.io | bfa1cf19a14da7cb13842fa0567c6c555d4abab4 | [
"CC-BY-3.0"
] | null | null | null | 205.27027 | 26,348 | 0.910632 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# Logistic Regression 3-class Classifier\n\n\nShow below is a logistic-regression classifiers decision boundaries on the\n`iris <https://en.wikipedia.org/wiki/Iris_flower_data_set>`_ dataset. The\ndatapoints are colored according to their labels.\n\n\n",
"_____no_output_____"
]
],
[
[
"print(__doc__)\n\n\n# Code source: Gaël Varoquaux\n# Modified for documentation by Jaques Grobler\n# License: BSD 3 clause\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn import linear_model, datasets\nimport pandas as pd\n\nmydata = pd.read_csv(\"dataset.csv\") \ndt = mydata.values\n\nX = dt[:, :2]\n\nY = dt[:, 3]\nY = Y.astype('int')\n\n# import some data to play with\n#iris = datasets.load_iris()\n#X = iris.data[:, :2] # we only take the first two features.\n#Y = iris.target\n\nh = .02 # step size in the mesh\n\nlogreg = linear_model.LogisticRegression(C=1e5)\n\n# we create an instance of Neighbours Classifier and fit the data.\nlogreg.fit(X, Y)\n\n# Plot the decision boundary. For that, we will assign a color to each\n# point in the mesh [x_min, x_max]x[y_min, y_max].\nx_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5\ny_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5\nxx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))\nZ = logreg.predict(np.c_[xx.ravel(), yy.ravel()])\n\n# Put the result into a color plot\nZ = Z.reshape(xx.shape)\nplt.figure(1, figsize=(4, 3))\nplt.pcolormesh(xx, yy, Z, cmap=plt.cm.Paired)\n\n# Plot also the training points\nplt.scatter(X[:, 0], X[:, 1], c=Y, edgecolors='k', cmap=plt.cm.Paired)\nplt.xlabel('Length_Data')\nplt.ylabel('Width_Data')\n\nplt.xlim(xx.min(), xx.max())\nplt.ylim(yy.min(), yy.max())\nplt.xticks(())\nplt.yticks(())\n\nplt.show()",
"Automatically created module for IPython interactive environment\n"
]
]
] | [
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f82777ad9e2d6b0f82a42a7ac72cee34f4051f | 760,193 | ipynb | Jupyter Notebook | session2/tut01.ipynb | elixir-luxembourg/julia-training | a1f378cd6b4f27ecf0c91f14980ad29b495ab0af | [
"CC-BY-4.0"
] | null | null | null | session2/tut01.ipynb | elixir-luxembourg/julia-training | a1f378cd6b4f27ecf0c91f14980ad29b495ab0af | [
"CC-BY-4.0"
] | 2 | 2021-04-22T08:03:55.000Z | 2021-04-22T09:53:09.000Z | session2/tut01.ipynb | elixir-luxembourg/julia-training | a1f378cd6b4f27ecf0c91f14980ad29b495ab0af | [
"CC-BY-4.0"
] | 1 | 2021-04-22T10:00:18.000Z | 2021-04-22T10:00:18.000Z | 116.755184 | 15,712 | 0.621372 | [
[
[
"empty"
]
]
] | [
"empty"
] | [
[
"empty"
]
] |
e7f8280c96c433ae9e470f02380d08c410e692ae | 52,382 | ipynb | Jupyter Notebook | models/miniscout/.ipynb_checkpoints/prop_fit-checkpoint.ipynb | cohen39/fixedwing_gazebo | 40fb8f94dcd16a5e1441eff7f22306e39ef02018 | [
"BSD-3-Clause"
] | 3 | 2020-10-07T15:04:59.000Z | 2022-03-29T01:29:23.000Z | models/miniscout/.ipynb_checkpoints/prop_fit-checkpoint.ipynb | cohen39/fixedwing_gazebo | 40fb8f94dcd16a5e1441eff7f22306e39ef02018 | [
"BSD-3-Clause"
] | 1 | 2021-10-07T09:46:10.000Z | 2021-10-07T09:46:10.000Z | models/miniscout/.ipynb_checkpoints/prop_fit-checkpoint.ipynb | cohen39/fixedwing_gazebo | 40fb8f94dcd16a5e1441eff7f22306e39ef02018 | [
"BSD-3-Clause"
] | 5 | 2020-04-11T00:02:46.000Z | 2020-10-07T15:04:58.000Z | 327.3875 | 21,308 | 0.930491 | [
[
[
"import pandas as pd\nimport re\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"data = {}\nheader = ['V', 'J', 'Pe', 'Ct', 'Cp', 'PWR', 'Torque', 'Thrust'] \nwith open('PER3_6x3.dat.txt', 'r') as f:\n for line in f.readlines():\n \n match_rpm = re.match('.*PROP.*RPM =\\s*([0-9]+)', line)\n if match_rpm is not None:\n rpm = int(match_rpm.group(1))\n data[rpm] = []\n continue\n\n match_data = re.match('(\\s*[-]?\\d+\\.\\d*\\s*)+', line)\n if match_data is not None:\n line_data = [float(v) for v in match_data.group(0).split()]\n data[rpm].append(line_data)\n if len(line_data) != 8:\n raise IOError(line)\n\n for k in data.keys():\n data[k] = pd.DataFrame(data[k], columns=header)",
"_____no_output_____"
],
[
"for k in [1000, 6000, 10000, 20000]:\n plt.plot(data[k].J, data[k].Ct, label=k)\n plt.legend()",
"_____no_output_____"
],
[
"ct_coef = np.polyfit(data[6000].J, data[6000].Ct, 4)\nJ = np.linspace(0,1)\nplt.plot(J, np.polyval(ct_coef, J))\nplt.plot(data[6000].J, data[6000].Ct, '.')\nplt.xlabel('J')\nplt.ylabel('Ct')\nprint(np.flip(ct_coef))",
"[ 0.09916565 0.00738079 -0.27449944 0.10678074 0.01674628]\n"
],
[
"cp_coef = np.polyfit(data[6000].J, data[6000].Cp, 4)\nJ = np.linspace(0,1)\nplt.plot(J, np.polyval(cp_coef, J))\nplt.plot(data[6000].J, data[6000].Cp, '.')\nplt.xlabel('J')\nplt.ylabel('Cp')\nprint(np.flip(cp_coef))",
"[ 0.04099138 0.02454836 -0.0241211 -0.13055477 0.04986827]\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7f82ecd7cdf2ed1b8b7402135b2a1eb5dbf623c | 20,223 | ipynb | Jupyter Notebook | ga_sim.ipynb | linea-it/ga_sim | 769adab09217e05215c081ec254cd46ee53021fa | [
"MIT"
] | null | null | null | ga_sim.ipynb | linea-it/ga_sim | 769adab09217e05215c081ec254cd46ee53021fa | [
"MIT"
] | 2 | 2022-01-26T13:45:15.000Z | 2022-02-08T20:11:09.000Z | ga_sim.ipynb | linea-it/ga_sim | 769adab09217e05215c081ec254cd46ee53021fa | [
"MIT"
] | null | null | null | 30.502262 | 528 | 0.548188 | [
[
[
"## ga_sim",
"_____no_output_____"
],
[
"This jn is intended to create simulations of dwarf galaxies and globular clusters using as field stars the catalog of DES. These simulations will be later copied to gawa jn, a pipeline to detect stellar systems with field's stars. In principle this pipeline read a table in data base with g and r magnitudes, subtract the extinction in each band, and randomize the positions in RA and DEC in order to avoid stellar systems in the FoV. The star clusters are inserted later, centered in each HP pixel with specific nside.\n\nTo complete all the steps you just have to run all the cells below in sequence.",
"_____no_output_____"
],
[
"Firstly, install the packages not available in the image via terminal. Restart the kernel and so you can run the cell bellow.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom astropy.coordinates import SkyCoord\nfrom astropy import units as u\nimport healpy as hp\nimport astropy.io.fits as fits\nfrom astropy.table import Table\nfrom astropy.io.fits import getdata\nimport sqlalchemy\nimport json\nfrom pathlib import Path\nimport os\nimport sys\nimport parsl\nfrom parsl.app.app import python_app, bash_app\nfrom parsl.configs.local_threads import config\nfrom time import sleep\nfrom tqdm import tqdm\nfrom ga_sim import (\n make_footprint,\n faker,\n join_cat,\n write_sim_clus_features,\n download_iso,\n read_cat,\n gen_clus_file,\n read_error,\n clus_file_results,\n join_cats_clean,\n split_files,\n clean_input_cat,\n clean_input_cat_dist\n)\n\nparsl.clear()\nparsl.load(config)\n",
"_____no_output_____"
]
],
[
[
"Below are the items of the configuration for field stars and simulations. A small description follows as a comment.",
"_____no_output_____"
]
],
[
[
"# Main settings:\nconfg = \"ga_sim.json\"\n\n# read config file\nwith open(confg) as fstream:\n param = json.load(fstream)\n\nage_simulation = 1.0e10 # in years\nZ_simulation = 0.001 # Assuming Z_sun = 0.0152\n\n# Diretório para os resultados\nos.system(\"mkdir -p \" + param['results_path'])\n\n# Reading reddening files\nhdu_ngp = fits.open(\"sample_data/SFD_dust_4096_ngp.fits\", memmap=True)\nngp = hdu_ngp[0].data\n\nhdu_sgp = fits.open(\"sample_data/SFD_dust_4096_sgp.fits\", memmap=True)\nsgp = hdu_sgp[0].data\n",
"_____no_output_____"
]
],
[
[
"Downloading the isochrone table with the last improvements from Padova.\nPrinting age and metalicity of isochrone downloaded. Try one more time in case of problems. Sometimes there is a problem with the connection to Padova.",
"_____no_output_____"
]
],
[
[
"download_iso(param['padova_version_code'], param['survey'], Z_simulation,\n age_simulation, param['av_simulation'], param['file_iso'])\n",
"_____no_output_____"
]
],
[
[
"Checking age and metalicity of the isochrone:",
"_____no_output_____"
]
],
[
[
"# Reading [M/H], log_age, mini, g\niso_info = np.loadtxt(param['file_iso'], usecols=(1, 2, 3, 26), unpack=True)\nFeH_iso = iso_info[0][0]\nlogAge_iso = iso_info[1][0]\nm_ini_iso = iso_info[2]\ng_iso = iso_info[3]\n\nprint('[Fe/H]={:.2f}, Age={:.2f} Gyr'.format(FeH_iso, 10**(logAge_iso-9)))\n\nmM_mean = (param['mM_max'] + param['mM_min']) / 2.\nprint(np.max(m_ini_iso[g_iso + mM_mean < param['mmax']]))\nmean_mass = (np.min(m_ini_iso[g_iso + mM_mean < param['mmax']]) +\n np.max(m_ini_iso[g_iso + mM_mean < param['mmax']])) / 2.\n\nprint('Mean mass (M_sun): {:.2f}'.format(mean_mass))\n",
"_____no_output_____"
],
[
"hpx_ftp = make_footprint(param['ra_min'], param['ra_max'], param['dec_min'], param['dec_max'],\n param['nside_ftp'], output_path=param['results_path'])\nprint(len(hpx_ftp))\n",
"_____no_output_____"
]
],
[
[
"Reading the catalog and writing as a fits file (to avoid read from the DB many times in the case the same catalog will be used multiple times).",
"_____no_output_____"
]
],
[
[
"RA, DEC, MAG_G, MAGERR_G, MAG_R, MAGERR_R = read_cat(\n param['vac_ga'], param['ra_min'], param['ra_max'], param['dec_min'], param['dec_max'],\n param['mmin'], param['mmax'], param['cmin'], param['cmax'],\n \"DES_Y6_Gold_v1_derred.fits\", 1.19863, 0.83734, ngp, sgp, param['results_path'])\n",
"_____no_output_____"
]
],
[
[
"The cells below reads the position, calculates the extinction using the previous function and <br>\ncorrect the aparent magnitude (top of the Galaxy), filter the stars for magnitude and color ranges, <br> \nand writes a file with the original position of the stars and corrected magnitudes.",
"_____no_output_____"
],
[
"## Simulation of dwarf galaxies and globular clusters\n\nIn fact, the dwarf galaxies and globular clusters are very similar in terms of stellar populations. Dwarf galaxies\nhave a half-light radius larger than globular clusters (given the amount of dark matter) with the same absolute magnitude. The code below simulates stars using a Kroupa or Salpeter IMF, and an exponential radius for the 2D distribution of stars. ",
"_____no_output_____"
],
[
"Generating the properties of clusters based on properties stated above. Writting to file 'objects.dat'.",
"_____no_output_____"
]
],
[
[
"RA_pix, DEC_pix, r_exp, ell, pa, dist, mass, mM, hp_sample_un = gen_clus_file(\n param['ra_min'],\n param['ra_max'],\n param['dec_min'],\n param['dec_max'],\n param['nside_ini'],\n param['border_extract'],\n param['mM_min'],\n param['mM_max'],\n param['log10_rexp_min'],\n param['log10_rexp_max'],\n param['log10_mass_min'],\n param['log10_mass_max'],\n param['ell_min'],\n param['ell_max'],\n param['pa_min'],\n param['pa_max'],\n param['results_path']\n)\n",
"_____no_output_____"
]
],
[
[
"## Dist stars\nReading data from magnitude and errors.\n\n",
"_____no_output_____"
]
],
[
[
"mag1_, err1_, err2_ = read_error(param['file_error'], 0.015, 0.015)\n",
"_____no_output_____"
]
],
[
[
"Now simulating the clusters using 'faker' function.",
"_____no_output_____"
]
],
[
[
"@python_app\ndef faker_app(N_stars_cmd, frac_bin, IMF_author, x0, y0, rexp, ell_, pa, dist, hpx, output_path):\n\n global param\n\n faker(\n N_stars_cmd,\n frac_bin,\n IMF_author,\n x0,\n y0,\n rexp,\n ell_,\n pa,\n dist,\n hpx,\n param['cmin'],\n param['cmax'],\n param['mmin'],\n param['mmax'],\n mag1_,\n err1_,\n err2_,\n param['file_iso'],\n output_path\n )\n\n\n# Diretório dos arquivo _clus.dat gerados pela faker.\nfake_clus_path = param['results_path'] + '/fake_clus'\n\nfutures = list()\n\n# Cria uma Progressbar (Opcional)\nwith tqdm(total=len(hp_sample_un), file=sys.stdout) as pbar:\n pbar.set_description(\"Submit Parsls Tasks\")\n\n # Submissão dos Jobs Parsl\n for i in range(len(hp_sample_un)):\n # Estimating the number of stars in cmd dividing mass by mean mass\n N_stars_cmd = int(mass[i] / mean_mass)\n # os.register_at_fork(after_in_child=lambda: _get_font.cache_clear())\n futures.append(\n faker_app(\n N_stars_cmd,\n param['frac_bin'],\n param['IMF_author'],\n RA_pix[i],\n DEC_pix[i],\n r_exp[i],\n ell[i],\n pa[i],\n dist[i],\n hp_sample_un[i],\n output_path=fake_clus_path\n )\n )\n\n pbar.update()\n\n# Progressbar para acompanhar as parsl.tasks.\nprint(\"Tasks Done:\")\nwith tqdm(total=len(futures), file=sys.stdout) as pbar2:\n # is_done é um array contendo True ou False para cada task\n # is_done.count(True) retorna a quantidade de tasks que já terminaram.\n is_done = list()\n done_count = 0\n while is_done.count(True) != len(futures):\n is_done = list()\n for f in futures:\n is_done.append(f.done())\n\n # Só atualiza a pbar se o valor for diferente.\n if is_done.count(True) != done_count:\n done_count = is_done.count(True)\n # Reset é necessário por que a quantidade de iterações\n # é maior que a quantidade de jobs.\n pbar2.reset(total=len(futures))\n # Atualiza a pbar\n pbar2.update(done_count)\n\n if done_count < len(futures):\n sleep(3)\n",
"_____no_output_____"
]
],
[
[
"Now functions to join catalogs of simulated clusters and field stars, and to estimate signal-to-noise ratio.",
"_____no_output_____"
]
],
[
[
"# Le os arquivos _clus.dat do diretório \"result/fake_clus\"\n# Gera o arquivo \"result/<survey>_mockcat_for_detection.fits\"\nmockcat = join_cat(\n param['ra_min'],\n param['ra_max'],\n param['dec_min'],\n param['dec_max'],\n hp_sample_un,\n param['survey'],\n RA,\n DEC,\n MAG_G,\n MAG_R,\n MAGERR_G,\n MAGERR_R,\n param['nside_ini'],\n param['mmax'],\n param['mmin'],\n param['cmin'],\n param['cmax'],\n input_path=fake_clus_path,\n output_path=param['results_path'])\nprint(mockcat)\n",
"_____no_output_____"
]
],
[
[
"If necessary, split the catalog with simulated clusters into many files according HP schema.",
"_____no_output_____"
]
],
[
[
"os.makedirs(param['hpx_cats_path'], exist_ok=True)\nipix_cats = split_files(mockcat, 'ra', 'dec',\n param['nside_ini'], param['hpx_cats_path'])\n",
"_____no_output_____"
],
[
"sim_clus_feat = write_sim_clus_features(\n mockcat, hp_sample_un, param['nside_ini'], mM, output_path=param['results_path'])\n",
"_____no_output_____"
]
],
[
[
"Merge both files in a single file.",
"_____no_output_____"
]
],
[
[
"clus_file_results(param['results_path'], \"star_clusters_simulated.dat\",\n sim_clus_feat, 'results/objects.dat')\n",
"_____no_output_____"
]
],
[
[
"## Plots\n\nA few plots to characterize the simulated clusters.",
"_____no_output_____"
]
],
[
[
"from ga_sim.plot import (\n general_plots,\n plot_ftp,\n plots_ang_size,\n plots_ref,\n plot_err,\n plot_clusters_clean\n)\n\ngeneral_plots(param['star_clusters_simulated'])\n",
"_____no_output_____"
]
],
[
[
"Plot footprint map to check area.",
"_____no_output_____"
]
],
[
[
"hpx_ftp = param['results_path'] + \"/ftp_4096_nest.fits\"\n\nplot_ftp(hpx_ftp, param['star_clusters_simulated'],\n mockcat, param['ra_max'], param['ra_min'], param['dec_min'], param['dec_max'])\n",
"_____no_output_____"
],
[
"# Diretório onde estão os arquivo _clus.dat\nplots_ang_size(param['star_clusters_simulated'], param['results_path'],\n param['mmin'], param['mmax'], param['cmin'], param['cmax'],\n param['output_plots'])\n",
"_____no_output_____"
],
[
"plots_ref(FeH_iso, param['star_clusters_simulated'], param['output_plots'])\n",
"_____no_output_____"
]
],
[
[
"Plotting errors in main magnitude band.",
"_____no_output_____"
]
],
[
[
"# Plots to analyze the simulated clusters.\nplot_err(mockcat, param['output_plots'])\n",
"_____no_output_____"
]
],
[
[
"## Removing stars close to each other\n\nNow, we have to remove stars that are not detected in the pipeline of detection of the survey. In principle, the software used by detect sources is SExtractor, which parameter deblend is set to blend sources very close to each other.\n\nTo remove sources close to each other, the approach below (or the function on that) read catalogs from ipixels (HealPixels).\nTo each star the distance to all sources are calculated. If the second minimum distance (the first one is zero, since it is the iteration of the stars with itself) is less than the distance defined as a parameter of the function, the star is not listed in the filtered catalog.\nThe function runs in parallel, in order to run faster using all the cores of node.\n\nFirstly, setting the string to read position of stars.",
"_____no_output_____"
]
],
[
[
"@python_app\ndef clean_input_cat_dist_app(file_name, ra_str, dec_str, min_dist_arcsec):\n\n clean_input_cat_dist(\n file_name,\n ra_str,\n dec_str,\n min_dist_arcsec\n )\n\n\nfutures = list()\n\n# Cria uma Progressbar (Opcional)\nwith tqdm(total=len(ipix_cats), file=sys.stdout) as pbar:\n pbar.set_description(\"Submit Parsls Tasks\")\n\n # Submissão dos Jobs Parsl\n for i in ipix_cats:\n futures.append(\n clean_input_cat_dist_app(\n i, param['ra_str'], param['dec_str'], param['min_dist_arcsec'])\n )\n\n pbar.update()\n\n# Espera todas as tasks Parsl terminarem\n# Este loop fica monitarando as parsl.futures\n# Até que todas tenham status done.\n# Esse bloco todo é opcional\n\nprint(\"Tasks Done:\")\nwith tqdm(total=len(futures), file=sys.stdout) as pbar2:\n # is_done é um array contendo True ou False para cada task\n # is_done.count(True) retorna a quantidade de tasks que já terminaram.\n is_done = list()\n done_count = 0\n while is_done.count(True) != len(futures):\n is_done = list()\n for f in futures:\n is_done.append(f.done())\n\n # Só atualiza a pbar se o valor for diferente.\n if is_done.count(True) != done_count:\n done_count = is_done.count(True)\n # Reset é necessário por que a quantidade de iterações\n # é maior que a quantidade de jobs.\n pbar2.reset(total=len(futures))\n # Atualiza a pbar\n pbar2.update(done_count)\n\n if done_count < len(futures):\n sleep(3)\n",
"_____no_output_____"
]
],
[
[
"After filtering stars in HealPixels, join all the HP into a single catalog called final cat.",
"_____no_output_____"
]
],
[
[
"ipix_clean_cats = [i.split('.')[0] + '_clean_dist.fits' for i in ipix_cats]\njoin_cats_clean(ipix_clean_cats,\n param['final_cat'], param['ra_str'], param['dec_str'])\n",
"_____no_output_____"
]
],
[
[
"Plot clusters comparing filtered and not filtered stars in each cluster. The region sampled is the center of the cluster where the crowding is more intense.</br>\nBelow the clusters with stars were filtered by max distance.",
"_____no_output_____"
]
],
[
[
"plot_clusters_clean(ipix_cats, ipix_clean_cats,\n param['nside_ini'], param['ra_str'], param['dec_str'], 0.01)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f831abf5986e7356bbbbaf2195a62fb2e2a310 | 106,200 | ipynb | Jupyter Notebook | Linear_Regression.ipynb | tejashrigadre/Linear_Regression | e37350cf51a4227affe85a8ea0e0e6f9815f9b7e | [
"MIT"
] | null | null | null | Linear_Regression.ipynb | tejashrigadre/Linear_Regression | e37350cf51a4227affe85a8ea0e0e6f9815f9b7e | [
"MIT"
] | null | null | null | Linear_Regression.ipynb | tejashrigadre/Linear_Regression | e37350cf51a4227affe85a8ea0e0e6f9815f9b7e | [
"MIT"
] | null | null | null | 106,200 | 106,200 | 0.822881 | [
[
[
"Let's see the simple code for Linear Regression.\nWe will be creating a model to predict weight of a person based on independent variable height using simple linear regression.\nweight-height dataset is downloaded from kaggle\nhttps://www.kaggle.com/sonalisingh1411/linear-regression-using-weight-height/data",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive')\n%cd /content/drive/My\\ Drive",
"Drive already mounted at /content/drive; to attempt to forcibly remount, call drive.mount(\"/content/drive\", force_remount=True).\n/content/drive/My Drive\n"
],
[
"%cd 'Colab Notebooks'",
"/content/drive/My Drive/Colab Notebooks\n"
]
],
[
[
"1. Simple Linear Regression with one independent variable",
"_____no_output_____"
],
[
"We will read the data file and do some data exploration.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd \nimport matplotlib.pyplot as plt\n",
"_____no_output_____"
],
[
"df = pd.read_csv('weight-height.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df['Gender'].unique()",
"_____no_output_____"
],
[
"df.corr()",
"_____no_output_____"
]
],
[
[
"We can see that there is high co-relation between height and weight columns.\nWe will use Linear Regression model from sklearn library",
"_____no_output_____"
]
],
[
[
"x = df['Height']\ny = df['Weight']",
"_____no_output_____"
]
],
[
[
"We will split the data into train and test datasets using sklearn preprocessing library",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(x,y,test_size=0.2, random_state=42)",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"X_train = X_train.to_numpy()\nX_train = X_train.reshape(-1,1)",
"_____no_output_____"
]
],
[
[
"reshape() is called to make X_train 2-dimesional that is row and column format",
"_____no_output_____"
]
],
[
[
"X_train.shape",
"_____no_output_____"
],
[
"X_test = X_test.to_numpy()\nX_test = X_test.reshape(-1,1)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression",
"_____no_output_____"
],
[
"model = LinearRegression()\nmodel.fit(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"model is created as instnace of LinearRegression. \nWith .fit() method, optimal values of coefficients (b0,b1) are calculated using existing input X_train and y_train.",
"_____no_output_____"
]
],
[
[
"model.score(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"The arguments to .score() are also X_train and y_train and it returns the R2 (coefficient of determination).",
"_____no_output_____"
]
],
[
[
"Intercept,coef = model.intercept_,model.coef_\nprint(\"Intercept is :\",Intercept, sep='\\n')\nprint(\"Coefficient/slope is :\",coef , sep='\\n')",
"Intercept is :\n-349.7878205824451\nCoefficient/slope is :\n[7.70218561]\n"
]
],
[
[
"Model attributes model.intercept_, model.coef_ give the value of (b01,b1)",
"_____no_output_____"
],
[
"Now, we will use trained model to predict on test data",
"_____no_output_____"
]
],
[
[
"y_pred = model.predict(X_test)",
"_____no_output_____"
],
[
"y_pred",
"_____no_output_____"
]
],
[
[
"We can use also slope-intercept of line y = y-intercept + slope * x to predict the values on test data. We will use model.intercept_ and model.coef_ value for predictiion",
"_____no_output_____"
]
],
[
[
"y_pred1 = Intercept + coef * X_test\ny_pred1",
"_____no_output_____"
]
],
[
[
"We can see output of both y_pred and y_pred1 is same.",
"_____no_output_____"
],
[
"We will plot the graph of predicted and actual values of weights using seaborn and matplotlib library",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nax = sns.regplot(x=y_pred, y=y_test,\n x_estimator=np.mean)",
"_____no_output_____"
]
],
[
[
"To clearly see the the plot, let's draw 20 samples from training dataset with actual weight values and plot it with predicted weight values for training dataset.\nThe red dots represent the actual weight values(20 samples drawn) and the green line represents the predcted weight values by the model. The vertical distance between red dot and the green line is the error which we have to minimize to best fit the model.",
"_____no_output_____"
]
],
[
[
"plt.scatter(X_train[0:20], y_train[0:20], color = \"red\")\nplt.plot(X_train, model.predict(X_train), color = \"green\")\nplt.title(\"Weight vs Height\")\nplt.xlabel(\"Height\")\nplt.ylabel(\"Weight\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"2. Multiple Linear Regressiom",
"_____no_output_____"
],
[
"A regression with 2 or more independet variables is multiple linear regression. \nWe will use same dataset to implemet multiple linear regression.\nThe 2 independent variables will be gender and height which be used to predict the weight.",
"_____no_output_____"
]
],
[
[
"x = df.drop(columns = 'Weight')\ny = df['Weight']",
"_____no_output_____"
],
[
"x.columns",
"_____no_output_____"
]
],
[
[
"Gender column is categorical. We can not use it directly as model can work only with numbers. We have to convert it to one-hot-encoding using pandas get_dummies() method. A new column will be create dropping earlier column . The new column contain values 1 and 0 for male and female respectively.",
"_____no_output_____"
]
],
[
[
"x = pd.get_dummies(x, columns = ['Gender'], drop_first = True)",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"print(x.shape)\nprint(y.shape)",
"(10000, 2)\n(10000,)\n"
]
],
[
[
"Rest of the steps will be same as simple linear regression.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(x,y,test_size=0.2, random_state=42)",
"_____no_output_____"
],
[
"X_train = X_train.to_numpy()\nX_train = X_train.reshape(-1,2)\n\nX_test = X_test.to_numpy()\nX_test = X_test.reshape(-1,2)",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"mulLR = LinearRegression()\nmulLR.fit(X_train,y_train)",
"_____no_output_____"
],
[
"mulLR.score(X_train,y_train)",
"_____no_output_____"
],
[
"Intercept,coef = mulLR.intercept_,mulLR.coef_\nprint(\"Intercept is :\",Intercept, sep='\\n')\nprint(\"Coefficient/slope is :\",coef , sep='\\n')",
"Intercept is :\n-244.69356793639193\nCoefficient/slope is :\n[ 5.97314123 19.34720343]\n"
]
],
[
[
"Coefficient array will have 2 values for gender and height respectively.",
"_____no_output_____"
]
],
[
[
"y_pred = mulLR.predict(X_test)\ny_pred",
"_____no_output_____"
]
],
[
[
"Alternate method : Predicting weight using coefficient and intercept values in equation",
"_____no_output_____"
]
],
[
[
"y_pred1 = Intercept + np.sum(coef * X_test, axis = 1)\ny_pred1",
"_____no_output_____"
]
],
[
[
"y_pred and y_pred1 both have same predicted values ",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nax = sns.regplot(x=y_pred, y=y_test,\n x_estimator=np.mean)",
"_____no_output_____"
]
],
[
[
"Above plot shows graph representing predicted and actual weight values on test dataset.",
"_____no_output_____"
],
[
"3. Polynomial Regression",
"_____no_output_____"
],
[
"We will use polynomial regression to find the weight using same dataset. Note that polynomial regression is the special case of linear regression.",
"_____no_output_____"
],
[
"Import class PolynomialFeatures from sklearn.preprocessing",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import PolynomialFeatures",
"_____no_output_____"
],
[
"x = df['Height']\ny = df['Weight']",
"_____no_output_____"
],
[
"transformer = PolynomialFeatures(degree = 2, include_bias = False)",
"_____no_output_____"
]
],
[
[
"We have to include terms like x2(x squared) as additional features when using polynomial regression.\nWe have to transform the inputfor that transformer is defined with degree (defines the degree of polynomial regression function) and include_bias decides whether to include bias or not.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(x,y,test_size=0.2, random_state=42)",
"_____no_output_____"
],
[
"X_train = X_train.to_numpy()\nX_train = X_train.reshape(-1,1)\n\nX_test = X_test.to_numpy()\nX_test = X_test.reshape(-1,1)",
"_____no_output_____"
],
[
"transformer.fit(X_train) ",
"_____no_output_____"
],
[
"X_trans = transformer.transform(X_train)",
"_____no_output_____"
]
],
[
[
"Above two lines of code can be fit into one line as below, both will give same output",
"_____no_output_____"
]
],
[
[
"x_trans = PolynomialFeatures(degree=2, include_bias=False).fit_transform(X_train)\n",
"_____no_output_____"
],
[
"X_transtest = PolynomialFeatures(degree=2, include_bias=False).fit_transform(X_test)",
"_____no_output_____"
]
],
[
[
"Each value in the first column is squared and stored in second column as feature ",
"_____no_output_____"
]
],
[
[
"print(x_trans)",
"[[ 61.39164365 3768.93390949]\n [ 74.6976372 5579.7370037 ]\n [ 68.50781491 4693.32070353]\n ...\n [ 64.3254058 4137.75783102]\n [ 69.07449203 4771.28544943]\n [ 67.58883983 4568.25126988]]\n"
]
],
[
[
"Create and fit the model",
"_____no_output_____"
]
],
[
[
"poly_LR = LinearRegression().fit(x_trans,y_train)",
"_____no_output_____"
],
[
"poly_LR.score(x_trans,y_train)",
"_____no_output_____"
],
[
"y_pred = poly_LR.predict(X_transtest)\ny_pred",
"_____no_output_____"
],
[
"Intercept,coef = mulLR.intercept_,mulLR.coef_\nprint(\"Intercept is :\",Intercept, sep='\\n')\nprint(\"Coefficient/slope is :\",coef , sep='\\n')",
"Intercept is :\n-244.69356793639193\nCoefficient/slope is :\n[ 5.97314123 19.34720343]\n"
]
],
[
[
"The score of ploynomial regression can slighly be better than linear regression due to added complexity but the high R2 scoe does not always mean good model. Sometimes ploynomial regression could lead to overfitting due to its complexity in defining the equation for regression.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f8382c8d263814fa224266896387c79edd9ab6 | 519,910 | ipynb | Jupyter Notebook | Model.ipynb | markorland/markorland | a14d75b0d75cdaaf49209c0cb9074e5d05992a0e | [
"MIT"
] | null | null | null | Model.ipynb | markorland/markorland | a14d75b0d75cdaaf49209c0cb9074e5d05992a0e | [
"MIT"
] | 1 | 2018-04-06T21:37:39.000Z | 2018-04-06T21:37:39.000Z | Model.ipynb | markorland/markorland | a14d75b0d75cdaaf49209c0cb9074e5d05992a0e | [
"MIT"
] | 2 | 2018-04-06T19:57:26.000Z | 2019-03-13T02:30:46.000Z | 43.178307 | 1,134 | 0.351297 | [
[
[
"import pandas as pd\nimport numpy as np\n\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.metrics.pairwise import cosine_similarity\nfrom nltk.corpus import stopwords\nfrom sklearn.decomposition import TruncatedSVD\n\nstop = stopwords.words('english')\n\nimport matplotlib.pyplot as plt\n\nplt.style.use('ggplot')\nplt.style.use('default')\n# plt.style.use('dark_background')\n%matplotlib inline",
"_____no_output_____"
],
[
"whiskey = pd.read_csv('./Scraping/Scraped_Data/master_whiskey.csv', encoding='latin1')\n\n# drop f's and no grades\nwhiskey = whiskey[(whiskey['grade'] != 'f') & (whiskey['grade'] != 'No Grade')].copy()\n\n# fill missing ages, vintages, region, abv\nwhiskey['age'].fillna('No Age', inplace=True)\nwhiskey['vint'].fillna('No Vint', inplace=True)\nwhiskey['region'].fillna('No Region', inplace=True)\nwhiskey['abv'].fillna('No Abv', inplace=True)\n\n# drop \"yrs\" for the age\nwhiskey['age'] = whiskey['age'].map(lambda x : x.split(' yrs')[0])\n\n# drop missing revies\nwhiskey = whiskey.dropna().copy()\n\n# convert letter grades to numbers\ngrade_replacement = {'a': 5, 'aminus': 4.5, 'bplus': 4.25, 'b': 4, 'bminus': 3.5, 'cplus': 3.25, 'c': 3, 'cminus': 2.5, 'dplus': 2.25, 'd': 2, 'dminus': 1.5}\n # {'A': 5, 'A-': 4.75, 'B+': 4.25, 'B': 4, 'B-': 3.75, 'C+': 3.25, 'C': 3, 'C-': 2.75, 'D+': 2.25, 'D': 2, 'D-': 1.75}\nwhiskey['grade'] = whiskey['grade'].map(grade_replacement)",
"_____no_output_____"
],
[
"beer = pd.read_csv('./Scraping/Scraped_Data/master_beer.csv', encoding='latin1')\n\n# remove reviews without text reviews\nbeer = beer[beer['r_text'].str.contains('No Review') == False]\nbeer = beer[beer['breakdown'].str.contains('look:') == True]\n\n# fill missing user name\nbeer['username'].fillna('Missing Username', inplace=True)\n# fill nan ibu's\nbeer['ibu'].fillna('No IBU', inplace=True)",
"_____no_output_____"
],
[
"# create id's for modeling\nwhiskey['id'] = 'w'\nbeer['id'] = 'b'",
"_____no_output_____"
],
[
"# creating all_reviews df\nbeer.rename({'r_text':'review', 'score_y':'user_rating'}, axis=1, inplace=True)\nwhiskey.rename({'w_name':'name', 'grade':'user_rating', 'w_type':'style'}, axis=1, inplace=True)\n\nall_reviews = beer[['name', 'review', 'username', 'user_rating', 'abv', 'style', 'id']].append(whiskey[['name', 'review', 'username', 'user_rating', 'abv', 'style', 'id']])",
"_____no_output_____"
]
],
[
[
"## Cleaning",
"_____no_output_____"
]
],
[
[
"beer.head()",
"_____no_output_____"
],
[
"beer.shape",
"_____no_output_____"
],
[
"beer = beer[beer['r_text'].str.contains('No Review') == False]",
"_____no_output_____"
],
[
"beer.shape",
"_____no_output_____"
],
[
"beer[beer['breakdown'].str.contains('look:') != True]['name'].value_counts()",
"_____no_output_____"
],
[
"beer = beer[beer['breakdown'].str.contains('look:') == True]",
"_____no_output_____"
],
[
"beer.shape",
"_____no_output_____"
],
[
"beer.isnull().sum()",
"_____no_output_____"
],
[
"beer['username'].fillna('Missing Username', inplace=True)",
"_____no_output_____"
],
[
"beer['ibu'].value_counts()",
"_____no_output_____"
],
[
"beer['ibu'].fillna('No IBU', inplace=True)",
"_____no_output_____"
],
[
"beer.isnull().sum()",
"_____no_output_____"
],
[
"whiskey.isnull().sum()",
"_____no_output_____"
],
[
"whiskey['age'].value_counts()",
"_____no_output_____"
],
[
"whiskey[whiskey['age'].isnull()].head()",
"_____no_output_____"
],
[
"whiskey['age'].fillna('No Age', inplace=True)",
"_____no_output_____"
],
[
"whiskey[whiskey['vint'].isnull()].head()",
"_____no_output_____"
],
[
"whiskey['vint'].fillna('No Vint', inplace=True)",
"_____no_output_____"
],
[
"whiskey[whiskey['region'].isnull()].head()",
"_____no_output_____"
],
[
"whiskey['region'].fillna('No Region', inplace=True)",
"_____no_output_____"
],
[
"whiskey['age'] = whiskey['age'].map(lambda x : x.split(' yrs')[0])",
"_____no_output_____"
],
[
"for i in whiskey['age']:\n i = i.split(' yrs')[0]\n print(i)",
"12\n12\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n5\n5\n16\n16\nNo Age\n21\n21\nNo Age\nNAS\nNAS\nNo Age\nNo Age\nNo Age\n14\n17\n15\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n11\n25\n15\n15\n15\nNo Age\nNo Age\n9\n18\n19\n17\n17\n32\n19\n19\n19\n19\n35\n15\n21\n21\n15\n26\n26\n26\n26\n20\nNo Age\n12\n12\n12\n12\n11\n11\nNo Age\nNo Age\n18\n18\n23\n23\n23\nNo Age\n40\n40\n23\n20\n20\nNo Age\n27\n30\n30\n30\n31\n31\n21\n21\n15\n14\n20\n8\n5\n5\n5\n5\n12\n12\n12\n12\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n3\n3\n3\n17\n17\n7\n2\nNo Age\nNo Age\nNo Age\n22\n22\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n7\n7\n7\n7\n23\n13\n13\n18\n18\n18\n18\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n20\n20\nNo Age\n13\n13\n10\n8\n12\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n24\n24\n24\n24\n14\n14\n19\n8\n8\n21\n21\n14\nNo Age\nNo Age\nNo Age\nNo Age\n14\n15\n15\n21\n19\n19\n8\n8\n8\nNo Age\nNAS\nNAS\nNAS\n21\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n37\nNo Age\nNo Age\nNo Age\n33\n33\n33\n33\n33\n19\n19\n23\n14\n14\n18\nNo Age\nNo Age\nNo Age\nNo Age\n7\nNo Age\nNo Age\nNo Age\n10\nNo Age\nNo Age\n25\n25\n14\n8\n8\n24\n24\n20\n7\nNo Age\n12\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n23\n23\n23\n14\n14\nNo Age\nNAS\nNAS\nNo Age\n4\nNo Age\nNo Age\nNo Age\n12\n10\n6\n10\n10\n10\n16\n16\n16\n16\n16\n15\n15\n15\n16\n16\n16\n16\n13\nNo Age\nNo Age\nNo Age\nNAS\nNAS\nNAS\nNAS\nNAS\nNAS\nNAS\nNo Age\n29\n29\n26\n26\n19\n25\n8\nNo Age\nNo Age\nNo Age\n8\n29\n29\n29\n21\n21\n16\n16\n18\n11\n12\n21\n21\n21\n21\n21\n21\n21\n30\n19\n19\n19\n19\n30\n14\n14\n25\n25\n7\n7\n7\n7\n27\n27\nNo Age\nNAS\nNAS\n50\n50\n50\n50\n50\n25\n25\n26\n26\nNo Age\nNo Age\nNo Age\nNo Age\nNo Age\n18\n36\n36\n36\n36\n36\n37\n37\n13\n10\n10\nNo Age\n10\n10\nNo Age\n14\nNo Age\nNo Age\n12\n18\n18\n18\n19\n21\n21\n21\n25\n25\n25\nNo Age\nNo Age\n12\n19\n5\n5\n5\nNAS\nNAS\n16\n16\n16\n16\nNo Age\nNo Age\nNo Age\n35\nNo Age\nNo Age\nNo Age\nNAS\nNAS\nNAS\nNo Age\n16\n18\n18\n18\n18\n8\n8\n8\n8\n20\n30\nNo Age\n10\n8\nNo Age\n7\n7\n7\nNo Age\nNo Age\n8\n2\n2\n3\n3\nNo Age\nNo Age\n2\n2\n2\nNo Age\nNo Age\nNo Age\nNo Age\n17\n17\n25\n"
],
[
"whiskey['age'].value_counts()",
"_____no_output_____"
],
[
"whiskey.isnull().sum()",
"_____no_output_____"
],
[
"whiskey2.isnull().sum()",
"_____no_output_____"
],
[
"whiskey = whiskey.dropna().copy()",
"_____no_output_____"
],
[
"whiskey.shape",
"_____no_output_____"
],
[
"whiskey['review'].dropna(inplace=True)",
"_____no_output_____"
],
[
"whiskey.shape",
"_____no_output_____"
],
[
"whiskey.head()",
"_____no_output_____"
],
[
"beer.head()",
"_____no_output_____"
],
[
"beer.rename({'r_text':'review', 'score_y':'user_rating'}, axis=1, inplace=True)\n\nwhiskey.rename({'w_name':'name', 'grade':'user_rating', 'w_type':'style'}, axis=1, inplace=True)",
"_____no_output_____"
],
[
"all_reviews[all_reviews['username'] == 'rodbeermunch']",
"_____no_output_____"
],
[
"all_reviews = beer[['name', 'review', 'username', 'user_rating', 'abv', 'style', 'id']].append(whiskey[['name', 'review', 'username', 'user_rating', 'abv', 'style', 'id']])\nall_reviews.head()",
"_____no_output_____"
],
[
"all_reviews['review'].isnull().sum()",
"_____no_output_____"
],
[
"all_reviews.shape",
"_____no_output_____"
],
[
"all_reviews = all_reviews.dropna().copy()",
"_____no_output_____"
]
],
[
[
"## Modeling",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics.pairwise import cosine_similarity",
"_____no_output_____"
],
[
"from sklearn.decomposition import TruncatedSVD",
"_____no_output_____"
],
[
"vect = CountVectorizer(ngram_range=(2,2), stop_words=stop, min_df=2)\nX = vect.fit_transform(all_reviews['review'])",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"svd = TruncatedSVD(n_components=5, n_iter=7, random_state=42)",
"_____no_output_____"
],
[
"X_svd = svd.fit_transform(X)",
"_____no_output_____"
],
[
"# import pickle\n\n# with open('C:/Users/Mark/Personal_GitHub/Portfolio/Capstone_What_Should_I_Drink/Pickle/svd.pkl', wb+) as f:\n# pickle.dump(X_svd, f)",
"_____no_output_____"
],
[
"X_svd.shape",
"_____no_output_____"
],
[
"cosine_similarity(X_svd)",
"_____no_output_____"
],
[
"import sys\n\nsys.getsizeof(X_svd) / 1000000000",
"_____no_output_____"
]
],
[
[
"## Model with grouped reviews",
"_____no_output_____"
]
],
[
[
"grouped_reviews = all_reviews.groupby('name')['review'].sum()",
"_____no_output_____"
],
[
"grouped_reviews.head()",
"_____no_output_____"
],
[
"from sklearn.metrics.pairwise import cosine_similarity",
"_____no_output_____"
],
[
"from sklearn.decomposition import TruncatedSVD",
"_____no_output_____"
],
[
"vect = CountVectorizer(ngram_range=(2,2), stop_words=stop, min_df=2)\nX = vect.fit_transform(grouped_reviews)",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"svd = TruncatedSVD(n_components=100, n_iter=7, random_state=42)",
"_____no_output_____"
],
[
"X_svd = svd.fit_transform(X)",
"_____no_output_____"
],
[
"X_svd.shape",
"_____no_output_____"
],
[
"# import pickle\n\n# with open('C:/Users/Mark/Personal_GitHub/Portfolio/Capstone_What_Should_I_Drink/Pickle/svd.pkl', wb+) as f:\n# pickle.dump(X_svd, f)",
"_____no_output_____"
],
[
"cos_sim = cosine_similarity(X_svd, X_svd)",
"_____no_output_____"
],
[
"cos_sim",
"_____no_output_____"
],
[
"cos_sim.shape",
"_____no_output_____"
],
[
"df_grouped_reviews = pd.DataFrame(grouped_reviews)\ndf_grouped_reviews.head()",
"_____no_output_____"
],
[
"df_grouped_reviews.index",
"_____no_output_____"
],
[
"pd.DataFrame(cos_sim)",
"_____no_output_____"
],
[
"df_cos_sim = pd.DataFrame(cos_sim, index=df_grouped_reviews.index)\ndf_cos_sim",
"_____no_output_____"
],
[
"df_cos_reviews = pd.concat([df_grouped_reviews, df_cos_sim], axis=1)\ndf_cos_reviews.head()",
"_____no_output_____"
],
[
"df_cos_reviews = df_cos_reviews.drop('review', axis=1)",
"_____no_output_____"
],
[
"df_cos_reviews.head()",
"_____no_output_____"
],
[
"df_cos_reviews.columns = df_cos_reviews.index",
"_____no_output_____"
],
[
"df_cos_reviews",
"_____no_output_____"
],
[
"all_reviews.head()",
"_____no_output_____"
],
[
"all_reviews_cosine = all_reviews.merge(df_cos_reviews, left_on='name', right_index=True)",
"_____no_output_____"
],
[
"all_reviews_cosine.head()",
"_____no_output_____"
],
[
"# all_reviews_cosine.to_csv('./Scraping/Scraped_Data/Data/all_reviews_cosine.csv', index=False)",
"_____no_output_____"
]
],
[
[
"## Recommender",
"_____no_output_____"
]
],
[
[
"all_reviews_cosine = pd.read_csv('./Scraping/Scraped_Data/Data/all_reviews_cosine.csv')",
"C:\\Users\\Mark\\Anaconda3\\lib\\site-packages\\IPython\\core\\interactiveshell.py:2728: DtypeWarning: Columns (4) have mixed types. Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"beer.head()",
"_____no_output_____"
],
[
"whiskey.head()",
"_____no_output_____"
],
[
"all_reviews_cosine.head()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7f84b07671306e7ed6ec8947fdb1d4536d7df25 | 108,247 | ipynb | Jupyter Notebook | src/recommendation/data_analysis/data_analysis_1.ipynb | odobenuskr/2019_capstone_FlexAds | e37708484d983326806c0d6a0c5ae7157ea57e59 | [
"MIT"
] | 12 | 2019-03-08T15:49:24.000Z | 2020-08-03T05:01:44.000Z | src/recommendation/data_analysis/data_analysis_1.ipynb | kookmin-sw/2019-cap1-2019_4 | e64aa4590ac58dca535024118ed4d4bd0b3eb699 | [
"MIT"
] | 63 | 2019-03-08T15:23:19.000Z | 2019-06-01T07:10:23.000Z | src/recommendation/data_analysis/data_analysis_1.ipynb | odobenuskr/2019_capstone_FlexAds | e37708484d983326806c0d6a0c5ae7157ea57e59 | [
"MIT"
] | 11 | 2019-03-10T02:12:24.000Z | 2021-04-02T07:16:01.000Z | 44.711689 | 18,344 | 0.502776 | [
[
[
"1) aisles.csv : aisle_id, aisle\n - 소분류\n \n2) departments.csv : department_id, department\n - 대분류\n \n2) order_products.csv : order_id, product_id, add_to_cart_order, reordered : train, prior\n - 주문id, 상품 id, 장바구니에 담긴 순서, 재구매 여부\n \n3) orders.csv : order_id, user_id, eval_set, order_number, order_dow, order_hour_of_day, day_since\n - 주문 id, 사용자 id, 데이터 구분, 주문 수량, 주문 요일, 주문 시간, 재구매까지 걸린 시간\n \n4) products.csv : product_id, product_name, aisle_id, department_id\n - 상품 id, 상품 이름, 소분류 id, 대분류 id\n \n> 내가 이 상품을 살것인가 추천하는 기준? 재구매가 몇번이상인지, 재구매까지 걸린 기간이 짧음\n\n> 그 외 추천 : 내가 사는 물건이랑 같은 소분류안에 있는 것, 내가 사는 물건과 겹치는 게 많은 사용자의 구매목록에서 내가 사지 않은 것\n",
"_____no_output_____"
]
],
[
[
"import numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\nimport matplotlib.pyplot as plt\nimport seaborn as sns\ncolor = sns.color_palette()\n\n%matplotlib inline",
"_____no_output_____"
],
[
"# data\n\ntrain = pd.read_csv(\"order_products__train.csv\")\nprior = pd.read_csv(\"order_products__prior.csv\")\norders_df = pd.read_csv(\"orders.csv\")\nproducts_df = pd.read_csv(\"products.csv\")\naisles_df = pd.read_csv(\"aisles.csv\")\ndepartments_df = pd.read_csv(\"departments.csv\")\nsubmit = pd.read_csv(\"sample_submission.csv\")\n",
"_____no_output_____"
],
[
"submit",
"_____no_output_____"
],
[
"# check orders.csv\n# 확인할만큼 괄호에 숫자 넣기 (default = 5)\n\norders_df.head(3)",
"_____no_output_____"
],
[
"train",
"_____no_output_____"
],
[
"prior",
"_____no_output_____"
],
[
"# check order_products__train.csv\n# 주문된 상품에 관한 정보\n\ntrain.head(3)",
"_____no_output_____"
],
[
"# 데이터 분류\n# eval_set이 기준 : 총 3개로 나뉘고 그 3개의 덩어리안에 데이터가 몇개씩 있는지 확인\n\ncnt_srs = orders_df.eval_set.value_counts()\nprint (cnt_srs)",
"prior 3214874\ntrain 131209\ntest 75000\nName: eval_set, dtype: int64\n"
],
[
"def get_unique_count(x):\n return len(np.unique(x))\n\n# eval_set이 같은 것끼리 그룹화\ncnt_srs = orders_df.groupby(\"eval_set\")[\"user_id\"].aggregate(get_unique_count)\n\nprint (cnt_srs)\n\n#총 206209명의 고객들이 있음",
"eval_set\nprior 206209\ntest 75000\ntrain 131209\nName: user_id, dtype: int64\n"
],
[
"#order number 중복 있음 : 유니크한 값 아니고 단지 \"구매 횟수\" 라는 것을 확인\n\ncnt_srs = orders_df.groupby(\"user_id\")[\"order_number\"].aggregate(np.max).reset_index()\ncnt_srs\n\n# userid - ordernumber\ncnt_srs = cnt_srs.order_number.value_counts()\ncnt_srs",
"_____no_output_____"
],
[
"# 요일 기준 : 요일당 주문 건수\n\ncnt_day = orders_df.order_dow.value_counts()\nprint (cnt_day)",
"0 600905\n1 587478\n2 467260\n5 453368\n6 448761\n3 436972\n4 426339\nName: order_dow, dtype: int64\n"
],
[
"plt.figure(figsize=(12,8))\nsns.countplot(x=\"order_dow\", data=orders_df, color=color[4])\nplt.ylabel('Count', fontsize=12)\nplt.xlabel('Day of week', fontsize=12)\nplt.xticks(rotation='vertical')\nplt.title(\"Frequency of order by week day\", fontsize=15)\nplt.show()",
"_____no_output_____"
],
[
"# 물건이 제일 많이 주문된 시간은 언제? \n\ncnt_hour = orders_df.order_hour_of_day.value_counts()\nprint (cnt_hour)",
"10 288418\n11 284728\n15 283639\n14 283042\n13 277999\n12 272841\n16 272553\n9 257812\n17 228795\n18 182912\n8 178201\n19 140569\n20 104292\n7 91868\n21 78109\n22 61468\n23 40043\n6 30529\n0 22758\n1 12398\n5 9569\n2 7539\n4 5527\n3 5474\nName: order_hour_of_day, dtype: int64\n"
],
[
"plt.figure(figsize=(12,8))\nsns.countplot(x=\"order_hour_of_day\", data=orders_df, color=color[1])\nplt.ylabel('Count', fontsize=12)\nplt.xlabel('Hour of day', fontsize=12)\nplt.xticks(rotation='vertical')\nplt.title(\"Frequency of order by hour of day\", fontsize=15)\nplt.show()",
"_____no_output_____"
],
[
"# 요일마다 어떤 시간에 제일 주문건수가 많은가?? \n\ngrouped_df = orders_df.groupby([\"order_dow\", \"order_hour_of_day\"])[\"order_number\"].aggregate(\"count\").reset_index()\nprint (grouped_df)",
" order_dow order_hour_of_day order_number\n0 0 0 3936\n1 0 1 2398\n2 0 2 1409\n3 0 3 963\n4 0 4 813\n5 0 5 1168\n6 0 6 3329\n7 0 7 12410\n8 0 8 28108\n9 0 9 40798\n10 0 10 48465\n11 0 11 51035\n12 0 12 51443\n13 0 13 53849\n14 0 14 54552\n15 0 15 53954\n16 0 16 49463\n17 0 17 39753\n18 0 18 29572\n19 0 19 22654\n20 0 20 18277\n21 0 21 14423\n22 0 22 11246\n23 0 23 6887\n24 1 0 3674\n25 1 1 1830\n26 1 2 1105\n27 1 3 748\n28 1 4 809\n29 1 5 1607\n.. ... ... ...\n138 5 18 24310\n139 5 19 18741\n140 5 20 13322\n141 5 21 9515\n142 5 22 7498\n143 5 23 5265\n144 6 0 3306\n145 6 1 1919\n146 6 2 1214\n147 6 3 863\n148 6 4 802\n149 6 5 1136\n150 6 6 3243\n151 6 7 11319\n152 6 8 22960\n153 6 9 30839\n154 6 10 35665\n155 6 11 36994\n156 6 12 37121\n157 6 13 37564\n158 6 14 38748\n159 6 15 38093\n160 6 16 35562\n161 6 17 30398\n162 6 18 24157\n163 6 19 18346\n164 6 20 13392\n165 6 21 10501\n166 6 22 8532\n167 6 23 6087\n\n[168 rows x 3 columns]\n"
],
[
"# 재구매까지 걸리는 시간별 구매량 \n# 30일이 제일 많고 26일이 제일 적음\n\ncnt_prior_order = orders_df.days_since_prior_order.value_counts()\nprint (cnt_prior_order)",
"30.0 369323\n7.0 320608\n6.0 240013\n4.0 221696\n3.0 217005\n5.0 214503\n2.0 193206\n8.0 181717\n1.0 145247\n9.0 118188\n14.0 100230\n10.0 95186\n13.0 83214\n11.0 80970\n12.0 76146\n0.0 67755\n15.0 66579\n16.0 46941\n21.0 45470\n17.0 39245\n20.0 38527\n18.0 35881\n19.0 34384\n22.0 32012\n28.0 26777\n23.0 23885\n27.0 22013\n24.0 20712\n25.0 19234\n29.0 19191\n26.0 19016\nName: days_since_prior_order, dtype: int64\n"
],
[
"train.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1384617 entries, 0 to 1384616\nData columns (total 4 columns):\norder_id 1384617 non-null int64\nproduct_id 1384617 non-null int64\nadd_to_cart_order 1384617 non-null int64\nreordered 1384617 non-null int64\ndtypes: int64(4)\nmemory usage: 42.3 MB\n"
],
[
"# 학습데이터 확인 > orders 랑 똑같음\n\ntrain.head()",
"_____no_output_____"
],
[
"# summary : reorder 가 횟수가 아니라 맞다, 아니다뿐이라 별 도움이 안 됨ㅎ... > 각 상품마다 재구매 기간의 평균을 구해보까..?\n\npd.set_option('display.float_format', lambda x: '%.3f' % x)\ntrain.describe()",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f84f15db71976cd9224bf32b8eeaa67de0a005 | 63,446 | ipynb | Jupyter Notebook | intro-to-tensorflow/intro_to_tensorflow.ipynb | ajmaradiaga/tf-examples | 48c795d2b324bfeccfcf2dd21502a19dfa287418 | [
"Apache-2.0"
] | null | null | null | intro-to-tensorflow/intro_to_tensorflow.ipynb | ajmaradiaga/tf-examples | 48c795d2b324bfeccfcf2dd21502a19dfa287418 | [
"Apache-2.0"
] | null | null | null | intro-to-tensorflow/intro_to_tensorflow.ipynb | ajmaradiaga/tf-examples | 48c795d2b324bfeccfcf2dd21502a19dfa287418 | [
"Apache-2.0"
] | null | null | null | 81.865806 | 35,636 | 0.786291 | [
[
[
"<h1 align=\"center\">TensorFlow Neural Network Lab</h1>",
"_____no_output_____"
],
[
"<img src=\"image/notmnist.png\">\nIn this lab, you'll use all the tools you learned from *Introduction to TensorFlow* to label images of English letters! The data you are using, <a href=\"http://yaroslavvb.blogspot.com/2011/09/notmnist-dataset.html\">notMNIST</a>, consists of images of a letter from A to J in different fonts.\n\nThe above images are a few examples of the data you'll be training on. After training the network, you will compare your prediction model against test data. Your goal, by the end of this lab, is to make predictions against that test set with at least an 80% accuracy. Let's jump in!",
"_____no_output_____"
],
[
"To start this lab, you first need to import all the necessary modules. Run the code below. If it runs successfully, it will print \"`All modules imported`\".",
"_____no_output_____"
]
],
[
[
"import hashlib\nimport os\nimport pickle\nfrom urllib.request import urlretrieve\n\nimport numpy as np\nfrom PIL import Image\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import LabelBinarizer\nfrom sklearn.utils import resample\nfrom tqdm import tqdm\nfrom zipfile import ZipFile\n\nprint('All modules imported.')",
"All modules imported.\n"
]
],
[
[
"The notMNIST dataset is too large for many computers to handle. It contains 500,000 images for just training. You'll be using a subset of this data, 15,000 images for each label (A-J).",
"_____no_output_____"
]
],
[
[
"def download(url, file):\n \"\"\"\n Download file from <url>\n :param url: URL to file\n :param file: Local file path\n \"\"\"\n if not os.path.isfile(file):\n print('Downloading ' + file + '...')\n urlretrieve(url, file)\n print('Download Finished')\n\n# Download the training and test dataset.\ndownload('https://s3.amazonaws.com/udacity-sdc/notMNIST_train.zip', 'notMNIST_train.zip')\ndownload('https://s3.amazonaws.com/udacity-sdc/notMNIST_test.zip', 'notMNIST_test.zip')\n\n# Make sure the files aren't corrupted\nassert hashlib.md5(open('notMNIST_train.zip', 'rb').read()).hexdigest() == 'c8673b3f28f489e9cdf3a3d74e2ac8fa',\\\n 'notMNIST_train.zip file is corrupted. Remove the file and try again.'\nassert hashlib.md5(open('notMNIST_test.zip', 'rb').read()).hexdigest() == '5d3c7e653e63471c88df796156a9dfa9',\\\n 'notMNIST_test.zip file is corrupted. Remove the file and try again.'\n\n# Wait until you see that all files have been downloaded.\nprint('All files downloaded.')",
"All files downloaded.\n"
],
[
"def uncompress_features_labels(file):\n \"\"\"\n Uncompress features and labels from a zip file\n :param file: The zip file to extract the data from\n \"\"\"\n features = []\n labels = []\n\n with ZipFile(file) as zipf:\n # Progress Bar\n filenames_pbar = tqdm(zipf.namelist(), unit='files')\n \n # Get features and labels from all files\n for filename in filenames_pbar:\n # Check if the file is a directory\n if not filename.endswith('/'):\n with zipf.open(filename) as image_file:\n image = Image.open(image_file)\n image.load()\n # Load image data as 1 dimensional array\n # We're using float32 to save on memory space\n feature = np.array(image, dtype=np.float32).flatten()\n\n # Get the the letter from the filename. This is the letter of the image.\n label = os.path.split(filename)[1][0]\n\n features.append(feature)\n labels.append(label)\n return np.array(features), np.array(labels)\n\n# Get the features and labels from the zip files\ntrain_features, train_labels = uncompress_features_labels('notMNIST_train.zip')\ntest_features, test_labels = uncompress_features_labels('notMNIST_test.zip')\n\n# Limit the amount of data to work with a docker container\ndocker_size_limit = 150000\ntrain_features, train_labels = resample(train_features, train_labels, n_samples=docker_size_limit)\n\n# Set flags for feature engineering. This will prevent you from skipping an important step.\nis_features_normal = False\nis_labels_encod = False\n\n# Wait until you see that all features and labels have been uncompressed.\nprint('All features and labels uncompressed.')",
"100%|██████████| 210001/210001 [00:21<00:00, 9650.17files/s]\n100%|██████████| 10001/10001 [00:01<00:00, 9583.57files/s]\n"
]
],
[
[
"<img src=\"image/Mean Variance - Image.png\" style=\"height: 75%;width: 75%; position: relative; right: 5%\">\n## Problem 1\nThe first problem involves normalizing the features for your training and test data.\n\nImplement Min-Max scaling in the `normalize_grayscale()` function to a range of `a=0.1` and `b=0.9`. After scaling, the values of the pixels in the input data should range from 0.1 to 0.9.\n\nSince the raw notMNIST image data is in [grayscale](https://en.wikipedia.org/wiki/Grayscale), the current values range from a min of 0 to a max of 255.\n\nMin-Max Scaling:\n$\nX'=a+{\\frac {\\left(X-X_{\\min }\\right)\\left(b-a\\right)}{X_{\\max }-X_{\\min }}}\n$\n\n*If you're having trouble solving problem 1, you can view the solution [here](https://github.com/udacity/deep-learning/blob/master/intro-to-tensorFlow/intro_to_tensorflow_solution.ipynb).*",
"_____no_output_____"
]
],
[
[
"# Problem 1 - Implement Min-Max scaling for grayscale image data\ndef normalize_grayscale(image_data):\n \"\"\"\n Normalize the image data with Min-Max scaling to a range of [0.1, 0.9]\n :param image_data: The image data to be normalized\n :return: Normalized image data\n \"\"\"\n # Implement Min-Max scaling for grayscale image data\n x_min = 0\n x_max = 255\n a = 0.1\n b = 0.9\n \n return a + (((image_data - x_min) * (b - a)) / (x_max-x_min))\n\n\n### DON'T MODIFY ANYTHING BELOW ###\n# Test Cases\nnp.testing.assert_array_almost_equal(\n normalize_grayscale(np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 255])),\n [0.1, 0.103137254902, 0.106274509804, 0.109411764706, 0.112549019608, 0.11568627451, 0.118823529412, 0.121960784314,\n 0.125098039216, 0.128235294118, 0.13137254902, 0.9],\n decimal=3)\nnp.testing.assert_array_almost_equal(\n normalize_grayscale(np.array([0, 1, 10, 20, 30, 40, 233, 244, 254,255])),\n [0.1, 0.103137254902, 0.13137254902, 0.162745098039, 0.194117647059, 0.225490196078, 0.830980392157, 0.865490196078,\n 0.896862745098, 0.9])\n\nif not is_features_normal:\n train_features = normalize_grayscale(train_features)\n test_features = normalize_grayscale(test_features)\n is_features_normal = True\n\nprint('Tests Passed!')",
"Tests Passed!\n"
],
[
"if not is_labels_encod:\n # Turn labels into numbers and apply One-Hot Encoding\n encoder = LabelBinarizer()\n encoder.fit(train_labels)\n train_labels = encoder.transform(train_labels)\n test_labels = encoder.transform(test_labels)\n\n # Change to float32, so it can be multiplied against the features in TensorFlow, which are float32\n train_labels = train_labels.astype(np.float32)\n test_labels = test_labels.astype(np.float32)\n is_labels_encod = True\n\nprint('Labels One-Hot Encoded')",
"Labels One-Hot Encoded\n"
],
[
"assert is_features_normal, 'You skipped the step to normalize the features'\nassert is_labels_encod, 'You skipped the step to One-Hot Encode the labels'\n\n# Get randomized datasets for training and validation\ntrain_features, valid_features, train_labels, valid_labels = train_test_split(\n train_features,\n train_labels,\n test_size=0.05,\n random_state=832289)\n\nprint('Training features and labels randomized and split.')",
"Training features and labels randomized and split.\n"
],
[
"# Save the data for easy access\npickle_file = 'notMNIST.pickle'\nif not os.path.isfile(pickle_file):\n print('Saving data to pickle file...')\n try:\n with open('notMNIST.pickle', 'wb') as pfile:\n pickle.dump(\n {\n 'train_dataset': train_features,\n 'train_labels': train_labels,\n 'valid_dataset': valid_features,\n 'valid_labels': valid_labels,\n 'test_dataset': test_features,\n 'test_labels': test_labels,\n },\n pfile, pickle.HIGHEST_PROTOCOL)\n except Exception as e:\n print('Unable to save data to', pickle_file, ':', e)\n raise\n\nprint('Data cached in pickle file.')",
"Data cached in pickle file.\n"
]
],
[
[
"# Checkpoint\nAll your progress is now saved to the pickle file. If you need to leave and comeback to this lab, you no longer have to start from the beginning. Just run the code block below and it will load all the data and modules required to proceed.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\n# Load the modules\nimport pickle\nimport math\n\nimport numpy as np\nimport tensorflow as tf\nfrom tqdm import tqdm\nimport matplotlib.pyplot as plt\n\n# Reload the data\npickle_file = 'notMNIST.pickle'\nwith open(pickle_file, 'rb') as f:\n pickle_data = pickle.load(f)\n train_features = pickle_data['train_dataset']\n train_labels = pickle_data['train_labels']\n valid_features = pickle_data['valid_dataset']\n valid_labels = pickle_data['valid_labels']\n test_features = pickle_data['test_dataset']\n test_labels = pickle_data['test_labels']\n del pickle_data # Free up memory\n\nprint('Data and modules loaded.')",
"Data and modules loaded.\n"
]
],
[
[
"\n## Problem 2\n\nNow it's time to build a simple neural network using TensorFlow. Here, your network will be just an input layer and an output layer.\n\n<img src=\"image/network_diagram.png\" style=\"height: 40%;width: 40%; position: relative; right: 10%\">\n\nFor the input here the images have been flattened into a vector of $28 \\times 28 = 784$ features. Then, we're trying to predict the image digit so there are 10 output units, one for each label. Of course, feel free to add hidden layers if you want, but this notebook is built to guide you through a single layer network. \n\nFor the neural network to train on your data, you need the following <a href=\"https://www.tensorflow.org/resources/dims_types.html#data-types\">float32</a> tensors:\n - `features`\n - Placeholder tensor for feature data (`train_features`/`valid_features`/`test_features`)\n - `labels`\n - Placeholder tensor for label data (`train_labels`/`valid_labels`/`test_labels`)\n - `weights`\n - Variable Tensor with random numbers from a truncated normal distribution.\n - See <a href=\"https://www.tensorflow.org/api_docs/python/constant_op.html#truncated_normal\">`tf.truncated_normal()` documentation</a> for help.\n - `biases`\n - Variable Tensor with all zeros.\n - See <a href=\"https://www.tensorflow.org/api_docs/python/constant_op.html#zeros\"> `tf.zeros()` documentation</a> for help.\n\n*If you're having trouble solving problem 2, review \"TensorFlow Linear Function\" section of the class. If that doesn't help, the solution for this problem is available [here](intro_to_tensorflow_solution.ipynb).*",
"_____no_output_____"
]
],
[
[
"# All the pixels in the image (28 * 28 = 784)\nfeatures_count = 784\n# All the labels\nlabels_count = 10\n\n# Set the features and labels tensors\nfeatures = tf.placeholder(tf.float32)\nlabels = tf.placeholder(tf.float32)\n\n# Set the weights and biases tensors\nweights = tf.Variable(tf.random_normal([features_count, labels_count]))\nbiases = tf.Variable(tf.zeros([labels_count]))\n\n### DON'T MODIFY ANYTHING BELOW ###\n\n#Test Cases\nfrom tensorflow.python.ops.variables import Variable\n\nassert features._op.name.startswith('Placeholder'), 'features must be a placeholder'\nassert labels._op.name.startswith('Placeholder'), 'labels must be a placeholder'\nassert isinstance(weights, Variable), 'weights must be a TensorFlow variable'\nassert isinstance(biases, Variable), 'biases must be a TensorFlow variable'\n\nassert features._shape == None or (\\\n features._shape.dims[0].value is None and\\\n features._shape.dims[1].value in [None, 784]), 'The shape of features is incorrect'\nassert labels._shape == None or (\\\n labels._shape.dims[0].value is None and\\\n labels._shape.dims[1].value in [None, 10]), 'The shape of labels is incorrect'\nassert weights._variable._shape == (784, 10), 'The shape of weights is incorrect'\nassert biases._variable._shape == (10), 'The shape of biases is incorrect'\n\nassert features._dtype == tf.float32, 'features must be type float32'\nassert labels._dtype == tf.float32, 'labels must be type float32'\n\n# Feed dicts for training, validation, and test session\ntrain_feed_dict = {features: train_features, labels: train_labels}\nvalid_feed_dict = {features: valid_features, labels: valid_labels}\ntest_feed_dict = {features: test_features, labels: test_labels}\n\n# Linear Function WX + b\nlogits = tf.matmul(features, weights) + biases\n\nprediction = tf.nn.softmax(logits)\n\n# Cross entropy\ncross_entropy = -tf.reduce_sum(labels * tf.log(prediction), reduction_indices=1)\n\n# Training loss\nloss = tf.reduce_mean(cross_entropy)\n\n# Create an operation that initializes all variables\ninit = tf.global_variables_initializer()\n\n# Test Cases\nwith tf.Session() as session:\n session.run(init)\n session.run(loss, feed_dict=train_feed_dict)\n session.run(loss, feed_dict=valid_feed_dict)\n session.run(loss, feed_dict=test_feed_dict)\n biases_data = session.run(biases)\n\nassert not np.count_nonzero(biases_data), 'biases must be zeros'\n\nprint('Tests Passed!')",
"Tests Passed!\n"
],
[
"# Determine if the predictions are correct\nis_correct_prediction = tf.equal(tf.argmax(prediction, 1), tf.argmax(labels, 1))\n# Calculate the accuracy of the predictions\naccuracy = tf.reduce_mean(tf.cast(is_correct_prediction, tf.float32))\n\nprint('Accuracy function created.')",
"Accuracy function created.\n"
]
],
[
[
"<img src=\"image/Learn Rate Tune - Image.png\" style=\"height: 70%;width: 70%\">\n## Problem 3\nBelow are 2 parameter configurations for training the neural network. In each configuration, one of the parameters has multiple options. For each configuration, choose the option that gives the best acccuracy.\n\nParameter configurations:\n\nConfiguration 1\n* **Epochs:** 1\n* **Learning Rate:**\n * 0.8\n * 0.5\n * 0.1\n * 0.05\n * 0.01\n\nConfiguration 2\n* **Epochs:**\n * 1\n * 2\n * 3\n * 4\n * 5\n* **Learning Rate:** 0.2\n\nThe code will print out a Loss and Accuracy graph, so you can see how well the neural network performed.\n\n*If you're having trouble solving problem 3, you can view the solution [here](intro_to_tensorflow_solution.ipynb).*",
"_____no_output_____"
]
],
[
[
"# Change if you have memory restrictions\nbatch_size = 256\n\n# Find the best parameters for each configuration\n\n#When epochs = 1, the best learning_rate is 0.5 with an accuracy of 0.7526666522026062\n\n#When multiple epochs \n#2 = 0.7515999674797058\n#3 = 0.7605332732200623\n#4 = 0.771733283996582\n#5 = 0.7671999335289001\nepochs = 4\nlearning_rate = 0.2\n\n### DON'T MODIFY ANYTHING BELOW ###\n# Gradient Descent\noptimizer = tf.train.GradientDescentOptimizer(learning_rate).minimize(loss) \n\n# The accuracy measured against the validation set\nvalidation_accuracy = 0.0\n\n# Measurements use for graphing loss and accuracy\nlog_batch_step = 50\nbatches = []\nloss_batch = []\ntrain_acc_batch = []\nvalid_acc_batch = []\n\nwith tf.Session() as session:\n session.run(init)\n batch_count = int(math.ceil(len(train_features)/batch_size))\n\n for epoch_i in range(epochs):\n \n # Progress bar\n batches_pbar = tqdm(range(batch_count), desc='Epoch {:>2}/{}'.format(epoch_i+1, epochs), unit='batches')\n \n # The training cycle\n for batch_i in batches_pbar:\n # Get a batch of training features and labels\n batch_start = batch_i*batch_size\n batch_features = train_features[batch_start:batch_start + batch_size]\n batch_labels = train_labels[batch_start:batch_start + batch_size]\n\n # Run optimizer and get loss\n _, l = session.run(\n [optimizer, loss],\n feed_dict={features: batch_features, labels: batch_labels})\n\n # Log every 50 batches\n if not batch_i % log_batch_step:\n # Calculate Training and Validation accuracy\n training_accuracy = session.run(accuracy, feed_dict=train_feed_dict)\n validation_accuracy = session.run(accuracy, feed_dict=valid_feed_dict)\n\n # Log batches\n previous_batch = batches[-1] if batches else 0\n batches.append(log_batch_step + previous_batch)\n loss_batch.append(l)\n train_acc_batch.append(training_accuracy)\n valid_acc_batch.append(validation_accuracy)\n\n # Check accuracy against Validation data\n validation_accuracy = session.run(accuracy, feed_dict=valid_feed_dict)\n\nloss_plot = plt.subplot(211)\nloss_plot.set_title('Loss')\nloss_plot.plot(batches, loss_batch, 'g')\nloss_plot.set_xlim([batches[0], batches[-1]])\nacc_plot = plt.subplot(212)\nacc_plot.set_title('Accuracy')\nacc_plot.plot(batches, train_acc_batch, 'r', label='Training Accuracy')\nacc_plot.plot(batches, valid_acc_batch, 'x', label='Validation Accuracy')\nacc_plot.set_ylim([0, 1.0])\nacc_plot.set_xlim([batches[0], batches[-1]])\nacc_plot.legend(loc=4)\nplt.tight_layout()\nplt.show()\n\nprint('Validation accuracy at {}'.format(validation_accuracy))",
"Epoch 1/4: 100%|██████████| 557/557 [00:03<00:00, 175.65batches/s]\nEpoch 2/4: 100%|██████████| 557/557 [00:03<00:00, 180.07batches/s]\nEpoch 3/4: 100%|██████████| 557/557 [00:03<00:00, 179.54batches/s]\nEpoch 4/4: 100%|██████████| 557/557 [00:03<00:00, 179.55batches/s]\n"
]
],
[
[
"## Test\nYou're going to test your model against your hold out dataset/testing data. This will give you a good indicator of how well the model will do in the real world. You should have a test accuracy of at least 80%.",
"_____no_output_____"
]
],
[
[
"### DON'T MODIFY ANYTHING BELOW ###\n# The accuracy measured against the test set\ntest_accuracy = 0.0\n\nwith tf.Session() as session:\n \n session.run(init)\n batch_count = int(math.ceil(len(train_features)/batch_size))\n\n for epoch_i in range(epochs):\n \n # Progress bar\n batches_pbar = tqdm(range(batch_count), desc='Epoch {:>2}/{}'.format(epoch_i+1, epochs), unit='batches')\n \n # The training cycle\n for batch_i in batches_pbar:\n # Get a batch of training features and labels\n batch_start = batch_i*batch_size\n batch_features = train_features[batch_start:batch_start + batch_size]\n batch_labels = train_labels[batch_start:batch_start + batch_size]\n\n # Run optimizer\n _ = session.run(optimizer, feed_dict={features: batch_features, labels: batch_labels})\n\n # Check accuracy against Test data\n test_accuracy = session.run(accuracy, feed_dict=test_feed_dict)\n\n\nassert test_accuracy >= 0.80, 'Test accuracy at {}, should be equal to or greater than 0.80'.format(test_accuracy)\nprint('Nice Job! Test Accuracy is {}'.format(test_accuracy))",
"Epoch 1/4: 100%|██████████| 557/557 [00:00<00:00, 1230.28batches/s]\nEpoch 2/4: 100%|██████████| 557/557 [00:00<00:00, 1277.16batches/s]\nEpoch 3/4: 100%|██████████| 557/557 [00:00<00:00, 1209.55batches/s]\nEpoch 4/4: 100%|██████████| 557/557 [00:00<00:00, 1254.66batches/s]"
]
],
[
[
"# Multiple layers\nGood job! You built a one layer TensorFlow network! However, you might want to build more than one layer. This is deep learning after all! In the next section, you will start to satisfy your need for more layers.",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
e7f858cf946562b5a147bccee1b1484494a13733 | 252,783 | ipynb | Jupyter Notebook | src/Analysis/bible_bitexts_analysis.ipynb | AlexJonesNLP/crosslingual-analysis-101 | 6cda1ad9d3f8133943cf736a554a646c865ebb4b | [
"MIT"
] | null | null | null | src/Analysis/bible_bitexts_analysis.ipynb | AlexJonesNLP/crosslingual-analysis-101 | 6cda1ad9d3f8133943cf736a554a646c865ebb4b | [
"MIT"
] | null | null | null | src/Analysis/bible_bitexts_analysis.ipynb | AlexJonesNLP/crosslingual-analysis-101 | 6cda1ad9d3f8133943cf736a554a646c865ebb4b | [
"MIT"
] | null | null | null | 58.623145 | 14,810 | 0.5257 | [
[
[
"### Dependencies",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport pingouin as pg\nimport seaborn as sns\nimport scipy.stats\nimport sklearn\nimport matplotlib.pyplot as plt\nfrom tqdm import tqdm",
"_____no_output_____"
]
],
[
[
"### Loading dataframes containing variables",
"_____no_output_____"
]
],
[
[
"# Loading the dataframes we'll be using\n\n# Contains the DEPENDENT variables relating to language PAIRS\nlang_pair_dv = pd.read_csv('/Data/Bible experimental vars/bible_dependent_vars_LANGUAGE_PAIR.csv')\n\n# Contains the INDEPENDENT variables relating to language PAIRS\nlang_pair_iv = pd.read_csv('/Data/bible_predictors_LANGUAGE_PAIR.csv')\n\n# Contains ALL variables relating to INDIVIDUAL languages\nindiv_lang_vars = pd.read_csv('/Data/bible_all_features_LANGUAGE.csv')",
"_____no_output_____"
],
[
"# Tallying zero-shot sub-cases\n\nprint('Simple zero-shot languages (LaBSE): {}'.format(sum(np.array(indiv_lang_vars['Total sentences (LaBSE)']==0))))\nprint('Simple zero-shot languages (LASER): {}'.format(sum(np.array(indiv_lang_vars['Total sentences (LASER)']==0))))\nprint('Double zero-shot language pairs (LaBSE): {}'.format(sum(np.array(lang_pair_iv['Combined sentences (LaBSE)']==0))))\nprint('Double zero-shot language pairs (LASER): {}'.format(sum(np.array(lang_pair_iv['Combined sentences (LASER)']==0))))",
"Simple zero-shot languages (LaBSE): 35\nSimple zero-shot languages (LASER): 45\nDouble zero-shot language pairs (LaBSE): 595\nDouble zero-shot language pairs (LASER): 990\n"
],
[
"# It's pretty helpful to combine the IVs and DVs for language pairs, as Pingouin prefers to work with \n# single dataframe objects\nmaster_pair = pd.concat([lang_pair_iv, lang_pair_dv], axis=1)",
"_____no_output_____"
],
[
"master_pair.corr()",
"_____no_output_____"
],
[
"pg.ancova(data=master_pair, \n dv='F1-score (LASER, average)', \n between='Same Genus?',\n covar=['Combined sentences (LASER)', \n 'Combined in-family sentences (LASER)',\n 'Combined in-genus sentences (LASER)'])",
"_____no_output_____"
],
[
"pg.partial_corr(data=master_pair,\n x='Phonological Distance (lang2vec)',\n y='Average margin score (LASER, average)',\n covar=['Combined sentences (LASER)',\n 'Combined in-family sentences (LASER)',\n 'Combined in-genus sentences (LASER)'])",
"_____no_output_____"
],
[
"double_zero_shot_labse = master_pair[np.array(master_pair['Combined sentences (LaBSE)'])==0]\ndouble_zero_shot_laser = master_pair[np.array(master_pair['Combined sentences (LASER)'])==0]",
"_____no_output_____"
],
[
"double_zero_shot_labse['Gromov-Hausdorff dist. (LaBSE, average)'] = -double_zero_shot_labse['Gromov-Hausdorff dist. (LaBSE, average)']\ndouble_zero_shot_labse['Gromov-Hausdorff dist. (LASER, average)'] = -double_zero_shot_laser['Gromov-Hausdorff dist. (LASER, average)']",
"/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \n"
],
[
"double_zero_shot_labse['Singular value gap (LaBSE, average)'] = -double_zero_shot_labse['Singular value gap (LaBSE, average)']\ndouble_zero_shot_laser['Singular value gap (LASER, average)'] = -double_zero_shot_laser['Singular value gap (LASER, average)']",
"/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:1: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \"\"\"Entry point for launching an IPython kernel.\n/usr/local/lib/python3.7/dist-packages/ipykernel_launcher.py:2: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n \n"
],
[
"double_zero_shot_labse = double_zero_shot_labse[['Same Family?', 'Same Genus?', \n 'Character-level Overlap (multiset Jaccard coefficient, Book of Matthew)',\n 'Token-level Overlap (multiset Jaccard coefficient, Book of John)',\n 'Same Word Order?', 'Same Polysynthesis Status?',\t'Geographic Distance (lang2vec)',\n 'Inventory Distance (lang2vec)',\t'Syntactic Distance (lang2vec)',\n 'Phonological Distance (lang2vec)', 'F1-score (LaBSE, average)',\t\n 'Gromov-Hausdorff dist. (LaBSE, average)',\n 'Singular value gap (LaBSE, average)',\n 'ECOND-HM (LaBSE, average)',\n 'Average margin score (LaBSE, average)', 'Language pair']]\n\ndouble_zero_shot_laser = double_zero_shot_laser[['Same Family?', 'Same Genus?', \n 'Character-level Overlap (multiset Jaccard coefficient, Book of Matthew)',\n 'Token-level Overlap (multiset Jaccard coefficient, Book of John)',\n 'Same Word Order?', 'Same Polysynthesis Status?',\t'Geographic Distance (lang2vec)',\n 'Inventory Distance (lang2vec)',\t'Syntactic Distance (lang2vec)',\n 'Phonological Distance (lang2vec)', 'F1-score (LASER, average)',\t\n 'Gromov-Hausdorff dist. (LASER, average)',\n 'Singular value gap (LASER, average)',\n 'ECOND-HM (LASER, average)',\n 'Average margin score (LASER, average)', 'Language pair']]",
"_____no_output_____"
],
[
"print(pg.anova(data=double_zero_shot_labse, dv='F1-score (LaBSE, average)', between='Same Word Order?'))\nprint(pg.anova(data=double_zero_shot_labse, dv='F1-score (LaBSE, average)', between='Same Polysynthesis Status?'))\nprint(pg.anova(data=double_zero_shot_labse, dv='F1-score (LaBSE, average)', between='Same Family?'))\nprint(pg.anova(data=double_zero_shot_labse, dv='F1-score (LaBSE, average)', between='Same Genus?'))",
"_____no_output_____"
],
[
"print(scipy.stats.pearsonr(double_zero_shot_labse['F1-score (LaBSE, average)'], \n double_zero_shot_labse['Syntactic Distance (lang2vec)']))",
"_____no_output_____"
],
[
"def corrUtilIO(corr: tuple, s1:str, s2:str):\n r, p = corr\n out = 'Correlation between {} and {}: {} | p-value: {}'.format(s1, s2, r, p)\n return out",
"_____no_output_____"
],
[
"print('Examining double-zero shot language pairs (LaBSE)')\nprint('--------------------------------------------------')\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_labse['F1-score (LaBSE, average)'], \n double_zero_shot_labse['Inventory Distance (lang2vec)']), \n 'F1-score', 'inventory distance'))\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_labse['Gromov-Hausdorff dist. (LaBSE, average)'], \n double_zero_shot_labse['Inventory Distance (lang2vec)']), \n 'Gromov-Hausdorff distance', 'inventory distance'))\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_labse['Singular value gap (LaBSE, average)'], \n double_zero_shot_labse['Inventory Distance (lang2vec)']), \n 'singular value gap', 'inventory distance'))\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_labse['ECOND-HM (LaBSE, average)'], \n double_zero_shot_labse['Inventory Distance (lang2vec)']), \n 'ECOND-HM', 'inventory distance'))\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_labse['Average margin score (LaBSE, average)'], \n double_zero_shot_labse['Inventory Distance (lang2vec)']), \n 'average margin score', 'inventory distance'))",
"Examining double-zero shot language pairs (LaBSE)\n--------------------------------------------------\nCorrelation between F1-score and inventory distance: -0.33724823383287633 | p-value: 2.7175319027258977e-17\nCorrelation between Gromov-Hausdorff distance and inventory distance: -0.0686797479579177 | p-value: 0.09418309896867325\nCorrelation between singular value gap and inventory distance: -0.2207380504269484 | p-value: 5.31897763102463e-08\nCorrelation between ECOND-HM and inventory distance: -0.3930150872874848 | p-value: 2.0514426878687908e-23\nCorrelation between average margin score and inventory distance: -0.3048283011613173 | p-value: 2.925467934356142e-14\n"
],
[
"X_to_regress_1 = ['Inventory Distance (lang2vec)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nX_to_regress_2 = ['Inventory Distance (lang2vec)', 'Character-level Overlap (multiset Jaccard coefficient, Book of Matthew)']",
"_____no_output_____"
],
[
"pg.linear_regression(X=double_zero_shot_labse[X_to_regress_2], y=double_zero_shot_labse['F1-score (LaBSE, average)'])",
"_____no_output_____"
],
[
"print('Examining double-zero shot language pairs (LASER)')\nprint('--------------------------------------------------')\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_laser['F1-score (LASER, average)'], \n double_zero_shot_laser['Inventory Distance (lang2vec)']), \n 'F1-score', 'inventory distance'))\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_laser['Gromov-Hausdorff dist. (LASER, average)'], \n double_zero_shot_laser['Inventory Distance (lang2vec)']), \n 'Gromov-Hausdorff distance', 'inventory distance'))\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_laser['Singular value gap (LASER, average)'], \n double_zero_shot_laser['Inventory Distance (lang2vec)']), \n 'singular value gap', 'inventory distance'))\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_laser['ECOND-HM (LASER, average)'], \n double_zero_shot_laser['Inventory Distance (lang2vec)']), \n 'ECOND-HM', 'inventory distance'))\nprint(corrUtilIO(scipy.stats.pearsonr(double_zero_shot_laser['Average margin score (LASER, average)'], \n double_zero_shot_laser['Inventory Distance (lang2vec)']), \n 'average margin score', 'inventory distance'))",
"Examining double-zero shot language pairs (LASER)\n--------------------------------------------------\nCorrelation between F1-score and inventory distance: -0.127669949469008 | p-value: 5.6142950992676594e-05\nCorrelation between Gromov-Hausdorff distance and inventory distance: 0.050291835337428856 | p-value: 0.11378722704645745\nCorrelation between singular value gap and inventory distance: -0.07911088506905038 | p-value: 0.012777024986595574\nCorrelation between ECOND-HM and inventory distance: 0.1440475335405774 | p-value: 5.355038479530176e-06\nCorrelation between average margin score and inventory distance: -0.09227268163614558 | p-value: 0.0036628574255727206\n"
],
[
"simple_zero_shot_labse = indiv_lang_vars[np.array(indiv_lang_vars['Total sentences (LaBSE)'])==0]\nsimple_zero_shot_laser = indiv_lang_vars[np.array(indiv_lang_vars['Total sentences (LASER)'])==0]\n\nsimple_zero_shot_labse = simple_zero_shot_labse.drop(['Total sentences (LaBSE)', 'Total in-family sentences (LaBSE)', \n 'Total in-genus sentences (LaBSE)', 'Total sentences (LASER)', \n 'Total in-family sentences (LASER)', 'Total in-genus sentences (LASER)',\n 'Average F1 (LASER)', 'Average G-H dist. (LASER)', 'Average SVG (LASER)',\n 'Average ECOND-HM (LASER)', 'Grand mean margin score (LASER)'], axis=1)\n\nsimple_zero_shot_laser = simple_zero_shot_laser.drop(['Total sentences (LaBSE)', 'Total in-family sentences (LaBSE)', \n 'Total in-genus sentences (LaBSE)', 'Total sentences (LASER)', \n 'Total in-family sentences (LASER)', 'Total in-genus sentences (LASER)',\n 'Average F1 (LaBSE)', 'Average G-H dist. (LaBSE)', 'Average SVG (LaBSE)',\n 'Average ECOND-HM (LaBSE)', 'Grand mean margin score (LaBSE)'], axis=1)",
"_____no_output_____"
],
[
"print('Running ANOVAs to check for omnibus group mean differences in the DVs for basic word order')\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average F1 (LaBSE)', between='Basic Word Order', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average G-H dist. (LaBSE)', between='Basic Word Order', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average SVG (LaBSE)', between='Basic Word Order', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average ECOND-HM (LaBSE)', between='Basic Word Order', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Grand mean margin score (LaBSE)', between='Basic Word Order', ss_type=3))\nprint('\\n')\nprint('Running ANOVAs to check for omnibus group mean differences in the DVs for polysyntheticity')\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average F1 (LaBSE)', between='Polysynthetic?', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average G-H dist. (LaBSE)', between='Polysynthetic?', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average SVG (LaBSE)', between='Polysynthetic?', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average ECOND-HM (LaBSE)', between='Polysynthetic?', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Grand mean margin score (LaBSE)', between='Polysynthetic?', ss_type=3))\nprint('\\n')\nprint('Running ANOVAs to check for omnibus group mean differences in the DVs for family')\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average F1 (LaBSE)', between='Family', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average G-H dist. (LaBSE)', between='Family', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average SVG (LaBSE)', between='Family', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average ECOND-HM (LaBSE)', between='Family', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Grand mean margin score (LaBSE)', between='Family', ss_type=3))\nprint('\\n')\nprint('Running ANOVAs to check for omnibus group mean differences in the DVs for genus')\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average F1 (LaBSE)', between='Genus', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average G-H dist. (LaBSE)', between='Genus', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average SVG (LaBSE)', between='Genus', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average ECOND-HM (LaBSE)', between='Genus', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Grand mean margin score (LaBSE)', between='Genus', ss_type=3))\nprint('\\n')\nprint('Running ANOVAs to check for omnibus group mean differences in the DVs for script')\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average F1 (LaBSE)', between='Script', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average G-H dist. (LaBSE)', between='Script', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average SVG (LaBSE)', between='Script', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Average ECOND-HM (LaBSE)', between='Script', ss_type=3))\nprint(pg.anova(data=simple_zero_shot_labse, dv='Grand mean margin score (LaBSE)', between='Script', ss_type=3))",
"Running ANOVAs to check for omnibus group mean differences in the DVs for basic word order\n Source ddof1 ddof2 F p-unc np2\n0 Basic Word Order 7 27 2.198701 0.066555 0.363071\n Source ddof1 ddof2 F p-unc np2\n0 Basic Word Order 7 27 1.396682 0.247187 0.265841\n Source ddof1 ddof2 F p-unc np2\n0 Basic Word Order 7 27 0.450401 0.861108 0.104561\n Source ddof1 ddof2 F p-unc np2\n0 Basic Word Order 7 27 2.199274 0.066492 0.363131\n Source ddof1 ddof2 F p-unc np2\n0 Basic Word Order 7 27 1.715491 0.147461 0.307842\n\n\nRunning ANOVAs to check for omnibus group mean differences in the DVs for polysyntheticity\n Source ddof1 ddof2 F p-unc np2\n0 Polysynthetic? 1 33 0.220212 0.641964 0.006629\n Source ddof1 ddof2 F p-unc np2\n0 Polysynthetic? 1 33 2.030424 0.163567 0.057962\n Source ddof1 ddof2 F p-unc np2\n0 Polysynthetic? 1 33 1.376524 0.249093 0.040043\n Source ddof1 ddof2 F p-unc np2\n0 Polysynthetic? 1 33 0.220054 0.642084 0.006624\n Source ddof1 ddof2 F p-unc np2\n0 Polysynthetic? 1 33 0.636075 0.430837 0.01891\n\n\nRunning ANOVAs to check for omnibus group mean differences in the DVs for family\n Source ddof1 ddof2 F p-unc np2\n0 Family 19 15 1.421432 0.247139 0.642918\n Source ddof1 ddof2 F p-unc np2\n0 Family 19 15 2.388181 0.046171 0.751555\n Source ddof1 ddof2 F p-unc np2\n0 Family 19 15 0.204183 0.999226 0.205486\n Source ddof1 ddof2 F p-unc np2\n0 Family 19 15 1.421729 0.247009 0.642966\n Source ddof1 ddof2 F p-unc np2\n0 Family 19 15 1.968022 0.093944 0.713699\n\n\nRunning ANOVAs to check for omnibus group mean differences in the DVs for genus\n Source ddof1 ddof2 F p-unc np2\n0 Genus 27 7 1.662188 0.250874 0.865071\n Source ddof1 ddof2 F p-unc np2\n0 Genus 27 7 14.154914 0.000689 0.982014\n Source ddof1 ddof2 F p-unc np2\n0 Genus 27 7 231758.080717 1.556980e-18 0.999999\n Source ddof1 ddof2 F p-unc np2\n0 Genus 27 7 1.662553 0.250771 0.865096\n Source ddof1 ddof2 F p-unc np2\n0 Genus 27 7 4.19455 0.028288 0.941789\n\n\nRunning ANOVAs to check for omnibus group mean differences in the DVs for script\n Source ddof1 ddof2 F p-unc np2\n0 Script 4 30 1.646355 0.188539 0.180001\n Source ddof1 ddof2 F p-unc np2\n0 Script 4 30 1.505646 0.225546 0.167189\n Source ddof1 ddof2 F p-unc np2\n0 Script 4 30 149.732352 2.301662e-19 0.9523\n Source ddof1 ddof2 F p-unc np2\n0 Script 4 30 1.646298 0.188553 0.179996\n Source ddof1 ddof2 F p-unc np2\n0 Script 4 30 2.587595 0.056838 0.256513\n"
],
[
"sns.barplot(simple_zero_shot_labse['Basic Word Order'], simple_zero_shot_labse['Average F1 (LaBSE)'])\nplt.ylabel('Meta-average F1 (LaBSE), zero-shot only', fontsize=12)\nplt.xlabel('Basic word order', fontsize=14)",
"/usr/local/lib/python3.7/dist-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
],
[
"sns.barplot(simple_zero_shot_laser['Basic Word Order'], simple_zero_shot_laser['Average F1 (LASER)'])\nplt.ylabel('Meta-average F1 (LASER), zero-shot only', fontsize=12)\nplt.xlabel('Basic word order', fontsize=14)",
"/usr/local/lib/python3.7/dist-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
],
[
"sns.barplot(simple_zero_shot_labse['Basic Word Order'], simple_zero_shot_labse['Average ECOND-HM (LaBSE)'])\nplt.ylabel('Meta-average ECOND-HM (LaBSE), zero-shot only', fontsize=11)\nplt.xlabel('Basic word order', fontsize=14)",
"/usr/local/lib/python3.7/dist-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
],
[
"pg.pairwise_tukey(data=simple_zero_shot_labse, dv='Average F1 (LaBSE)', between='Basic Word Order')",
"/usr/local/lib/python3.7/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"pg.pairwise_tukey(data=simple_zero_shot_laser, dv='Average F1 (LASER)', between='Basic Word Order')",
"_____no_output_____"
]
],
[
[
"### Experimenting with sklearn models for feature selection\n\n",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.model_selection import cross_val_score\nfrom itertools import chain, combinations # Used for exhaustive feature search",
"_____no_output_____"
],
[
"# The model we'll use to choose the best features for predicting F1-score for LaBSE\nlabse_f1_model = LinearRegression()",
"_____no_output_____"
],
[
"# All the possible pair-centric LaBSE IVs\nlabse_pair_iv = ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)',\n 'Combined in-genus sentences (LaBSE)', 'Same Family?', 'Same Genus?',\n 'Character-level Overlap (multiset Jaccard coefficient, Book of Matthew)',\n 'Token-level Overlap (multiset Jaccard coefficient, Book of John)',\n 'Same Word Order?', 'Same Polysynthesis Status?', \n 'Geographic Distance (lang2vec)', 'Syntactic Distance (lang2vec)',\n 'Phonological Distance (lang2vec)', 'Inventory Distance (lang2vec)']\nX_pair_labse = master_pair[labse_pair_iv]\n\n# The first DV we'll look at\ny_pair_f1_labse = master_pair['F1-score (LaBSE, average)']",
"_____no_output_____"
],
[
"# Exhaustive feature search on language pair features\ndef getBestFeatures(model, X, y, score_method):\n FOLDS = 10\n n_features = X.shape[1]\n all_subsets = chain.from_iterable(combinations(range(n_features), k) for k in range(n_features+1))\n\n best_score = -np.inf\n best_features = None\n for subset in all_subsets:\n if len(subset)!=0: # Search over all non-empty subsets of features \n score_by_fold = sklearn.model_selection.cross_validate(model, \n X.iloc[:, np.array(subset)], \n y, \n cv=FOLDS, \n scoring=score_method)['test_score']\n #scoring='neg_mean_squared_error')\n\n # Convert R2 to adjusted R2 to take into account the number of predictors\n def adjustedR2(r2, n, p):\n num = (1-r2)*(n-1)\n denom = n-p-1\n adj_r2 = 1 - (num/denom)\n return adj_r2\n \n if score_method=='r2':\n # Compute the adjusted R2 instead\n n_subset_features = len(subset)\n # Fraction of data used for training during CV\n train_frac = (FOLDS-1) / FOLDS # e.g. with 10 folds, we use 9/10 of the data for training\n sample_size = round(train_frac*X.shape[0])\n score_by_fold = list(map(lambda r2: adjustedR2(r2,sample_size,n_subset_features), score_by_fold)) #[adjustedR2(r2, n_subset_features, sample_size) for r2 in score_by_fold]\n\n score = np.average(score_by_fold)\n\n # If score is current optimum . . .\n if score > best_score:\n best_score, best_features = score, subset # . . . flag it as such\n print('Score: {} Features: {}'.format(best_score, [X.columns[i] for i in best_features]))\n\n best_features = [X.columns[i] for i in best_features] # Return just the best features\n return best_features",
"_____no_output_____"
],
[
"labse_pair_f1_best_features = getBestFeatures(model=labse_f1_model, \n X=X_pair_labse, \n y=y_pair_f1_labse,\n score_method='r2') # really adjusted R2",
"Score: 0.04128646949432839 Features: ['Combined sentences (LaBSE)']\nScore: 0.17838707077008623 Features: ['Combined in-family sentences (LaBSE)']\nScore: 0.22443671542909233 Features: ['Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nScore: 0.2941476121717451 Features: ['Combined sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nScore: 0.3153931783656644 Features: ['Combined in-family sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nScore: 0.3386625149111925 Features: ['Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?']\nScore: 0.3492214395987304 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nScore: 0.3872478837547058 Features: ['Combined sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?']\nScore: 0.4009222646126024 Features: ['Combined in-family sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?']\nScore: 0.4257946237905684 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?']\nScore: 0.4280964281852465 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?', 'Geographic Distance (lang2vec)']\n"
],
[
"# Repeating the same process for LASER\n\n# All the possible pair-centric LASER IVs\nlaser_pair_iv = ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)',\n 'Combined in-genus sentences (LASER)', 'Same Family?', 'Same Genus?',\n 'Character-level Overlap (multiset Jaccard coefficient, Book of Matthew)',\n 'Token-level Overlap (multiset Jaccard coefficient, Book of John)',\n 'Same Word Order?', 'Same Polysynthesis Status?', \n 'Geographic Distance (lang2vec)', 'Syntactic Distance (lang2vec)',\n 'Phonological Distance (lang2vec)', 'Inventory Distance (lang2vec)']\nX_pair_laser = master_pair[laser_pair_iv]\n\n# The first DV we'll look at (for LASER)\ny_pair_f1_laser = master_pair['F1-score (LASER, average)']",
"_____no_output_____"
],
[
"laser_f1_model = LinearRegression()",
"_____no_output_____"
],
[
"laser_pair_f1_best_features = getBestFeatures(model=laser_f1_model, \n X=X_pair_laser, \n y=y_pair_f1_laser, \n score_method='r2')",
"Score: -0.01978130854908604 Features: ['Combined sentences (LASER)']\nScore: 0.25089903587019724 Features: ['Combined in-family sentences (LASER)']\nScore: 0.2781519844206934 Features: ['Combined in-family sentences (LASER)', 'Same Family?']\nScore: 0.31903495057038245 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nScore: 0.3316275968804237 Features: ['Combined in-family sentences (LASER)', 'Same Family?', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nScore: 0.3482534147350469 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?']\nScore: 0.35438798384031955 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Syntactic Distance (lang2vec)']\nScore: 0.36074765639270434 Features: ['Combined in-family sentences (LASER)', 'Same Family?', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?']\nScore: 0.36195359973421626 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Geographic Distance (lang2vec)']\nScore: 0.3660700225060908 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Syntactic Distance (lang2vec)']\nScore: 0.3689909119141931 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?', 'Syntactic Distance (lang2vec)']\nScore: 0.3694833894592186 Features: ['Combined in-family sentences (LASER)', 'Same Family?', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?']\nScore: 0.3703835734557118 Features: ['Combined in-family sentences (LASER)', 'Same Family?', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Syntactic Distance (lang2vec)']\nScore: 0.37823186943463705 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Syntactic Distance (lang2vec)']\nScore: 0.3797975436317183 Features: ['Combined in-family sentences (LASER)', 'Same Family?', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Syntactic Distance (lang2vec)']\nScore: 0.3800551193435479 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Geographic Distance (lang2vec)', 'Syntactic Distance (lang2vec)']\nScore: 0.38065200958413714 Features: ['Combined in-family sentences (LASER)', 'Same Family?', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Geographic Distance (lang2vec)', 'Syntactic Distance (lang2vec)']\nScore: 0.38200560070891776 Features: ['Combined in-family sentences (LASER)', 'Same Family?', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Geographic Distance (lang2vec)', 'Syntactic Distance (lang2vec)', 'Phonological Distance (lang2vec)']\n"
],
[
"# Overlapping best predictors\nset(laser_pair_f1_best_features)&set(labse_pair_f1_best_features)",
"_____no_output_____"
],
[
"# Checking out the best predictors for the other DVs\n\n# LaBSE\ny_pair_gh_labse = master_pair['Gromov-Hausdorff dist. (LaBSE, average)']\ny_pair_svg_labse = master_pair['Singular value gap (LaBSE, average)']\ny_pair_econdhm_labse = master_pair['ECOND-HM (LaBSE, average)']\ny_pair_avgmarg_labse = master_pair['Average margin score (LaBSE, average)']\nlabse_gh_model, labse_svg_model, labse_econdhm_model, labse_avgmarg_model = LinearRegression(), LinearRegression(), LinearRegression(), LinearRegression()\n\n# LASER\ny_pair_gh_laser = master_pair['Gromov-Hausdorff dist. (LASER, average)']\ny_pair_svg_laser = master_pair['Singular value gap (LASER, average)']\ny_pair_econdhm_laser = master_pair['ECOND-HM (LASER, average)']\ny_pair_avgmarg_laser = master_pair['Average margin score (LASER, average)']\nlaser_gh_model, laser_svg_model, laser_econdhm_model, laser_avgmarg_model = LinearRegression(), LinearRegression(), LinearRegression(), LinearRegression()",
"_____no_output_____"
],
[
"# LaBSE best feature selection\nprint('Getting best features for LaBSE, GH')\nlabse_pair_gh_best_features = getBestFeatures(labse_gh_model, X_pair_labse, y_pair_gh_labse, 'r2')\nprint('Getting best features for LaBSE, SVG')\nlabse_pair_svg_best_features = getBestFeatures(labse_svg_model, X_pair_labse, y_pair_svg_labse, 'r2')\nprint('Getting best features for LaBSE, ECOND-HM')\nlabse_pair_econdhm_best_features = getBestFeatures(labse_econdhm_model, X_pair_labse, y_pair_econdhm_labse, 'r2')\nprint('Getting best features for LaBSE, avg. margin score')\nlabse_pair_avgmarg_best_features = getBestFeatures(labse_avgmarg_model, X_pair_labse, y_pair_avgmarg_labse, 'r2')\n\n# LASER best feature selection\nprint('Getting best features for LASER, GH')\nlaser_pair_gh_best_features = getBestFeatures(laser_gh_model, X_pair_laser, y_pair_gh_laser, 'r2')\nprint('Getting best features for LASER, SVG')\nlaser_pair_svg_best_features = getBestFeatures(laser_svg_model, X_pair_laser, y_pair_svg_laser, 'r2')\nprint('Getting best features for LASER, ECOND-HM')\nlaser_pair_econdhm_best_features = getBestFeatures(laser_econdhm_model, X_pair_laser, y_pair_econdhm_laser, 'r2')\nprint('Getting best features for LASER, avg. margin score')\nlaser_pair_avgmarg_best_features = getBestFeatures(laser_avgmarg_model, X_pair_laser, y_pair_avgmarg_laser, 'r2')",
"Getting best features for LaBSE, GH\nScore: -0.0413396886380951 Features: ['Combined sentences (LaBSE)']\nScore: -0.021350223866934324 Features: ['Combined in-family sentences (LaBSE)']\nScore: -0.01679224278785668 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)']\nScore: -0.002414935334796575 Features: ['Combined in-family sentences (LaBSE)', 'Same Word Order?']\nScore: 0.0003457227233038096 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Same Word Order?']\nScore: 0.0042619988612207175 Features: ['Combined in-family sentences (LaBSE)', 'Same Word Order?', 'Same Polysynthesis Status?']\nScore: 0.006178065339854944 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Same Word Order?', 'Same Polysynthesis Status?']\nScore: 0.007388992714442766 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Geographic Distance (lang2vec)']\nGetting best features for LaBSE, SVG\nScore: -13.442845600864931 Features: ['Combined sentences (LaBSE)']\nScore: -12.91560970563965 Features: ['Same Family?']\nScore: -12.70894889458604 Features: ['Same Genus?']\nGetting best features for LaBSE, ECOND-HM\nScore: -0.07663792223141372 Features: ['Combined sentences (LaBSE)']\nScore: 0.17450005561272933 Features: ['Combined in-family sentences (LaBSE)']\nScore: 0.18470724634286412 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)']\nScore: 0.18710971735580179 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Combined in-genus sentences (LaBSE)']\nScore: 0.19515508854609553 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Same Polysynthesis Status?']\nScore: 0.1972223430662685 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Combined in-genus sentences (LaBSE)', 'Same Polysynthesis Status?']\nScore: 0.19857398579111657 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Combined in-genus sentences (LaBSE)', 'Same Family?', 'Same Polysynthesis Status?']\nGetting best features for LaBSE, avg. margin score\nScore: 0.003132645432362735 Features: ['Combined sentences (LaBSE)']\nScore: 0.08877712075136177 Features: ['Combined in-family sentences (LaBSE)']\nScore: 0.11039046088757831 Features: ['Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nScore: 0.13403592045753307 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)']\nScore: 0.17985617121967118 Features: ['Combined sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)']\nScore: 0.21704669662166767 Features: ['Combined in-family sentences (LaBSE)', 'Same Polysynthesis Status?']\nScore: 0.22581187960281346 Features: ['Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?']\nScore: 0.24842980585512003 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Same Polysynthesis Status?']\nScore: 0.27335157866345083 Features: ['Combined sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?']\nScore: 0.297625168503587 Features: ['Combined sentences (LaBSE)', 'Combined in-family sentences (LaBSE)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Polysynthesis Status?']\nGetting best features for LASER, GH\nScore: -0.04232319615283292 Features: ['Combined sentences (LASER)']\nScore: -0.011643389665897275 Features: ['Combined in-family sentences (LASER)']\nScore: -0.011590823223872415 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)']\nScore: -0.011067803063791825 Features: ['Combined in-family sentences (LASER)', 'Same Family?']\nScore: 0.009656949432331452 Features: ['Combined in-family sentences (LASER)', 'Same Word Order?']\nScore: 0.01582487308917795 Features: ['Combined in-family sentences (LASER)', 'Same Word Order?', 'Same Polysynthesis Status?']\nScore: 0.01741500169028971 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?']\nScore: 0.01914508611124365 Features: ['Combined in-family sentences (LASER)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Geographic Distance (lang2vec)']\nScore: 0.02014953773989433 Features: ['Combined in-family sentences (LASER)', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Geographic Distance (lang2vec)']\nScore: 0.020236921735400204 Features: ['Combined in-family sentences (LASER)', 'Same Genus?', 'Token-level Overlap (multiset Jaccard coefficient, Book of John)', 'Same Word Order?', 'Same Polysynthesis Status?', 'Geographic Distance (lang2vec)']\nGetting best features for LASER, SVG\nScore: -2.1239321109011433 Features: ['Combined sentences (LASER)']\nScore: -2.087700196995328 Features: ['Combined in-family sentences (LASER)']\nScore: -2.03830090855165 Features: ['Combined in-genus sentences (LASER)']\nScore: -2.037664077793555 Features: ['Same Family?']\nScore: -2.0058829314780606 Features: ['Same Word Order?']\nScore: -1.9890764864614519 Features: ['Combined in-genus sentences (LASER)', 'Same Word Order?']\nGetting best features for LASER, ECOND-HM\nScore: -0.08398775179803149 Features: ['Combined sentences (LASER)']\nScore: 0.033989541419546 Features: ['Combined in-family sentences (LASER)']\nScore: 0.05607357917655924 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)']\nScore: 0.07199849287006928 Features: ['Combined in-family sentences (LASER)', 'Same Polysynthesis Status?']\nScore: 0.09467553484494366 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Polysynthesis Status?']\nScore: 0.10073009738796554 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Family?', 'Same Polysynthesis Status?']\nScore: 0.10155085237237542 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Polysynthesis Status?', 'Syntactic Distance (lang2vec)']\nScore: 0.11599358877094387 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Family?', 'Same Polysynthesis Status?', 'Syntactic Distance (lang2vec)']\nScore: 0.1165912748426974 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Family?', 'Same Polysynthesis Status?', 'Syntactic Distance (lang2vec)', 'Phonological Distance (lang2vec)']\nGetting best features for LASER, avg. margin score\nScore: -0.08558564615220951 Features: ['Combined sentences (LASER)']\nScore: 0.028976489862599188 Features: ['Combined in-family sentences (LASER)']\nScore: 0.0623230762576779 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)']\nScore: 0.06384934429762971 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Family?']\nScore: 0.07024430593135979 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Word Order?']\nScore: 0.07052342218821048 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Polysynthesis Status?']\nScore: 0.08030722587494216 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Syntactic Distance (lang2vec)']\nScore: 0.08734223706788617 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Polysynthesis Status?', 'Syntactic Distance (lang2vec)']\nScore: 0.08791708013904954 Features: ['Combined sentences (LASER)', 'Combined in-family sentences (LASER)', 'Same Polysynthesis Status?', 'Syntactic Distance (lang2vec)', 'Phonological Distance (lang2vec)']\n"
]
],
[
[
"### Applying PCA as an additional feature selection tool",
"_____no_output_____"
]
],
[
[
"pca = sklearn.decomposition.PCA(n_components=5)\nlabse_pair_pca = pca.fit_transform(X_pair_labse)",
"_____no_output_____"
],
[
"labse_pair_pca.shape",
"_____no_output_____"
]
],
[
[
"### PCR",
"_____no_output_____"
]
],
[
[
"# Implement principal component regression (PCR)\ndef PCR(model, X, y, n_components, score_method):\n FOLDS = 10\n pca = sklearn.decomposition.PCA(n_components=n_components)\n X_pca = pca.fit_transform(X)\n score_by_fold = sklearn.model_selection.cross_validate(model, \n X_pca, \n y, \n cv=FOLDS, \n scoring=score_method)['test_score']\n # Convert R2 to adjusted R2 to take into account the number of predictors\n def adjustedR2(r2, n, p):\n num = (1-r2)*(n-1)\n denom = n-p-1\n adj_r2 = 1 - (num/denom)\n return adj_r2\n \n if score_method=='r2':\n # Compute the adjusted R2 instead\n n_subset_features = X.shape[1]\n # Fraction of data used for training during CV\n train_frac = (FOLDS-1) / FOLDS # e.g. with 10 folds, we use 9/10 of the data for training\n sample_size = round(train_frac*X.shape[0])\n score_by_fold = list(map(lambda r2: adjustedR2(r2,sample_size,n_subset_features), score_by_fold)) #[adjustedR2(r2, n_subset_features, sample_size) for r2 in score_by_fold]\n\n score = np.average(score_by_fold) \n return score",
"_____no_output_____"
],
[
"def optimizeComponentsPCR(X, y, score_method):\n score_list = []\n for n in range(1, X.shape[1]+1):\n lr_model = LinearRegression()\n score_n = PCR(lr_model, X, y, n, score_method)\n score_list.append(score_n)\n print('Number of components: {} | Score: {}'.format(n, score_n))\n return max(enumerate(score_list), key=lambda x: x[1])[0]+1",
"_____no_output_____"
],
[
"# Computing the optimal number of components for predicting each of our DVs (LaBSE)\n\nlabse_best_components = []\n\nprint('Getting best number of components for predicting F1-score (LaBSE)')\nres1 = optimizeComponentsPCR(X_pair_labse, y_pair_f1_labse, 'r2')\nprint('Optimal components: {}'.format(res1))\nlabse_best_components.append(res1)\n\nprint('Getting best number of components for predicting G-H dist. (LaBSE)')\nres2 = optimizeComponentsPCR(X_pair_labse, y_pair_gh_labse, 'r2')\nprint('Optimal components: {}'.format(res2))\nlabse_best_components.append(res2)\n\nprint('Getting best number of components for predicting SVG (LaBSE)')\nres3 = optimizeComponentsPCR(X_pair_labse, y_pair_svg_labse, 'r2')\nprint('Optimal components: {}'.format(res3))\nlabse_best_components.append(res3)\n\nprint('Getting best number of components for predicting ECOND-HM (LaBSE)')\nres4 = optimizeComponentsPCR(X_pair_labse, y_pair_econdhm_labse, 'r2')\nprint('Optimal components: {}'.format(res4))\nlabse_best_components.append(res4)\n\nprint('Getting best number of components for predicting avg. margin score (LaBSE)')\nres5 = optimizeComponentsPCR(X_pair_labse, y_pair_avgmarg_labse, 'r2')\nprint('Optimal components: {}'.format(res5))\nlabse_best_components.append(res5)\n\nprint('\\nAverage best number of components (LaBSE): {}'.format(np.average(labse_best_components)))",
"Getting best number of components for predicting F1-score (LaBSE)\nNumber of components: 1 | Score: 0.17972379521571577\nNumber of components: 2 | Score: 0.1901240819549382\nNumber of components: 3 | Score: 0.21476442655085473\nNumber of components: 4 | Score: 0.29956825492307526\nNumber of components: 5 | Score: 0.34289235673145446\nNumber of components: 6 | Score: 0.3421048165496681\nNumber of components: 7 | Score: 0.3372982915953129\nNumber of components: 8 | Score: 0.3360520687440106\nNumber of components: 9 | Score: 0.3335706883430518\nNumber of components: 10 | Score: 0.3312733377591113\nNumber of components: 11 | Score: 0.4039158484777078\nNumber of components: 12 | Score: 0.4075171131971723\nNumber of components: 13 | Score: 0.4070012098494723\nOptimal components: 12\nGetting best number of components for predicting G-H dist. (LaBSE)\nNumber of components: 1 | Score: -0.023450041867626937\nNumber of components: 2 | Score: -0.022016428319914638\nNumber of components: 3 | Score: -0.019884421939913976\nNumber of components: 4 | Score: 0.00909835522611423\nNumber of components: 5 | Score: 0.004125602923978655\nNumber of components: 6 | Score: 0.003271217259553949\nNumber of components: 7 | Score: -0.003200684233527129\nNumber of components: 8 | Score: -0.0035266503736736342\nNumber of components: 9 | Score: -0.016886078448594754\nNumber of components: 10 | Score: -0.020666874055134076\nNumber of components: 11 | Score: -0.01911219759191459\nNumber of components: 12 | Score: -0.021771350806071576\nNumber of components: 13 | Score: -0.021459883412738846\nOptimal components: 4\nGetting best number of components for predicting SVG (LaBSE)\nNumber of components: 1 | Score: -15.327637555806144\nNumber of components: 2 | Score: -15.170055032880992\nNumber of components: 3 | Score: -15.348415463605608\nNumber of components: 4 | Score: -15.723437710804806\nNumber of components: 5 | Score: -20.138578085396354\nNumber of components: 6 | Score: -20.22582664045378\nNumber of components: 7 | Score: -26.17349375339247\nNumber of components: 8 | Score: -26.338045762239894\nNumber of components: 9 | Score: -27.895656205308597\nNumber of components: 10 | Score: -30.258586287075996\nNumber of components: 11 | Score: -30.659528153335394\nNumber of components: 12 | Score: -33.59505881116864\nNumber of components: 13 | Score: -33.5589871805668\nOptimal components: 2\nGetting best number of components for predicting ECOND-HM (LaBSE)\nNumber of components: 1 | Score: 0.17240243596218333\nNumber of components: 2 | Score: 0.17070721517851442\nNumber of components: 3 | Score: 0.18531565361127683\nNumber of components: 4 | Score: 0.18461839643528005\nNumber of components: 5 | Score: 0.19525640859385313\nNumber of components: 6 | Score: 0.1983754682800799\nNumber of components: 7 | Score: 0.17397518755381663\nNumber of components: 8 | Score: 0.17525965113784145\nNumber of components: 9 | Score: 0.16983126022206396\nNumber of components: 10 | Score: 0.1656266328722466\nNumber of components: 11 | Score: 0.11960482469443368\nNumber of components: 12 | Score: 0.0718231184334466\nNumber of components: 13 | Score: 0.06989622254615739\nOptimal components: 6\nGetting best number of components for predicting avg. margin score (LaBSE)\nNumber of components: 1 | Score: 0.09010179162968665\nNumber of components: 2 | Score: 0.10532237744780584\nNumber of components: 3 | Score: 0.13163965015930104\nNumber of components: 4 | Score: 0.21540326700058202\nNumber of components: 5 | Score: 0.2556903557196278\nNumber of components: 6 | Score: 0.25530312961692075\nNumber of components: 7 | Score: 0.2500554679730965\nNumber of components: 8 | Score: 0.24878856597548843\nNumber of components: 9 | Score: 0.24146731977725172\nNumber of components: 10 | Score: 0.23949006569759934\nNumber of components: 11 | Score: 0.26486129986939516\nNumber of components: 12 | Score: 0.26686805094549276\nNumber of components: 13 | Score: 0.26614060820906227\nOptimal components: 12\n\nAverage best number of components (LaBSE): 7.2\n"
],
[
"# Computing the optimal number of components for predicting each of our DVs (LASER)\n\nlaser_best_components = []\n\nprint('Getting best number of components for predicting F1-score (LASER)')\nres1 = optimizeComponentsPCR(X_pair_laser, y_pair_f1_laser, 'r2')\nprint('Optimal components: {}'.format(res1))\nlaser_best_components.append(res1)\n\nprint('Getting best number of components for predicting G-H dist. (LASER)')\nres2 = optimizeComponentsPCR(X_pair_laser, y_pair_gh_laser, 'r2')\nprint('Optimal components: {}'.format(res2))\nlaser_best_components.append(res2)\n\nprint('Getting best number of components for predicting SVG (LASER)')\nres3 = optimizeComponentsPCR(X_pair_laser, y_pair_svg_laser, 'r2')\nprint('Optimal components: {}'.format(res3))\nlaser_best_components.append(res3)\n\nprint('Getting best number of components for predicting ECOND-HM (LASER)')\nres4 = optimizeComponentsPCR(X_pair_laser, y_pair_econdhm_laser, 'r2')\nprint('Optimal components: {}'.format(res4))\nlaser_best_components.append(res4)\n\nprint('Getting best number of components for predicting avg. margin score (LASER)')\nres5 = optimizeComponentsPCR(X_pair_laser, y_pair_avgmarg_laser, 'r2')\nprint('Optimal components: {}'.format(res5))\nlaser_best_components.append(res5)\n\nprint('\\nAverage best number of components (LASER): {}'.format(np.average(laser_best_components)))",
"Getting best number of components for predicting F1-score (LASER)\nNumber of components: 1 | Score: 0.25132995699234767\nNumber of components: 2 | Score: 0.24533497663419532\nNumber of components: 3 | Score: 0.24429672952637901\nNumber of components: 4 | Score: 0.32625652037710645\nNumber of components: 5 | Score: 0.324452619728815\nNumber of components: 6 | Score: 0.33632049976740347\nNumber of components: 7 | Score: 0.3330385036809088\nNumber of components: 8 | Score: 0.3315329782183334\nNumber of components: 9 | Score: 0.34004006789883046\nNumber of components: 10 | Score: 0.3454269658345027\nNumber of components: 11 | Score: 0.37131678954138614\nNumber of components: 12 | Score: 0.36917892682023223\nNumber of components: 13 | Score: 0.36910899491731164\nOptimal components: 11\nGetting best number of components for predicting G-H dist. (LASER)\nNumber of components: 1 | Score: -0.015035956926025273\nNumber of components: 2 | Score: -0.015123503675341422\nNumber of components: 3 | Score: -0.013855788261972046\nNumber of components: 4 | Score: 0.018119577453560652\nNumber of components: 5 | Score: 0.012091670481161932\nNumber of components: 6 | Score: 0.011570117389545654\nNumber of components: 7 | Score: 0.006528029298617133\nNumber of components: 8 | Score: 0.005809074326431041\nNumber of components: 9 | Score: -0.0020434895764344873\nNumber of components: 10 | Score: -0.007913653166674573\nNumber of components: 11 | Score: -0.004456867117794383\nNumber of components: 12 | Score: -0.007335936437130874\nNumber of components: 13 | Score: -0.006800213465730199\nOptimal components: 4\nGetting best number of components for predicting SVG (LASER)\nNumber of components: 1 | Score: -2.092335835039092\nNumber of components: 2 | Score: -2.0953046899998853\nNumber of components: 3 | Score: -2.1048317282868187\nNumber of components: 4 | Score: -2.2438478980664636\nNumber of components: 5 | Score: -2.462178615394362\nNumber of components: 6 | Score: -2.529091627679765\nNumber of components: 7 | Score: -3.8417726735413944\nNumber of components: 8 | Score: -3.88208279102322\nNumber of components: 9 | Score: -3.62729747769029\nNumber of components: 10 | Score: -3.7558858385761056\nNumber of components: 11 | Score: -3.7403916673114375\nNumber of components: 12 | Score: -4.192159341439939\nNumber of components: 13 | Score: -4.171736703881545\nOptimal components: 1\nGetting best number of components for predicting ECOND-HM (LASER)\nNumber of components: 1 | Score: 0.02406136741858822\nNumber of components: 2 | Score: 0.038590178900507324\nNumber of components: 3 | Score: 0.053384939847624935\nNumber of components: 4 | Score: 0.07516391377271717\nNumber of components: 5 | Score: 0.08662986056238306\nNumber of components: 6 | Score: 0.09745978213582872\nNumber of components: 7 | Score: 0.0916740163540157\nNumber of components: 8 | Score: 0.09169259044772156\nNumber of components: 9 | Score: 0.10009273047292475\nNumber of components: 10 | Score: 0.10446411769286446\nNumber of components: 11 | Score: 0.08806274092131895\nNumber of components: 12 | Score: 0.08274294196038093\nNumber of components: 13 | Score: 0.0881484428879068\nOptimal components: 10\nGetting best number of components for predicting avg. margin score (LASER)\nNumber of components: 1 | Score: 0.020245403773229077\nNumber of components: 2 | Score: 0.03262116728466258\nNumber of components: 3 | Score: 0.05925128565550993\nNumber of components: 4 | Score: 0.07978972709241317\nNumber of components: 5 | Score: 0.07751119669949094\nNumber of components: 6 | Score: 0.07687859192950443\nNumber of components: 7 | Score: 0.06303077147862242\nNumber of components: 8 | Score: 0.0609031941020155\nNumber of components: 9 | Score: 0.07182133673477151\nNumber of components: 10 | Score: 0.06734702514129629\nNumber of components: 11 | Score: 0.032973086274687646\nNumber of components: 12 | Score: -0.012399648033380761\nNumber of components: 13 | Score: -0.02237236910831223\nOptimal components: 4\n\nAverage best number of components (LASER): 6.0\n"
],
[
"# Perform ablation analysis to see how removing each predictor individually affects the regression fit\ndef ablateLinReg(X, y, score_method):\n FOLDS = 10\n n_features = X.shape[1]\n ablation_feature_diffs = {}\n\n model = LinearRegression()\n\n # Convert R2 to adjusted R2 to take into account the number of predictors\n def adjustedR2(r2, n, p):\n num = (1-r2)*(n-1)\n denom = n-p-1\n adj_r2 = 1 - (num/denom)\n return adj_r2\n\n # Getting baseline score using all the features\n score_by_fold = sklearn.model_selection.cross_validate(model,\n X,\n y,\n cv=FOLDS,\n scoring=score_method)['test_score']\n if score_method=='r2':\n # Compute the adjusted R2 instead\n N = n_features-1\n # Fraction of data used for training during CV\n train_frac = (FOLDS-1) / FOLDS # e.g. with 10 folds, we use 9/10 of the data for training\n sample_size = round(train_frac*X.shape[0])\n score_by_fold = list(map(lambda r2: adjustedR2(r2, sample_size, N), score_by_fold)) \n baseline_score = np.average(score_by_fold) \n \n\n # We'll drop each of the features one-by-one and see how the fit (adjusted R2) of the model changes\n for i in range(n_features):\n dropped_feature = X.columns[i]\n X_ablated = X.drop(columns=dropped_feature) # Ablated feature space\n score_by_fold = sklearn.model_selection.cross_validate(model, \n X_ablated, \n y, \n cv=FOLDS, \n scoring=score_method)['test_score']\n \n if score_method=='r2':\n # Compute the adjusted R2 instead\n N = n_features-1\n # Fraction of data used for training during CV\n train_frac = (FOLDS-1) / FOLDS # e.g. with 10 folds, we use 9/10 of the data for training\n sample_size = round(train_frac*X.shape[0])\n score_by_fold = list(map(lambda r2: adjustedR2(r2, sample_size, N), score_by_fold)) \n \n score_diff = baseline_score - np.average(score_by_fold)\n # The higher the score_diff, the more important that feature is\n ablation_feature_diffs[dropped_feature] = score_diff\n\n # Return dictionary sorted in descending order\n ablation_feature_diffs = {k: v for k, v in sorted(ablation_feature_diffs.items(), key=lambda item: item[1], reverse=True)}\n for k,v in zip(ablation_feature_diffs.keys(), ablation_feature_diffs.values()):\n print('Dropped feature: {} | Score difference: {}'.format(k, v))\n print('\\n')\n return ablation_feature_diffs",
"_____no_output_____"
],
[
"print('LaBSE F1-score ablation experiment')\nlabse_f1_ablation = ablateLinReg(X_pair_labse, y_pair_f1_labse, 'r2')\nprint('LaBSE GH dist. ablation experiment')\nlabse_gh_ablation = ablateLinReg(X_pair_labse, y_pair_gh_labse, 'r2')\nprint('LaBSE SVG ablation experiment')\nlabse_svg_ablation = ablateLinReg(X_pair_labse, y_pair_svg_labse, 'r2')\nprint('LaBSE ECOND-HM ablation experiment')\nlabse_econdhm_ablation = ablateLinReg(X_pair_labse, y_pair_econdhm_labse, 'r2')\nprint('LaBSE avg. margin score ablation experiment')\nlabse_avgmarg_ablation = ablateLinReg(X_pair_labse, y_pair_avgmarg_labse, 'r2')\n\nprint('LASER F1-score ablation experiment')\nlaser_f1_ablation = ablateLinReg(X_pair_laser, y_pair_f1_laser, 'r2')\nprint('LASER GH dist. ablation experiment')\nlaser_gh_ablation = ablateLinReg(X_pair_laser, y_pair_gh_laser, 'r2')\nprint('LASER SVG ablation experiment')\nlaser_svg_ablation = ablateLinReg(X_pair_laser, y_pair_svg_laser, 'r2')\nprint('LASER ECOND-HM ablation experiment')\nlaser_econdhm_ablation = ablateLinReg(X_pair_laser, y_pair_econdhm_laser, 'r2')\nprint('LASER avg. margin score ablation experiment')\nlaser_avgmarg_ablation = ablateLinReg(X_pair_laser, y_pair_avgmarg_laser, 'r2')",
"LaBSE F1-score ablation experiment\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: 0.08709077917140412\nDropped feature: Same Polysynthesis Status? | Score difference: 0.07801459067082295\nDropped feature: Combined in-family sentences (LaBSE) | Score difference: 0.021541212157679013\nDropped feature: Combined sentences (LaBSE) | Score difference: 0.017490943900182687\nDropped feature: Geographic Distance (lang2vec) | Score difference: 0.0011430479674968685\nDropped feature: Phonological Distance (lang2vec) | Score difference: 0.0001029806808426903\nDropped feature: Same Genus? | Score difference: -0.0001531678426618388\nDropped feature: Same Family? | Score difference: -0.0007611213982388065\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.0008741335340861633\nDropped feature: Combined in-genus sentences (LaBSE) | Score difference: -0.0018140830062930435\nDropped feature: Same Word Order? | Score difference: -0.0018827690611165626\nDropped feature: Syntactic Distance (lang2vec) | Score difference: -0.002441537556928719\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -0.013319129609875913\n\n\nLaBSE GH dist. ablation experiment\nDropped feature: Same Word Order? | Score difference: 0.006897161648695593\nDropped feature: Same Polysynthesis Status? | Score difference: 0.00645739364107778\nDropped feature: Geographic Distance (lang2vec) | Score difference: 0.0023524478594463827\nDropped feature: Combined in-family sentences (LaBSE) | Score difference: 0.0008682008555018113\nDropped feature: Same Genus? | Score difference: -2.759898914258402e-05\nDropped feature: Combined sentences (LaBSE) | Score difference: -0.00030655642704104785\nDropped feature: Same Family? | Score difference: -0.0008277636397534191\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: -0.0009677888537195992\nDropped feature: Combined in-genus sentences (LaBSE) | Score difference: -0.0013085775103701472\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.0038018125687619628\nDropped feature: Phonological Distance (lang2vec) | Score difference: -0.005744979994610077\nDropped feature: Syntactic Distance (lang2vec) | Score difference: -0.007565268961590954\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -0.008039777900211409\n\n\nLaBSE SVG ablation experiment\nDropped feature: Geographic Distance (lang2vec) | Score difference: 0.5141641599718483\nDropped feature: Combined sentences (LaBSE) | Score difference: 0.22714934044942225\nDropped feature: Same Genus? | Score difference: -0.016431399398932456\nDropped feature: Combined in-genus sentences (LaBSE) | Score difference: -0.03781069649957658\nDropped feature: Same Family? | Score difference: -0.11725021464062024\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: -0.39791610304009595\nDropped feature: Phonological Distance (lang2vec) | Score difference: -0.8385217008515014\nDropped feature: Same Word Order? | Score difference: -1.9244485135487892\nDropped feature: Same Polysynthesis Status? | Score difference: -2.1738540974039644\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -2.534763936260454\nDropped feature: Combined in-family sentences (LaBSE) | Score difference: -2.8475637580025968\nDropped feature: Syntactic Distance (lang2vec) | Score difference: -3.113730680884963\nDropped feature: Inventory Distance (lang2vec) | Score difference: -3.22421194282796\n\n\nLaBSE ECOND-HM ablation experiment\nDropped feature: Combined in-family sentences (LaBSE) | Score difference: 0.05063046779669611\nDropped feature: Combined sentences (LaBSE) | Score difference: 0.024817862287948897\nDropped feature: Same Polysynthesis Status? | Score difference: 0.013061189424213152\nDropped feature: Same Genus? | Score difference: 7.79825257350708e-06\nDropped feature: Same Word Order? | Score difference: -0.00010921185526750754\nDropped feature: Combined in-genus sentences (LaBSE) | Score difference: -0.00022050860585579035\nDropped feature: Same Family? | Score difference: -0.00043902324188749287\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -0.0005549923298013015\nDropped feature: Syntactic Distance (lang2vec) | Score difference: -0.0006484675761729564\nDropped feature: Geographic Distance (lang2vec) | Score difference: -0.005292842536381986\nDropped feature: Phonological Distance (lang2vec) | Score difference: -0.008499506025806527\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.041923091467889875\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: -0.0543895933454894\n\n\nLaBSE avg. margin score ablation experiment\nDropped feature: Same Polysynthesis Status? | Score difference: 0.07679985887940755\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: 0.034876410633977506\nDropped feature: Combined sentences (LaBSE) | Score difference: 0.024301586963855176\nDropped feature: Combined in-family sentences (LaBSE) | Score difference: 0.007719308465425179\nDropped feature: Same Genus? | Score difference: -1.3773338356448672e-05\nDropped feature: Geographic Distance (lang2vec) | Score difference: -8.434771939935803e-05\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.0007721091921477075\nDropped feature: Same Family? | Score difference: -0.001094174462460018\nDropped feature: Phonological Distance (lang2vec) | Score difference: -0.0018141949070326624\nDropped feature: Combined in-genus sentences (LaBSE) | Score difference: -0.0018694720877838744\nDropped feature: Same Word Order? | Score difference: -0.0031653663381369657\nDropped feature: Syntactic Distance (lang2vec) | Score difference: -0.003227684390767427\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -0.01814723784528116\n\n\nLASER F1-score ablation experiment\nDropped feature: Combined in-family sentences (LASER) | Score difference: 0.03532655308145333\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: 0.034090201192904346\nDropped feature: Same Word Order? | Score difference: 0.014845703435949376\nDropped feature: Syntactic Distance (lang2vec) | Score difference: 0.009062735090504759\nDropped feature: Same Polysynthesis Status? | Score difference: 0.006155983489184336\nDropped feature: Geographic Distance (lang2vec) | Score difference: 0.0016916191473048126\nDropped feature: Phonological Distance (lang2vec) | Score difference: 0.001103256677490616\nDropped feature: Same Family? | Score difference: 0.000568760502462462\nDropped feature: Combined in-genus sentences (LASER) | Score difference: 0.00015885817914013112\nDropped feature: Same Genus? | Score difference: -0.0007391347953327743\nDropped feature: Combined sentences (LASER) | Score difference: -0.000841200574562273\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.0025014959703905104\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -0.006197126425025512\n\n\nLASER GH dist. ablation experiment\nDropped feature: Combined in-family sentences (LASER) | Score difference: 0.01170852720237383\nDropped feature: Same Polysynthesis Status? | Score difference: 0.011516445324771418\nDropped feature: Same Word Order? | Score difference: 0.010489577791007787\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: 0.002804860489116035\nDropped feature: Geographic Distance (lang2vec) | Score difference: 0.0027856004947055205\nDropped feature: Combined sentences (LASER) | Score difference: 6.921745205095125e-05\nDropped feature: Same Genus? | Score difference: 1.4123918417818793e-05\nDropped feature: Combined in-genus sentences (LASER) | Score difference: -0.0006598227178345791\nDropped feature: Same Family? | Score difference: -0.0021272974244777917\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.003883400899434042\nDropped feature: Phonological Distance (lang2vec) | Score difference: -0.004149404810619562\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -0.007408686284347976\nDropped feature: Syntactic Distance (lang2vec) | Score difference: -0.010804680206020845\n\n\nLASER SVG ablation experiment\nDropped feature: Geographic Distance (lang2vec) | Score difference: 0.12210076492366895\nDropped feature: Same Word Order? | Score difference: 0.045996229455272264\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: 0.024431252872374465\nDropped feature: Same Genus? | Score difference: 0.0006070374917470645\nDropped feature: Combined sentences (LASER) | Score difference: -0.0012463497644237265\nDropped feature: Combined in-genus sentences (LASER) | Score difference: -0.025368497825972725\nDropped feature: Phonological Distance (lang2vec) | Score difference: -0.0494573144482775\nDropped feature: Same Family? | Score difference: -0.068461744711942\nDropped feature: Syntactic Distance (lang2vec) | Score difference: -0.1629783877770743\nDropped feature: Same Polysynthesis Status? | Score difference: -0.16455936176583563\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -0.32840567146623023\nDropped feature: Combined in-family sentences (LASER) | Score difference: -0.4006159427286615\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.5136894728137866\n\n\nLASER ECOND-HM ablation experiment\nDropped feature: Combined in-family sentences (LASER) | Score difference: 0.05376929029901581\nDropped feature: Same Polysynthesis Status? | Score difference: 0.04303776283631127\nDropped feature: Combined sentences (LASER) | Score difference: 0.018473247825301464\nDropped feature: Syntactic Distance (lang2vec) | Score difference: 0.01436651882626154\nDropped feature: Same Family? | Score difference: 0.009733927772574924\nDropped feature: Phonological Distance (lang2vec) | Score difference: 0.0023229796708655326\nDropped feature: Geographic Distance (lang2vec) | Score difference: -0.00048052019916516864\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: -0.0005638277784352774\nDropped feature: Same Genus? | Score difference: -0.0007712784773083181\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.0011656812150869916\nDropped feature: Combined in-genus sentences (LASER) | Score difference: -0.0013382095941681382\nDropped feature: Same Word Order? | Score difference: -0.004028024792243651\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: -0.016029288917307788\n\n\nLASER avg. margin score ablation experiment\nDropped feature: Combined sentences (LASER) | Score difference: 0.030704686799008576\nDropped feature: Combined in-family sentences (LASER) | Score difference: 0.028208706676016784\nDropped feature: Same Polysynthesis Status? | Score difference: 0.0015596997047992028\nDropped feature: Same Word Order? | Score difference: 0.0013753483275052783\nDropped feature: Character-level Overlap (multiset Jaccard coefficient, Book of Matthew) | Score difference: 0.0013201462157613336\nDropped feature: Geographic Distance (lang2vec) | Score difference: 0.0008338622100955949\nDropped feature: Syntactic Distance (lang2vec) | Score difference: 0.0007432489929740799\nDropped feature: Combined in-genus sentences (LASER) | Score difference: 7.327060421214587e-06\nDropped feature: Same Genus? | Score difference: -0.0015927878852523975\nDropped feature: Same Family? | Score difference: -0.0026651532178667003\nDropped feature: Phonological Distance (lang2vec) | Score difference: -0.015857708281955263\nDropped feature: Token-level Overlap (multiset Jaccard coefficient, Book of John) | Score difference: -0.019002520856765303\nDropped feature: Inventory Distance (lang2vec) | Score difference: -0.05307703403547537\n\n\n"
],
[
"# Let's see how important each feature is, on average, according to the ablation experiments\n\n# LaBSE\nfeature_orders_in_ablation_labse = {}\nfor idx, item in enumerate(labse_f1_ablation.keys()):\n feature_orders_in_ablation_labse[item] = [idx]\nfor idx, item in enumerate(labse_gh_ablation.keys()):\n feature_orders_in_ablation_labse[item].append(idx)\nfor idx, item in enumerate(labse_svg_ablation.keys()):\n feature_orders_in_ablation_labse[item].append(idx)\nfor idx, item in enumerate(labse_econdhm_ablation.keys()):\n feature_orders_in_ablation_labse[item].append(idx)\nfor idx, item in enumerate(labse_avgmarg_ablation.keys()):\n feature_orders_in_ablation_labse[item].append(idx)\n\nfor k in feature_orders_in_ablation_labse: \n feature_orders_in_ablation_labse[k] = np.average(feature_orders_in_ablation_labse[k])\n\n# LASER\nfeature_orders_in_ablation_laser = {}\nfor idx, item in enumerate(laser_f1_ablation.keys()):\n feature_orders_in_ablation_laser[item] = [idx]\nfor idx, item in enumerate(laser_gh_ablation.keys()):\n feature_orders_in_ablation_laser[item].append(idx)\nfor idx, item in enumerate(laser_svg_ablation.keys()):\n feature_orders_in_ablation_laser[item].append(idx)\nfor idx, item in enumerate(laser_econdhm_ablation.keys()):\n feature_orders_in_ablation_laser[item].append(idx)\nfor idx, item in enumerate(laser_avgmarg_ablation.keys()):\n feature_orders_in_ablation_laser[item].append(idx)\n\nfor k in feature_orders_in_ablation_laser: \n feature_orders_in_ablation_laser[k] = np.average(feature_orders_in_ablation_laser[k])",
"_____no_output_____"
],
[
"# Sort the average feature order lists\nfeature_orders_in_ablation_labse = sorted(feature_orders_in_ablation_labse.items(), key=lambda item: item[1])\nfeature_orders_in_ablation_laser = sorted(feature_orders_in_ablation_laser.items(), key=lambda item: item[1])",
"_____no_output_____"
],
[
"feature_orders_in_ablation_labse",
"_____no_output_____"
],
[
"feature_orders_in_ablation_laser",
"_____no_output_____"
]
],
[
[
"Taking a look at the loadings of the first principal components",
"_____no_output_____"
]
],
[
[
"pca = sklearn.decomposition.PCA(n_components=7)\nX_pair_labse_pca = pca.fit_transform(X_pair_labse)",
"_____no_output_____"
],
[
"pca_labse_loadings = pd.DataFrame(pca.components_.T, columns=['PC1', 'PC2', 'PC3', 'PC4', 'PC5', 'PC6', 'PC7'], index=X_pair_labse.columns)\npca_labse_loadings",
"_____no_output_____"
],
[
"pca = sklearn.decomposition.PCA(n_components=6)\nX_pair_laser_pca = pca.fit_transform(X_pair_laser)\n\npca_laser_loadings = pd.DataFrame(pca.components_.T, columns=['PC1', 'PC2', 'PC3', 'PC4', 'PC5', 'PC6'], index=X_pair_laser.columns)\npca_laser_loadings",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7f85f0b04440ba7a74917756621654dd0d4d101 | 8,103 | ipynb | Jupyter Notebook | notebooks/08.Make.generalized.ufuncs.ipynb | IsabelAverill/Scipy-2017---Numba | 4d74aba75ca8072c616c46fdc27ce78777f35f84 | [
"CC-BY-4.0"
] | 149 | 2016-06-28T21:57:25.000Z | 2022-01-09T00:03:09.000Z | notebooks/08.Make.generalized.ufuncs.ipynb | IsabelAverill/Scipy-2017---Numba | 4d74aba75ca8072c616c46fdc27ce78777f35f84 | [
"CC-BY-4.0"
] | 9 | 2017-06-11T21:20:59.000Z | 2018-10-18T13:57:30.000Z | notebooks/08.Make.generalized.ufuncs.ipynb | IsabelAverill/Scipy-2017---Numba | 4d74aba75ca8072c616c46fdc27ce78777f35f84 | [
"CC-BY-4.0"
] | 68 | 2016-06-30T00:26:57.000Z | 2021-12-28T18:50:38.000Z | 22.571031 | 251 | 0.506109 | [
[
[
"## Generalized ufuncs",
"_____no_output_____"
],
[
"We've just seen how to make our own ufuncs using `vectorize`, but what if we need something that can operate on an input array in any way that is not element-wise?\n\nEnter `guvectorize`. \n\nThere are several important differences between `vectorize` and `guvectorize` that bear close examination. Let's take a look at a few simple examples.",
"_____no_output_____"
]
],
[
[
"import numpy\nfrom numba import guvectorize",
"_____no_output_____"
],
[
"@guvectorize('int64[:], int64, int64[:]', '(n),()->(n)')\ndef g(x, y, result):\n for i in range(x.shape[0]):\n result[i] = x[i] + y",
"_____no_output_____"
]
],
[
[
"* Declaration of input/output layouts\n* No return statements",
"_____no_output_____"
]
],
[
[
"x = numpy.arange(10)",
"_____no_output_____"
]
],
[
[
"In the cell below we call the function `g` with a preallocated array for the result.",
"_____no_output_____"
]
],
[
[
"result = numpy.zeros_like(x)\nresult = g(x, 5, result)\nprint(result)",
"_____no_output_____"
]
],
[
[
"But wait! We can still call `g` as if it were defined as `def g(x, y)`\n\n```python\nres = g(x, 5)\nprint(res)\n```\n\nWe don't recommend this as it can have unintended consequences if some of the elements of the `results` array are not operated on by the function `g`. (The advantage is that you can preserve existing interfaces to previously written functions).",
"_____no_output_____"
]
],
[
[
"@guvectorize('float64[:,:], float64[:,:], float64[:,:]', \n '(m,n),(n,p)->(m,p)')\ndef matmul(A, B, C):\n m, n = A.shape\n n, p = B.shape\n for i in range(m):\n for j in range(p):\n C[i, j] = 0\n for k in range(n):\n C[i, j] += A[i, k] * B[k, j]",
"_____no_output_____"
],
[
"a = numpy.random.random((500, 500))",
"_____no_output_____"
],
[
"out = matmul(a, a, numpy.zeros_like(a))",
"_____no_output_____"
],
[
"%timeit matmul(a, a, numpy.zeros_like(a))",
"_____no_output_____"
],
[
"%timeit a @ a",
"_____no_output_____"
]
],
[
[
"And it also supports the `target` keyword argument",
"_____no_output_____"
]
],
[
[
"def g(x, y, res):\n for i in range(x.shape[0]):\n res[i] = x[i] + numpy.exp(y)\n \ng_serial = guvectorize('float64[:], float64, float64[:]', \n '(n),()->(n)')(g)\ng_par = guvectorize('float64[:], float64, float64[:]', \n '(n),()->(n)', target='parallel')(g)",
"_____no_output_____"
],
[
"%timeit res = g_serial(numpy.arange(1000000).reshape(1000, 1000), 3)\n%timeit res = g_par(numpy.arange(1000000).reshape(1000, 1000), 3)",
"_____no_output_____"
]
],
[
[
"## [Exercise: Writing signatures](./exercises/08.GUVectorize.Exercises.ipynb#Exercise:-2D-Heat-Transfer-signature)",
"_____no_output_____"
],
[
"What's up with these boundary conditions?\n\n```python\nfor i in range(I):\n Tn[i, 0] = T[i, 0]\n Tn[i, J - 1] = Tn[i, J - 2]\n\n for j in range(J):\n Tn[0, j] = T[0, j]\n Tn[I - 1, j] = Tn[I - 2, j]\n```\n\nWe don't pass in `Tn` explicitly, which means Numba allocates it for us (thanks!) but it's allocated using `numpy.empty_like` so if we don't touch every value in `Tn` in the function, those empty values will stick around and cause trouble. \n\nSolutions? The one above, or pass it in explicitly after doing something like `Tn = Ti.copy()`",
"_____no_output_____"
],
[
"## [Exercise: Remove the vanilla loops](./exercises/08.GUVectorize.Exercises.ipynb#Exercise:-2D-Heat-Transfer-Time-loop)",
"_____no_output_____"
],
[
"The example above loops in time outside of the `vectorize`d function. That means it's looping in vanilla Python which is not the fastest thing in the world. \n\nMove the time loop inside the function.",
"_____no_output_____"
],
[
"## Demo: Why not `jit` the `run_ftcs` function?",
"_____no_output_____"
],
[
"Because, at the moment, it won't work. (bummer).",
"_____no_output_____"
]
],
[
[
"@guvectorize('float64[:,:], float64[:,:]', '(n,n)->(n,n)')\ndef gucopy(a, b):\n I, J = a.shape\n for i in range(I):\n for j in range(J):\n b[i, j] = a[i, j]",
"_____no_output_____"
],
[
"from numba import jit",
"_____no_output_____"
],
[
"@jit\ndef make_a_copy():\n a = numpy.random.random((25,25))\n b = gucopy(a)\n \n return a, b",
"_____no_output_____"
],
[
"a, b = make_a_copy()\nassert numpy.allclose(a, b)",
"_____no_output_____"
],
[
"make_a_copy.inspect_types()",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
e7f8689dd28d7f50e540774ccbad95d3953e4c47 | 13,660 | ipynb | Jupyter Notebook | git_utils.ipynb | liaa-3r/utilidades-colab | 7c29ded202145b16b63a64687ebbb46e61c41e9f | [
"MIT"
] | null | null | null | git_utils.ipynb | liaa-3r/utilidades-colab | 7c29ded202145b16b63a64687ebbb46e61c41e9f | [
"MIT"
] | null | null | null | git_utils.ipynb | liaa-3r/utilidades-colab | 7c29ded202145b16b63a64687ebbb46e61c41e9f | [
"MIT"
] | null | null | null | 35.115681 | 184 | 0.525037 | [
[
[
"# Git_Utils",
"_____no_output_____"
],
[
"## Instruções:\nPara clonar um repositório, primeiro copie o url no GitHub ou no GitLab e insira no campo `REMOTE`.\n\nO formato deve ser, conforme o caso:\n```\nhttps://github.com/<nome_da_organizacao>/<nome_do_projeto>.git\n``` \nou \n```\nhttps://gitlab.com/<nome_da_organizacao>/<nome_do_subgrupo>/<nome_do_projeto.git\n```\n\n\nEm seguida, verifique se os campos `GIT_CONFIG_PATH` e `PROJECTS_PATH` correspondem aos caminhos no seu Drive para o arquivo de configuração do git e para a pasta de projetos.\n\nPor fim, execute a célula.\n\n**Atenção: o arquivo de configuração do git deve ter ao menos três linhas, na seguinte ordem:** \n```\nemail\nuser\naccess_token\n```\n**Para instruções sobre como obter tokens de acesso pessoal no GitHub e no GitLab, veja os guias oficiais:**\n\n+ [GitHub](https://help.github.com/pt/github/authenticating-to-github/creating-a-personal-access-token-for-the-command-line#creating-a-token);\n+ [GitLab](https://docs.gitlab.com/ee/user/profile/personal_access_tokens.html).",
"_____no_output_____"
]
],
[
[
"REPO_HTTPS_URL = 'https://gitlab.com/liaa-3r/sinapses/ia-dispositivos-legais.git'\nGIT_CONFIG_PATH = 'C:\\\\Users\\\\cmlima\\\\Desenvolvimento\\\\LIAA-3R\\\\config'\nPROJECTS_PATH = 'C:\\\\Users\\\\cmlima\\\\Desenvolvimento\\\\LIAA-3R\\\\projetos'\nACTION = \"pull\"\nBRANCH = 'master'\nCOMMIT_MESSAGE = \"\" \n\nimport os, re\nimport ipywidgets as widgets\nfrom ipywidgets import Layout\nfrom IPython.display import display, clear_output\n\nclass bcolors:\n HEADER = '\\033[95m'\n OKBLUE = '\\033[94m'\n OKGREEN = '\\033[92m'\n WARNING = '\\033[93m'\n FAIL = '\\033[91m'\n ENDC = '\\033[0m'\n BOLD = '\\033[1m'\n UNDERLINE = '\\033[4m'\n\nw_repo_https_url = widgets.Text(value=REPO_HTTPS_URL, description='REPO', disabled=False, layout=Layout(width='90%'))\nw_git_config_path = widgets.Text(value=GIT_CONFIG_PATH, description='CONFIG', disabled=False, layout=Layout(width='90%'))\nw_projects_path = widgets.Text(value=PROJECTS_PATH, description='PROJECT', disabled=False, layout=Layout(width='90%'))\n\nw_action = widgets.Dropdown(\n options=['commit-pull-push', 'commit', 'pull', 'push', 'clone'],\n value='pull',\n description='ACTION',\n disabled=False,\n layout=Layout(width='50%')\n)\n\nw_branch = widgets.Text(value=BRANCH, description='BRANCH', disabled=False)\n\nw_commit_message = widgets.Textarea(\n value='',\n placeholder='seja breve e objetivo(a)...',\n description='COMMIT',\n disabled=False,\n layout=Layout(width='90%')\n)\n\nw_execute_button = widgets.Button(\n description='executar',\n disabled=False,\n button_style='success',\n icon='play-circle'\n)\nw_exit_button = widgets.Button(\n description='sair',\n disabled=False,\n button_style='',\n icon='',\n layout=Layout(align_self='flex-end', margin=\"0 5px 0 0\")\n)\n\nform = widgets.VBox([\n w_repo_https_url, \n w_git_config_path, \n w_projects_path,\n widgets.HBox([w_action, w_branch]),\n w_commit_message,\n widgets.HBox([w_exit_button, w_execute_button], layout=Layout(align_self='flex-end', margin=\"20px 10% 0 0\"))\n], layout=Layout(width='90%', display='flex', align_items='flex-start', justify_content='flex-start'))\n\ndef print_error(message):\n print()\n print(bcolors.FAIL + 'O script não pôde ser concluído.')\n print(bcolors.FAIL + bcolors.BOLD + 'Erro: ' + message)\n\ndef is_valid_url(url):\n return re.match(\"^https:\\/\\/(.+\\/){1,}(.+)\\.git$\", url)\n\ndef repo_exists(path):\n if os.path.isdir(path):\n %cd {path}\n output = !git rev-parse --is-inside-work-tree 2>/dev/null || echo 0\n return output != '0'\n return False\n\ndef git_is_set():\n token = !git config user.password\n return len(token) > 0\n\ndef is_github(url):\n return 'https://github.com' in url\n\ndef get_credentials(path, url):\n file_path = os.path.join(path, 'github_config.txt' if is_github(url) else 'gitlab_config.txt') \n if not os.path.isfile(file_path):\n raise Exception('Arquivo de configuração não localizado.')\n with open(file_path, 'r') as file:\n email = file.readline()\n user = file.readline()\n token = file.readline()\n return (email,user,token)\n\ndef clone(url, root_path, token):\n %cd {root_path}\n if not is_github(url):\n url = 'https://oauth2:' + token + '@gitlab.com' + url.replace('https://gitlab.com', '') \n !git clone {url}\n path = os.path.join(root_path, re.search(\"([^\\/]*)\\.git$\", url).group(1))\n %cd {path}\n %ls\n print('remote:')\n !git remote -v\n\ndef pull(branch, url, token):\n if not is_github(url):\n remote = 'https://oauth2:' + token + '@gitlab.com' + url.replace('https://gitlab.com', '')\n !git pull {remote} {branch}\n\ndef push(branch, url, token):\n if is_github(url):\n remote = 'https://' + token + '@github.com' + url.replace('https://github.com', '')\n else:\n remote = 'https://oauth2:' + token + '@gitlab.com' + url.replace('https://gitlab.com', '')\n !git push {remote} {branch}\n\ndef commit(message):\n if len(message) == 0:\n message = 'Atualizado via git_utils'\n !git add .\n !git commit -m '{message}'\n\ndef clear_all(b):\n form.close()\n clear_output()\n \ndef wait():\n w_wait_button = widgets.Button(\n description='Clique para concluir o script, limpando o output',\n disabled=False,\n layout=Layout(align_self='center', margin=\"0 5px 0 0\")\n )\n w_wait_button.on_click(clear_all)\n display(w_wait_button)\n \ndef exit(b):\n form.close()\n clear_output()\n print(bcolors.OKBLUE + bcolors.BOLD + 'Script encerrado pelo usuário...')\n\ndef execute(b):\n\n print(bcolors.OKBLUE + bcolors.BOLD + 'iniciando...\\n')\n print(bcolors.ENDC + 'reunindo parâmetros...')\n \n try:\n \n if not is_valid_url(w_repo_https_url.value):\n raise Exception('Remoto inválido.')\n\n repo_url = w_repo_https_url.value\n project_name = re.search(\"([^\\/]*)\\.git$\", repo_url).group(1)\n projects_path = w_projects_path.value\n config_path = w_git_config_path.value\n repo_path = os.path.join(projects_path, project_name)\n branch = w_branch.value\n action = w_action.value\n commit_message = w_commit_message.value\n\n user, email, token = get_credentials(config_path, repo_url)\n\n if not repo_exists(repo_path) and action != 'clone':\n raise Exception('O repositório local não foi localizado. Você deve primeiro cloná-lo.')\n\n print()\n\n if not git_is_set():\n print('configurando o git...')\n git_config(config_path, repo_url)\n print()\n\n if action == 'clone':\n print('clonando repositório...')\n clone(repo_url, projects_path, token)\n elif action == 'pull':\n print('atualizando repositório local (pull)...')\n pull(branch, repo_url, token)\n elif action == 'push':\n print('atualizando repositório remoto (push)...')\n push(branch, repo_url, token)\n elif action == 'commit':\n print('iniciando commit...')\n commit(commit_message)\n elif action == 'commit-pull-push':\n print('iniciando sequência...')\n commit(commit_message)\n pull(branch, repo_url, token)\n push(branch, repo_url, token)\n else:\n raise Exception('A ação selecionada não está implementada.')\n \n except Exception as error:\n print_error(str(error))\n \n else:\n print()\n print(bcolors.OKGREEN + bcolors.BOLD + 'Script concluído.')\n \n finally:\n print()\n wait()\n \ndisplay(form)\n\nw_execute_button.on_click(execute)\nw_exit_button.on_click(exit)\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code"
]
] |
e7f86d8c7a7db1ade3b318f7d9456146f5757aa5 | 33,437 | ipynb | Jupyter Notebook | day03.ipynb | BaoZiAKmeng/BaoZiAKmeng | 717759ddc12f617ba80fe21aa50679876a8c727f | [
"Apache-2.0"
] | null | null | null | day03.ipynb | BaoZiAKmeng/BaoZiAKmeng | 717759ddc12f617ba80fe21aa50679876a8c727f | [
"Apache-2.0"
] | null | null | null | day03.ipynb | BaoZiAKmeng/BaoZiAKmeng | 717759ddc12f617ba80fe21aa50679876a8c727f | [
"Apache-2.0"
] | null | null | null | 27.725539 | 71 | 0.538595 | [
[
[
"file='D:\\\\zzs\\\\kaifangX.txt'\nopen_file=open(file,mode='r',encoding='gbk',errors = 'ignore')\nc_list=[]\nfor i in range(0,1000):\n line=open_file.readline()\n strip_line=line.strip('\\n')\n split_line=line.split(',')\n try:\n e_mail=split_line[9]\n except:\n print()\n print(e_mail)\nprint(c_list)",
"[email protected]\[email protected]\n\n\[email protected]\[email protected] \[email protected]\[email protected]\n\n\[email protected] \[email protected] \[email protected]\[email protected]\[email protected] \[email protected]\n\n\[email protected] \[email protected] \[email protected] \[email protected]\[email protected] \[email protected] \[email protected] \[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected] \[email protected]\[email protected]\[email protected]\n\n\n\n\[email protected]\[email protected] \[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\n\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected] \[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected] \[email protected] \[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n-\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\[email protected]\n[]\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
e7f880463d797a7f3becfe584b668b3114d9389a | 4,611 | ipynb | Jupyter Notebook | Assignments/answers/Lab_3-answers.ipynb | unmeshvrije/python-for-beginners | d8943130bfd2499a458d92d5f6db97170fd53810 | [
"Apache-2.0"
] | 7 | 2019-08-13T15:36:50.000Z | 2021-09-09T20:37:21.000Z | Assignments/answers/Lab_3-answers.ipynb | unmeshvrije/python-for-beginners | d8943130bfd2499a458d92d5f6db97170fd53810 | [
"Apache-2.0"
] | 2 | 2019-07-04T08:30:38.000Z | 2019-07-16T13:44:45.000Z | Assignments/answers/Lab_3-answers.ipynb | unmeshvrije/python-for-beginners | d8943130bfd2499a458d92d5f6db97170fd53810 | [
"Apache-2.0"
] | 4 | 2019-07-29T10:57:24.000Z | 2021-03-17T15:02:36.000Z | 22.062201 | 234 | 0.481891 | [
[
[
"# 1. Zipping Lists",
"_____no_output_____"
]
],
[
[
"import string\n\nfirst_example_list = [c for c in string.ascii_lowercase]\nsecond_example_list = [i for i in range(len(string.ascii_lowercase))]",
"_____no_output_____"
],
[
"def zip_lists(first_list, second_list):\n new_list = []\n for i in range(min(len(first_list), len(second_list))):\n new_list.append(first_list[i])\n new_list.append(second_list[i])\n return new_list\n\nprint(zip_lists(first_example_list, second_example_list))",
"['a', 0, 'b', 1, 'c', 2, 'd', 3, 'e', 4, 'f', 5, 'g', 6, 'h', 7, 'i', 8, 'j', 9, 'k', 10, 'l', 11, 'm', 12, 'n', 13, 'o', 14, 'p', 15, 'q', 16, 'r', 17, 's', 18, 't', 19, 'u', 20, 'v', 21, 'w', 22, 'x', 23, 'y', 24, 'z', 25]\n"
]
],
[
[
"# 2. Age Differences",
"_____no_output_____"
]
],
[
[
"example_people = [(16, \"Brian\"), (12, \"Lucy\"), (18, \"Harold\")]",
"_____no_output_____"
],
[
"def age_differences(people):\n for i in range(len(people) - 1):\n first_name = people[i][1]\n first_age = people[i][0]\n \n second_name = people[i + 1][1]\n second_age = people[i + 1][0]\n \n if first_age > second_age:\n difference = first_age - second_age\n print(\"{} is {} years older than {}.\".format(first_name, difference, second_name))\n \nage_differences(example_people)",
"Brian is 4 years older than Lucy.\n"
]
],
[
[
"# 3. Remove the Duplicates",
"_____no_output_____"
]
],
[
[
"example_doubled_list = [1, 1, 2, 3, 3, 4, 3]",
"_____no_output_____"
],
[
"def remove_doubles(doubled_list):\n no_doubles = []\n for number in doubled_list:\n if number not in no_doubles:\n no_doubles.append(number)\n return no_doubles\n\nprint(remove_doubles(example_doubled_list))",
"[1, 2, 3, 4]\n"
]
],
[
[
"# 4. Only the Duplicates",
"_____no_output_____"
]
],
[
[
"first_example_list = [1, 2, 3, 4]\nsecond_example_list = [1, 4, 5, 6]",
"_____no_output_____"
],
[
"def get_duplicates(first_list, second_list):\n duplicates = []\n for number in first_list:\n if number in second_list:\n duplicates.append(number)\n return duplicates\n\nprint(get_duplicates(first_example_list, second_example_list))",
"[1, 4]\n"
]
],
[
[
"# 5. Count the Duplicates",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
e7f882fe6d8e0a44129ffbdfc91e2c8c35221a58 | 3,627 | ipynb | Jupyter Notebook | PRELIM_EXAM.ipynb | Singko25/Linear-Algebra-58020 | bd282e9f17eddc4422f37e0dc7664a322bd3197c | [
"Apache-2.0"
] | null | null | null | PRELIM_EXAM.ipynb | Singko25/Linear-Algebra-58020 | bd282e9f17eddc4422f37e0dc7664a322bd3197c | [
"Apache-2.0"
] | null | null | null | PRELIM_EXAM.ipynb | Singko25/Linear-Algebra-58020 | bd282e9f17eddc4422f37e0dc7664a322bd3197c | [
"Apache-2.0"
] | null | null | null | 22.388889 | 237 | 0.384891 | [
[
[
"<a href=\"https://colab.research.google.com/github/Singko25/Linear-Algebra-58020/blob/main/PRELIM_EXAM.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"##SAAVEDRA",
"_____no_output_____"
],
[
"##QUESTION 1",
"_____no_output_____"
]
],
[
[
"import numpy as np \nC = np.eye(4)\nprint(C)",
"[[1. 0. 0. 0.]\n [0. 1. 0. 0.]\n [0. 0. 1. 0.]\n [0. 0. 0. 1.]]\n"
]
],
[
[
"##QUESTION 2",
"_____no_output_____"
]
],
[
[
"import numpy as np\nC = np.eye(4)\nprint('C = ')\nprint(C)\n\narray = np.multiply(2,C)\nprint('Doubled = ')\nprint(array)",
"C = \n[[1. 0. 0. 0.]\n [0. 1. 0. 0.]\n [0. 0. 1. 0.]\n [0. 0. 0. 1.]]\nDoubled = \n[[2. 0. 0. 0.]\n [0. 2. 0. 0.]\n [0. 0. 2. 0.]\n [0. 0. 0. 2.]]\n"
]
],
[
[
"##QUESTION 3\n",
"_____no_output_____"
]
],
[
[
"import numpy as np \nA = np.array([2,7,4])\nB = np.array([3,9,8])\n\ncross = np.cross(A,B)\nprint(cross)",
"[20 -4 -3]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f887ac20e7609ebd07c45d581889af8a30c5ff | 5,982 | ipynb | Jupyter Notebook | Mean & SD.ipynb | sharlenechen0113/Real-Estate-Price-Prediction | 374bf0b5ac11a04672fdb6354320bb1466602938 | [
"MIT"
] | null | null | null | Mean & SD.ipynb | sharlenechen0113/Real-Estate-Price-Prediction | 374bf0b5ac11a04672fdb6354320bb1466602938 | [
"MIT"
] | null | null | null | Mean & SD.ipynb | sharlenechen0113/Real-Estate-Price-Prediction | 374bf0b5ac11a04672fdb6354320bb1466602938 | [
"MIT"
] | null | null | null | 33.049724 | 257 | 0.533266 | [
[
[
"<a href=\"https://colab.research.google.com/github/sharlenechen0113/Real-Estate-Price-Prediction/blob/main/Mean%20%26%20SD.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"_____no_output_____"
],
[
"import torch\nimport torch.nn as nn\nimport torch.optim as optim\nimport torch.nn.functional as F\nimport torchvision.datasets as dset\nimport torchvision.transforms as T\nfrom torch.utils.data import TensorDataset\nfrom torch.utils.data import DataLoader\nfrom torch.utils.data import sampler\n\nimport numpy as np\nimport pandas as pd\nfrom PIL import Image\nfrom sklearn import preprocessing, metrics, model_selection",
"_____no_output_____"
],
[
"USE_GPU = True\n\ndtype = torch.float # we will be using float throughout this tutorial\n\nif USE_GPU and torch.cuda.is_available():\n device = torch.device('cuda')\nelse:\n device = torch.device('cpu')\n\n# Constant to control how frequently we print train loss\nprint_every = 100\n\nprint('using device:', device)",
"_____no_output_____"
],
[
"train_mean = 0.0\ntrain_std = 0.0\nval_mean = 0.0\nval_std = 0.0",
"_____no_output_____"
],
[
"district_list = ['villages_towns_Sanxia', 'villages_towns_Sanzhi',\n 'villages_towns_Sanchong', 'villages_towns_Zhonghe',\n 'villages_towns_Zhongshan', 'villages_towns_Zhongzheng',\n 'villages_towns_Wugu', 'villages_towns_Xinyi', 'villages_towns_Neihu',\n 'villages_towns_Bali', 'villages_towns_Beitou',\n 'villages_towns_Nangang', 'villages_towns_Tucheng',\n 'villages_towns_Shilin', 'villages_towns_Datong', 'villages_towns_Daan',\n 'villages_towns_Wenshan', 'villages_towns_Xindian',\n 'villages_towns_Xinzhuang', 'villages_towns_Songshan',\n 'villages_towns_Banqiao', 'villages_towns_Linkou',\n 'villages_towns_Shulin', 'villages_towns_Yonghe',\n 'villages_towns_Xizhi', 'villages_towns_Taishan',\n 'villages_towns_Tamsui', 'villages_towns_Shenkeng',\n 'villages_towns_Ruifang', 'villages_towns_Wanhua',\n 'villages_towns_Wanli', 'villages_towns_Luzhou',\n 'villages_towns_Gongliao', 'villages_towns_Jinshan',\n 'villages_towns_Shuangxi', 'villages_towns_Yingge']\nbuilding_material = ['building_materials_RC', 'building_materials_RB',\n 'building_materials_brick', 'building_materials_steel',\n 'building_materials_SRC', 'building_materials_PRX',\n 'building_materials_other_material']",
"_____no_output_____"
],
[
"FILE = '/content/drive/MyDrive/SC201_Final_Project/Data/final_data_taipei.csv'",
"_____no_output_____"
],
[
"# standardize data\n# train_mean = 154170.694\n# train_std = 79570.40139\ndata = pd.read_csv(FILE)\ndata = data[data.unit_price != 0]\ndata = data[data.unit_price != 2211457]\nprint(data.count())\n\nsd = {}\nmean = {}\ncolumns = ['zoning', 'total_floors', 'floors_area', 'unit_price', 'unit_berth_price', 'total_berth_price', 'main_building_area', 'auxiliary_building_area', 'balcony_area', 'building_age']\n\nfor column in columns:\n print(column)\n sd_each = data[column].std()\n # sd[column] = sd_each\n\n mean_each = data[column].mean()\n # mean[column] = mean_each\n\n data[column] = (data[column] - mean_each)/sd_each\n\ndata.to_csv('/content/drive/MyDrive/SC201_Final_Project/Data/new_data_taipei.csv', encoding=\"utf_8_sig\", index=False)\n\n",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f8892e33fc355afc152aec09b6c576c020446b | 1,391 | ipynb | Jupyter Notebook | Hackerrank/3. conditional statements.ipynb | Anna-MarieTomm/Learn_Python_with_Anna-Marie | e1d7b0f95674a91b1f30acd8923e0fc54f823182 | [
"MIT"
] | null | null | null | Hackerrank/3. conditional statements.ipynb | Anna-MarieTomm/Learn_Python_with_Anna-Marie | e1d7b0f95674a91b1f30acd8923e0fc54f823182 | [
"MIT"
] | null | null | null | Hackerrank/3. conditional statements.ipynb | Anna-MarieTomm/Learn_Python_with_Anna-Marie | e1d7b0f95674a91b1f30acd8923e0fc54f823182 | [
"MIT"
] | null | null | null | 19.871429 | 49 | 0.403307 | [
[
[
"def conditions(n): \n for n in range(1,101): \n if (n % 2 != 0 and n<21):\n print(\"Weird\")\n \n elif (n %2 == 0 and 2<= n<=5): \n print (\"Not Weird\")\n\n elif (n % 2 == 0 and 6<= n<=20):\n print(\"Weird\")\n\n elif (n % 2==0 and 2<n):\n print (\"Not Weird\")\n\n else: \n print(\"Not Weird\")\n return \n\n \nconditions(30)\n\n",
"Weird\n"
]
]
] | [
"code"
] | [
[
"code"
]
] |
e7f88df687e6275008d1fc995ab9b3e4ec116390 | 42,676 | ipynb | Jupyter Notebook | jupyter/BLOOMBERG/SektorEksternal/script/SektorEksternal1_3.ipynb | langpp/bappenas | f780607192bb99b9bc8fbe29412b4c6c49bf15ae | [
"Apache-2.0"
] | 1 | 2021-03-17T03:10:49.000Z | 2021-03-17T03:10:49.000Z | jupyter/BLOOMBERG/SektorEksternal/script/SektorEksternal1_3.ipynb | langpp/bappenas | f780607192bb99b9bc8fbe29412b4c6c49bf15ae | [
"Apache-2.0"
] | null | null | null | jupyter/BLOOMBERG/SektorEksternal/script/SektorEksternal1_3.ipynb | langpp/bappenas | f780607192bb99b9bc8fbe29412b4c6c49bf15ae | [
"Apache-2.0"
] | 1 | 2021-03-17T03:12:34.000Z | 2021-03-17T03:12:34.000Z | 85.011952 | 2,221 | 0.645257 | [
[
[
"#IMPORT SEMUA LIBARARY",
"_____no_output_____"
],
[
"#IMPORT LIBRARY PANDAS\nimport pandas as pd\n#IMPORT LIBRARY UNTUK POSTGRE\nfrom sqlalchemy import create_engine\nimport psycopg2\n#IMPORT LIBRARY CHART\nfrom matplotlib import pyplot as plt\nfrom matplotlib import style\n#IMPORT LIBRARY BASE PATH\nimport os\nimport io\n#IMPORT LIBARARY PDF\nfrom fpdf import FPDF\n#IMPORT LIBARARY CHART KE BASE64\nimport base64\n#IMPORT LIBARARY EXCEL\nimport xlsxwriter ",
"_____no_output_____"
],
[
"#FUNGSI UNTUK MENGUPLOAD DATA DARI CSV KE POSTGRESQL",
"_____no_output_____"
],
[
"def uploadToPSQL(columns, table, filePath, engine):\n #FUNGSI UNTUK MEMBACA CSV\n df = pd.read_csv(\n os.path.abspath(filePath),\n names=columns,\n keep_default_na=False\n )\n #APABILA ADA FIELD KOSONG DISINI DIFILTER\n df.fillna('')\n #MENGHAPUS COLUMN YANG TIDAK DIGUNAKAN\n del df['kategori']\n del df['jenis']\n del df['pengiriman']\n del df['satuan']\n \n #MEMINDAHKAN DATA DARI CSV KE POSTGRESQL\n df.to_sql(\n table, \n engine,\n if_exists='replace'\n )\n \n #DIHITUNG APABILA DATA YANG DIUPLOAD BERHASIL, MAKA AKAN MENGEMBALIKAN KELUARAN TRUE(BENAR) DAN SEBALIKNYA\n if len(df) == 0:\n return False\n else:\n return True",
"_____no_output_____"
],
[
"#FUNGSI UNTUK MEMBUAT CHART, DATA YANG DIAMBIL DARI DATABASE DENGAN MENGGUNAKAN ORDER DARI TANGGAL DAN JUGA LIMIT\n#DISINI JUGA MEMANGGIL FUNGSI MAKEEXCEL DAN MAKEPDF",
"_____no_output_____"
],
[
"def makeChart(host, username, password, db, port, table, judul, columns, filePath, name, subjudul, limit, negara, basePath):\n #TEST KONEKSI DATABASE\n try:\n #KONEKSI KE DATABASE\n connection = psycopg2.connect(user=username,password=password,host=host,port=port,database=db)\n cursor = connection.cursor()\n #MENGAMBL DATA DARI TABLE YANG DIDEFINISIKAN DIBAWAH, DAN DIORDER DARI TANGGAL TERAKHIR\n #BISA DITAMBAHKAN LIMIT SUPAYA DATA YANG DIAMBIL TIDAK TERLALU BANYAK DAN BERAT\n postgreSQL_select_Query = \"SELECT * FROM \"+table+\" ORDER BY tanggal ASC LIMIT \" + str(limit)\n \n cursor.execute(postgreSQL_select_Query)\n mobile_records = cursor.fetchall() \n uid = []\n lengthx = []\n lengthy = []\n #MELAKUKAN LOOPING ATAU PERULANGAN DARI DATA YANG SUDAH DIAMBIL\n #KEMUDIAN DATA TERSEBUT DITEMPELKAN KE VARIABLE DIATAS INI\n for row in mobile_records:\n uid.append(row[0])\n lengthx.append(row[1])\n if row[2] == \"\":\n lengthy.append(float(0))\n else:\n lengthy.append(float(row[2]))\n\n #FUNGSI UNTUK MEMBUAT CHART\n #bar\n style.use('ggplot')\n \n fig, ax = plt.subplots()\n #MASUKAN DATA ID DARI DATABASE, DAN JUGA DATA TANGGAL\n ax.bar(uid, lengthy, align='center')\n #UNTUK JUDUL CHARTNYA\n ax.set_title(judul)\n ax.set_ylabel('Total')\n ax.set_xlabel('Tanggal')\n \n ax.set_xticks(uid)\n #TOTAL DATA YANG DIAMBIL DARI DATABASE, DIMASUKAN DISINI\n ax.set_xticklabels((lengthx))\n b = io.BytesIO()\n #CHART DISIMPAN KE FORMAT PNG\n plt.savefig(b, format='png', bbox_inches=\"tight\")\n #CHART YANG SUDAH DIJADIKAN PNG, DISINI DICONVERT KE BASE64\n barChart = base64.b64encode(b.getvalue()).decode(\"utf-8\").replace(\"\\n\", \"\")\n #CHART DITAMPILKAN\n plt.show()\n \n #line\n #MASUKAN DATA DARI DATABASE\n plt.plot(lengthx, lengthy)\n plt.xlabel('Tanggal')\n plt.ylabel('Total')\n #UNTUK JUDUL CHARTNYA\n plt.title(judul)\n plt.grid(True)\n l = io.BytesIO()\n #CHART DISIMPAN KE FORMAT PNG\n plt.savefig(l, format='png', bbox_inches=\"tight\")\n #CHART YANG SUDAH DIJADIKAN PNG, DISINI DICONVERT KE BASE64\n lineChart = base64.b64encode(l.getvalue()).decode(\"utf-8\").replace(\"\\n\", \"\")\n #CHART DITAMPILKAN\n plt.show()\n \n #pie\n #UNTUK JUDUL CHARTNYA\n plt.title(judul)\n #MASUKAN DATA DARI DATABASE\n plt.pie(lengthy, labels=lengthx, autopct='%1.1f%%', \n shadow=True, startangle=180)\n \n plt.axis('equal')\n p = io.BytesIO()\n #CHART DISIMPAN KE FORMAT PNG\n plt.savefig(p, format='png', bbox_inches=\"tight\")\n #CHART YANG SUDAH DIJADIKAN PNG, DISINI DICONVERT KE BASE64\n pieChart = base64.b64encode(p.getvalue()).decode(\"utf-8\").replace(\"\\n\", \"\")\n #CHART DITAMPILKAN\n plt.show()\n \n #MENGAMBIL DATA DARI CSV YANG DIGUNAKAN SEBAGAI HEADER DARI TABLE UNTUK EXCEL DAN JUGA PDF\n header = pd.read_csv(\n os.path.abspath(filePath),\n names=columns,\n keep_default_na=False\n )\n #MENGHAPUS COLUMN YANG TIDAK DIGUNAKAN\n header.fillna('')\n del header['tanggal']\n del header['total']\n #MEMANGGIL FUNGSI EXCEL\n makeExcel(mobile_records, header, name, limit, basePath)\n #MEMANGGIL FUNGSI PDF\n makePDF(mobile_records, header, judul, barChart, lineChart, pieChart, name, subjudul, limit, basePath) \n \n #JIKA GAGAL KONEKSI KE DATABASE, MASUK KESINI UNTUK MENAMPILKAN ERRORNYA\n except (Exception, psycopg2.Error) as error :\n print (error)\n\n #KONEKSI DITUTUP\n finally:\n if(connection):\n cursor.close()\n connection.close()",
"_____no_output_____"
],
[
"#FUNGSI MAKEEXCEL GUNANYA UNTUK MEMBUAT DATA YANG BERASAL DARI DATABASE DIJADIKAN FORMAT EXCEL TABLE F2\n#PLUGIN YANG DIGUNAKAN ADALAH XLSXWRITER",
"_____no_output_____"
],
[
"def makeExcel(datarow, dataheader, name, limit, basePath):\n #MEMBUAT FILE EXCEL\n workbook = xlsxwriter.Workbook(basePath+'jupyter/BLOOMBERG/SektorEksternal/excel/'+name+'.xlsx')\n #MENAMBAHKAN WORKSHEET PADA FILE EXCEL TERSEBUT\n worksheet = workbook.add_worksheet('sheet1'))\n #SETINGAN AGAR DIBERIKAN BORDER DAN FONT MENJADI BOLD\n row1 = workbook.add_format({'border': 2, 'bold': 1})\n row2 = workbook.add_format({'border': 2})\n #MENJADIKAN DATA MENJADI ARRAY\n data=list(datarow)\n isihead=list(dataheader.values)\n header = []\n body = []\n \n #LOOPING ATAU PERULANGAN, KEMUDIAN DATA DITAMPUNG PADA VARIABLE DIATAS\n for rowhead in dataheader:\n header.append(str(rowhead))\n \n for rowhead2 in datarow:\n header.append(str(rowhead2[1]))\n \n for rowbody in isihead[1]:\n body.append(str(rowbody))\n \n for rowbody2 in data:\n body.append(str(rowbody2[2]))\n \n #MEMASUKAN DATA DARI VARIABLE DIATAS KE DALAM COLUMN DAN ROW EXCEL\n for col_num, data in enumerate(header):\n worksheet.write(0, col_num, data, row1)\n \n for col_num, data in enumerate(body):\n worksheet.write(1, col_num, data, row2)\n \n #FILE EXCEL DITUTUP\n workbook.close()",
"_____no_output_____"
],
[
"#FUNGSI UNTUK MEMBUAT PDF YANG DATANYA BERASAL DARI DATABASE DIJADIKAN FORMAT EXCEL TABLE F2\n#PLUGIN YANG DIGUNAKAN ADALAH FPDF",
"_____no_output_____"
],
[
"def makePDF(datarow, dataheader, judul, bar, line, pie, name, subjudul, lengthPDF, basePath):\n #FUNGSI UNTUK MENGATUR UKURAN KERTAS, DISINI MENGGUNAKAN UKURAN A4 DENGAN POSISI LANDSCAPE\n pdf = FPDF('L', 'mm', [210,297])\n #MENAMBAHKAN HALAMAN PADA PDF\n pdf.add_page()\n #PENGATURAN UNTUK JARAK PADDING DAN JUGA UKURAN FONT\n pdf.set_font('helvetica', 'B', 20.0)\n pdf.set_xy(145.0, 15.0)\n #MEMASUKAN JUDUL KE DALAM PDF\n pdf.cell(ln=0, h=2.0, align='C', w=10.0, txt=judul, border=0)\n \n #PENGATURAN UNTUK UKURAN FONT DAN JUGA JARAK PADDING\n pdf.set_font('arial', '', 14.0)\n pdf.set_xy(145.0, 25.0)\n #MEMASUKAN SUB JUDUL KE PDF\n pdf.cell(ln=0, h=2.0, align='C', w=10.0, txt=subjudul, border=0)\n #MEMBUAT GARIS DI BAWAH SUB JUDUL\n pdf.line(10.0, 30.0, 287.0, 30.0)\n pdf.set_font('times', '', 10.0)\n pdf.set_xy(17.0, 37.0)\n \n #PENGATURAN UNTUK UKURAN FONT DAN JUGA JARAK PADDING\n pdf.set_font('Times','',10.0) \n #MENGAMBIL DATA HEADER PDF YANG SEBELUMNYA SUDAH DIDEFINISIKAN DIATAS\n datahead=list(dataheader.values)\n pdf.set_font('Times','B',12.0) \n pdf.ln(0.5)\n \n th1 = pdf.font_size\n \n #MEMBUAT TABLE PADA PDF, DAN MENAMPILKAN DATA DARI VARIABLE YANG SUDAH DIKIRIM\n pdf.cell(100, 2*th1, \"Kategori\", border=1, align='C')\n pdf.cell(177, 2*th1, datahead[0][0], border=1, align='C')\n pdf.ln(2*th1)\n pdf.cell(100, 2*th1, \"Jenis\", border=1, align='C')\n pdf.cell(177, 2*th1, datahead[0][1], border=1, align='C')\n pdf.ln(2*th1)\n pdf.cell(100, 2*th1, \"Pengiriman\", border=1, align='C')\n pdf.cell(177, 2*th1, datahead[0][2], border=1, align='C')\n pdf.ln(2*th1)\n pdf.cell(100, 2*th1, \"Satuan\", border=1, align='C')\n pdf.cell(177, 2*th1, datahead[0][3], border=1, align='C')\n pdf.ln(2*th1)\n \n #PENGATURAN PADDING\n pdf.set_xy(17.0, 75.0)\n \n #PENGATURAN UNTUK UKURAN FONT DAN JUGA JARAK PADDING\n pdf.set_font('Times','B',11.0) \n data=list(datarow)\n epw = pdf.w - 2*pdf.l_margin\n col_width = epw/(lengthPDF+1)\n \n #PENGATURAN UNTUK JARAK PADDING\n pdf.ln(0.5)\n th = pdf.font_size\n \n #MEMASUKAN DATA HEADER YANG DIKIRIM DARI VARIABLE DIATAS KE DALAM PDF\n pdf.cell(50, 2*th, str(\"Negara\"), border=1, align='C')\n for row in data:\n pdf.cell(40, 2*th, str(row[1]), border=1, align='C')\n pdf.ln(2*th)\n \n #MEMASUKAN DATA ISI YANG DIKIRIM DARI VARIABLE DIATAS KE DALAM PDF\n pdf.set_font('Times','B',10.0)\n pdf.set_font('Arial','',9)\n pdf.cell(50, 2*th, negara, border=1, align='C')\n for row in data:\n pdf.cell(40, 2*th, str(row[2]), border=1, align='C')\n pdf.ln(2*th)\n \n #MENGAMBIL DATA CHART, KEMUDIAN CHART TERSEBUT DIJADIKAN PNG DAN DISIMPAN PADA DIRECTORY DIBAWAH INI\n #BAR CHART\n bardata = base64.b64decode(bar)\n barname = basePath+'jupyter/BLOOMBERG/SektorEksternal/img/'+name+'-bar.png'\n with open(barname, 'wb') as f:\n f.write(bardata)\n \n #LINE CHART\n linedata = base64.b64decode(line)\n linename = basePath+'jupyter/BLOOMBERG/SektorEksternal/img/'+name+'-line.png'\n with open(linename, 'wb') as f:\n f.write(linedata)\n \n #PIE CHART\n piedata = base64.b64decode(pie)\n piename = basePath+'jupyter/BLOOMBERG/SektorEksternal/img/'+name+'-pie.png'\n with open(piename, 'wb') as f:\n f.write(piedata)\n \n #PENGATURAN UNTUK UKURAN FONT DAN JUGA JARAK PADDING\n pdf.set_xy(17.0, 75.0)\n col = pdf.w - 2*pdf.l_margin\n widthcol = col/3\n #MEMANGGIL DATA GAMBAR DARI DIREKTORY DIATAS\n pdf.image(barname, link='', type='',x=8, y=100, w=widthcol)\n pdf.set_xy(17.0, 75.0)\n col = pdf.w - 2*pdf.l_margin\n pdf.image(linename, link='', type='',x=103, y=100, w=widthcol)\n pdf.set_xy(17.0, 75.0)\n col = pdf.w - 2*pdf.l_margin\n pdf.image(piename, link='', type='',x=195, y=100, w=widthcol)\n pdf.ln(2*th)\n \n #MEMBUAT FILE PDF\n pdf.output(basePath+'jupyter/BLOOMBERG/SektorEksternal/pdf/'+name+'.pdf', 'F')",
"_____no_output_____"
],
[
"#DISINI TEMPAT AWAL UNTUK MENDEFINISIKAN VARIABEL VARIABEL SEBELUM NANTINYA DIKIRIM KE FUNGSI\n#PERTAMA MANGGIL FUNGSI UPLOADTOPSQL DULU, KALAU SUKSES BARU MANGGIL FUNGSI MAKECHART\n#DAN DI MAKECHART MANGGIL FUNGSI MAKEEXCEL DAN MAKEPDF",
"_____no_output_____"
],
[
"#DEFINISIKAN COLUMN BERDASARKAN FIELD CSV\ncolumns = [\n \"kategori\",\n \"jenis\",\n \"tanggal\",\n \"total\",\n \"pengiriman\",\n \"satuan\",\n]\n\n#UNTUK NAMA FILE\nname = \"SektorEksternal1_3\"\n#VARIABLE UNTUK KONEKSI KE DATABASE\nhost = \"localhost\"\nusername = \"postgres\"\npassword = \"1234567890\"\nport = \"5432\"\ndatabase = \"bloomberg_sektoreksternal\"\ntable = name.lower()\n#JUDUL PADA PDF DAN EXCEL\njudul = \"Data Sektor Eksternal\"\nsubjudul = \"Badan Perencanaan Pembangunan Nasional\"\n#LIMIT DATA UNTUK SELECT DI DATABASE\nlimitdata = int(8)\n#NAMA NEGARA UNTUK DITAMPILKAN DI EXCEL DAN PDF\nnegara = \"Indonesia\"\n#BASE PATH DIRECTORY\nbasePath = 'C:/Users/ASUS/Documents/bappenas/'\n#FILE CSV\nfilePath = basePath+ 'data mentah/BLOOMBERG/SektorEksternal/' +name+'.csv';\n#KONEKSI KE DATABASE\nengine = create_engine('postgresql://'+username+':'+password+'@'+host+':'+port+'/'+database)\n\n#MEMANGGIL FUNGSI UPLOAD TO PSQL\ncheckUpload = uploadToPSQL(columns, table, filePath, engine)\n#MENGECEK FUNGSI DARI UPLOAD PSQL, JIKA BERHASIL LANJUT MEMBUAT FUNGSI CHART, JIKA GAGAL AKAN MENAMPILKAN PESAN ERROR\nif checkUpload == True:\n makeChart(host, username, password, database, port, table, judul, columns, filePath, name, subjudul, limitdata, negara, basePath)\nelse:\n print(\"Error When Upload CSV\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f8986f826814239427115987a412edf0820475 | 184,668 | ipynb | Jupyter Notebook | fe588/fe588_introduction.ipynb | bkoyuncu/notes | 0e660f46b7d17fdfddc2cad1bb60dcf847f5d1e4 | [
"MIT"
] | 191 | 2016-01-21T19:44:23.000Z | 2022-03-25T20:50:50.000Z | fe588/fe588_introduction.ipynb | onurboyar/notes | 2ec14820af044c2cfbc99bc989338346572a5e24 | [
"MIT"
] | 2 | 2018-02-18T03:41:04.000Z | 2018-11-21T11:08:49.000Z | fe588/fe588_introduction.ipynb | onurboyar/notes | 2ec14820af044c2cfbc99bc989338346572a5e24 | [
"MIT"
] | 138 | 2015-10-04T21:57:21.000Z | 2021-06-15T19:35:55.000Z | 67.668743 | 36,670 | 0.785085 | [
[
[
"Install Python. \n\njupyter --no-browser\n\nIn the favorite browser, type\nhttp://localhost:8888 (or the port that is assigned)\n\n\nBasic usage of jupyter notebooks.\n- create a newdocument by clicking the New Notebook\n- start typing code in the shaded textbox\n- execute the code",
"_____no_output_____"
]
],
[
[
"\nx = 0.1\nN = 3\n\na = 1\nb = 0\nc = -1\n\nprint('f(' + str(x) + ') = ' + str(a*x**2 + b*x + c))\n\n\n\n",
"f(0.1) = -0.99\n"
],
[
"a = 1\nb = 1\nprint(a*b,a*(b+1),a*(b+2),a*(b+3))\n\na = 2\nprint(a*b,a*(b+1),a*(b+2),a*(b+3))\n \na = 3\nprint(a*b,a*(b+1),a*(b+2),a*(b+3))\n \na = 4\nprint(a*b,a*(b+1),a*(b+2),a*(b+3))\n ",
"(1, 2, 3, 4)\n(2, 4, 6, 8)\n(3, 6, 9, 12)\n(4, 8, 12, 16)\n"
]
],
[
[
"Fibionacci Series\n",
"_____no_output_____"
]
],
[
[
"N = 0\n\na_1 = 1\na_2 = 0\n\nx = 1\nif N>0:\n print('x_' + str(0) + ' = ' + str(x) )\n\n \nfor i in range(1,N):\n x = a_1 + a_2\n print('x_' + str(i) + ' = ' + str(x) )\n a_2 = a_1\n a_1 = x\n ",
"_____no_output_____"
],
[
"l = -1\nr = 1\ndelta = 0.1\nsteps = (r-l)/delta+1\n\nprint '-'*20\nprint('| '),\nprint('x'),\nprint('| '),\nprint('3*x**2 + 2*x + 3'),\nprint('| ')\n\nfor i in range(0,int(steps)):\n x = l+i*delta\n print '-'*20\n print('| '),\n print(x),\n print('| '),\n print(3*x**2 + 2*x + 3),\n print('| ')\n",
"--------------------\n| x | 3*x**2 + 2*x + 3 | \n--------------------\n| -1.0 | 4.0 | \n--------------------\n| -0.9 | 3.63 | \n--------------------\n| -0.8 | 3.32 | \n--------------------\n| -0.7 | 3.07 | \n--------------------\n| -0.6 | 2.88 | \n--------------------\n| -0.5 | 2.75 | \n--------------------\n| -0.4 | 2.68 | \n--------------------\n| -0.3 | 2.67 | \n--------------------\n| -0.2 | 2.72 | \n--------------------\n| -0.1 | 2.83 | \n--------------------\n| 0.0 | 3.0 | \n--------------------\n| 0.1 | 3.23 | \n--------------------\n| 0.2 | 3.52 | \n--------------------\n| 0.3 | 3.87 | \n--------------------\n| 0.4 | 4.28 | \n--------------------\n| 0.5 | 4.75 | \n--------------------\n| 0.6 | 5.28 | \n--------------------\n| 0.7 | 5.87 | \n--------------------\n| 0.8 | 6.52 | \n--------------------\n| 0.9 | 7.23 | \n--------------------\n| 1.0 | 8.0 | \n"
],
[
"def f(x):\n return 3*x**2 + 2*x + 3\n\nl = -1\nr = 1\ndelta = 0.1\n\nsteps = (r-l)/delta\n\nfor i in range(0,int(steps)):\n x = l+i*delta\n print x,\n print f(x)",
"-1.0 4.0\n-0.9 3.63\n-0.8 3.32\n-0.7 3.07\n-0.6 2.88\n-0.5 2.75\n-0.4 2.68\n-0.3 2.67\n-0.2 2.72\n-0.1 2.83\n0.0 3.0\n0.1 3.23\n0.2 3.52\n0.3 3.87\n0.4 4.28\n0.5 4.75\n0.6 5.28\n0.7 5.87\n0.8 6.52\n0.9 7.23\n"
],
[
"def f(r, T, S_0):\n return S_0*(1+r)**T\n\ninterest_rate = 0.12\nT = 10\nS_0 = 100\n\nl = 1\nr = T\ndelta = 1\n\nsteps = (r-l)/delta\n\nfor i in range(0,int(steps)):\n T = l+i*delta\n print T,\n print f(interest_rate, T, S_0)",
"1 112.0\n2 125.44\n3 140.4928\n4 157.351936\n5 176.23416832\n6 197.382268518\n7 221.068140741\n8 247.596317629\n9 277.307875745\n"
]
],
[
[
"Arrays, lists",
"_____no_output_____"
]
],
[
[
"a = [1,2,5,7,5, 3.2, 7]\nnames = ['Ali','Veli','Fatma','Asli']\n\n#for s in names:\n# print(s)\n\nprint(names[3])\n\nprint(len(names))",
"Asli\n4\n"
],
[
"for i in range(len(names)-1,-1,-1):\n print(names[i])\n \nfor i in range(len(names)):\n print(names[len(names)-i])\n",
"Asli\nFatma\nVeli\nAli\n"
],
[
"for n in reversed(names):\n print(n)",
"Asli\nFatma\nVeli\nAli\n"
]
],
[
[
"Average and standard deviation\n",
"_____no_output_____"
]
],
[
[
"#x = [0.1,3,-2.1,5,12,3,17]\nx = [1,-1,0]\n\ns1 = 0.0\nfor a in x:\n s1 += a\n\nmean = s1/len(x)\n\ns2 = 0.0\nfor a in x:\n s2 += (a-mean)**2\n\nvariance = s2/len(x)\n\nprint('mean = '),\nprint(mean)\nprint('variance = '),\nprint(variance)\n",
"mean = 0.0\nvariance = 0.666666666667\n"
]
],
[
[
"Find the minimum in an array",
"_____no_output_____"
]
],
[
[
"a = [2,5,1.2, 0,-4, 3]\nmn = a[0]\n\nfor i in range(1,len(a)):\n if a[i]<mn:\n mn = a[i]\n\nprint(mn)",
"-4\n"
],
[
"a.sort()",
"_____no_output_____"
],
[
"a.append(-7)",
"_____no_output_____"
],
[
"v = a.pop()",
"_____no_output_____"
],
[
"a.reverse()",
"_____no_output_____"
],
[
"v = a.pop(0)",
"_____no_output_____"
],
[
"a.sort",
"_____no_output_____"
],
[
"a = 5",
"_____no_output_____"
],
[
"a.bit_length",
"_____no_output_____"
]
],
[
[
"Homework: Value counts given an array of integers",
"_____no_output_____"
]
],
[
[
"a = [5, 3, 1, 1, 6, 3, 2]\n\nua = []\n\nfor j in a:\n found = False\n for i in ua:\n if j==i:\n found = True;\n break;\n if not found:\n ua.append(j)\n \nprint(ua) \n \n\nfor i in ua:\n s = 0\n for j in a:\n if i==j:\n s = s+1\n\n print(i, s)\n\n\n\n\n",
"[5, 3, 1, 6, 2]\n(5, 1)\n(3, 2)\n(1, 2)\n(6, 1)\n(2, 1)\n"
],
[
"a = [5, 3, 1, 1, 6, 3, 2]\n\nca = []\nsum = 0\n\nfor j in a:\n ca.append(sum)\n sum += j\n\nca.append(sum)\n\nprint(ca)\n#ca = [0, 5, 8, 9, 10, 16, 19, 21]\n",
"[0, 5, 8, 9, 10, 16, 19, 21]\n"
],
[
"a = [3, 6, 7, 2]\n\n#oa = [3, 6, 7, 2, 2, 7, 6, 3]\n\noa = a\nfor i in reversed(a):\n oa.append(i)\n\noa\n\noa = a\nfor i in range()\n",
"_____no_output_____"
],
[
"a = [3, 4, 6]\n\noa = list(a)\noa = a\n\nprint(a)\n\nfor i in range(1,len(a)+1):\n# print(a[-i])\n oa.append(a[-i]) \n \noa",
"[3, 4, 6]\n"
],
[
"a+list(reversed(a))",
"_____no_output_____"
],
[
"a0 = 0\na = [2, 6, 3, 1, 4, 8, 3, 5, 5]\n\nprev = a0\n\nInc = []\nDec = []\nfor i in a:\n Inc.append(i>prev)\n Dec.append(i<prev)\n prev = i\n\nprint(Inc)\nprint(Dec)\n\n#Inc = [True, True, False, False, True, True, False, True, False]\n#Dec = [False, False, True, True,False, False, True, False, False]\n\n\n",
"[True, True, False, False, True, True, False, True, False]\n[False, False, True, True, False, False, True, False, False]\n"
]
],
[
[
"Generate random walk in an array",
"_____no_output_____"
]
],
[
[
"import random\n\nN = 10\nmu = 0\nsig = 1\n\nx = 0\na = [x]\nfor i in range(N):\n w = random.gauss(mu, sig)\n x = x + w\n a.append(x)\n\nprint(a)\nlen(a)",
"[0, 0.07173293905450501, -0.3652340160453349, -0.07610430577230803, -1.4172015782500376, -0.31469586619290335, -1.4458834127459201, -0.7189045208807692, 0.9895551731951309, 0.1012103597338051, -1.0353093339238497]\n"
]
],
[
[
"List Comprehension",
"_____no_output_____"
]
],
[
[
"\n\nN = 100\nmu = 0\nsig = 1\n\na = [random.gauss(mu, sig) for i in range(N)]\n\n\nfor i in range(len(a)-1):\n a[i+1] = a[i] + a[i+1]",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pylab as plt\n\nplt.plot(a)\nplt.show()",
"/Users/cemgil/anaconda/envs/py27/lib/python2.7/site-packages/matplotlib/font_manager.py:273: UserWarning: Matplotlib is building the font cache using fc-list. This may take a moment.\n warnings.warn('Matplotlib is building the font cache using fc-list. This may take a moment.')\n"
]
],
[
[
"Moving Average",
"_____no_output_____"
]
],
[
[
"# Window Lenght\nW = 20\n\ny = []\nfor i in range(len(a)):\n s = 0\n n = 0\n for j in range(W):\n if i-j < 0:\n break;\n \n s = s + a[i-j]\n n = n + 1\n y.append(s/n)\n\nplt.plot(a)\nplt.plot(y)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"Moving average, second version",
"_____no_output_____"
]
],
[
[
"# Window Lenght\nW = 20\n\ny = []\ns = 0\nn = 0\nfor i in range(len(a)):\n s = s + a[i]\n if i>=W:\n s = s - a[i-W]\n else:\n n = n + 1\n \n y.append(s/n)\n\nplt.plot(a)\nplt.plot(y)\nplt.show()\n",
"_____no_output_____"
],
[
"def mean(a):\n s = 0\n for x in a:\n s = s+x\n return s/float(len(a))\n \ndef var(a):\n mu = mean(a)\n s = 0\n for i in range(len(a)):\n s = s+ (a[i]-mu)**2\n return float(s)/len(a)\n \na = [3,4,1,2]\n \nprint(mean(a))\nprint(var(a))\n",
"2.5\n1.25\n"
]
],
[
[
"Mean and Variance, online calculation",
"_____no_output_____"
]
],
[
[
"def mean(a):\n mu = 0.0\n for i in range(len(a)):\n mu = i/(i+1.0)*mu + 1.0/(i+1.0)*a[i]\n return mu\n\na = [3,4,1,2]\n\n#print(a)\nprint(mean(a))",
"2.5\n"
]
],
[
[
"Implement the recursive formula for the variance\n",
"_____no_output_____"
]
],
[
[
"for i in range(1,len(a)+1):\n print(i)",
"1\n2\n3\n4\n"
],
[
"a = [i**2 for i in range(10)]\na",
"_____no_output_____"
],
[
"st = 'if'\n\nif st == 'if':\n print('if')\nelif st == 'elif':\n print('elif')\nelse:\n print('not elif')",
"if\n"
],
[
"if x<10 and x>3:\n ",
"_____no_output_____"
],
[
"for i in range(10):\n if i%2:\n continue\n \n print i\n ",
"0\n2\n4\n6\n8\n"
],
[
"x",
"_____no_output_____"
],
[
"del x",
"_____no_output_____"
],
[
"from math import exp\nimport math as m\n\n\n\nx = 3\n\nexp(x)\n\nm.sin(x)",
"_____no_output_____"
],
[
"if x == 3:\n print 'x'",
"x\n"
],
[
"i = 0\nwhile i<10:\n i+=1\n print(i*10)",
"10\n20\n30\n40\n50\n60\n70\n80\n90\n100\n"
],
[
"%matplotlib inline\nimport matplotlib.pylab as plt\n\nx = [z/10. for z in range(-20,21)]\nx2 = [z**2 for z in x]\nx3 = [z**3 for z in x]\nsinx = [math.sin(z) for z in x]\n\nplt.plot(x,x)\nplt.plot(x,x2)\nplt.plot(x,sinx)\nplt.show()\n\n",
"_____no_output_____"
],
[
"import numpy as np\n\nx = [z for z in np.arange(-2,2,0.1)]\n\nx",
"_____no_output_____"
],
[
"x = [2,3]\n\ny = (2,3)",
"_____no_output_____"
],
[
"x[0] = 3\n\n\ny = (z for z in range(5))",
"_____no_output_____"
],
[
"for i in y:\n print(i)",
"0\n1\n2\n3\n4\n"
],
[
"import random\n\nmath.pi*(2*random.random()-1)",
"_____no_output_____"
],
[
"def unif():\n return 2*random.random() - 1\n\nN = 100\n\n# Generate N points in the square and store in a list\npoints = [[unif(), unif()] for i in range(N) ]\n\n# For each point check if it is in the circle and count the total number\n# plot points in the circle as blue\n# plot points outside as red\n\ncount = 0\npx_in = []\npy_in = []\npx_out = []\npy_out = []\nfor x in points:\n if x[0]**2 + x[1]**2< 1:\n count += 1\n px_in.append(x[0])\n py_in.append(x[1])\n else:\n px_out.append(x[0])\n py_out.append(x[1])\n \n\nprint(4.0*count/N)\n\nplt.plot(px_in, py_in,'.b')\nplt.plot(px_out, py_out,'.r')\nplt.show()\n",
"_____no_output_____"
],
[
"def unif():\n return 2*random.random() - 1\n\nN = 1000000\n\n\ncount = 0\nfor i in range(N):\n x = [unif(), unif(), unif()]\n if x[0]**2 + x[1]**2 + x[2]**2 < 1:\n count += 1\n\nprint(8*float(count)/N)\nprint(4.0/3.0*math.pi)\n",
"4.190776\n4.18879020479\n"
],
[
"for x in points:\n print(x)",
"[0.5669673838721772, 0.8780780353618138]\n[0.27412079600315264, 0.6816166464306519]\n[0.5328030431009356, 0.2771763253798074]\n[0.823868172070928, -0.5271300068861811]\n[0.8956904454811281, -0.5074709824240229]\n[0.3592475026139912, 0.9804238195673225]\n[-0.14259791062631044, 0.3106098012654106]\n[-0.31312787419231314, 0.3527459657040679]\n[0.1898388211757973, 0.2483440273290023]\n[0.8936854598353037, -0.9638186968466502]\n"
],
[
"y = [3,4,5]\nx = ['a','b','d']\n\nfor u,v in zip(x, y):\n print(u,v)",
"('a', 3)\n('b', 4)\n('d', 5)\n"
],
[
"import sys\n\nsys.stdout",
"_____no_output_____"
],
[
"a = [3,4,5]\na = map(lambda x: x**2, a)\n\nreduce(lambda a,b: a*b, a)\n\n",
"_____no_output_____"
],
[
"ln = 'twinkle tinkle little star'\n\nln.split()",
"_____no_output_____"
],
[
"lst = [1,3,4]\n\nlst.insert(2,4)\nlst",
"_____no_output_____"
],
[
"import sys\n\nN = int(raw_input().strip())\nlst = []\nfor line in sys.stdin:\n tok = line.split()\n if tok[0] == 'insert':\n i = int(tok[1])\n val = int(tok[2])\n lst.insert(i,val)\n elif tok[0] == 'print':\n print(lst)\n elif tok[0] == 'remove':\n val = int(tok[1])\n lst.remove(val)\n elif tok[0] == 'append':\n val = int(tok[1])\n lst.append(val)\n elif tok[0] == 'sort':\n lst.sort()\n elif tok[0] == 'pop':\n lst.pop()\n elif tok[0] == 'reverse':\n lst.reverse()\n else:\n print('none')\n \n ",
"_____no_output_____"
],
[
"p = raw_input('Enter Price ')\nC = raw_input('Enter Capital ')\nprint 'Number of Items'\nprint float(C)/int(p)",
"Enter Price 12\nEnter Capital 34\nNumber of Items\n2.83333333333\n"
],
[
"\nc = {'A': 3, 'B': 7, 'C': [2,3], 'D': 'Adana'}\n#print(c)\n\nc['D']\n",
"_____no_output_____"
]
],
[
[
"Catalog",
"_____no_output_____"
]
],
[
[
"def fun(x, par):\n print(x, par['volatility'])\n\nparams = {'volatility': 0.1, 'interest_rate': 0.08}\n\nsig = params['volatility']\nr = params['interest_rate']\n\nfun(3, params)\n\n",
"(3, 0.1)\n"
],
[
"plate = {'Istanbul':34}\ncity = 'Istanbul'\nprint 'the number plate for', city,'is', plate[city]",
"the number plate for Istanbul is 34\n"
],
[
"plate = {'Istanbul':34, 'Adana': '01', 'Ankara': '06', 'Izmir': 35, 'Hannover': 'H'}\ncities = ['Adana', 'Ankara','Istanbul','Izmir','Hannover']\nfor city in cities:\n print('the number plate for', city,'is', plate[city])",
"('the number plate for', 'Adana', 'is', '01')\n('the number plate for', 'Ankara', 'is', '06')\n('the number plate for', 'Istanbul', 'is', 34)\n('the number plate for', 'Izmir', 'is', 35)\n('the number plate for', 'Hannover', 'is', 'H')\n"
],
[
"for i in plate.keys():\n print i",
"Ankara\nAdana\nHannover\nIzmir\nIstanbul\n"
],
[
"plate.has_key('Balikesir')",
"_____no_output_____"
],
[
"plate['Eskisehir'] = 26\n",
"_____no_output_____"
],
[
"for i in sorted(plate.keys()):\n print i, plate[i]",
" Adana 01\nAnkara 06\nEskisehir 26\nHannover H\nIstanbul 34\nIzmir 35\n"
],
[
"students = {273: {'Name': 'Ali', 'Surname': 'Yasar', 'Gender': 'M'}}\nstudents[395] = {'Name': 'Ayse', 'Surname': 'Oz', 'Gender': 'F'}\nstudents[398] = {'Name': 'Ayse', 'Surname': 'Atik', 'Gender': 'F'}\nstudents[112] = {'Name': 'Ahmet', 'Surname': 'Uz', 'Gender': 'M'}\nstudents[450] = {'Name': 'Veli', 'Surname': 'Gez', 'Gender': 'M'}\nstudents[451] = {'Name': 'Taylan', 'Surname': 'Cemgil', 'Gender': 'U'}\n",
"_____no_output_____"
],
[
"for i in students:\n if students[i]['Gender'] is 'F':\n print students[i]['Name']",
"Ayse\n"
],
[
"counts = {'M': 0, 'F': 0}\n\nfor i in students:\n G = students[i]['Gender']\n if counts.has_key(G):\n counts[G] += 1\n else:\n counts[G] = 1\n\ncounts",
"_____no_output_____"
],
[
"counts = {}\n\nfor i in students:\n G = students[i]['Name']\n if counts.has_key(G):\n counts[G] += 1\n else:\n counts[G] = 1\n\ncounts\n",
"_____no_output_____"
]
],
[
[
"Tuples (Immutable Arrays, no change possible after creation)",
"_____no_output_____"
]
],
[
[
"a = ('Ankara', '06')\n\na.count('Istanbul')",
"_____no_output_____"
],
[
"%matplotlib inline\n\nimport numpy as np\nimport matplotlib.pylab as plt\n\nx = np.arange(-2,2,0.1)\n\nplt.plot(x,x)\nplt.plot(x,x**2)\nplt.plot(x,np.sin(x))\nplt.show()\n\n",
"_____no_output_____"
]
],
[
[
"Numpy arrays versus matrices",
"_____no_output_____"
]
],
[
[
"A = np.random.rand(3,5)\nx = np.random.rand(5,1)\nprint(A.dot(x))\n\n",
"[[ 0.69119365]\n [ 1.25372192]\n [ 1.58909165]]\n"
],
[
"A = np.mat(A)\nx = np.mat(x)\nprint(A*x)",
"[[ 0.69119365]\n [ 1.25372192]\n [ 1.58909165]]\n"
],
[
"a = np.mat(np.random.rand(3,1))\nb = np.mat(np.random.rand(3,1))\n\nprint(a)\nprint(b)\n\na.T*b",
"[[ 0.08673755]\n [ 0.0456282 ]\n [ 0.71475827]]\n[[ 0.27890196]\n [ 0.64473462]\n [ 0.90795421]]\n"
],
[
"N = 1000\nD = 3\n\nX = np.random.rand(N, D)\n\nmu = X.mean(axis=0, keepdims=True)\n\n#print(mu)\nprint((X - mu).T.dot(X-mu)/(N-1.))",
"[[ 0.0824769 -0.00162642 0.00029607]\n [-0.00162642 0.08155178 -0.00138188]\n [ 0.00029607 -0.00138188 0.08613698]]\n"
],
[
"np.cov(X.T)",
"_____no_output_____"
],
[
"print(np.mat(np.arange(1,11)).T*np.mat(np.arange(1,11)))",
"[[ 1 2 3 4 5 6 7 8 9 10]\n [ 2 4 6 8 10 12 14 16 18 20]\n [ 3 6 9 12 15 18 21 24 27 30]\n [ 4 8 12 16 20 24 28 32 36 40]\n [ 5 10 15 20 25 30 35 40 45 50]\n [ 6 12 18 24 30 36 42 48 54 60]\n [ 7 14 21 28 35 42 49 56 63 70]\n [ 8 16 24 32 40 48 56 64 72 80]\n [ 9 18 27 36 45 54 63 72 81 90]\n [ 10 20 30 40 50 60 70 80 90 100]]\n"
]
],
[
[
"B&S with Monte Carlo Call and Put pricing, use a catalog and numpy, avoid using for loops",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef European(Param, S0=1., T=1., Strike=1.,N=10000 ):\n '''\n Price_Call, Price_Put = European(Param, S0, T, Strike,N)\n \n Param: Market parameters, a catalog with fields \n Param['InterestRate'] : Yearly risk free interest rate\n Param['Volatility'] : \n S0 : Initial asset price\n T : Time Period (in Years)\n Strike: Strike Price\n N : Number of Monte Carlo Samples\n '''\n W = np.sqrt(T)*np.random.standard_normal(N)\n ST = S0*np.exp(T*(Param['InterestRate']-0.5*Param['Volatility']**2) + Param['Volatility']*W)\n CT = np.maximum(ST-Strike, 0)\n PT = np.maximum(Strike-ST, 0)\n\n Price_C = CT.mean()*np.exp(-Param['InterestRate']*T)\n Price_P = PT.mean()*np.exp(-Param['InterestRate']*T)\n return Price_C, Price_P\n\ndef Lookback(Param, S0=1., T=1., Strike=1., Steps=12, N=10000 ):\n '''\n Price_Call, Price_Put = Lookback(Param, S0, T, Strike, Steps, N)\n \n Param: Market parameters, a catalog with fields \n Param['InterestRate'] : Yearly risk free interest rate\n Param['Volatility'] : \n S0 : Initial asset price\n T : Time Period (in Years)\n Strike: Strike Price\n Steps : Number of steps to monitor the stock price\n N : Number of Monte Carlo Samples\n '''\n \n Tstep = T/Steps\n Smax = S0*np.ones(N)\n Smin = S0*np.ones(N)\n St = S0*np.ones(N)\n \n for t in range(Steps):\n W = np.sqrt(Tstep)*np.random.standard_normal(N)\n St = St*np.exp(Tstep*(Param['InterestRate']-0.5*Param['Volatility']**2) + Param['Volatility']*W)\n Smax = np.maximum(St, Smax)\n Smin = np.minimum(St, Smin)\n\n CT = np.maximum(Smax-Strike, 0)\n PT = np.maximum(Strike-Smin, 0)\n\n Price_C = CT.mean()*np.exp(-Param['InterestRate']*T)\n Price_P = PT.mean()*np.exp(-Param['InterestRate']*T)\n return Price_C, Price_P\n\ndef Asian(Param, S0=1., T=1., Strike=1., Steps=12, N=10000 ):\n '''\n Price_Call, Price_Put = Asian(Param, S0, T, Strike, Steps, N)\n \n Param: Market parameters, a catalog with fields \n Param['InterestRate'] : Yearly risk free interest rate\n Param['Volatility'] : \n S0 : Initial asset price\n T : Time Period (in Years)\n Strike: Strike Price\n Steps : Number of steps to monitor the stock price\n N : Number of Monte Carlo Samples\n '''\n \n Tstep = T/Steps\n Smean = np.zeros(N) \n St = S0*np.ones(N)\n \n for t in range(Steps):\n W = np.sqrt(Tstep)*np.random.standard_normal(N)\n St = St*np.exp(Tstep*(Param['InterestRate']-0.5*Param['Volatility']**2) + Param['Volatility']*W)\n i = t+1\n Smean = (i-1)*Smean/i + St/i\n \n CT = np.maximum(Smean-Strike, 0)\n PT = np.maximum(Strike-Smean, 0)\n\n Price_C = CT.mean()*np.exp(-Param['InterestRate']*T)\n Price_P = PT.mean()*np.exp(-Param['InterestRate']*T)\n return Price_C, Price_P\n\n\ndef FloatingLookback(Param, S0=1., T=1., Steps=12, N=10000 ):\n '''\n Price_Call, Price_Put = FloatingLookback(Param, S0, T, Steps, N)\n \n Param: Market parameters, a catalog with fields \n Param['InterestRate'] : Yearly risk free interest rate\n Param['Volatility'] : \n S0 : Initial asset price\n T : Time Period (in Years)\n Steps : Number of steps to monitor the stock price\n N : Number of Monte Carlo Samples\n '''\n \n Tstep = T/Steps\n Smax = S0*np.ones(N)\n Smin = S0*np.ones(N)\n St = S0*np.ones(N)\n \n for t in range(Steps):\n W = np.sqrt(Tstep)*np.random.standard_normal(N)\n St = St*np.exp(Tstep*(Param['InterestRate']-0.5*Param['Volatility']**2) + Param['Volatility']*W)\n Smax = np.maximum(St, Smax)\n Smin = np.minimum(St, Smin)\n\n CT = np.maximum(St-Smin, 0)\n PT = np.maximum(Smax-St, 0)\n\n Price_C = CT.mean()*np.exp(-Param['InterestRate']*T)\n Price_P = PT.mean()*np.exp(-Param['InterestRate']*T)\n return Price_C, Price_P\n\n \n\nParam = {'Volatility': 0.25, 'InterestRate': 0.11}\nPrice_C, Price_P = European(Param, S0=100, T=1.0, Strike=100)\nprint 'European\\nCall= ', Price_C,'\\n','Put = ', Price_P\n\nPrice_C, Price_P = Asian(Param, S0=100, T=1.0, Strike=100, Steps=1000)\nprint 'Asian\\nCall= ', Price_C,'\\n','Put = ', Price_P\n\nPrice_C, Price_P = Lookback(Param, S0=100, T=1.0, Strike=100, Steps=1000)\nprint 'Lookback\\nCall= ', Price_C,'\\n','Put = ', Price_P\n\nPrice_C, Price_P = FloatingLookback(Param, S0=100, T=1.0, Steps=1000)\nprint 'FloatingLookback\\nCall= ', Price_C,'\\n','Put = ', Price_P\n",
"European\nCall= 15.5726197769 \nPut = 5.13556380233\nAsian\nCall= 8.16477817074 \nPut = 3.17271035914\nLookback\nCall= 25.6819276647 \nPut = 12.5838549789\nFloatingLookback\nCall= 23.0385882044 \nPut = 15.3296952253\n"
]
],
[
[
"Next week Assignment:\n\n\nConsolidate all pricing methods into one single function avoiding code repetitions.\n",
"_____no_output_____"
]
],
[
[
"def OptionPricer(type_of_option, Params):\n '''\n Price_Call, Price_Put = OptionPricer(type_of_option, Param, S0, T, Strike, Steps, N)\n type_of_option = 'European'\n 'Asian', 'Lookback', 'FloatingLookback'\n Param: Parameter catalog with fields \n Param['InterestRate'] : Yearly risk free interest rate (default: 0.11)\n Param['Volatility'] : scalar or array of length steps (default: 0.11)\n Param['S0'] : Initial asset price (default: 1)\n Param['T '] : Time Period (in Years) (default: 1)\n Param['Strike']: Strike Price (default: 1)\n Param['Steps'] : Number of steps to monitor the stock price (default: 12)\n Param['N'] : Number of Monte Carlo Samples (default: 1000)\n '''",
"_____no_output_____"
],
[
"# Some test cases\npar = {'Volatility': [0.01,0.01,0.01,0.03,0.03], 'InterestRate': 0.11}\nOptionPricer('Asian', par)",
"_____no_output_____"
]
],
[
[
"Next week: Kalman Filtering (Learn Numpy and matplotlib)",
"_____no_output_____"
]
],
[
[
"th = 0.5\nA = np.mat([[np.cos(th), np.sin(th)],[-np.sin(th), np.cos(th)]])\n\nx = np.mat([1,0]).T\nx = np.mat([[1],[0]])\n\nfor t in range(10):\n x = A*x + 0*np.random.randn(2,1)\n print(x)\n\n\n",
"[[ 2.18797242]\n [-0.08841785]]\n[[ 2.91934019]\n [-0.83328774]]\n[[ 2.60784679]\n [-2.60834758]]\n[[ 0.55056669]\n [-4.53854234]]\n[[-1.83571934]\n [-4.13840597]]\n[[-3.21258182]\n [-1.98156118]]\n[[-4.7905288]\n [-1.6219334]]\n[[-4.95505597]\n [ 0.89512108]]\n[[-5.46246567]\n [ 4.18711598]]\n[[-2.32141535]\n [ 5.0248705 ]]\n"
],
[
"name = raw_input(\"What is your name? \")\nprint name",
"What is your name? Ali\nAli\n"
],
[
"\ndef fun(x):\n print x,\n\nx = map(fun,range(1,10+1))\n\nx = map(fun,range(1,10+1))\n",
"1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10\n"
],
[
"def myfun(x,y):\n return x*y\n\ndef myfun2(x,y):\n return x+y\n\ndef funn(f, x, y): \n return f(x,y)\n\n\nprint funn(myfun2, 3, 5)\n \nl = [1,2,3,4]\n\ndef power(x):\n return 2**x\n\nr = sum(map(power, l))\n\nprint(r)",
"8\n30\n"
],
[
"s = 'insert 0 5'\n\nu = s.split(' ')\nprint(u)",
"['insert', '0', '5']\n"
],
[
"l = [1,2,3]\nl.pop",
"_____no_output_____"
],
[
"N = int(raw_input())\n\nl = []\n\nfor i in range(N):\n s = raw_input()\n items = s.split(' ')\n cmd = items[0]\n if cmd == 'insert':\n pos = int(items[1])\n num = int(items[2])\n l.insert(pos, num)\n \n if cmd == 'print':\n print l\n if cmd == 'remove':\n num = int(items[1])\n l.remove(num)\n if cmd == 'append':\n num = int(items[1])\n l.append(num)\n if cmd == 'sort':\n l.sort()\n if cmd == 'pop':\n l.pop()\n if cmd == 'reverse':\n l.reverse()",
"_____no_output_____"
],
[
"N = 2\nl = '1 2'.split(' ')\nl2 = [int(i) for i in l[0:N+1]]\nc = tuple(l2)\nprint hash(c)",
"3713081631934410656\n"
],
[
"48*21",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f89897c33ba9ddf36c080d2f45683ea8ed2f87 | 74,655 | ipynb | Jupyter Notebook | docs/tutorials/TCalc_tutorial.ipynb | Bhavesh012/Telescope-Calculator | 049cebdcc023df0375438c54ffbd4ba256e3f121 | [
"MIT"
] | 4 | 2021-06-25T21:22:04.000Z | 2021-06-29T10:00:08.000Z | docs/tutorials/TCalc_tutorial.ipynb | Bhavesh012/Telescope-Calculator | 049cebdcc023df0375438c54ffbd4ba256e3f121 | [
"MIT"
] | null | null | null | docs/tutorials/TCalc_tutorial.ipynb | Bhavesh012/Telescope-Calculator | 049cebdcc023df0375438c54ffbd4ba256e3f121 | [
"MIT"
] | 1 | 2021-09-28T05:18:36.000Z | 2021-09-28T05:18:36.000Z | 138.25 | 20,130 | 0.849293 | [
[
[
"# uncomment the bottom line to install the package\n# !pip install TCalc==1.1.1",
"_____no_output_____"
]
],
[
[
"Importing classes from the `tcalc` module",
"_____no_output_____"
]
],
[
[
"from TCalc.tcalc import eyepiece, telescope, barlow_lens, focal_reducer",
"_____no_output_____"
]
],
[
[
"To quickly access the docstring, run `help(classname)`",
"_____no_output_____"
]
],
[
[
"help(eyepiece)",
"Help on class eyepiece in module TCalc.tcalc:\n\nclass eyepiece(builtins.object)\n | eyepiece(f_e, fov_e=50)\n | \n | Class representing a single eyepiece\n | Args:\n | f_e: focal length of the eyepiece (mm) \n | fov_e: field of view of the eyepiece (deg). Defaults to 50 degrees.\n | \n | Methods defined here:\n | \n | __init__(self, f_e, fov_e=50)\n | Initialize self. See help(type(self)) for accurate signature.\n | \n | ----------------------------------------------------------------------\n | Data descriptors defined here:\n | \n | __dict__\n | dictionary for instance variables (if defined)\n | \n | __weakref__\n | list of weak references to the object (if defined)\n\n"
]
],
[
[
"For an example, let's try to have estimate the specifications of Celestron's 8 SE telescope.",
"_____no_output_____"
]
],
[
[
"c8 = telescope(D_o=203.2, f_o=2032, user_D_eye=None, user_age=22) # adding configuration of 8in scope\nomni_40 = eyepiece(40, 52) # defining 40 mm eyepiece\nomni_25 = eyepiece(25, 52) # defining 25 mm eyepiece",
"Focal Ratio:'10.0'\nTrue Focal Ratio:'10.0'\n"
],
[
"# adding eyepiece to the telescope\nc8.add_eyepiece(omni_40, id='omni_40', select=True)\nc8.add_eyepiece(omni_25, id='omni_25', select=True)",
"_____no_output_____"
],
[
"# listing all the added eyepieces in a table format\nc8.list_eyepiece()",
"\n Currently included eyepieces:\n Name Focal Length FOV\n -------------- -------------- --------------\n 'omni_40' 40 mm 52 degrees \n 'omni_25' 25 mm 52 degrees \n\n The currently selected eyepiece is 'omni_25'\n\n\n Additional optical parts available:\n Name Type Power\n -------------- -------------- --------------\n\n No optical part is selected\n\n"
],
[
"# listing overall configuration of the telescope\nc8.say_configuration() # remember this with 25 mm eyepiece",
"\n The telescope has the following layout:\n Aperture diameter: 203.2 mm\n Focal length: 2032 mm, corresponding to a focal ratio of 10.0\n\n In good atmospheric conditions, the resolution of the telescope (Dawes limit) is 0.6 arcseconds\n By wavelength, the resolution is\n 400 nm (blue): 0.5 arcsec\n 550 nm (green): 0.7 arcsec\n 700 nm (red): 0.9 arcsec\n\n The maximum possible magnification factor is 406.4\n This means the minimum compatible eyepiece focal length is 5.0 mm\n\n The minimum magnification factor and corresponding maximum eyepiece focal length depend on the diameter of the observer's eye.\n For a telescope user with an eye diameter of 7 mm (apropriate for an age around 25 years):\n The minimum magnification factor is 29.0\n This means the maximum compatible eyepiece focal length is 406.4 mm\n\n The faintest star that can be seen by this telescope is 13.5 mag\n\n The currently selected eyepiece is 'omni_25', which has the following layout:\n Focal length: 25 mm\n Field of view: 52 degrees\n\n With this eyepiece:\n The magnification factor is 81.3. This is compatible with the telescope limits.\n The true field of view is 1 degrees\n The exit pupil diameter is 2.5 mm\n\n The faintest surface brightness that can be seen by this telescope is 12.50\n\n"
],
[
"# selecting different eyepiece\nc8.select_eyepiece('omni_40')\nc8.say_configuration()",
"\n The telescope has the following layout:\n Aperture diameter: 203.2 mm\n Focal length: 2032 mm, corresponding to a focal ratio of 10.0\n\n In good atmospheric conditions, the resolution of the telescope (Dawes limit) is 0.6 arcseconds\n By wavelength, the resolution is\n 400 nm (blue): 0.5 arcsec\n 550 nm (green): 0.7 arcsec\n 700 nm (red): 0.9 arcsec\n\n The maximum possible magnification factor is 406.4\n This means the minimum compatible eyepiece focal length is 5.0 mm\n\n The minimum magnification factor and corresponding maximum eyepiece focal length depend on the diameter of the observer's eye.\n For a telescope user with an eye diameter of 7 mm (apropriate for an age around 25 years):\n The minimum magnification factor is 29.0\n This means the maximum compatible eyepiece focal length is 406.4 mm\n\n The faintest star that can be seen by this telescope is 13.5 mag\n\n The currently selected eyepiece is 'omni_40', which has the following layout:\n Focal length: 40 mm\n Field of view: 52 degrees\n\n With this eyepiece:\n The magnification factor is 50.8. This is compatible with the telescope limits.\n The true field of view is 1 degrees\n The exit pupil diameter is 4.0 mm\n\n The faintest surface brightness that can be seen by this telescope is 32.00\n\n"
],
[
"# calling individual functions\nc8._compute_focal_ratio()",
"Focal Ratio:'10.0'\nTrue Focal Ratio:'10.0'\n"
],
[
"# adding additional optical parts\nreducer = focal_reducer(.5) # defining focal reducer of 0.5x\nbarlow = barlow_lens(2) # defining barlow lens of 2x ",
"_____no_output_____"
],
[
"c8.add_optic(reducer,'reducer 1', select=True) # adding reducer to the telescope\nc8.add_optic(barlow,'barlow 1', select=False) # adding barlow to the telescope\n\n#if the magnifications limits get reached then warning will be printed. ",
"Focal Ratio:'5.0'\nTrue Focal Ratio:'10.0'\nNote: The magnification produced by this eyepiece is not compatible with the telescope.\n"
],
[
"c8.add_optic(reducer,'reducer 1', select=False) \nc8.add_optic(barlow,'barlow 1', select=True) ",
"Focal Ratio:'20.0'\nTrue Focal Ratio:'10.0'\n"
],
[
"# printing configuration again with barlow lens\nc8.say_configuration()",
"\n The telescope has the following layout:\n Aperture diameter: 203.2 mm\n Focal length: 2032 mm, corresponding to a focal ratio of 10.0\n 'barlow 1', a Barlow lens, has been added to the optical path. This increases the focal length by 2\n This results in\n Focal length: 4064 mm, corresponding to a focal ratio of 20.0\n\n In good atmospheric conditions, the resolution of the telescope (Dawes limit) is 0.6 arcseconds\n By wavelength, the resolution is\n 400 nm (blue): 0.5 arcsec\n 550 nm (green): 0.7 arcsec\n 700 nm (red): 0.9 arcsec\n\n The maximum possible magnification factor is 406.4\n This means the minimum compatible eyepiece focal length is 10.0 mm\n\n The minimum magnification factor and corresponding maximum eyepiece focal length depend on the diameter of the observer's eye.\n For a telescope user with an eye diameter of 7 mm (apropriate for an age around 25 years):\n The minimum magnification factor is 29.0\n This means the maximum compatible eyepiece focal length is 406.4 mm\n\n The faintest star that can be seen by this telescope is 13.5 mag\n\n The currently selected eyepiece is 'omni_40', which has the following layout:\n Focal length: 40 mm\n Field of view: 52 degrees\n\n With this eyepiece:\n The magnification factor is 101.6. This is compatible with the telescope limits.\n The true field of view is 1 degrees\n The exit pupil diameter is 2.0 mm\n\n The faintest surface brightness that can be seen by this telescope is 8.00\n\n"
]
],
[
[
"You can notice that if used a *2x barlow lens* on a *40mm eyepiece*, the brightness of the object will be decresead by **4 times!**\n\nThis way you can simulate different scenarios and find out which accesories are optimal for your purpose. This will save you both time and money on costly accesories! ",
"_____no_output_____"
],
[
"For advanced users, the plot functionality provides the plots of `resolution performance`, `maginfication_limits` and `eyepiece_limits`.",
"_____no_output_____"
]
],
[
[
"c8.show_resolving_power()\nc8.show_magnification_limits()\nc8.show_eyepiece_limits()",
"_____no_output_____"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
e7f8b71b1d94abd4dc0f01583abb7b2ab80e8d20 | 12,076 | ipynb | Jupyter Notebook | py/chat/tmasma_demo.ipynb | bakfire07/courseproject | 59546a66301f9485c4926e294f59ebac60dd1c0e | [
"MIT"
] | null | null | null | py/chat/tmasma_demo.ipynb | bakfire07/courseproject | 59546a66301f9485c4926e294f59ebac60dd1c0e | [
"MIT"
] | null | null | null | py/chat/tmasma_demo.ipynb | bakfire07/courseproject | 59546a66301f9485c4926e294f59ebac60dd1c0e | [
"MIT"
] | null | null | null | 35.41349 | 141 | 0.427211 | [
[
[
"# %load tmasma_bot.py\n'''\nThis is an implementation of ELiza. It is taken from https://www.smallsurething.com/implementing-the-famous-eliza-chatbot-in-python/\nThe code has been modified to adapt for the course\n'''\n\nimport re\nimport random\n \nreflections = {\n \"am\": \"are\",\n \"was\": \"were\",\n \"i\": \"you\",\n \"i'd\": \"you would\",\n \"i've\": \"you have\",\n \"i'll\": \"you will\",\n \"my\": \"your\",\n \"are\": \"am\",\n \"you've\": \"I have\",\n \"you'll\": \"I will\",\n \"your\": \"my\",\n \"yours\": \"mine\",\n \"you\": \"me\",\n \"me\": \"you\"\n}\n \npsychobabble = [\n [r'I need (.*)',\n [\"Why do you need {0}?\",\n \"Would it really help you to get {0}?\",\n \"Are you sure you need {0}?\"]],\n \n [r'Why don\\'?t you ([^\\?]*)\\??',\n [\"Do you really think I don't {0}?\",\n \"Perhaps eventually I will {0}.\",\n \"Do you really want me to {0}?\"]],\n \n [r'Why can\\'?t I ([^\\?]*)\\??',\n [\"Do you think you should be able to {0}?\",\n \"If you could {0}, what would you do?\",\n \"I don't know -- why can't you {0}?\",\n \"Have you really tried?\"]],\n \n [r'I can\\'?t (.*)',\n [\"How do you know you can't {0}?\",\n \"Perhaps you could {0} if you tried.\",\n \"What would it take for you to {0}?\"]],\n \n [r'I am (.*)',\n [\"Did you come to me because you are {0}?\",\n \"How long have you been {0}?\",\n \"How do you feel about being {0}?\"]],\n \n [r'I\\'?m (.*)',\n [\"How does being {0} make you feel?\",\n \"Do you enjoy being {0}?\",\n \"Why do you tell me you're {0}?\",\n \"Why do you think you're {0}?\"]],\n \n [r'Are you ([^\\?]*)\\??',\n [\"Why does it matter whether I am {0}?\",\n \"Would you prefer it if I were not {0}?\",\n \"Perhaps you believe I am {0}.\",\n \"I may be {0} -- what do you think?\"]],\n \n [r'What (.*)',\n [\"Why do you ask?\",\n \"How would an answer to that help you?\",\n \"What do you think?\"]],\n \n [r'How (.*)',\n [\"How do you suppose?\",\n \"Perhaps you can answer your own question.\",\n \"What is it you're really asking?\"]],\n \n [r'Because (.*)',\n [\"Is that the real reason?\",\n \"What other reasons come to mind?\",\n \"Does that reason apply to anything else?\",\n \"If {0}, what else must be true?\"]],\n \n [r'(.*) sorry (.*)',\n [\"There are many times when no apology is needed.\",\n \"What feelings do you have when you apologize?\"]],\n \n [r'Hello(.*)',\n [\"Hello... I'm glad you could drop by today.\",\n \"Hi there... how are you today?\",\n \"Hello, how are you feeling today?\"]],\n \n [r'I think (.*)',\n [\"Do you doubt {0}?\",\n \"Do you really think so?\",\n \"But you're not sure {0}?\"]],\n \n [r'(.*) friend (.*)',\n [\"Tell me more about your friends.\",\n \"When you think of a friend, what comes to mind?\",\n \"Why don't you tell me about a childhood friend?\"]],\n \n [r'Yes',\n [\"You seem quite sure.\",\n \"OK, but can you elaborate a bit?\"]],\n \n [r'(.*) computer(.*)',\n [\"Are you really talking about me?\",\n \"Does it seem strange to talk to a computer?\",\n \"How do computers make you feel?\",\n \"Do you feel threatened by computers?\"]],\n \n [r'Is it (.*)',\n [\"Do you think it is {0}?\",\n \"Perhaps it's {0} -- what do you think?\",\n \"If it were {0}, what would you do?\",\n \"It could well be that {0}.\"]],\n \n [r'It is (.*)',\n [\"You seem very certain.\",\n \"If I told you that it probably isn't {0}, what would you feel?\"]],\n \n [r'Can you ([^\\?]*)\\??',\n [\"What makes you think I can't {0}?\",\n \"If I could {0}, then what?\",\n \"Why do you ask if I can {0}?\"]],\n \n [r'Can I ([^\\?]*)\\??',\n [\"Perhaps you don't want to {0}.\",\n \"Do you want to be able to {0}?\",\n \"If you could {0}, would you?\"]],\n \n [r'You are (.*)',\n [\"Why do you think I am {0}?\",\n \"Does it please you to think that I'm {0}?\",\n \"Perhaps you would like me to be {0}.\",\n \"Perhaps you're really talking about yourself?\"]],\n \n [r'You\\'?re (.*)',\n [\"Why do you say I am {0}?\",\n \"Why do you think I am {0}?\",\n \"Are we talking about you, or me?\"]],\n \n [r'I don\\'?t (.*)',\n [\"Don't you really {0}?\",\n \"Why don't you {0}?\",\n \"Do you want to {0}?\"]],\n \n [r'I feel (.*)',\n [\"Good, tell me more about these feelings.\",\n \"Do you often feel {0}?\",\n \"When do you usually feel {0}?\",\n \"When you feel {0}, what do you do?\"]],\n \n [r'I have (.*)',\n [\"Why do you tell me that you've {0}?\",\n \"Have you really {0}?\",\n \"Now that you have {0}, what will you do next?\"]],\n \n [r'I would (.*)',\n [\"Could you explain why you would {0}?\",\n \"Why would you {0}?\",\n \"Who else knows that you would {0}?\"]],\n \n [r'Is there (.*)',\n [\"Do you think there is {0}?\",\n \"It's likely that there is {0}.\",\n \"Would you like there to be {0}?\"]],\n \n [r'My (.*)',\n [\"I see, your {0}.\",\n \"Why do you say that your {0}?\",\n \"When your {0}, how do you feel?\"]],\n \n [r'You (.*)',\n [\"We should be discussing you, not me.\",\n \"Why do you say that about me?\",\n \"Why do you care whether I {0}?\"]],\n \n [r'Why (.*)',\n [\"Why don't you tell me the reason why {0}?\",\n \"Why do you think {0}?\"]],\n \n [r'I want (.*)',\n [\"What would it mean to you if you got {0}?\",\n \"Why do you want {0}?\",\n \"What would you do if you got {0}?\",\n \"If you got {0}, then what would you do?\"]],\n \n [r'(.*) mother(.*)',\n [\"Tell me more about your mother.\",\n \"What was your relationship with your mother like?\",\n \"How do you feel about your mother?\",\n \"How does this relate to your feelings today?\",\n \"Good family relations are important.\"]],\n \n [r'(.*) father(.*)',\n [\"Tell me more about your father.\",\n \"How did your father make you feel?\",\n \"How do you feel about your father?\",\n \"Does your relationship with your father relate to your feelings today?\",\n \"Do you have trouble showing affection with your family?\"]],\n \n [r'(.*) child(.*)',\n [\"Did you have close friends as a child?\",\n \"What is your favorite childhood memory?\",\n \"Do you remember any dreams or nightmares from childhood?\",\n \"Did the other children sometimes tease you?\",\n \"How do you think your childhood experiences relate to your feelings today?\"]],\n \n [r'(.*)\\?',\n [\"Why do you ask that?\",\n \"Please consider whether you can answer your own question.\",\n \"Perhaps the answer lies within yourself?\",\n \"Why don't you tell me?\"]],\n \n [r'quit',\n [\"Thank you for talking with me.\",\n \"Good-bye.\",\n \"Thank you, that will be $150. Have a good day!\"]],\n \n [r'(.*)',\n [\"Please tell me more.\",\n \"Let's change focus a bit... Tell me about your family.\",\n \"Can you elaborate on that?\",\n \"Why do you say that {0}?\",\n \"I see.\",\n \"Very interesting.\",\n \"{0}.\",\n \"I see. And what does that tell you?\",\n \"How does that make you feel?\",\n \"How do you feel when you say that?\"]]\n]\n \n \ndef reflect(fragment):\n tokens = fragment.lower().split()\n for i, token in enumerate(tokens):\n if token in reflections:\n tokens[i] = reflections[token]\n return ' '.join(tokens)\n \n \ndef analyze(statement):\n for pattern, responses in psychobabble:\n match = re.match(pattern, statement.rstrip(\".!\"))\n if match:\n response = random.choice(responses)\n return response.format(*[reflect(g) for g in match.groups()])\n \n \ndef main():\n print \"TMASMA Bot> Hello. How are you feeling today? \\n\"\n \n while True:\n statement = raw_input(\"You> \")\n print \"TMASMA Bot> {} \\n\".format( analyze(statement))\n \n if statement == \"quit\":\n break\n \n \n",
"_____no_output_____"
],
[
"main()",
"TMASMA Bot> Hello. How are you feeling today? \n\nYou> I am feeling good\nTMASMA Bot> How long have you been feeling good? \n\nYou> long back\nTMASMA Bot> Please tell me more. \n\nYou> i have been bored for 1 hour\nTMASMA Bot> you have been bored for 1 hour. \n\nYou> yes \nTMASMA Bot> Why do you say that yes? \n\nYou> Just because I want to say this!\nTMASMA Bot> I see. And what does that tell you? \n\nYou> It doesn't tell a shit\nTMASMA Bot> Why do you say that it doesn't tell a shit? \n\n"
]
]
] | [
"code"
] | [
[
"code",
"code"
]
] |
e7f8bff25e1ef8f23be56cceb2d9cbd342973e7b | 28,891 | ipynb | Jupyter Notebook | week-6/sc_6.ipynb | mattDevigili/dms-smm695 | c1789c5667e2854449bd49cf0290e5cee149bdcf | [
"MIT"
] | 34 | 2020-05-14T05:04:34.000Z | 2021-11-07T20:37:04.000Z | week-6/sc_6.ipynb | mattDevigili/dms-smm695 | c1789c5667e2854449bd49cf0290e5cee149bdcf | [
"MIT"
] | null | null | null | week-6/sc_6.ipynb | mattDevigili/dms-smm695 | c1789c5667e2854449bd49cf0290e5cee149bdcf | [
"MIT"
] | 23 | 2020-05-21T10:24:10.000Z | 2022-03-24T23:37:30.000Z | 23.916391 | 236 | 0.537122 | [
[
[
"# Week 6 - SMM695\n\nMatteo Devigili\n\nJune, 28th 2021\n\n[_PySpark_](https://spark.apache.org/docs/latest/api/python/index.html#): during this lecture, we will approach Spark through Python\n\n<img src=\"images/_1.png\" width=\"20%\">",
"_____no_output_____"
],
[
"**Agenda**:\n1. Introduction to Spark\n1. Installing PySpark\n1. PySpark Basics\n1. PySpark and Pandas\n1. PySpark and SQL\n1. Load data from your DBMS",
"_____no_output_____"
],
[
"# Introduction to Spark\n\n**Big Data Challenge**:\n\n* Cost of storing data has dropped\n* The need for parallel computation has increased\n\n\n**Note**: [IBM Blue Gen\\L](https://www.ibm.com/ibm/history/ibm100/us/en/icons/bluegene/)",
"_____no_output_____"
],
[
"**What is [Apache Spark](https://spark.apache.org)**?\n\n> \"Apache Spark is a unified computing engine and a set of libraries for parallel data processing on computer clusters\"\n\n[Chambers and Zaharia 2018](#references)",
"_____no_output_____"
],
[
"**Programming Languages Supported**:\n<img src=\"images/_0.png\" width=\"50%\">",
"_____no_output_____"
],
[
"**Spark's philosophy**:\n\n* *Unified*: Spark offers a large variety of data analytics tools\n* *Computing Engine*: Spark focuses on computing, not on storage\n* *Libraries*: Spark has different libraries to perform several tasks",
"_____no_output_____"
],
[
"**Apache Spark Libraries**:\n\n* *Spark SQL*\n* *Spark Streaming*\n* *Spark MLlib*\n* *Spark GraphX*\n\n[Third-party projects](https://spark.apache.org/third-party-projects.html)",
"_____no_output_____"
],
[
"**Spark Application**:\n\n| Component ||Role |\n|----|----|---|\n| *Spark Driver*| | Execute user-defined tasks |\n| *Cluster Manager* | | Manage workers nodes|\n| *Executors* | | Execute tasks |\n\n<img src=\"images/_5.png\" width=80%>",
"_____no_output_____"
],
[
"**From Python to Spark code and back**:\n\n\n\nSource: _Bill Chambers, Matei Zaharia 2018_ (p. 23)",
"_____no_output_____"
],
[
"# Installing PySpark\n\nThere are several ways to set-up PySpark on your local machine. Here, two methods are discussed:\n* Pure-python users: \n```python\npip install pyspark\n```\n* Conda users:\n```python\nconda install pyspark\n```\nFurther info at [Spark Download page](https://spark.apache.org/downloads.html).",
"_____no_output_____"
],
[
"## Requirements\n\nPay attention to the following:\n\n>Spark runs on Java 8/11\n\nCheck java version running on your machine. Type the following on your terminal:\n```python\njava -version\n```\n\nIf you are running a different Java version, install java 8/11! Check out [Spark Downloading info](https://spark.apache.org/docs/latest/#downloading).",
"_____no_output_____"
],
[
"# PySpark - Basics",
"_____no_output_____"
],
[
"## Libraries",
"_____no_output_____"
]
],
[
[
"#to create a spark session object\nfrom pyspark.sql import SparkSession\n\n# functions\nimport pyspark.sql.functions as F\n\n# data types\nfrom pyspark.sql.types import *\n\n# import datetime \nfrom datetime import date as dt",
"_____no_output_____"
]
],
[
[
"* More info on **Functions** at these [link-1](https://spark.apache.org/docs/latest/api/python/pyspark.sql.html#module-pyspark.sql.functions) & [link-2](https://spark.apache.org/docs/2.3.0/api/sql/index.html#year)\n* More info on **Data Types** at this [link](https://spark.apache.org/docs/latest/sql-ref-datatypes.html)",
"_____no_output_____"
],
[
"## Opening a Session\n\nThe **SparkSession** is a driver process that enables:\n\n* to control our Spark Application\n* to execute user-defined manipulations\n\nCheck this [link](https://spark.apache.org/docs/latest/api/python/pyspark.sql.html#pyspark.sql.SparkSession) for further reference.",
"_____no_output_____"
]
],
[
[
"# to open a Session\nspark = SparkSession.builder.appName('last_dance').getOrCreate()",
"_____no_output_____"
]
],
[
[
"**Spark UI**\n\n<img src=\"images/_6.png\" width=60%>\n\nThe spark UI is useful to monitor your application. You have the following tabs:\n\n* *Jobs*: info concerning Spark jobs\n* *Stages*: info on individual stages and their tasks\n* *Storage*: info on data that is currently in our spark application\n* *Environment*: info on configurations and current settings of our application\n* *Executors*: info on the executors that run our application\n* *SQL*: refers to both SQL and DataFrames",
"_____no_output_____"
]
],
[
[
"spark",
"_____no_output_____"
]
],
[
[
"## Create Dataframe\n\nIn order to create a dataframe from scratch, we need to:\n1. Create a schema, passing:\n * Column names\n * Data types\n1. Pass values as an array of tuples",
"_____no_output_____"
]
],
[
[
"# Here, I define a schema\n# .add(field, data_type=None, nullable=True, metadata=None)\n\nschema = StructType().add(\"id\", \"integer\", True).add(\"first_name\", \"string\", True).add(\n \"last_name\", \"string\", True).add(\"dob\", \"date\", True)\n\n'''\nschema = StructType().add(\"id\", IntegerType(), True).add(\"first_name\", StringType(), True).add(\n \"last_name\", StringType(), True).add(\"dob\", DateType(), True)\n'''\n\n# Then, I can pass some values\ndf = spark.createDataFrame([(1, 'Michael', \"Jordan\", dt(1963, 2, 17)),\n (2, 'Scottie', \"Pippen\", dt(1965, 9, 25)),\n (3, 'Dennis', \"Rodman\", dt(1961, 5, 16))],\n schema=schema)\n\n# Let's explore Schema structure\ndf.printSchema()",
"_____no_output_____"
],
[
"# We can also leverage on functions to create a new column\ndf=df.withColumn('age', F.year(F.current_date()) - F.year(df.dob))\n\ndf.show()",
"_____no_output_____"
]
],
[
[
"**Transformations**\n\n* Immutability: once created, data structures can not be changed\n* Lazy evaluation: computational instructions will be executed at the very last",
"_____no_output_____"
],
[
"**Actions**\n\n* view data\n* collect data\n* write to output data sources",
"_____no_output_____"
],
[
"# PySpark and Pandas",
"_____no_output_____"
],
[
"## Load a csv",
"_____no_output_____"
],
[
"Loading a csv file from you computer, you need to type:\n* Pands:\n * db = pd.read_csv('path/to/movies.csv')\n* Pyspark:\n * df = spark.read.csv('path/to/movies.csv', header=True, inferSchema=True)\n\nHere, we will import a csv directly from GitHub. Data are provided by [FiveThirtyEight](https://github.com/fivethirtyeight)\n[<img src=\"images/_2.png\" width=\"50%\">](https://fivethirtyeight.com/features/the-dollar-and-cents-case-against-hollywoods-exclusion-of-women/)",
"_____no_output_____"
]
],
[
[
"# import pandas\nimport pandas as pd\n\n# import SparkFiles\nfrom pyspark import SparkFiles\n\n# target dataset\nurl = 'https://raw.githubusercontent.com/fivethirtyeight/data/master/bechdel/movies.csv'",
"_____no_output_____"
],
[
"# loading data with pandas\ndb = pd.read_csv(url)\n\n# loading data with pyspark\nspark.sparkContext.addFile(url)\ndf = spark.read.csv(SparkFiles.get('movies.csv'), header=True, inferSchema=True)",
"_____no_output_____"
]
],
[
[
"## Inspecting dataframes",
"_____no_output_____"
]
],
[
[
"# pandas info\ndb.info()",
"_____no_output_____"
],
[
"# pyspark schema\ndf.printSchema()",
"_____no_output_____"
],
[
"# pandas fetch 5\ndb.head(5)",
"_____no_output_____"
],
[
"# pyspark fetch 5\ndf.show(5)\n\ndf.take(5)",
"_____no_output_____"
],
[
"# pandas filtering:\ndb[db.year == 1970]",
"_____no_output_____"
],
[
"# pyspark filtering:\ndf[df.year == 1970].show()",
"_____no_output_____"
],
[
"# get columns and data types\nprint(\"\"\"\nPandas db.columns:\n===================\n{}\n\nPySpark df.columns:\n===================\n{}\n\nPandas db.dtype:\n===================\n{}\n\nPySpark df.dtypes:\n===================\n{}\n\n\"\"\".format(db.columns, df.columns, db.dtypes, df.dtypes), flush = True)",
"_____no_output_____"
]
],
[
[
"## Columns",
"_____no_output_____"
]
],
[
[
"# pandas add a column\ndb['newcol'] = db.domgross/db.intgross\n\n# pyspark add a column\ndf=df.withColumn('newcol', df.domgross/df.intgross)",
"_____no_output_____"
],
[
"# pandas rename columns\ndb.rename(columns={'newcol': 'dgs/igs'}, inplace=True)\n\n# pyspark rename columns\ndf=df.withColumnRenamed('newcol', 'dgs/igs')",
"_____no_output_____"
]
],
[
[
"## Drop",
"_____no_output_____"
]
],
[
[
"# pandas drop `code' column\ndb.drop('code', axis=1, inplace=True)\n\n# pyspark drop `code' column\ndf=df.drop('code')",
"_____no_output_____"
],
[
"# pandas dropna()\ndb.dropna(subset=['domgross'], inplace=True)\n\n# pyspark dropna()\ndf=df.dropna(subset='domgross')",
"_____no_output_____"
]
],
[
[
"## Stats",
"_____no_output_____"
]
],
[
[
"# pandas describe\ndb.describe()",
"_____no_output_____"
],
[
"# pyspark describe\ndf.describe(['year', 'budget']).show()",
"_____no_output_____"
]
],
[
[
"# Pyspark and SQL",
"_____no_output_____"
]
],
[
[
"# pyspark rename 'budget_2013$'\ndf=df.withColumnRenamed('budget_2013$', 'budget_2013')",
"_____no_output_____"
],
[
"# Create a temporary table \ndf.createOrReplaceTempView('bechdel')\n\n# Run a simple SQL command\nsql = spark.sql(\"\"\"SELECT imdb, year, title, budget FROM bechdel LIMIT(5)\"\"\")\nsql.show()",
"_____no_output_____"
],
[
"# AVG budget differences\nsql_avg = spark.sql(\n \"\"\"\n SELECT \n binary, \n COUNT(*) AS count, \n format_number(AVG(budget),2) AS avg_budget, \n format_number((SELECT AVG(budget) FROM bechdel),2) AS avg_budget_samp,\n format_number(AVG(budget_2013),2) AS avg_budget2013,\n format_number((SELECT AVG(budget_2013) FROM bechdel),2) AS avg_budget2013_samp\n FROM bechdel GROUP BY binary\n \"\"\"\n)\n\nsql_avg.show()",
"_____no_output_____"
]
],
[
[
"# Load data from DBMS",
"_____no_output_____"
],
[
"To run the following you need to restart the notebook.",
"_____no_output_____"
]
],
[
[
"# to create a spark session object\nfrom pyspark.sql import SparkSession",
"_____no_output_____"
]
],
[
[
"## PostgreSQL",
"_____no_output_____"
],
[
"To interact with postgre you need to:\n \n* Download the *postgresql-42.2.22.jar file* [here](https://jdbc.postgresql.org/download.html)\n* Include the path to the downloaded jar file into SparkSession()",
"_____no_output_____"
]
],
[
[
"# Open a session running data from PostgreSQL\nspark_postgre = SparkSession \\\n .builder \\\n .appName(\"last_dance_postgre\") \\\n .config(\"spark.jars\", \"/Users/matteodevigili/py3venv/dms695/share/py4j/postgresql-42.2.22.jar\") \\\n .getOrCreate()",
"_____no_output_____"
],
[
"spark_postgre",
"_____no_output_____"
],
[
"# Read data from PostgreSQL running at localhost\ndf = spark_postgre.read \\\n .format(\"jdbc\") \\\n .option(\"url\", \"jdbc:postgresql://localhost:5432/pagila\") \\\n .option(\"dbtable\", \"film\") \\\n .option(\"user\", \"dms695\") \\\n .option(\"password\", \"smm695\") \\\n .option(\"driver\", \"org.postgresql.Driver\") \\\n .load()\n\ndf.printSchema()",
"_____no_output_____"
],
[
"# get some stats\ndf.describe(['release_year', 'rental_rate', 'rental_duration']).show()",
"_____no_output_____"
],
[
"# Create a temporary table \ndf.createOrReplaceTempView('film')\n\n# Run a simple SQL command\nsql = spark_postgre.sql(\"\"\"SELECT title, release_year, length, rating FROM film LIMIT(1)\"\"\")\nsql.show()",
"_____no_output_____"
]
],
[
[
"## MongoDB",
"_____no_output_____"
],
[
"For further reference check the [Python Guide provided by Mongo](https://docs.mongodb.com/spark-connector/current/python-api/) or the [website for the mongo-spark connector](https://spark-packages.org/package/mongodb/mongo-spark).",
"_____no_output_____"
]
],
[
[
"# add path to Mongo\nspark_mongo = SparkSession \\\n .builder \\\n .appName(\"last_dance_mongo\") \\\n .config(\"spark.mongodb.input.uri\", \"mongodb://127.0.0.1/amazon.music\") \\\n .config(\"spark.mongodb.output.uri\", \"mongodb://127.0.0.1/amazon.music\") \\\n .config('spark.jars.packages', 'org.mongodb.spark:mongo-spark-connector_2.12:3.0.1') \\\n .getOrCreate()",
"_____no_output_____"
],
[
"spark_mongo",
"_____no_output_____"
],
[
"# load data from MongoDB\ndf = spark_mongo.read.format(\"mongo\").load()\n\ndf.printSchema()",
"_____no_output_____"
],
[
"# get some stats\ndf.describe(['overall', 'unixReviewTime']).show()",
"_____no_output_____"
],
[
"# Create a temporary table \ndf.createOrReplaceTempView('music')\n\n# Run a simple SQL command\nsql = spark_mongo.sql(\"\"\"SELECT asin, date, helpful, overall, unixReviewTime FROM music LIMIT(1)\"\"\")\nsql.show()",
"_____no_output_____"
]
],
[
[
"# References\n\n* Bill Chambers, Matei Zaharia 2018,[\"Spark: The Definitive Guide\"](https://www.oreilly.com/library/view/spark-the-definitive/9781491912201/) <img src=\"images/_3.png\" width=\"20%\">\n* Pramod Singh 2019, [\"Learn PySpark: Build Python-based Machine Learning and Deep Learning Models\n\"](https://www.ibs.it/learn-pyspark-build-python-based-libro-inglese-pramod-singh/e/9781484249604) <img src=\"images/_4.png\" width=\"18%\">",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
e7f8da404c99c3383fdd9b0912b46e1ec3ebb5f7 | 179,625 | ipynb | Jupyter Notebook | python/algorithmia/algorithmia.ipynb | Giorat/pocketbotanist | 1d918bcead6cbe2822f9c04c42a62f315aabceb9 | [
"MIT"
] | 3 | 2018-06-08T05:44:16.000Z | 2019-02-27T19:28:04.000Z | python/algorithmia/algorithmia.ipynb | riccardogiorato/pocketbotanist | 1d918bcead6cbe2822f9c04c42a62f315aabceb9 | [
"MIT"
] | 8 | 2018-06-08T06:43:45.000Z | 2019-05-08T06:37:28.000Z | python/algorithmia/algorithmia.ipynb | riccardogiorato/pocketbotanist | 1d918bcead6cbe2822f9c04c42a62f315aabceb9 | [
"MIT"
] | null | null | null | 1,009.129213 | 174,247 | 0.953837 | [
[
[
"!pip install tensorflow\n!pip install Algorithmia\n!pip install numpy",
"_____no_output_____"
],
[
"import Algorithmia\nimport tensorflow as tf\nimport os\nimport numpy as np \nimport base64\nfrom io import BytesIO\nfrom PIL import Image\nfrom matplotlib.pyplot import imshow\nimport json\n\n# Login to Algorithmia API to fetch data\nclient = Algorithmia.client('sim+lHZ2+0fW+jdBFWnX6U2EaMA1')",
"_____no_output_____"
],
[
"graph_def = tf.GraphDef()\nlabels = []\n# data://riccardogiorato/modeltf_test/labels.txt\nlabels_filename = client.file(\"data://riccardogiorato/modeltf_test/labels.txt\").getFile().name\n# data://riccardogiorato/modeltf_test/model.pb\nfilename = client.file(\"data://riccardogiorato/modeltf_test/model.pb\").getFile().name\n\n# Import the TF graph\nwith tf.gfile.FastGFile(filename, 'rb') as f:\n graph_def.ParseFromString(f.read())\n tf.import_graph_def(graph_def, name='')\n\n# Create a list of labels.\nwith open(labels_filename, 'rt') as lf:\n for l in lf:\n labels.append(l.strip())\n\noutput_layer = 'loss:0'\ninput_node = 'Placeholder:0'\n\ndef decode_base64(data):\n \"\"\"\n Decode base64, padding being optional.\n\n :param data: Base64 data as an ASCII byte string\n :returns: The decoded byte string.\n\n \"\"\"\n data = data.encode('UTF-8')\n missing_padding = len(data) % 4\n if missing_padding != 0:\n data += b'='* (4 - missing_padding)\n return base64.decodestring(data)\n\n# API calls will begin at the apply() method, with the request body passed as 'input'\ndef apply(input):\n\n encoded_image = input.split(\",\")[1]\n \n image_bytes = BytesIO(decode_base64(encoded_image))\n im = Image.open(image_bytes)\n colors = np.array(im).T\n bgr_image = np.array([colors[0],colors[1],colors[2]]).transpose()\n\n with tf.Session() as sess:\n prob_tensor = sess.graph.get_tensor_by_name(output_layer)\n predictions, = sess.run(prob_tensor, {input_node: [bgr_image] })\n highest_probability_index = np.argmax(predictions)\n \n return(json.dumps({'label': labels[highest_probability_index],'value': np.float64(round(predictions[highest_probability_index],8))})) ",
"_____no_output_____"
],
[
"# test out the local Algorithmia future function on a sample image\napply(\"data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAOMAAADjCAYAAAB3jqNMAAAgAElEQVR4Xoy8B7RdZbX+/Vtr7bV7Ob2kJycnPSQhgVBC6EgLXS4KoheviggKXhsqKhas1/+9dgUVRKRdQRAQEOkQII30Xs/J6W33svZa6xtz7nNy8Y7/941vMTJIztlllfeZ85nPfOZrNC7o9G08fKfK2MgIC5YuIV/OkU2PUZdKgO8yqX0Ks+d00t3TQ9WAxrp6jpu/kBdffwPTDPL8008xZ+Z0qnjkcSmXy3iehxyG42L6BgEjgOEZOIUylUKRcDRCxfNI9/VjxsOEwkHCsTCBQIA5c+awbds2KvjM7pjDnDlzyeeKpAeHWPv3fxBrqKdSKmHhEQ6HmT59OoVykVK1jBGwiCUS9PX0UFdXx+jAIKZh0NDQQDKVYvlJKzn++OMp5yssWLCAt99+mztuv51Vp5/Fug0bsMMhKpUKAcPENgwMH8aGh7n6hutpnTyJfDZLyffIZDL4vg+ehxMIcXTfAXZv304iGiMSjzHU18+ylSfS0NKsnxGLxTAMA991mTx5MpV8gVQyycJFC/jL40/w5BN/BUwq5RDffmktzxUiOG4SN2Bj2gZj+IQwsPAJJAx29ReohCJYwSqBkE0mnQMjjOEb+FUP0zfxHANcDyNq4lselh3QZyKHb+up67nh+RhetfYL18O2LAKBCs11UapBFycKUQNMw8M3fTlNKqZHPBigwfOZbNuMdaVx8hWiZZfc/n7oM7lkxQLuuf2T5If3EUsGcf0ShXyWqe0NdHd3630eOHiASF0dxfQYJ5xxBuddewVepcJvfvZzhnt6+dLXv8HTf3maoG/iV1xGsyPMmjWL119/XZ+R4XkEg0Fc1yU9PMyyE04gGo3S3t7Ou1u3MXv2bA4fPkwikWD7ls1Uq1V97Q033MCu3dvZsmWLPhs5l0gkomumsbGR/r5BRkdH9fVtbW20t7YyMjKC4zj6/+FsRtd5S0sL/f39rLlsDS+uWwthi5bWZoZHBnUtm6ZJxAphGQZ9R7oImhalSpmG2dNJV0uETUO/t+p7GMkZ7X40EVfwDBw+DOEwnXPmUCjk8XB0AbW3t+mXdnZ2snv3biqFgn7J5z//eZ7ftoXGlma++qEbWDJvPmnHoeg4OG5VAT5yoBcMWRQuRiyKgUldJMqZZ57JjM7ZZCplNry1lk985MN87Zt3EAgF9TvlJviGQf/+bj73jTvIVaoEAwEGevtIxuP0HD1KMZ+lUCiwY8cO5s2fy+EjBwlFo5ScCtVyBVduZEsL+3bvYda0DvyAxfkXXsiq1adxcP8hprS20ZRM8f7LLqfo+1TxoVzm9Y0buficc2lrbGIkm+EfWzbwx3vv482XXmb54uPoHxnGN00y+Zye47YtW5g0aZKC851XXyMaClMYTXPJddeRampk7csvc9mFFxGMhDm0/wBNrS0UKxVmzunkoQcfpm1SC2bVZ1JjGwca5+Gefx29o2DH40QtC8eoYtoWZcvF9T26ekewo23kI0VCqTCFcgnTDYDv4ZUsAp6luKoiCHIxgxaeIVgX5NUO05IfgMQT3/UwPbP2Gr8CloUZcklEbOJhH6wq2YBPNBSk3jKo+C4B3yUS8OmIhIh7UPY8XBzagiFaqlUaLZMtL+5iZngmbzz0U5767U9pbJmMH61gBExMv4IVCRBJpYjYQSq5LP09R/nJfffw/AsvaPD884MP0RqNE7LCjAyNkB5JU/CKZIeHCQZDGjSS0biCZd++faRSKZqbm/U5CGAloJdKJQYGBrj88st58A/3s2r1anp7e3Xd9A/0EQqF6OjoUMAKiAWMyWSSgaEhDfKytmStV3yHYrFIuVDADoeJxaNMnTKNXTv2KuAlIOzYtxO7Kcbs+XMZ6uvRdSyATKYaSQai7H13K0bYothYC4ryHtu2cT0D1zIxQu0Nfl1jg4JRskzXgQNgmLS2t5IvZPTkipUS5513nkaCGTNm8NzTTzNv3jz6+vponjuHyJR2Fs2YxU+/fReV4TTRWIyy5/Iv136Q/Rt3cdtttzE4OEg8HscKhnA8l1AkolHJdSoU0qP8+uc/IxgJaoSQyCXZ8XBXl2bTO7/xTd54Zz0SNPp7ejUrlkslwsEAPT09+tk+HhWnhGnbCpLpU6Zy6OBBIqEQoYBNABvPMllz6aXMmt1B0A4TwGDZwkWctOIEYg0NOKUSbfPmcMrZZ/LYgw+xZ9t2HvzjA4TrEmxYt46H7/8jUydNZtmK5ZQrFQWinG99fb0+cMuyOHr0KBecfQ4Dvf3c/8cHOPWcs+mcNo17f3M3VMo0trZxwUUXsWLliQrgV158idxgnue37+fiW7/IkRlL2Z6GhJ0iVy5hhQIEoiEs26BklkiEI2zb2k+0OYU9NUymUsA3fMKBGE4B3GwV/ACmpDzLxzN9BXINaO8BY6AGxlqc9JW9ePIew4OACQGPSMQiHPCIx01yroMftWkUfDtVQpZBMGxSHzBoBZLyVgOiQIsPkUoJu2Dg9pbp297PFRd3cuaCU5lcH8S1yzX2ZBkY8SiG4VP1qyQTcQqVIgPDQwqMudNmMHCki0rBoZjJUS45+JYriRm3XNFsQ9XTzxJQCRAFmAIAAY6AYf/+/bXsFIkwe+YsXS8f+chHuOaaazjSdZjt27fzne98R5lONptVQAuYzz73XIaGhhSkuVyOquEeA5ecWzKV4NDBwzhlj6lTp8qd5ISTl/Ps6y/iWB6tzY2k02kF3KT2qaSsCM//+Um+/l/f58f3/Cf1TY0a0Cuu8MkAlmlitM6f5cuCkhOQKLJo0SK+9O+fY8fO7dx7328544wzePr5ZzVayO/kpCO2zZEjRxQ071tzCaPRIKOVEu3xJBv+9gLDR3so5vLkqw5m3tOLkZOaNm0aF15xBctWncLwwCBUHLr27OSpvz6BZRmMpMf0JsqNFergeB7lbIG+rh4+/+WvUjZh/569hGybUrFIpVrW18tNHOrvo+pWCIRCmhnlxh+QBwEKRrfsEU7E6ejs5NoPXUc6k2fa5MmEDZM1a9YQMAPM7uxk0onLONBzlLNPP4PjFy3mhcefYN3aNzWaymIWij5vzhxa2tpI53OYgYBSY6FBQmHkfIS27Nu+ixUrT8KwQ0xpbuFP995HJBYjHAzq4pHzksWSLw5TKgT5yuMvs7VxBocKEbJlHyMYomQZSv0dw1VgBcMB+g4OEzCTBOuquJODWFGhsVDJulRyJpSEdnoSTzUrGpYUIbXDkEw5kRnHGatSbX3LOGe15X0mhMAKgB00CQUgFISM4RI1TZqqVRIhGzMEtukr0FtdaMKnzTRJASkfQpUKTVaQ7esGSMYbMEdGWLUgxsLJrbTM6KSQGWbanBkMVwqUAxCNhImEbYKRCOmxMQV9GINqyeXQth1E6hqJhC2l+lTdWqni12hef28vp6xapfdfACnrVYK0ZDa5z/JMLltziSYcSQIC0EWLFypgJOE8+eSTrFq1itdee00p7t7de/jMv3+Whx56SF8bTkR07ct9kvVpBUyWLlnG66+u1X8n4nUsXbaYx5/9C5GGBHMWdOqakcCweM5CCkMZ0pkcHcsW8PqWN/XnJbeKFQiQsGKEfQNj2nFz/UyxoL8UcAloWusaONrTjS/0yDTJO2VN888884xmxHAgoCn8rbfeYv6sTlZdfgnv9hyh4Lm4+RKr5i3kK5+8mURdiq5dXYyMjeiFCNjGHIeqZVIcGuGMFSu48opLqFSLVFyHwaFRpQ9yw+QGtrS3M9Ddq/XC+RetwY2GGezrp1qp6O/L1bLSWQkiAsZsZlQphGTeyW3t7Nu7VzOjUypjuCb1Lc1MmjKFj33i4/SPpnELRWZOncY1V13FyqXL+fVjj3LK5Wt4/xVXsGHDBoaPdNO7eTsB29IHPJpOa/arlkrkMhmWrzpVo6IXsPSBSOYPBIPE4nHaG5v54Q9+xMpTVisY7//t77SWdISO+JLJbPoPHODGz34eY/pCwqdfySvDntIVV7I7JmXHJ22aCAHEAq/sYeYNbNui6GWJzk9StdCHitSHZQ88C9syCQR85G3FUhXDCuDLwx4vC5WmKhh9vY96BHwh6fhBwDLwTQ/LMjEtFOzJIBScCl4oQCpgEDEErB4ByydiWESAMFVmEGA+Lik3QLQKCdenyTV4a+cAh/uhIVElhM20+kbsfA9nL5/CJdddidVchx2wKBRzWpu/8eprVHJ5wraNCBWG49HZOZdXn3yMhvZ2iU2MDg/TmKpXtiYZUEopyWzCVEQLOO644xSosp6EsobtICeeeCJ/+MMfatkyGmZ0eIR///znWL58OTfddJOuJaGOEsDy6QwfvP5DvPjii5S9iq41yYpaArgO8+ctIBKK61qJR+tYML8TP+izfttG0k5OyzpZ9y2RFLs2buPDN97If7/0LJGkpeffWlfP2y+9JoRVk4LRvmCq72BoZpz4kx8d0wuSL5ZI85Wf/AfPbd7Axaedzo2XX8UJy5ZRl0zSPnmyZoBEfR2zV59CIRSgsa6BgZ5elnbO5effvov97+7iqmuvZfHy47nk/AtobmvHdl3qQ2GCVoATTl1+7Hu7+vsoVSp6o+SGTGpr08Jaiv3swBj3Pfgwjz72Z40mru9T8SoKXjnH/FhaM7AEFHk/YZuxkSHCshCrVYwKxOpSdM6dy0du+FcO9/Uy0j9Ae3MLX/vyl/nxT36BEwzwq4f/xGmrTmXJwoU8/d+P8fQ9vyfa0Kj3QjKwBCvL8/nMbbfx9sYN9PT2ctLKldjBoNakuVKBRUuOY2w0ze9++3suvfAq/vH6BkYHRhkeOMjxK1cxrW0yo9k8c5Yu5YEnn+ahvz/B37YMc9CMkbZDpEs+ZctU0SbvG1Ql01kmzmhGAdQwJYUT8imEakDyPB+/UlEgErQww4bSTGPE1xod39By0Xd9rU8EdUZI6Kmr9Nd1K5iWpYHGDwj4a+C07Br1FNEmYBuEAoYGumDIUhosGdM0XEKep88riEtbxeWUYJCGKjS6Bg0uREoO4aDNy7srZG2ThGtSF4JItcSpnVFWLTmOSUun4cfiSs2b6+rZtPYtmuIJDMvCCAVrDMcKsHRGBw898IAylGw6zcMP/Ekz3fPPP0+l4uhzEkDJWj58+JAKLdVSkUgkqmwtGArxuc99jru++S1S9XX6POW53nXXXcr2/vznP9Pa2sqhQ4dUwBEWI1gwI7b+P5/P631KJKN0d/dw/LLlWvK88/Y65s+fr+Lcuo3rIW5Q11xHNVfBKsPOzVu5+PqreXvDOma3TWLta29g2Jbet4nqwajvaPGFvsmilkWsXx5PaJQXVMu/b/zyF9k22Kt0r1oq89rDf6YxFmfmjBl0zupg87atnHzxBXQVcziGqVlCKMbuDZuIFw0+/+nPsGTFcv762OMcONLFvp07+O/7/sCZZ51OtpjVrCygO9RzVBeC0D05SckgkoGFYgjgTll5Oq2T2olIdDJNCpWCnqNSw+6jevPkQch19KdHiIaD2BgaXQujeexohCvf/35ZQUTr64iHwhiux2OPPMKtt32OXz34R3KGTzwRZeO69UQDNsd3zmXTWxuUnmQHBvjozTfr3x9+9FH9Lqn7uru6uONrX6PquiQa6+gdHqJUdpgzbz4P/OYhvnnff7MrU8U2TCpOFcu0CQaCVFyPMadMOj1Ae1sbhVCYV4/m6Q/FKDhQxCPnmf8DRkcAVyTSFMEJevjB8D+D0bAgZBAImZrd7AyUSy6m/FzqLNfHcGtg9KgQjQcpV8pab0t2r4FRNGoReFTHQd4qNanQXgGjlABxSZOAMlopTcdrRTsM5WyJU2M27VhM9X2aqz6JikdTBNYe8hl0pK40SQib8ITOljm68S1eeeURhsjheFUakik2rn2LoNBqyySWTOoaCRkmW95Yq8Hv0N59uk5yY2mWLVumtZ2o7rJWJEMKRSybLgQsLSMO7d6LXYWVK1cqNZWS6flnn9VySIKSAPKyyy7jb3/7mz5fEZAEqFJPfvzjH6di1Fij3CNZX2NjQxy3ZBme65PLFqivq+kuotAfPHSA/Uf3k2prJB6M0n20n6GDB1l5xmm8/dabREJhDRbyvRMFhJYR0Wn1frlaOSa9S1qvT6VUnBFqJjRm76HDysFnT5nK5FkzOX3Vaq33/uv//B9OO2M1Czrn8Pe33mThOWcwJFFJqJbjMLW1jYe+/V/c+q8f5b4HHuAXv/gFw6NjPHDfvQwP9pPNpjFEYDBNvbE79+/TukoCg0Q4iYZNTU0KMrnQzGiRGz72bxw6fFgzqGEben4TmVHeJ3/kQgWMwYBJMhLFlAsvVClWHRVPZnbMIpxKkh0ZVSUvEYnwzPPPkw6aDGYz2KahWVZaG6cdv4JHfno3q849VyPjnj176BsZ1igbCga13pCja9cuPnHrrcydO4e8VyWTL1D24ZW127j9d/ezqccjWDWpAEWpdzAxTYOSP8aUxhQhz2C44tEjCy7tkhcwui4ZyTcSOk0D33AJBDyIeLghUeBqoDjWRvJN7LiPFTSQRGmMepiGieeOA9HztR1QGc1DTNiDqDeO9jfMYBBDRARZvJaFYFYA6fkepiVPAsJBKEkwCcsvRIG1qBY8/LKn2dEM24rMjiRM9j06qiVmh4I0Vy1idpltXT7VUEQiAdWch1v0yPZnmdsU4lOXHsfMhS1UqWJhUEhnmNzWRmZ4hEAsyoEDB7A9X4OWBFhhTXt27VaQSr0uzz2RSOq6kTUsa+LAwBGEmQsZqOYLlIaySjUvvPBCFdrkqvbu3XtM8CkWCuzdt08BJc9106ZNCu5LLrmE4dyYrtGJdkUoHCCfL7Ji+Qns3LGb5qYWbYkINRUmt3HzO4QakjQkG9m1dw+LFi1k29tvEU3Gj9Xt//OXcTHtnGvX+J5fy0SiHslC1qwkPTHf14scHE5rj6+lqYlgIk7KCDHUP0BdcxPZUpZFsztJtbbApBaq0YjWPZK5TM+nriiK2iB9w0Ns3rwZI19maLCfBQvmqU5gBy29cZIZD/f2aM0lFy0AFaFGLl5usGTAatkgHIty3JIlqsYKGCXKySE0Vc5VXifvz1SKhGyLSMDWtkr37sM0T5us0nbbpHbmLF7Evp276Jg+g9bGRq790AeZf8G59I2OEHKrygykp1hJZ1k8qZODBw9y+OBBZs2ezVAhj+l5eE6VquPQ2tbGwX37iNpBCpk0D/7lcXqGhti6azel9tnMveQaBvyEcj6r4jNUrSKEU1Z7KGmR8FxawxYl32VvweBI1ifrmOSrHoWqRalcxpKoLNkpaOAFqzhhqAZqKsw/gTFZxQyYlPIujHoYmtosfM8jZlvkjwxCIERoivQwfeyQBDQIazoEx/XJl4p4VriW8kyhyMpViYSgkC8Qqo/WAF7w8UqGFK2ETIOyWSUp4prv0Zgb45z2emZ4Lm1ykpbB4V6PYAjKrkF+tIpfCdPfnaEhmOfN336BYOkwq885jUq5rMHuiSeeYKx/gIVLl+jaFBYjvWcRwQqZrK6ZCy64gHfeeUeTxfz5C7QHKIesmZfeepFkU6NSS6/i4GYc/V0xm+Oyq66kVCjw7LPPahYUkMrxm9/8hl/+8pe88tLL3HTzp/jFz37OJZddylub1unakkNea5jS3wwTtEMUC2Wlq5Lp5JwWzJvH3559iqaZk2humcRQbx8fvPYDfP9H36dUKhKWKPeew1C1DYzO0xb6suDlC6Q3I6lYJGLJjKJGCcrNQFBRL6lfQBKzI0pP9m3ewYLL38e/3fBRvvLpW7nsyitoXDSP3v5+zY5yYgKM8089gxl1Tbz56hts3riJSibLnV/7Oh+88nJSjUl9XVdXF1OmzdK/mxikR0ePRSHh4XI+Bcm6pTK3feHz7NizWynDxHeMjGaU1sr5SmBIF/IsmN3JM398gGef+ztzFsyn5Ll890c/ZFZnJ8FokIHuo1x03vu0GfvpT3+aU66+jJfWvcHMlnYt9ruOHNHouXj2PLZt3KyCSN6pPbSAaerDFHHIdT0VC3bt2kVdKkm5kKHi2Pzsd/dQOfECXhopaIYj5BMqGgyXK4zZQYyATXQKZEsuyaJBxDTpAYZzDm7FwleqalLOeYRsk5LpEQ4aGLYIPRZO7RkeO6yYUEmRA2C0J4dRDmHYAW3B4Pr4QxnMiE/D3DpGy7W3CTORzGhbWl0SdH29NkmYo2UHXxaKbxIwXJqCAUbTDpV6W7wg+pkirgTG2ZYkcPkqq+qrOaEjaFLa38OqmY1Y+QqGb1MfC9M76lLIVghbYZwxDzc9xI4Hv0LS2UtjW2NNQPE8bUO4XlWfs/xb1mZbU7NmK2EoUtsJg5GALOVMLBzVn7377rtaQgwX01RzBYLJeE1H8Dxd5/I5hdEMH/vYx/X9Dz7wJ6LxmP58bGSUvzz5BB/+8IeJxuNUqg5jmQxusYgZqfUF5Sg7BRpaWhSAKn6WDDpmzMQpl1UMevQvj1F2qyTr6pg6pZ0tm9+lrb2FTDatGV5inzCQXLFANVvSQGicc92FvgBmeHhY071cWGNzsy5yOXHNTLGabC/pXzh2Q6qR41ev4s19u0hLL8yHSYkUZ5x0Mg8+8SSJVIpgOEQ6m8UNmiRDMbq37iRhBmlqaeHKSy4lGQxx9QUXUN/WoFlYM7EVUok6Kr3IQpGKUCTT1EAgEU8XFeiNkhpBHCXykORc8+LsKRbJjI7R2NrC2+vWccdXvsL7zjtPb/L9v/s9f3nmaTrmzWX6rFl0zO3g0J69LJwzl3gwxE2fuJGOVSey5eBuwmaAukSSXDar2bmtrolNL79KqL6JUDRWWxgBm5H+fgKycFxfa0etPfJ54qmkLoax/QPcM5bhr4czOIkEedMlYgbIFj16JavaQZpnQHfaoeqaKgxVpG4v+IR9S0Ho+CZO2iVoW5LgMHGxwob2TKXak0PorgYI+b0nxgWPYjqHZcW09eLJOpTyiTz1k6NkfY+KyLDvAaMIRCLABqqegjHpQb4Kw0X5lpr6V2cHGMu4uA21ha2CkX7u/xzVqoctUcKAhAd2qcrUYECzY6PlEfUNugbTeNiYrkl5sIBZKdNR2MKr93+D/pGjXHHFFRr4BXQVp6xgFBDIeiznC8ydO1d1CekRau/asvS1p6w8Wd8na/iN117DSkVqIBw/pCUy0YiXZ1gey3PKqlNVgf3b088QiUURqiq31ZUWRjSiwplQONdz9DsnWIgdCypzFDorGCnnXM3m4tSpmQRc9m3bxuUfvp5XX3lR63Lh5xLbpkyZov8Sg4strbhykZz0RVdeepo+0QkxQlSkSCKub5CaUTLWSy+/poA4euSIhr5p7VM578rLGZNnoc4Ok0QsRnFkjEvedzGe7/PMc88qqMu2r83afes2UT46yIyZM5V29B7p4tWnn8ISsWH8ECn/+BUrOP+iC5UurHv+RdpmTNOHIAX3oe4uFRhGu49y1TUf0JsnQJQbPDqSJhEKs/GddSxZtpT7H3yYYqXIX/78GDu2bdPPyxcLnHHB+SxasoQFxy3g3XfWccapq8gOj/DVL93OmOVhNCdJhqXOhKlTptDT3a0LbuakqWxYvwnTrkVHcX+4TlWzo2UHVT6f6NmFwinGysMU9xX5wf4uNuZdSjGTQbGR+TV186jjkvENOtoDDDseg56B7XoKMqGYVjWAXZEaDfysj42LYQYolwqk6qMUPB933EUjaqr0acXKZlZMvIpHIipiHBTzJaxYGLfq0dJqkvUL+IEQJU/cIaYuEnHjVGseAYLj/495hvb/HQ+GsmWk2DUJ4lWq0BKo+ei03BQK+z/9SwW4ZEoPrLKvPcF6z2W6AbOiAVKGSV+6SCwZJZv2CVfkHCtcPBPu/JflTJtSr/dXWkaS/WTxTgBK1oGcowg2Qlvl58Le5O+SJKplR9mdAPSBBx7ASoSOPRMB0f8iEpokpFU30QoREAtFvuYDH+CNN97g9/fdpxRVSqJMdky/b8Ic41qeBmAxe0iiitpJ2ppb8OQcLYtoKsGWHdupa2ggFo8wbfpUyhUpx8o41NpIgUi41ipzy5p4jBMvWeVPZEA5KSmMX1v3DgcOHVIeLj8bHk1rVhQeLp7KoG9xcNN2/uOB37H9wD68oM2kmTMUYDOapmhqrm9s5IUXXiDYECcYidK9bQe/+Oq36O0+yqdv/6L24kaOdlMsZo5lPN812btvLz/77T387Pe/5eJTVzO5vZ3bPnYjsxYtUIud1KwCgBs+dL0W363NLXrzX3/9Td54+RV1ahw4eICxcpGdO3dyy8c/oX7AUCKudeuHb7hBs2MwFmLn1m2cd/oZHN67j+9/69sYdXFalswlMzxKJBiivq7uGBibG5rYumkr4XhCI6HcRLHniYpbX9+MGRhvqfui8vpcfOU5PPf6If79kb+yvewxYjuMhWzyIib4JiM+jFRdZkagZMOQaWGIw8S2KUvjXmhkyRMTKEautgirjhhlR2ic2UrW8XHHM6LY/qQ1IcHALIunFkrZghaB0USYctUn4BuU+/sx66NKj92oi1kXrXlNpcco6VNo6jgY5fVRV5r60DtSxC2Lz9JWoHn1NYVV3C8CxIma51gG0oISjKKrPllRTRP5IvNjESbHw1SrLoOZPOFYkkDBpVhyuGJRmL/eeRNdu95SEArVk+uR5rqq8+MmCa/qahklLESUfmlPSHkQMC1aW9v0ucl6fumll0g7eX1WAqD/GxhT0aQCSRLPySefzKYNG9QkIN8rAf57P/gBjz76KJs2v4sdtmuinmRoz6Nj7ixtf8i5qdunajOvc472PoWJpZoa6B8eYvrMmRzpOoQdDOB4DuFIiKppaGZUfUayr1vRNWycdMVZfrpvUD9QLrJULDNQyeDLG8YftjH+oCQyyAekwlEyI6MEDZPe7m6C8Rjf+/kvGXUcBo7W/H5Kdxsb6Zg5i207tnPRmWdz56dvY+0rrzC7cw4fv/lTdMzp4HO33ar1ZbFcJhGNazM8EO2So4IAACAASURBVA5hNtZx3IknqECyZ/sOli5dSu/uA6x9800NEH7FYd/6TYTjcS648EJ+/4d79WZJvSDZ/JOf+pR+v1Cu4cFBvYmS/W+8+WbqJrcTjoY0WFxz2eXkh0b43l3fJ1Wf5JyPX8dfHnlUr0EEITkiwYhmFmkNVMo1v64wAgFIemAQowypZqErIYJ2hJF0jjXnnk7zKedjnnEJ+yzoMas4QZOyRHPDVxGj7LgwNMisuW0MSYPeMKnmx32ilZpN1Ch5WCUDt+Qq+L28T9grUI5buF5YF6WcjzToBR+uZK6qCa5E8SqU8kSaUvrzkGvhCEWVZBY18EMGZsxQ07gkbIWjZFdpW7gmMWGiVZ981aesPUsLX9JlSDzppr6n5jT/56P2OXL+AkafQMUnXHJpKZdI+g7TW5ooVYoEojHscs2B11EPv7zuNLy+bRTHxtQ8LgASdbdaLKkA5ZbKNLS1MnPmTP2dJIhDR44oKxGqOWPqlFqd6bra5J9YvwpqaUlY1DSJceoqr5PS7Hvf+x6f/tTNzJo2U3Eg2VYAJa8T5VXMLnLfrKCFdB6SDSmy42LOBG0tZUssW3oihw910dE5Q9f/WH6MeQvn887aN2hsalJVV7JsX3+vnqMKQZ5PQ2sLIQnGUxbP8nsO9+iFTaC8Zd5kBceEh9SRmzoeLeQETdMiFY7oZIIsdrmgqZNncN11H2KsWNZoNDHRIF5O+f1P/+PHJKJRtdSdecqpnHfhheSGhmiY3MYnPvlJXn39NTZu3UKrFOi9vdzy+c8RaW/hj3/6E7PnzVUb24F3t3P6qlX85ic/U/Flaec8Xn7xJda/u0kLY4mQn7nxk6RamrXBL+dayObUsSPZVI4PXn+99vYamur1AU1raaUhEuNr3/gmpXyWU6+7gueffob2SZP0gUtNMDY8ps1zUSbzuZpbKe9UlKqKqud1D7JwxSl0D4/R1D6F/Zu2ct6F53L2TV/g0IzZHLEM0uJgCUGfAWOiWspCl+yRzbNoaowDJcjLhIXUW2IvFXoqfZCCD3kPr1jFCBqYuSBufz+Ni1sZHqwpJyIETNBDiZ+eUyFoBymVqxgBA1++CJdQIKJtEjGbBwIWjlsi2BhGPOZS+MkaFboqhy0WVfm761EYzUEhrIiVtokbEopV87uqn/V/H1JPymsFkHKNZRe76BEdy1BveExvatDvTiTj+JLtXY+Z0Sq/uu5UYtkamCZ6cJ4ppvSaYCb13LQZs/WZyPoKR4J0d3epniAtBWlJybMREU80EDX+v/cQ1I9nWO2pjoM0NzrGkmXLqJYcNX2PjY0pmEWFlfNYu3Ytl1xxud7LdC5DVSONr+tj4jzdfIVly0/m3U1biMZDWuJ5lsdIelSjjWQ+ISESNA7s3nWMXcl5yJ9UIoFx6Qev8CWyqCBiWWzcuJGmzknax4snk7j4ZLIFRbG2PKSgNSxK+TwJoQTlivbbhrr7dTzKLzkE47UpEGmFCLAkop1y+mpK8hnhEG+//Cp33nknt9/+ZTZueZeL1qxh+OhRrv7Yv/LCSy8y0tev0w37DhzgkSceJ+tWSTU30Xu4m4WLF/Pu5s06fvSDr32TxlCUjRvW861vf5PnnvwrbdOn6Q3y7YBGN8msIuoYrksxn+fsCy5g5qIFjIwNqwLXLL0pw+IrX7+ThGUy6bTlbHz1FRqmTtMMLH3Ogd4ByiWpTS39v/YWgzaO2AfFlHywn+5dRyHVzrPbt+AbFv2797G2d4z0ivlk4jEFYjYIRwyPMcukKuZsbcLDnJTJkar0H72aZa1q1MAoVFX0hJyLnylCJCHyKgwNEW5tojRhbxsfV5PgYxgubm6UaEOCQtEmGDKVFvpacwZrAoT8KTriroCmqPYN3ZCjbSXJdrpkRQgS2iroLjnYJcgNFrBCsXEwBhTAAsYJ7+s/LXy139UseRJ5AkWDWKZAQvyYpRxN7fWUZRwvHJOhEGaGKjz35Q8wsuutfwKjb+rsCThV5syejUut3aWBtpCnv69b/dMyefH0k08ou5M6TtiRMK7/NzDKmpd7IWtemEWlWCKfznHdddfpuvjBD36gdavQ2K7DR/juf/yQV15/hZdefRk0wNWo74RO4BYcli5bya6dezhu6UJNbuveXUeyPqUZVdt1rkdTYyMH9+7Ra5BDvl/YYGNzE0bL1Da/7NeeaqouQTBgUTEkctYkcXEmJNvatF0h0x1Dw8NaD8qClykHcdqIK0eUQKdYpjCY5owL31dzzO/dy1BPv2ZGSf8yTRG2QnpT5fd79u3js5/9rHpFf3333Qylx7j7nntobW/XG/LO3/9BsrWFN995mxfeXsvb69eRaG7SED63o4PXX3yJ+bM7+c2P/5MIZi2SiRMsGCQcj+lDPHP16dwvExM+nHnuOezZt5errr6agWpBa4XeA4dYNn8B3/3ud/U6ZqxaqX3D7Zu3HHNJWL5HMBzRmUyvWPNnWhGbsXQRv2Dw3Tu/w6xF5wlueHPrITo6pmC7RT5y/uVc+8YL9ISg3yiTSwYYCFnHso82v4U+Fgs0TYqSGTWpJkyKw2UigRDlvIefMwhUDZyuMYJtdThZEXQKWBHp9dUAM7EgsDxC1QDlkX3Uz5nJaNojFJEaVPiugWnY6qoRLIpAL+0LJz+kKnG5WiJQH6YaqinYYl4RR5H4YEXmMdwy4YJNdtTEixiYwXHdRtqYsq4MHfZRu54QWkmY8hmmUl0IORDOVckPDDG3PkUobjNqmESqMqdp0m5XeP32aygd3KRe5QAVHC8EXo1WOvk802bOwrSklk8wMjRINBZWNifgu+WWW3jhhb9TLhYZHRtSuporSPFdO1R8MWqZbILliQYykWSyY2NMapmkrRHJjKKyPvHE46qoN7c2U/YdTjrpJP0OAZpY72T9i6ldsFIYzLJy9ZmaGcv5MeYvXUS6kMUPGARDtrbAxobTWIZJbmxMQSjnrpbE8YBgHH/yCv/oQD9Xvf9KzjhzNd/61jepb2pQ8Eiql0bnz3/1az524ydIj4wwqWOWZiqRZaVua4gnOPDmRqy6KB0zZtDddVTfOzHjVRjL6+doT1Ciz2hWb07n3DnasD3v/PP58X/+H7Zu385xS5eyefs2DQJSs5byBU5YvpxXnnqGu37+Uy675BJ++rt7VIgRIAn4Tj1xJTOaWzn7hJNontyuUTosQpPnks/lOPesszUoSPQRwUeu4Utf+Qr7s0MqkR/etYdzVp3GHV/+CulclmI0hBkK6kyiPLSLL76Yo0P9tE2dRjSepP9oL4c3b2OskKVUDRJxEvzk7ocZdBKMFsvkChU62hOkiwWGXIOXyTHQnGA4ZeLGA4y+J1oLKCRaWhWXiEyf7MrQctIURkT5K5k4OR+/aGBUfLz9/QQ7WnAzJl6p5lXVzPMeMAoYAtIucAcw4zEdRw6HA5TELK4mU8kC415WobJWCDvokR9IQySG1RDEjdXAKDdSXqv9aRnRsgyye/uw7CZc28SQP/I7sczJoPL/BqMAoFoDo6iuoSpESz6VdI5kOU9zSyPD2IR8aXIYtIc8nv/omTTmD5ATQ7qkVE88sIZmFaGhApxEPEW5lCdkB9i8aQO+U2HpypWqKSQScV555RXGMsPaEhnsrxkAJsBoihqltsAaDfZ98dmG9PNlRnbn1h3ce++93HHHHZxx1jmcfc6Z3PXd75BKJWozmDI0kc8rLT7WqBf9oFpl9Wln0d87THdXD45bZPXpqzjQc4iS62g5I2u5XKiQz+TV2TXhA5fzEMYpADdCyYg/rWOWznaJJ7OlpRkXTzOiIFesQe9s2Mg1135QF7osaKGJbqDm0fPFR2pa5EZG1ZDd2NCkANZ2w8AAkydNV+BIpFowdx79R/v07yJHS5oXKpxLp0k2NymIJKNJc18GcMWylIon6D94CDsR5/jTT+PA7j30bN5G54L5/OhnP+Wtd97hjY3ruf7K9zOlpZXzzzxL60UjHNIo51YcpcriulCbVKHAh66/nvYl85S/p/sGtD969ZVXYYVDVMW109igooA8/L5Dh/joHV+iIpRXvKx2kO0bNpJO56lLTefa913PtGQnQ44MzXqUrDB7hyv4tkvQ9zjYGePdUJluET1CQbzxptxEbxDHxfYtzK0lInuysCRJvjmA4YtwAdWSh10wcXYeJLJkBtWMofRVaj8Bo4oc4/1XscKJ97VuSpixwRJ2MCzzxlTLUuPIVLzMH9dIpUx2yCRHMACFgyOQTEKdWRtIlIWnvW3xpPra+vArBra0MhwoFKoqs+rirhl8MMazY+09hmbtgFej22IKiDhiGBd6XKFORhMMg1wwroCVaf2kUea2hRE+PLuJaMqnHAiBX9WZUyk35HkIw2pqbKGvt1uN5z3dR5jRMYsVK1boOhUwSpn1/At/U7NANl1QwUbeL+KeZMYJA4EAIF5f61Eq3RSHUlBmT2pgnT51FvsP7OW01avYsGGdTsfIIYlGjmgkpt0FEXtEIAwYQeZ2LkT0lcNd+5k3fw6ZSk5utBoBBIxC22LhGNnRUf0c+W45L3EAqSKbaEr5Mo3efbSLhsa62oS/4WvWkBeow8A3VEb/+je+QUN9vSpP4r8UuijTCrLIew4dVkO2qLHaEzx0SIvYQ9v3Mn12h/Z/ZBaxub7p2PYFcjLnvu99fOa2W1k8fz6nnXsu+w8f0oUmlqxcJs2t3/g6oWSCZ95+Ux/GaDbDeWedTdS0uO/H/0XPW+tZeuKJ/Oq39/DqSy9z3llncebpZ+CFbL3Ig9u287FPfUr7j9LqkOtZvXo1zQtns2TJEoa7e7jo7HM4fdVpNE5q10Uy5JS0RyrXpRMk0RAdK5YzKA8tFCIYjbFk7hLc4RAfPvciXn+9m9bZU/DdKn1Zhx1Vk2zEIho3OdrsMNoYokeEG2nP/bMTSicpkp5B5skjWMMu7rwUkUn1OlVRlG0uhKZmTarb9hA7oZPK/wcYQzMtSlWHSMyg2O+o8aKSFVm2JpcKGCeAKw4ZV7ZDASqHsxCPvgeMIvup+I4VMBSMnhR4kt0CkB3+/wdGU2tfoc4mtuORVGNuhSQOpVKZITdENBAm7MpYlcFsI8O673+JYtfzlAwxPHgE3KoGc6GiCxcuJD2Wxa2W2b9tK9//8Y/44r9/lh/99KfKvg4dOqig/dND99PaJj0/Q51b8hylDeLKf45Dg9g6g0HybuEYVRSBKGjWalFZI8loSte/2NgeeeQhivJe11UTutSjoVD42PYyGtT7R2lvnUoqWc+2He/S2FRPIB48Bka/6tJU38zo0CgjAwOqR0hS0kAgqqrMxSan1Izi5Uyexkmt4+m7ZhuSRqp8kTxEAcfQ0R4e+uMDXPH+q5kyczrFalnpjFysmLmFO4uNTFoCAki5EblKWRuhOv6yeAkiAQsnFyDKe6T9ICARoInoJ6qTuleGhjnnSzczUiwwpbGJqW3tvPTOW7ROn0Y2YKhzIR6wmTV5Cse3T+WWy67BSWe48tLLuOa6a5kybRonLTteM674T5vb27T3JA9BKMK/ffZWgvUpRrt7uOSMs7h0zRraJk3SrH7TFz/Pm9u3sHPbdmK2UFaLkVKJNR/+MOu7DlOfaqbv0GHu++497NkMYTFnRx2cQJB/bD5KJt5AMWWSi8NA2GWsOcpoBDVAKPd77yGqoAimjx/V/VNIxNXTZtVFpDuBWfGwRiUzbqVu1XzGBoX2BZT6iUVNxBn5O5MCWM0ublHSlLhwpA9oitucY+nY+J8s6otRXd4nSfNIHpJRfAFgnUUw7OPITgCS9ZSvSp9SsGkQtkzyfdXafjrj9FRqRvkzMeFR2z6gRlPl/1HHV+dNuFjBKFdoT8UZ7hkjk6tik6Rk5pkctNn8j9f58VfP57Nz2gg1VWsOHQddI9LzlsOybUJGla2bNpBsaAKnRHNjIzfecjMvvfEazbOmct+9d9MkA8B2XDOjgEoNA7at61n3OBIRyK5R8mM1t2/Q2txMMhankM3qWpSALfOPkYYkjbOmEwiHObJ9J5YV0AFz2eNIANXb3cPh3UdZumAZ+/dtJRIPYCaiRGIRXMdVR9mMyTM4vHufzPXrOpPP15abU9IMacTak36+VFA1SvhwTZGrqTwCKuHTLa1t2uiUglU2ePrdvffxH//5Y7q6juhimLggVercmvgjgBL0Zwp5pYtHDhxUUC+et7gm7x44UBuLCUldE9ZhUInaMsQs6V9GXe740ffpWLiAwa5ucoUCZ551Fmdfdglr9++mZ2RYASmAEpoQzlX4wsdu5Hc//DF/+tkvVA1MjFuV0pJNzz+fZ//6V+xoFCeX51ePPEhfPsued7fwgTWXsubcc2mdOlXP5YoPfoDD6RFef/VVXYQys3fSqtP47+ee4yOf/xw7unuxygZf+cg32b+hQEHm+WLQny8QmdZClzSk6ywGzBJdIQFjjJEIOLYYy/4XGD1qYHymi2mdU+nOlPECHpY8RDHSixo5BM6OHbScvoCBXk8XqSeUsVzVbSjsiE1ohknF9tQYIM9DTPXkhBb+38GoGdKRDbJ8jN6aMd+Lmnh1PlbIwBPngA4Z17yrE2A0REwaLqvnVVRCYVF+UFSz2m4d8lp3HIyqpLoQk0RaqSoYA1WXXP+gTIIp3Y3VNWFbDidObeW5h/7GmktPwXrhHl558icM9Bzlquv/TYd3j6mWlTJde7eTK5WIS3+xqUH7ju+79BJGKHP/X/6bWDTM8PrNzJ2/WAN/f18fqbo6tQbKdU5kSs+oKJAmlE0xfctWHiK2JGMxTUZyiHo+lB1l8pzZDI6MsG/bduxIlLAMLseiNLe2qrDXc2iQya3TWPvCX5m1dB4Fv0p9Qx3FvJgCXNqb2tm3bScL5szXz/zLY48TCMpODeNG8VBz1E811Po2gtLalEQtrcshzctwXZ32bHR2DciNpHGcMvF4jLlz5ymwJpqpMqIj6VzrwXhcWyOSIc8591zeevMtyqN55fdCYQWwUoOKHK0RrFSbqRR6KR7DkZEx3SojYgUoypgPBtl8jrLjqIH31PPO5bpP3cjeo91053IaLGY3tnDRSady93/+hCcfeJBYKKzUTQKLfMeEpC17+sgMpmTnc086hTXvO18piESszOAQv3z0IT554ydobm2jtb2J1pY2jvQPsPfQIc7+2MfIlDyuOOmjZA5G8X2beF2MnFXGaYnQXylQbQjiNAc4aHn0Jk2GQv8MxolmsdQqshtP6YktNM07jqFSGULSz7NwpV6T/aHGDJyd+5l6Ric9h2X/F5H7LWlM4svIvVEicVyMouPj+VWiMZucWHzy4sjx8SRDSlYZ3/emJvoIPaoNHNtjJs6hYZIzm8jE3JoRYDwzKiCl1qpthKczhaUBqUeDCkShfoFEULfokLURwNfNvYQWO64UrD4x1yBQdkhUfZoTYfZs3kP+SJpwNKXG8LjrcGDtZoJVlxPOWs2i0Vf52Rcv5ZYvfpn7/nC/tpfUXSS9xtEhbr31k0ydOYPrb/goTj7D1++8k/35NI+89DcCoqKXK+Tf2kKqqVkprmgPGvhVBa3o2pZgFE/ENOlIEpJD9lD6zC23sPXdzezbvfvY/KIay50ig6OjpBobtAZ0KlW1usleT4KbyS1t/P251zlu0XJco0B332GssE1SdncoO9ptSA+lNYAO9Qxw0ZqLNSg89thjuiuiJsGpC2f4MtAph1AB+eJyuTYpPWHgPjg4oDY0cRGIEBKVG10skM2kdYZMaIBsICUq09jwiF6gXlylomk/WyrqfjZzZ8/hzb+/opRUsqOcgGRAmdRWe1nI1hsilEIAXspUCMVk+0GfUFuzzuJhB9QNr1nRMHUAWFZJ07TpfOdnP2Ff71HdKUwiz3XnnM+1513A1q1bVXySACANYd2waHSMPzz7NF19vUxvauG2T96k1yvBx3I9Vp1/HudeeAG/u/deKm4JQ8zgLZMYzefZ23OIj37xToaPJPDHptBYNx3LDhGZGqTS5DNm2PjNAUaj0BVw6ElaZOLmP9HUY2B0ZcrCxNvYTSjWQsGoAVFAIAql5Xg4PQXCYx6VhIlFXDdBsoSK5n08ccJISpoy3hdMGArIatoCyZKyn44/PjD8nqRsyd41VYOqzCfmArT3uXQXBjA6myBmYAQDtfpWe4mipQjrqU1qGGPlmkNALIAS1cMBeSyquE6A0fNlsyVPhZyI48sIJkkXDq1fz/Tps+neMaC7MCycNY137n+EyY2zGMmNsfrMlZS2/Y4LFkf40pe/yfTZ05Vhac3nuuTTQ9x887/x9bu+wxXv/xceuvtuvnDH7fzXk3/GSwS1rVVnhOl94U2ijbWd3mRtqWAybjaXwFwWkTEc1l7ikX37WLR8mQo2xZEMF6y5kGIupyCVQCD1qOy3pP1It4oZDnF4xx4yubw+p6XLllGsOAz0Z+mcdxz7D+8kmgpR9V1ClollWGoj7dp3WEWtkFUbExThSbSUP95/P+F4BGPqcTN9GXVtTNUpZcwW8thyIzHIFCv0DQ4zta6JoUKGUF1M+3h18YTu2jWlqZmR0VG9SZFwWCnqhINBLkA9gQa6R4lkx6VLj6dYqLBp3XodkZLtFaXZJdsVSJbs6e7S9+hWeQMDhKMxNm/eoiqZfEeqsZWi7PcSCemkglMqEE8m9CbLXjelQpnegUFeevMtXn3jdRzfp6WtlcNvvMPP7vpebZuY8SgofcfPfO2rLDlhhW5u9cWbbtabnRkbo62lVjt/+Wt36DYds6bPrO1f43kMjY6wa9t2tm7fzZb9ZZac/AkO78op1aibbhFom0QmZlBKGgSnhdlaLbPt4C5aTj+OnEzKjzfTJ4QUURKlyV/NujSWAqTL+s/aIdMhVaj05YiFkxSlkVkFL1MkKspwScwNHkbYxmwEN+JiRixVPF2hqNL6EHuc7Bwlx3sZsuyVKj8TkaVUIeYFyb+wlZY1i8k5UAr76jjRQQCVYWvjUmoK7y3qNgBGIqTzWjL6KIf6WqW1VDMRkZNMrS0Ol7DjMisRYsv6HUwN1DN8dBhXDOuPv8D0xSsoVEoUK1nswhGanPX07vsH55xzDtvWbSeVipAvF9RgHbFh0C0Rr0vo9i2RKjTMm8W7h/Yo+KuYtBpB4iN5DNtWkUQSi7A0CcSSbCRJyCEGlJqjrPbHdcoKPgFhx/TJqthKLSeBfWRsrHYLDUNZliSE0REZOHbVIlmVnqYd4sRTzmFv127qWsKaMSWICPuoZHP0HehWFVZ2gpsYvbr00kuJR+LcfffdtT1wWprrVc3RreXEw2eHiNoRlq8+ndknr+RLV1/HpI4ZdB/YRfOC+Vrgdu3bj621SUX3/RBwyQOTNodkHnG0y43QaFKpsHjxYvqHhil6Bs0NDWxfv5FUNEIkWtvISTJla3NTbXOfeLzmPzVMjUqS6YrFEsl4HdligdbJkwmEZHetirZDhHfnfY+UbHJkWASqaDP/yZf/wctb32VG+yS+fu2/csNNn+TRvz9LDo/RA4cZ2LmHR599RrfJ+MJHP64ZWx7W0d5erWFlAyrxujZLLRmNqjopSq8tqpsRwIq18ddnNhMLLqb7aIWrblrA/rEq/b5Boc4iH4OhkMvBdIlQR4w9Unc7sgdqbXhbDmGDMg9pV0xCo1CojINRfi1zgY6PM1BUSbwoLVoBo8y/eQZBI4QTrM0ienGHaqvsVGeoDU12iauKRUfAM76P6v8eWxDyqlS15GA5BtMDAXo2DVNaFMKMxmpWOu0zSgk+zlMlAffK/F0AQzayCdY2rJLTnQBjRKyx7wGjIdm3UGJWKsHhbfvJ7e0lnmxRcSy3aSdxO673Y6yQ4d+uXsX6p38CY/t567VXmDxpqjb3W5uTZEZ7qQbE4B6n6JQIBSymtU/m7T07cCM1d5TpGSxqaidaqGqLbP369ZoZRYndvHGj7qs7UX8Ku5qYbxTWJN0EKZuEPk5pbyY9Nqr6xYR1TtbGBOMrVMp6bwcGhhgeGqntlVv1WbFiNZt3b2J6Z+sx7UTAKG61be+8SzyWICa7WIxvcSNs8pr3X1ObUFl65gq/UCxycKCPaz/0IU49bim//vkvWf/G28Qbm3jiH//gyrPPZqyUJ96U0t282yZPYvP6DboplexfKXKxCBOdHR3qV53w68mWd+ILlV6M3Oy+gUF+effv+dIdX9X+XtQyOeHE5XqjnnvuOdpaavteyg2R10fjCf0sqfOkVXJg1yGa2poZ6h/krAvP5fUNb2sLQqKv7twdieJGItimTXVwhMZkihs/eysXnHomP/3xD/nu977H6svWsK23m7GjvcQzBTZu2MDDT/yFL3zqFr3xIjaJOXnFCSfw4j/+wec/++9qoRNDsjxcGcOyAkGCkVTN7ROoI2TP4vbbH+Ku33+Mo2MO+YTNURyyUZtiCIYtePWp56k7/1TGUiHd3mLisAxPptwIyRYWPRUCZlDYpYJe6zTpTAxXFHjlrIwn+fglFyouhh/Etx2tpQMpB39SVLsYYi9zsyJnWlTLtX1URVgR2jnR33RFuhagSvYUOuv5xAjgPN/LpAsmcbBSS3PS0Ff7lxrJZdMyF68rD6EwZiqk3yfVg7DmY2D0XXzTYiiTxxJzSLlCREa4DJtM7wDDe/vF3MrU9lZ63t0CI0USSMYq0z65hTYrx1mnrqCuPka26NAQqfLpC+dz2qqFdDnDeJGgMhEBoxSrQ7J3rOUpGFtiKYY3bueWGz/FU089pQlBWVq5wvlrLtbRKMlWEzvATbQyRATauXkzdjKmyaW/5whTpk5W/UJEINm3RlibvE+EHUkKmXSWctmht6cPaV14WJy06jx2H95JfWvNmSOHCDjix370vgepOq6yGjkHUYmlTHv1pVe58sorMWYe3+lLcSl+zZUnn8y3b71dN8iV5qWMDU2bPI1Nm9frHilSZy0+42T29x5l5rTpOl4kvlNxIAggReAp9I/SNLlFF7UYOPdz7QAAIABJREFUB8QUIEqtRKChgSF2bd/DrV/7sg7/vv3a60ppCukiay6/iO4jh49thCUzazt21ab5BZgalfyAAlO2Zr/6X66mYbLszF3WloRsCmTFo/QPj9ASq+Oqiy7m5z/4oYbsqbNqu4lHrQB7Dh9k9qknqYmgZ/suptc3cvMtt/DxD32I5o5ZGhWFzlxx6WVKryQhFDNZnnr8cc686MLaSM3OHfSPFNVbmS1WeOGBvTz10Cv8dvPv6RstYU2ro1eyb8Rkd85jOJJnppdQZXVXyvingVfTlJ1RpUluERz2sKuy3YZs6yDDxiZVYVQjZWzJghNgLEozXGx1snW/jHIZeKEyNIW0+V7JiWopUx1mrckvmVHAGKiJIMqAx6OBL5MjksbKssNCAO/F3VCMEz5vhqq/6oYSlEm/0ZBWgIdzMIMRT+InzNq+Oe8Bo3RvItrXsBgUt5VsuCoLsOoyPRRj+1N/J9A2k0g4qgtfdpmbJzsKvrOXmBnQbUDDyLC1i+OWmdkxhcxQDwunhNn1yp94c/3DpCa3UCkVWDB/Lms3biBXdYgnIsqmT5l/HG/86c98/KabVZz77a9/QzAaURDJ1otXf/ADPPLwwzoULoZ56fFFhRGFw8xbOE/LI+0iuCXdIiOfydI5p1Nbc+/1k5rj/25saGbP7r0qFO090kV9Qzu9w72EErWtP4QxuGJ0iCc546RV/OiuHxJJ1LLzhOHggvMuUOeQMfekhb4dCCoVLI1mtZ+iozrjJlgpNCcAIT87dfVqdvQcUV4dkqDqODUXSLVmxA5YNpMnTaKQy9Ha0KhCi7xWLuT/YewtoOQq07XRZ+9yr2q3tMXdE2KEACHBLcgZ3IMPBJsZgsNhBOYMg8sAgwQdyOAQCJCQQIS4dpJOu3e5y77reXdVn7n/unfdW2uxAiHprt71vd9rj7AUbe/qxO133okPv/xcuFxfvvwWXD4nwsEIpk6b/L+HJZcT9e5C78l+NpXWh0pE0bNschX5hF2SyWZgc3vl7956/Q147A/3wl1cKpC6wsvpcaHI5Ra5fvLJykcOF8XwVEs7jvT2iMp4aVWV8ALDwUHEOvt0WQnqFWbS2N/RjrPPO0+GSXs3/Iy5x52Iux58FCX19TCaS/HPt7/HpCUTYfUWw68BfRrQZgD2J7KI2TRovCWdwF6KGplNQ4BsA8veBDcQKTjSFtgTGYQ14oO5aNfLP2MoK9A4BhWB5RqXb/QukYyVFWEp1W6A6mRpmoZBwKJ630jYnCgkyDDmPxTFC21pQaFMlAZU1LqA5n/sQcmMUfD7OOQxwuRSkTFogiqyKhriTQOw1JXCYOejMUChrYACMBCppUOtVCrS9YSiyJgtggJyZJOYUurAj5/vAmxWeQZEX/FsNbg86N6yB7acCqfVBpOiy3ZYjQaU15RATQdFxeC0yaVYtqQG446bjaC/D+PHj8OhtmYRrGLlNLluBDZ/sBoumxsPPvwIero6RfOUOGe+KLvCs0j0V6H0ZGvGeQbfSyqbkqDlObLYLUK+ZknJ1Rmze0H6n0FuMdnzw86kfH0yh3hWJ8yeCX84CINJk8SRjsZQYvHgl3U/y4d53oXnSyZktuVZLkz3mbAkM3Z26No2BSIm0QhCxmRPSM+L/GSVb5Qg7qRZ1VEygSDMJtMQIr2vuxvjxk3QqS41NTiwe4/sdjiQYUrm2oAp/vDRZixddg5a+3oxqrIaL/3xb7B6bFLSTpo4cYiCVaCc8IFxGlZWXq1LbESj8vW+/u5b6eVkgGN3Suqn4tsDK1fiL0/8VSZfQ3INwQBqR42STJrMZIQCxUuDl8iG9esxe/48XRUtl0NgsA8xfxhWi024l3wOl199NW6/9VaZ6OY8TixecipOOv1s7D7SjK7eCH5cuwsvvP8ieoM5xCw2hKxZtKsGdKY0DCopJEwWdGfiyDrNSBMuZlRlj2iLaSgOK4jm0giHkphV78S+/hAiRjfSyQRMsEqwxnqTwroQ7RlmM/asRiBryEKxKDA4VGSUtFyGHODIbIbKbZxUcbr9/xKMhf5JESyoETaLAk8M6Hx/H4oWjcVgPKIjeOwmwJxEqdeGviOtcIwYhixvBuon51S5mG3039AAV9KEfbv2STb2jR0jIucMxhrFiLb93RhMxCUYKc/CZ15qtCJ+qBUlDo+srZw2ixjCsJ82WgGPXYHPbsQpIw24f8UynHDhmVj11uuyWqltrJMLNjbgR8eWHTDCCKfBguXLrxcwOYEeXJMxaXCAV5DO4Plm5uKvzGoUt27taB3iMnLdQJC8kZC1UAgLj5kvAUzlcQJavB7f0DyEMUKK1cMPP4yxM6YNBaMEdiaLpfNOwKq3VknLxaw7rGqYnHG2Y3z+mVRaRNKUouFlWiwalzcl2SeVEt2Xwr/z9/mD8KXTVmIorx8mlCaqe3OKykkoNUiZDT1urxzyTT//LJmos70LT/ztSay46XZUN9Zg2pQp2Ll7N077rwuwbssmXfuybwCBtg7kqDaeyciahD8IHZ34/6kCTeHZFXfcIwpz7CtZAjOLbdn2qygL5EjTMRqFaU01OKtHz5SFCVhS1TCspExuRQYpBZTDAT9GjB8vB4DcTY7rCRxu3bEdJ11xuQguf7vmO7jqh2H+3LkYP3wE1GgCe5oOwuctgWp2IGe2IJVR0dQcwiU3X4Hdh4OIFbkQt6noUkSgDRFDFsmEBsVmRCgHxHJASMsibTLAlckitrMXxeUlcLtMOPTTdiy8cAp+2NUDi7NMWjqSlzMD+pjVYLRAoc4ML0qzhjTSoB5LjlU8xYZzumBxIhSHwj9r0A1tdJG4/1UQL3SthWBk52rgJUG5x2QOKUrye0oRdfJSZiMKGN05Mbvx+AzoiPFWUHSUDXmfihHaYAw9uw4ANh/sXhf4zK3FHnmuHkow9g4g0BFFwkQ5SFXcwtg+eIm0ae1CsdMrAtWZTBJ2owFOmxVKOgKvQ0WtW8U/7zpViNgH/Z1IxsOYOXMGwokI/H396DtyFJaUBrNigMNsxylLThZtmQ0bNgz5oFBZrlDJ8Vzw3zks5Jnj7KGju0OyHKf7xLF2d3fCPzgoXEMOuNjfvfzyy7KPZnLg8IVzEZ5HQvdoCtU4cTwSGQ4WdTWIRCiM9GAMsYi+Qxc1w6x+KdBOgMRl9kJkCinehhKNjagoQufHvkSZM4MxyP4zGEV4J5nESacsxY4D++VmEIArW45IVCamqVgCm7/fAHe5Tw4FG2B+YyJrnnrqKQmUSDSGWYuOxcVXX4UPvvgMRR4vPn72RdFr5eFg5iOonC+Wtm2tHVi+fDnsLrf0dE8//bS8jwJYgCLKVHUmI6OouBgPPPQQVtx5l/w8hQxv9bmR46WTyciHQM2e/kBAjHeq6utFU5WHhiPrusY6jJgwEZ98/gWe+vOT2L13r/y/xlEjYbVYxFqOG7WMZoDF7qahHjr7s+hOJFA2ajqa0xpCahYBh0m0bjIWDYmBGFwmB1I5A+jvFVaBqKKhyGJA/7ZOlJWXIJNMwAoDOo+0onRWLcKUJlSt0LIGGAMZaAkNuawiaA/Z85Hc6LTAUuREzqwHGlkSibDeWoj2qVCH/r+CUZNgNhmzyKVMwp8s1gzo+7EdjsYqRM2smnJQHVmkOg+hZMJYBImSI7KG8v05BS0btsFWNExokizvuGMjkNVa7IXJoMGDFAZ37IeasCJpNUsw2ly6hIk9lUO6dxA+u1tWVFo2DYJ6OMCyKTmUum1YMM6JG+dUYPF5S7CvrwXxeBAOG5+Nzhzq2n9YlAM5ILFYbJgzY7b0sqzKKMfIc1UIRj4bnu+SoiLs3Por3njvXdx9990ynedTZenYM6iz/UePHAWH3YKW/S1i/rR5c16yUclJFuUZZOaluPP0WbPQGw7AZKfFA0kJCRE7ywRiONx0eOgiMBvM8nNHQmGceNJiKVkljpzDvARjyO1QEH+lLmUhnfP3C7cJfyAe7lQgjiXLzkRHTw86ezoxtnE4fvpmLeaeeBwO5I1GGMgia2DVwdbz5um+FNt+/RVNR45InX75NVejJeQHrBZMGTMWT9z2e9i9DunLOKW1OzxDw5/rrrtOxIMZqAxwPoS9u/bigw8+kGksHxSX+6zrmf6/Xbt2iKvGh0ssIVn/3Ovx/RDAwBcfKKF+HJVX1dUJQIEQO0o2UO3rqosuxut/ew6btmxBe28P9h08gN5MRtgRMJqhGkxIambsbmqHaitC48z5CLurETCa0GtOI0zjGUKuOqNQNAfiZk3WO8TXxs3UElYQae9DRZHuSpTLpeDzmrF/xx74GuuRdFoQI8jaZEa0KwpVs+qUn0qrQE5FIFzsKDR4VQX9nTGoBpsIVImUI4crnITK8j5Pjypwh2WHqItZsc3k5ZlLEOGTBRIG2AYyiO8fBGwGGEttyDiCKB5RiSg9N3lBpNKospmwf/0hmBwlyHBgJCRGGvcYoVhzsPucsJlU1FhUbPtgLeAsBSONomcmp0MyqzurINTTD6/VCZPBKJQjlneUonBbDXAqBswc7sJ9S+txyn+dgK0HdqG8rAjPP/0U5o4ZA1dFmUyDC9A2f0cPps6cJmsRng0mAa7aIgmdccFzKVq9sbicG2arKdOm4um//g/qRulULfq4yOopPxFFMovzzjtP+kIOedZ89RWMNrMELMtd6ghfcsM1+OSrT6UtEEX+QASL5yxALp7GZ599JkEuTI1QSES1qHrB+Qd/j/GguGp9GvVdCsLFQjnJY/gKMhx8cwUBIAYk/fISwQiWnnsWmpr24q3XXsf5550ncnN2r0cOeGEMnI0ndHhbZ6dQl779bo3A4dpaWnC4uRmfbFiHm353t/goEja14es1oispUpFEmhgMmDt3rgSlr1wvM3nbsWTYt3ufBDhLZ8qwd3d0CBCbHiBd3d0ScIW+NxGNy63LkkZcjDjc0TRcePHFeOfNN2G0W0XNPJXJYPikCWjt6xlyjTplzkK88Pe/y8Rx5KRJ2LZ9Ow4eOYqtO3aKs1BHLIlIFvj8u03421vP4eNtcXRkDRiwKAgbcii2WVDemcBA2oxeJQNSZyMmFSmbrhNsTukShrL3UyGOTNRHbd++HeNOnIIjfWkBMLgtZvjbgyJ5765xIaSlxT3LmM3JsCPQkyDhSPfXIAOf+FLBi+YdqYiYye83ZeXPnkyXARXonaBr0iqyBKfHycg3I7U3AKvXjUSqG+XzqxAi5SvDQMzBRorZ3jYYTB6ktbw+q0FXFqdxjslugN1tgzUegtIaxEB3EFkTq5UMKobVIJZJw6qYYEtm0Xm0VXappMwxIzqMJmlzzZm0aJZOrnXjlduW4ZTFtagb3YilixfhzJNPgtut7yhZLfNzjQ0EcPXya7H6449xx4q7ZIDDs/Ptt99iMKRbtFHWUTIyFZU5rKbwMTVu8gp/uWwWwxvrJXtyMMRgqyirlGRFxs8zzzyDkrJiXLBsGZxFPvywcQNGNQ7Hex99IC1DIp2WwCspKkM2GEV1CaFyX0kpTBDBJRddJHvFH3/8cejzoO6u4qjxaIY8dUS0XTglzU+UCqVqAYrE20QTOI9VZDdYcl5343VY/+P3eOPtt7Bw8YmobqiXQDpy6JBssC0ep9TVpJ1c+JsLUVZZjueeeUZoTry11mzbgsrhDWhuOgQzjNj55RpUVVaCzlhUR2MwcvDDgL55xe3ytTm8ocL37BmzZSnPh0StTXoi8Pf54dCY5qGHHpIHyZ/LZjSj/UizEJrra2uRU1W8++670nvW1NWiqrZGF74dGEBrc7NM/LwV+s4x6Y/oLBGnE3WNDRgY6MeBnbtlR/fIw4/i5ttvweZ9+7D3cD/+582PsfCGFeizV6IzqSDE9UM6hxMcRrQGgENZDUkoGODtzCmoRYWVO76cIsOPBDQkTQpSmRRsFrP0Qr6R9aJ2V1JlQzxKHIoJSR5kq67WRhGrRCgJzcBA1AnE4s34/yMYC+LWWWY0CUYjtFQGpqQCQ0RFotkvOjm+EXbETGkoVBanAp3YtWk4tGE/DMXDqOYqSB3Fpsi6gyuQymqPDKRHqjls+HgjDA4fLHS0ovM0eY8i42KCO6fiwJ59wlW0W62iAk/bBdHODYRhUXKYPboWFQP78NXqP2DLtk048/STsfXnDYilExKEPCcyM/D7UVtZJZe/zeqQ4KHlHz1CVYJDotH/1QPOB6OQjbWcVEg8A7x0R08YiwO792HEmJHC3KBMJ6Vp7r33Xjx8/wPwVZXCSDJCaQmuv+dOaIEwSqvKseJ3dwhSi+d10oQpaNvXhMGuPiFKVNfVyvn7ZPVquQD4fguJjv+ulDSWawywgvwABZeYQZhRePiE00gnYE4gyepQDDAoRhFimj1vLn7Z/DOmTJ6MvsAATl52Ftat/0HKKO7ACCbmTcTjw8NPsG0gGcMcLjpXf4rxdQ1I2qwYM226LE4JFWr0FOHNZ57T5fkM+s1FlA9fPICUPtCJpC4R16UlOG8ZBhUha8yIo8aMkYdK5boPVr0jH1YqmpBJ2Pfff4+XX3wRA36/TNr4QdH5lzdcwdOjtKgYqsko9nJEgCXDEUSTCZGIpyzliOlTJOH09/aJ4QmrhNbNWzHmmIWYNPtYLLn6VnyfsmDTEcLKdJLyDIsKUyaD/RkFUUVF0GxERAFSHKoAKKWddIyW3YrYxnHyx+dGREl4sA/e2lKEYjF4PHaRsaCgVcxPJAxJ3pzOqDqaRvaJBHdzz1AQOdZJwPowK/8rIWssUbmjJERPyYp3Ri6ek4ktgy3dn4ElncPocS7EjBkYWBfHiEtPwmQkGgnYu68DUB06ZYNDHgLcqbeTy6Kk1oFqm4ZtD74P66Qp8kyRjMNXWiFnyOAwoqjUB3Mc2LZ1N4oqfbAabTLh5nSWlnDFVjtiYT+mNtSiOHQYq56/BmedNgPPP/+ysHL4vjn9F95pHhPNSkk8FDPABRdcgI8++kjOMa0ZKDRVAJ3znPMcsWcjK6i8skImm4R0EoHFs1tIUJz0Uz6SL3E9HugXiNwpZ56BmsYGLJkzB+dcsAxjp00Smh8Fll1mB9yqBZFASPRZDx0+gIHB/qFsyOpRbCtSugOcUjuhQQv6I3nqlCJvjlZb/OGYVQRNk9Fl5Zhio+EITly8RFI96/Bde3ajob4WsWxKaHZ2t1W4bhyOEOcqC2HKnxv1gULGpAijfkT1MOz9ZTPOOPVMHDzaKtOvH39Yh97Dzeg9chS5eAIZAUb+r8ZLAVxNDZFlF16IWbPnDjGvKcvwxddfYczYsdiyYzsmjZ8gq5JXnnse3Udb8eJLL0vpzEESHzalFfhwmVGDwQBUh+67JwOPPK2G71uclJM02TGKR8K4GdPRm4xJWU+tTcKtoomMoD80jaapbgT2dOKKv7yAL9CIQYNXnLkqM2Es9DqwKZRCjC5GqoIoLQLyzH8lGkeD24nucEz3KjGbhJgqmk5KBjlZuhuQ5PCGdZwMcHS1hcKLKw0Oazj8FGpTQXefAAJOVRkk+WgU4WIRqOHAICv26WaLIstpIn2CXfzGWd3EM5pGvLMDvmGlGFbJfVwOkXgaHf1+GK0ufRJNdI+RVn5GUd8urjSiwWXDpte+htlRA8Vhhd1qRE1xKbJ+Pwh24A53ECnMm16Hf73zE8p8dTAQ7M3eNRaXvtFnsSDS14V6rwP2wAF89My1WHLKdPy8daee4Qw6kEHWCPkHIbQ/tj1GqwQBLeuJxiHQpLO1TdQkRE7U75eBDAWtmGio7VSg/rEdEkEzAjBoB66qcv6JlGHwFtfWiD19sc+Hrz/5FCNHjUJz21GMmzUNlY21aKeZcDCB1qYjqKXd+O7d8Jb6xOuTXBe+Ckv/WDitM0mqx9ZqyXhmaOrIZlZk4/KoF1mOpjNoa27FsccvlDfo8xaLvAENKgnUptQdHCZMmj0daVNOPhyuG5LpFAb9fdDMJj0QOSjgQTMZBQFRX1eHM+aegA/eelemgOctOx97du0S6cRn//KELG//U4GrwAbhe2Mm9PpKBI3DPRGHPuIX2XwEwZAujEyqS2PNMPy8/icsO/tc8dNoPXpU9lssd/n1ZPxMzRVulf+PF12QSQGj9APfP6lc3qpKwOOUm4wfNBknNqMNgWgMJrNdBj9KSEP/QBQrP9uCl3aEEFXtMGfi+E29CxvbQ0gZTYiZjIiKDbn+tTORKMbUuHGoLSYcuXgehZMht5BrQhre5IBwJA6DTTelye/2hy4sJcWg4HoipYNeRYBGPvX8r/kfkAGch8dxT2xXFNgMlJ40o+VgCGrWrZOD1RyMKQWJjggUsxUGlfIUGShUl5K9pQE51QiNyB76OXIIxCBPhDB2QhnUwwM4tPYoRsybBrtZH/jYFcCHHPoHu9HXH4Kpaji8RTas+ewrFNeMFflFOodlKZbNcr69BeOqStG9ZzsavVFMHh7GI3fdCE9FlUhriEdIfkfOz5utFSs6JpJ923fLIafoGVul2++4Q3o5voSrmMthoK9flNn4a+/gAE499VRpqWSIk6cRCiTTbMbgwAAuvewyOffkvL74/PMyg2BGZnIijveYE6kyYUBjbR3inQNY/dG/MWXKVImXuvoaRGMRYdX8pxBVJKhbGCje2mLN5aDchu4YzGBk3c4g4BsWaFA8IZMkonS4h3nzjbeHGPOZHFXiiOIHJi04BimbIvU3kTFcxts9VmQ0DYPRiPAKOWWTHs5hRzKVwZkLlyA3GIbTZMWhI83CEWttOoTB1nb0dfQOcc/0EkvPlHz4Uv5mNMmMpMP8fuVKgfMxCNs6O7Bx0yZxA+rv7MIbr76GXFpXoC4AdAsGJhLsKrPE/0H6FSsxRXpGKpIzYHg4hk+cIJmRfaqUzskkjJoRHd09sKtWhI+24qqVj2Ln5l+weX0bFv5zDfZ3pRHNqVCbDuDEORPRH4tLL5lQDUhSsp9AbIrrxpMC0NZdGRhEqv6+JBhJ+tZgtKkIJLIiB58y53OBWIAr8LlI+NXRbYTF5Xf/egQSLSXWcHl2O6tKmuRogIlCw3GguzkImNwC/lZSXJMo0KjZGqYNHDNeCkaTKplax61yAqTD7MTlWH5V4C3KYmyVE79+tAvlo8dCS8ZRaTChhqZJuRQmjHQiHgJcxcAPTSEkUzl8/cNOVI+mrXdcdtY+BiTFi5MRpFoPwRjogRbchaXHFeGlJx8XOzw6QYlcRn6iSdcwZjS++Ll6rE5xJr7/wQekdaJCeOHs8O84bDbJdgyEwf4BvPDKy3jyySflzLc1H8W8hceKOzcDmwH3yCOPiArd0qVLcf311+O0U09F/cgREif889RTnXbsXBzubpd1y9lzj8cjj/w3Jk+fLmoWRcUeuayowlgQ1eZ7LfZWiJOWMuPYSVprWwBpLlppvUbPxHhWvgHfaMAfwKy5s2QUS1ymbk+mo274MuZLyXg6JcLETYke6VlYJrFcylp1MSCPwykyekkthVAuI9nXbLDi9BPPxjtPPoOLzj8b3YN+XHzpZfjqiy+QDITw6rMvwetwSlXGr1/gVw4t8/OGKEJmzmQQ7B1AUWWZDG7cZaVYtWoVxo4chVGNjVh+xTVCDI2KFoqmMxv+U1ktb74ju7p86ceMSSk+Yjaz1KPJZjB5wRxZf/BZcYXCPSh70mggLoMtt8uLGdMXYN2336Fzfzdw0v1YetMl2B2II9geh9LdhHPOmIzBYAY5kxV9IQ0B7gZVE1raOnHM+Cp09AxCyXHqaETWnkGOatPUgRWpCwVpKhgSg0p+cUYT0a1sR7f0c77hdniG+WRSmTVRp0XfORIULuytHKFqHAHlPyNNQbide2LdrIYlIp2oqHhu5iwjnIDJTqW8JHJiEcdeMymlMg8WZeuzBsrUm5FDElabCWoshkAkiobyaljTGkqzCiocSTSkuMICLlxwMxxzT0MOacQzAYjEeUklRk6ZhIwhAxssMKb5mSagJFTMn+LD+jdWI9u3Bnt/ek1YNCwn5XIWGfWcUN8KVmv8/Arqa7ws//znP8vwpaevD++99y5sHo9kJp9Pd+ZmFuTfoSxj/aiRQgQ+vPsQvl//Ay66+GKhzZE3+9nna3Dyyadi0cK5UvZySsug4jngNqC1s1NMlzjH+P7rb3DnnXfi0YcfwYRJEyVzSins88Hptug9rcGg6+gYDbo63MnnnqA1HemU4KONMocoHQebBRa2/8B+aUTvvf9e+Yasw1le9vTrLAzJUvlg5OKb06+TLj0HuzpbkHPzm9F/Ioex8+fIcGTr6k9RPZrKzwaEkgnxfPBaivDULffgjZdfQATAzv37cMkll+DDt95GfUmV2LWRCcIdEb9nwbtRbr98tuQDYRnBZpg/FP9MoLcft9/3B6z5+hssXLAA/V29+OKrr/53YpbXHC0EXmGyxf+m9AK/NgdCFHPmop3BSHja2NnToZhNMtFlv8hgjEdiggcdPHgUdz78ONo7B7B986+Iq8OBurNwtKMZsy65EPt6emHJOuFNduGUBWPQEQgiqroR0BQEMgp6+gOIBPw4ZkIDeigzaOK80iiyFjmLnr01g4IkLzl6MBpy8iu3E6U2oKO5A0nKlfe3YcZZs9BjYIWSQlpkp1QYiODhoIDDHiUNc9aEgZYQjFknNYJ1Y1H2y7xkE0YkY5TVJ+hUNCClBGZ2zrGktRhk+EMDG3quMvj94T6oRS5B3xDVVaJZUMqdbhIoQhZ1xhj+tvwPKF1wPvoJ1MuRPG6RnSO1ZQw2VdZWNKShL6aTezxFw9H1X6FeVXBo2/PwuJO6PWAsJu81k9UZQ2xTiI0tZEUGGP/hdJxrDVo5cODy/jvv6P04F/9e3RaAzI6CSnlpWZmY65a5S3DppZfiiSefRFdvD0I9fjz4178J8+iNp/+Gk087Rf7uyy+9BKfLJTo8b61aJQx+9qg7tv4qbmlMDJSRZFvFNQkDkMZFnLaKgZLJBH9Uz87KuZecLpbiTocb76x6D1dOOeKBAAAgAElEQVRfeR0O7j0gUB0G34033oiGEQ3ywwsQQNPkzRVeWYrQmkxY++13mDNzFlS7gnFzZ8FSU47eVFTIoKrRgP3BAZx07RVoqKjGC/feD5B7ZrNh8vhp+PSRZ1BttOLG++5EfyQsZSM5k/6WLmz84UchM1NLhw+fNwkfsmS1fBbjLcjdKF/84aT0yP968umnSamxdMkp+Oezz8FVWqrfmuKdocuD/GcJzL8/srFRBjuffvYZbrz5Zlx34y1Ikz0fi6J+2iRRyiNkqqBQ0N3WC6fNhQvO+S/xCPzrww9j3MxjcGSwEpaxl0LVFPiPHIZ3RBXGTxyPkoZqHNz6AxYvnYFg2oL2WBp91PZSrGjt70cmHMCY4TWIRgJwestFJS4u5lWqBEzCoIl2aVYEcvRBAKuHpIXSKIpcJoH+CHKD3YBdxZiJjYJ37/cHkY5nMKyyGK07euDvycLkKpLsYlNssKaB3h82AsEInLPnIMK9id0se0OWpNQCslqMyPj7Ee/qkL7UWV8Lg9OMmJqBpdSNrE1FmjtLTUFRCnDFc/DEUihKmTCjQcXjt74Ic/0EpIxcRzDQyc63IJlOYO5xM5GKhRDuC4qTs92kYFpDFQ5/9m8ML4vh2T9dCaNXH7RRKd7ONqq/T0pwlvSlxSVyNliaMpns3bcX77333pAszESKS735Bkw2m2TWuKqz93kRi44qS20o8FptKHcVg8TfF158UdqecJ8fU044QQybUj09OPvcc/D7u+6SS4QTW0I89+3Zg+OPW4QD+/YJi5996ttvvy3Tf55bglISsbjQ0kpLdWQaLwEO7Pj9lVPPW6zFk2F0tPWI1Fz9sJEo8nqlduYfFm9zm66oJuiBaBQDAU7D9H4lk9EELP7qSy/jtzfdhGg6BrvZiP5EBAuuuhitgQ7pvSgMHDcbUDN/noyRq0vL4KEQ7Zr1ePiMy3DxcSdJY3Pno/dj069bccPy6/Gvt9/HgV270dHSKmY47Gn5QxWY2gSrF4KJB7BQdvLPiWJdMIhgNCK9ZFtHF2ZOmYqP3v9AegUelv+nYOTPGBv0Cy2ru6dbgAFU3GYw8uw4q8pgc7t02f/eXnku/t4QUtEUbr3pNpme7du9FV0DA6iZcDmO2o5DKkv2vYZs1A+3wwt/Xyewbzde2Xofth/MoD2WRUc0i4xixVF/P8LkKkb9qPXZMG50A9oDSSTA3tIgK4+UziZDTiWvXQ9GrmAM2TiyBiuSXDNwxkNz14E4suGUmMcIgpymi5pJVBOCsSSsHjMq00Dzt/vgHt4oJTwFmVKclNaWIUfVOA5oaBtAe6xsEg6fF2nqQ3BAxEGRKQNHqRdZK5BUMnCwnOYFnwJs8Sy8iSzskQwmlWp47g+vQhk1XYiSqizasyLNYfd54Sp2YvgwH3LJHLRYCiNqS/HW/f+NpePKsWf3O/B370CMSlyUdEkkZPdLwadK+qV4vWg9dFiCkQMcwS6rqiQUnhnS4giTPNB0EO9++KGcj6hR11SV50ekmUrCtgqP0Yz9v+yUvvj8Cy/EJ59/htPOOgWHD7fJJDUWC+JPjz4hfWNhzkIKFquOEfUNUqVxvcZSlhUhZU+ZoTlo5Ln5/PPVMBqtKC33yXulcZS0YCcvO05j3R8NJ5BNK2g60CxlANM18Zp8oy6x8dJHvDz8gVBEyhDutpgVk6Ew/vgXaljeAYfLKdwwDnFCfj/Ouf1qEZNq6e/F3l+34Df//SgOdLaL90VDUSlGJVX8z52/Q7Q3AGdpiXgrllRXgUzq0087U27p3/72t8JzY2Hq5qSX43NOIllC5qle/wkKL2RI/h4fFi+UNH0VoWDrhg06XMmiS7tLVhRJCb1PZl3P8pR8OgLIeTi5DBYD0VIvLF4X7C4nBsMRqJpRIFudB5pxwXmXYNrkSbj7luVwVNfB7TRi8RXP4fPuKkRDKaSjKSnLsnEVxmwaXi2H/h27YKnScN0jN2NTez9iJhtimgHdoRCSmRzSwQgw0I3jZo+H4nOgJ2UA8dmaWd/nxTlUMugKj9lMABaDBRnFJnYccXorWviZGcVJKjIYRiZJbiGRR/yp0zCajJhVbcWG5zfC4BuBbDqC2pkNIrwlhjseK+A0iUAWpCylC1VGL13ZQ4sJYw6qnZqfFmgWTlM1ycxUEKfhqhJPwpVW4UmacOYE4J6rXoFaOUHv9ZSs6NsaDUkY7DZU19YC6YisCxrKXNj67qtQB5qBns1o2vMDPJVliKbi0ubUVlfnK6WEtCUEa/v7+iSzFYZz7GP5/9gX8uJ8/MFHRZTqthW3Yxihj6oOeZMeU9FALagCXZD6rtxDNjcfgb2ygkskFBd5MaaxEU37d8NiVBGJs9+zIEljpWBUWraaknKcdcbZOHrkCN56402pCqdMmyY9I98Dz9xll14k8o/c71JShuLWPKPKqecfLzbiRoMZnZ1d8LiK8O2a7+Fyu6U34ht12HRPgAI2NZZIie6H2EurqjTP1Fq58uqrBNA9a8ZMOIu8UioqWhaBoK4fAo6tS4swdtoUHNi7D4H9zahqHIZAOKzLqyRz4qkw58QTMHf+fFicbllFHDtvPq5ddgG8RR4YqUSd0ie1Bfzsf5apQ3AvsUnTickCqVuwUFyqxo4ZI3hWArIL1LBoIICLLr4EN910k47iaW3BvPnz8c2334pOCjMwx+D1oxsRyabg9nnR1dcPt8WFga5uHDtvIb5+7yM88fen8c83X8Huw62Yf9wCNMy5GRvjY8DRdSLM/qUPhqTO/SQ/jhmEtgMDXbtxyxOXY0s3RB3On0whnEiK4jfLKTXgR2OFE7Xj69ETTiGSpY+jGQZrEsU2G6rtBnz0/HdQzRrOu/4EHB5MYCBlRJbTWg58VMrYG9AfCEONW2BOmZHNJDC3zopvH1sH2MtEVIrM+WRiEOrwKmkzkmaCNfLOp6JORToWqdD6aoRs+2wqBqvPhQwnsFZF9n78jFhBUw3Oyj8dSqLGZEFtJoIPXt2FsJgnUckuDaPVBLOSQuXwkVAsBKQb4DFGge7tmAA/FsypR7L3KK677ho4ikqFcExaE4cs7Nnod8jdOPs+gv/p3yikXiYOgyZZkcHI3/vDHfeIWdFPGzdg4y+/DAWj3vLowSj6NsmkbBR4xtgysR/2WB1wOexw2Uxw2SxwiMSiRYxnd+zahf7OfkydMAWRfj/sVgeaDx8e0hKeMGmSvAeKrPFlsZpkyMPy9blnn5W9u7znm++5WuMhZdomqoLp1Gp24pfNm2CwWHQ2BzlyzET5pjiVyQkincEooF5+A5NZSgf+IDGqZlG4qrwMCmUWOTHSNPh8HiTDgSH7ZM5+wqm4ZEnu8oyq7ntAGUZ+ncsuuRwXX3kFrnjwXly5/Dp44gmsuPwqNO/dL3jV/lBwSBGaEKoCfpYgci7rC2x2/gydbZ1irkOGx+49e/DTN9/JoOgfL74oyJY1X30t6gTffPMN/vH6a4jGYrIEZn/MDMlgHDd1gqDySyvK0dPbD434zVQGdcPqsPL39+G0E47HglMWw1Zaja/XfIn/fvoLvLjdDIPqRTKiIeIPIpfOSKYiVpZ9K3Vq3JoJoaY1uP2N3+ODJj9CtEww2SR5ROIpxEIpKFlVANPhwV4YnEZYHAbEjvQANg/slhIoWVLYKF+RRkg7gJNXnIi97VFkLVYJSpbYDmMChmQYk+tLsfnD/Ti0IQRTcS00UwbZMrLhjVICKx5KGGaREVkLVQeQi+8G2ZFZHdvL8thulsm5rBgoNW4gTkEPRnMO8NDwJsf1SQZ1ahIfrvwQ1tqxspIqZEbNpMFOCfxMEmMnVWLOpAokNn6H7Z+9juXXXgzFmMKX//4EP/y4TiCSZrtRSklWZDGS1s2625igxaIx3cE4L8CdyCZl1cH/x4u3p7UTf1i5UriUjz3+OLx1ZfoQiDvFdFKYJgVgOEH1ooonExXOsAxwUss2EcOsaVPBxVY6p6HXH8DR9k50tfbCZrQiGYzguGMXibmTKNYPDGDVu+/KUIfUQF6u/YN9Q6LMPOfcTnClqCxlmZoXE5a6ldSeUBo9vX3wFBcLVCwSDIn2JF9m8veSmaFgZCDyFiE07P6VK3HFFVcI0sHlcQtaweVwCkCXPSO5guHObkycOgm7tu+ElZo6XL5mMpg0bRrWfvedlMd0KGZw9rd04S/PPI0fDu+HocQHq8eOopwBJVkD/vrQIzoLgXIg8bj4RRKs/tT/PIXlNyzH519/NTQd5HCnzFeCpsOHsfKhB7Fz1y4cM2mqUL5++G6trG34gRBRRFTOyDGjEQqHpSeJxHUhZwb6+GkT0RMYQGXtMHS0d4pNdn1FJW675Va8+tqb6Pf349dfNsLkq0BJkR1de3tx25pB/LIvhq6jA9TwlkEUS+JENAlTzoh4KiErH2vShNjBX/FfT9+AHYMxDHBgRMRNKgctFRNZxXgsKe8pHUnAIvtJAgIdOrg7k4QWjussDToRtx/FjAumoXy4S+QW7eznetP49+OvA5YRUEvqYCpyIqmmxRJOtRqQ4y7Rqk/42D+BrYGqA9i5thAxY7NRTGdkXsBsQh4khyB23QjHzJUHCcbUMKLHBkHfuSjGuRz49O+H0BGn9aBH9HRUYw4ZUxrjqz1Y0FiJYOsmvHnvJQLKuP2uWzFu4mQcbWnC3TffgPJhFRgMDcBp90g1Nm3qVLmwW1qOCq75n2+/JWePlRwxz9u3bEX1iLohdI7IiHb149YVt4uOERPQ9iP7JSjYdvT19VDsQ0fFyPhBv8olGBUNFrMVFrMRbpsFw4cNE9C+1eHEnv0HcbStA9m0imkTp2Lnpq3o6+iRZMTqMhwMYdb8ubBazbKF4LyDajmMtYLqAMnKMsBZfM7xWnV5sUTm0CJcU3GouRWJbA4aKS1m3TJ79MiR6OroxMYfNgomk7ozCpXNDAZpZnmQCQzgF+YN8OD9D2D0pEk4cPAAyuh/fvQoJs6cidbWFiSSMaSyaeEo8oF0tbTgTy88i7suvwYldZWCa00GkxgcHMC7a9fgrueeQuPIGnQcbsM3//gQdaqCqvoahNJJUROg5AclDrhLuuGaa/HF55+LzAJ7RgpTxZNpIT831NXLTTpy0gRcf/U1SESictGQlEycKtHzFVXVkhlZnnLXaDGaEIxFMZHB6O+XKV4gFIZDsaBj/0Fs3rkDMydORkltPZLZHCwOffPuNvlwZE8GngsehNMzDHFmmHhalOVsFg4ZIqIckEzSzFXvz1P+LiDUixOuvwh+Rw6HYiFkKAzN3SBL05wqVumioKq4UGZ3YO8XP8GQNaBi3jTJHlQxkEqEkoCpKEKBXsG/OlU3DG43Atk0VIdOwVKcBglG7roIuVOsBigWA3I04+GQSEvDYNbg8thgMSmIRSgmrROKU2kCJii2nIZqN8FsUmXNYbOmocVNcKpACVIYWLcTLVsHkbOWSnlsNFgQy8XhtCbx0JXHY+XyZUgd3Y0yNxBLp4bs6xlA8+bOwqo3/ynKXBbq+kKv0HheCWUjVpmXwddr1uCkJYtljzpz9ixZzq+8/345/CwJ2YI8//zzci456WRGJCzuT399ElHaTNTXIKkkheys+1PqJAWZKTDTG1XUVFUh3NePRTNn6wOfXA4tXZ3Ys38fkpQbMVrQ29whmOdF8xfJ3523cD7aeruQICVLFNrlbhx6SRCCZyAJ5cSzF2lmNScTH8KACg2tyWrH9l37BBvJZS+zX2V5uZSjxx5zLFbc8lu4vMVIRmNIxeOoqq5GZ3sHvvt+rexTyJ5gZpy/6Hi5eTgE8fp8Ig7LAGNZkqNBIBkLRUXykG59+D6x2v4n5fnDUelf2Vu1NR3GXW++jN0tB0RJACkN993yW4w1O1FaU41wLoO64Y0IDgxKP8ags3o9qB45XEbcg719aGttByip5/bIQpk26OXDqvH0U3+Xsov7VLLCeZlkcpoEI/sLlrsMRi76G0c3gl6WhNvRzz4Xy+CPjzyKaTNnYN78Y1FcUiZ7PbvLAYvJgNZdTVi/oxNv/9CGNS1+ZCxlsKa9EoQiLACT/Dv772RCXzwrkbR4QJRZjGj99WcsueVyFNW4hfvIvTzbNgp1Hz3Sh+aPfoKpohJGq03kUTU1hVxvL1BZAaPdhkwipl/wIgXOCbIGR205EoY0NKsJOSMnxXZB1LAsJo2KJGNiYrOGjOwRS8psCEUZ3GYpPWkLbqZ/CiHLRiPSBsoWZ2B2GGAxa7CpOTiyKoqsMUwudeK5216G3ToWBqMdYQaUWYXdksZvThiLb158AE3rPobVksLIhjr0BgPwD/Rj4cKFEpD8PDqPHMXKhx+Ew2nFPXffjWPnzMWP33+PQCgkxqaffvKJsOQZOP7AoCQCloZUeCN+lxbhHJYUdJh48ZJ5wb35C889h9ffeEP8QTUzdWBNUjJKMArPU5+ycjZhMxnEW6Ol6RCOnT5TTJ4oublhy2aRcokkUjK9Drb1YjDgx/DGEfJ32YNW1tVANRNnnJW+9P8pGGV+cdzpCzSnVbf9Zm3Nkox2YSS67dnXBLPDjqxqxDGzZoFwo8ryCvS29+KG62/E7MnT5QdubWlBbc0w8Vzk1Oj444/HsmXLBG1w5rLz4HC7RKump6MDJWVlCAT8YjXGH5jE5oIc49U334DthgRykRiOHzMB3/y4Di0cV/uD6GtvxYWPrkRfVxesLje8FhuWzluEe65ZDqfdLotTIlGohsYsLno2JqOUwYSaZaw2eEuKYUlnUWwwo3nPPrz67iqRSuCQgeuTJ554QlcRiydE0oMXiFhlpzOYPGM6tu/ZjtLqCvlglXgaZ51yJuYumI+33noLG3/eBIvVLkyGNNFCyODTjz7FE0/+GZ99cwj3fvsr/v1VE1J+m4zyybQI+EPiN8H/NhmtcounE2mkuWynWDM/E4MJLpOGwVQUSadJAs/l8SGbU0Qr1mQxCwdTSisuGynklu/j+QwItWONSgYKmfCaMQPVpsLp8SGZY4ZUkeMumMwPBqFJQzyTgq/aAlLrEvG0/KqkorCnYzh2TBneeehf1I3ASeeeClcl0NyThdFuQGUZqwGg+8fD+ObPb8E2ZiGylLXgJZFV4FMCuPSkGdjz04dY8/wDKKmzIRyNomFYPYbX1eCGq5ajuLpKLkbyVAkh8zjduPKqK5BLROTSXn7lVcJl5SEnb5bSix//619yDidPmSQSLSTyPvbYY+gbGMB9990nswD2ghygMLAI2GAlx6nt8muvw+nnno0HH3sYp599GgLBIIpKS5AgNYY43/y03mE2yvnp7uxCsc2OeTNnyeCRXEYO+QIRWr5psKomlBSVorOtQ+9Fo1GU19bAQD9LI1UXaDybt3/Pg1iotCHxP2XRdM1rN8ohLCv2SsB5vEXykGiI2nTwEAZj+nJ03txj5YAsOuZYvP3WKnz35Rr4Skvw3HPPSTl4/vnnw+5wyhcODA7qpcOiRWgcO1boQFNnzsBn/1oNG3sORUNKy8jivrArzIajOOueWzGYTkKxGsVaPGcyyhL/oosuwkfvvy/9AG+ucCSG4qIyuaX2fLMWX67+WAZOSWIuWSqqtGTLl4Iwoq+nR/daSCQkU27cuwutvT3SG1kocltcifriUmz59VeR9WB237hxg6Six19/FWectARTJo7F+OlTEO0LoqayGmecepow/z9e/YkcDE73CuDz+1b+AR9/tBrff/0Z2oJp3P/pIPYdTiGXcCAYjKDf3wWnywNNdSMSjiISiYIACrI/RG0vP8E1GK3IdB7BaXefjZ/9A8h05xBVskinaTKqq/iJTCYV/cTzLZ8JKW2psqfmOkLfwfIz5DpCS6Vgd3sR4xQ8l0ZtwzD0K2nZWzpLLbC4zMgYYjBlDcgEoqh1FaEyAaz+75dgqZuJrOJARs0KcJx9Xzqpe1WQzuVWuT9UEaaCA+8BswXjh9ei/fsv0f79gzB4InA4fEjxUBJPS6JBKAibxYCBvh6Ul5TjsquuxqiRY5CIBvHCc89gz8EDMm3k5UjPS6LAeru6JONfd+MNuPrqq7Ft2zYBYlx44YWCeCEFiqUfg48DPZa17BWZdDhjeOCBBxAaCODZF56Dq6wIqsWI9iOHZB1BVs8lV12Oju5uVA6rkL6epbDb40N3dy+smopTjjsOnf19oocUzrty69VtDjazVYaeA529MJosYmREO3Fq/ujQO13kjT8TQRp08+a8Qhk1e5xWU14kMCeP0wG3y4FEUncl4g9DtLm1qESMPjzuIrmR9m3eJZmjsa4RoUhEmBOvvPKK7oGe0wQHWBh6UD28vKoKew814aJLLxFDyndefw3xeAxJ2un8h0WaTTUh5B/E+SvvwLc7t2B4dbVMeb0ej9xsdfX1goagn8LAYAAV5dUSZOcevxhXL1qMCbNmIIwcqisq0d/VgWgkjm3btouGDgOFi2BO1/jPQDoplw37wsH+foyrqkO9r1iC8edftwoQe9y0qagZORyb1v+EB+68G3VV5Rg7Ywq6DrTgzTfflAAkQsnl9ooOCksrftCnLF0qCmKUe0j6/dg5qOGxzwbR3sISxYxDh1swddZEqAYVO/d1QMuqUqoyIEnDkgBjv5pIyLDCGIki0bQV5758M37aF0AsbRKaEmUuCjLxsnbi+DU/dODnl+LqIG+Bx99nOcrLgn+HfavdZobXakX74UNAd4vITpaVFkl1k0nnEOzpAwYCok7uq5sKf9Av01WQZ0qz3DxDxGDS1x7URB7m9KBn6y543S6EAv2ItbcCiTQmTB6BA2ufgSXbgqydEL2YHMzqsiopAQ/t3gmz045UIIgftmzDe+9/iEXz52DZ6afCVV6mX9qkVecNmvhsxAQ1HJQdHQ/zb2+9Wc4JPwdOLpkhWeq+v+o9mGxmFJWWy+fD/090zMUX/gY2mxUH25rx0muvwOV2SBVES/Frll8rWfKXrT8jEo/KxcHKJxSKQIkmcf7ZZ+FIaws279gu4JJc3pG6IOxGwTN/d78QnOPJKIqqKoYMeK3c2eZtxPm51JZXy+WvlI+p1oqL3CjyesDWdVh11RA8qEBNWbdpK4qLSzCicbSQc7PxDH539+/w+KOPo6OtHet/3ig3Eg8nG3p+owJaRvRzFNqGj5ZDtvjEk/DMn/4Iu9uFlJKVcrLwMluoBg4Eenpx7m3L0RYJSMa2OHSlbwZ548gRcgHwxolFk8IjIzPj8VvuQp3JjGHjx4j7sN2kwqCa0NbWjj8+/mccam0R6FHBaYsK0D63W2y8Tz3lFLQcOYqqklJ5K9t37cI/31mF1Wu+xqvvvA1rMoMLzjoLpZWlMDpt6NpxAO/++99CyeLt5nC6pUfmh8zbmWY/lFTgoTZlTfj+UD/ueH0/+tuyiIT8WHjcfDR39kAzmdDRFUZZWQV6uvp1sHIkjmg8LigfXhIepwfhSALeTBremgSSY+oQhVtWCsmsHriFF33+Cp+ZBCPScmALdB1CGAorKitvbJbCuazsBmtcdNSNou+oH2bVipQhKbQPm8GMcCCsl1kUTs6moFBig6hZskZk32iAlknJXKHGbMeeNethtZhhIkKHCuTkROYyqC82osgSwo/vPg57SRxOJ9cTSYwbM0rcgV0OG+bPPgbzFp0IX1EJxgyvx8lLFsNdXCS4Ui7QBfCT97Dgzz1rzmwRAC7xFaG/rR1vvPMGLrn4EumVx00cJ+UoPTu4h/xp42bJiGzHmC0vv+QyrLznHplAV44dLntTtjhUGaDZKpX2VeKC8/1jW3sngsEwXIoJ1153jVR+v27fprfl+WDkZSC0QyWHTDyD5GAANaNHyp8Zkjw16EwTkWmkb2Q2J2dTNHConDxx/HjYrEaUlxTJ6HrIwy6XQ68/hGAgJHW/QIuOduDVV17FsrOXCfh6yQknClpdxKqyOfFf54fOB0CCcQEdc9uKFdi7Zx96WlswefIkvPj6K1LrFxb1GWrxWMh1NCAaGMTwqZMwfuZ0xMxGGOxW1FRWIRDWuYqTp0zD2rXrUFlWJr/f3HQEz9/9AEr4gxZT01Kvy6kWlg1FddQJ/zHqFwUDnGVOsG9AF/qly5ICfeo7MIBzzlsmpOjf//73mD5pEsZMnIgM0ghGQuhv7RFEPi8H9o/r1m+Qcqi3uRn3/+lPMl2+d8UKeCsrYTNYkfWUYem1q/Dh55txw3Xn4Y2312PEpDHoD8TgK69Ae1uHVB0EuqfTGtxFRfDHIgiFo0ikMrIyMGVySB3ZhzNvOQeb43HESdlQ9CxXeGXyCBL+t0xmlfTQnkwuPTIu8npGDptDZgMmkckALG4gkdDlKzq6emBKuTjaFKfhdDQBpJNiKcARKxUBhASQ92MkbdluMgo7v2vteii+Mpi5o+TAxmSQw0aDGpNcBmkcP6Me/3jgYtSNdUExJ9Dd0YniYh/8A31Cn/rN5VehvKIKXocVK267FUabVUpUktYzaV1oWDSSMhn4Sosl2/FgJwMDIuJ8/m+WCemhAPpg78nPB4pV+lFWMRzsNB0+gksvvBDX33AdxkydhEQqLtKj3GMWVZQKLS2VS8LldcvfiSdSUqaOqqqF3eeW5MM+lI8hkw9GBhmrTA7DTLSCdbj0dYlAR/PDoLzHh7QjXAtxNsF/99SVaEQfUISnsrwM9dU1qKmpQCgcgINlQzoJh9WDDT+sQ2lZFfYfahKjyPqGERhWUYXl1yyX5XlvnkdG1M4QAiKdwfmXX40X/v4PINgHT7kdwcEEZs6Yhs3bt8LhdcLm1D3zCumdkg1csvMHjKW47M7isuuXQy32Im3ggUmIfgoX2cfNORarV6+WEoUDiUyvH4/ddgeqjBZ4a6tlfaKwhyGTPd8zMVPzgyHMjbcRFeP4QdKQhv/dcegQ5p1yMkaPGIl9e/di97btqBs5Ah3dXRg9epQMfObNOAZNBw4g5A8IjcZbUSqooBMWLcKDK+/D3t27MYVKA8R48lubTLj8+r/DPRq9gFAAACAASURBVPUyvP/Rt/CWlsDpK4JqdumOtSazSC9QliJJZQFeDlQ2j1EqUxMQBaEVhlQOkYF9WHLN6dg5EEIqa4KReNVUFvFsDg67WTRhCeWTXWEuK7e6yUY1dj1rOp1Ub9CgZjKSBTJaBiZFRdpAnCmjnm7BaditJqRiOQTbB+FUbEgncrBoKoL9vfBYrIIV5udD/wGzzQrVH4ItlUVvRycyJEpS8tBgpNkdTKqCZCQARx7QblczaKyw4ptXfouquhQoom216GgvclAj0YQIjL337tv48qs1utoCByksVTMJwX4yaIlxTrBipg6rlkUmGcMpJy0R1BaNaAjZGwyEpMXi3yWXk59/++EWPPXCMzhm6kyZfbD9ufOeuzFuwgQpvakmntOyMv1nRma/d7jlEFTFhFgoC6/HIV+bdMPe3h4Bz8cFqGGSJCbVCjVocznZsycSGZ1zadOFurlnpL4tn44pqyAcDKNheMP/PRi58ypyu1FS4kVjYwPMbGo72lBSVI4Du/eKcrSQXal2VlEli/QPX3kDxZWV0ktKBqSjrcEgWEGuCXJxDfUnXIyOUAqewVb4m3fA6VZRUU8LgLhIxvPGkGVoHu1QyJRE6JAWQ5dak8+D0mFVIjBk8roRJ6QsDXiKfJg6fTo+W/05KmqqMdDRiT/fdhcaS0tR5vEiTiQOb9W8FQE/DI69w36/TCGpDkDMoDGdw0B/H37p7cAVZ50rB5SSevRaCMR0nz5a263/+mucsex8GXSRutN08CCOmT8XqseFjZt1Ueb33l6F5pajOHjkCJKxFN58+Rl0Nccx/NSnkLOVYNK0qbIuiEZz4vFHe4UMcb60eSPoIpkU7mX3QD8sZptk30g0hORgL6oqytG58yecde/l2NTFzKcgGtSHP5RsZEtgobB0XsmAwZhRsnC7XUIM5rNQuTejZAVLKigkdgg3kl43VP9m76dl9Vvbyl4pB7Qf7IE5Z4YWj0kfS8I4idckAFvjWfTu3S8ADMpuFiohDm9Eh1RayoxooXLZbyUdLZHCohkleOfPpyGT0i9EZvl5c+YINpqHmno1hLqJazZ9MQgkAHDq6afjb0//HcU+D4xFbmQyKVRWl6NhWA38vCxcLqToWuZyoLOLgzsH/MEQKquqhpg/zJwePttIHFdcfiWKvMUio3LyaUtQUl6Cypoq2TH3cjBXUqyvO7JmjKqbIRuApraf0NZO5+60aKRyW8BzXICMMosXeJWkp3E9Ris4YRelM1A0Ba2HmrHz1x1o2t+EM848Qw9G3QeJAHwV2WQSXo8bVdUVmDBhPAdkCIeiCA4G0NzeBW9lGQaTcVSVV6K2tAKfvL8ayZgOuJVanrdxvo/hr/VlpQgnvbCOmYNwRkFjkRGbVj0PW4kLmXQc3hrvUKklXEKm9EIfxOEF9MN51jlno6F+ON546y3UjB2Ncy65CNv37BXsIFEpo0aOxdqfN8rOz5cBWjf8grf+/qzIYBD3yk1KITty0vvpx6sFJcQbzelx4+WXXsHKp57Gjj3b8OSKu+VAcNAjOitmMxYsWCB9IHsNqgZw2knhqp/XrcPc44/Dlp075D2x3/vkzbd1zpyiYc6SpTBGQ6hwl2DJdW/gna92o7i0Bom0EUdbu2G3upGg6YyqZ+uUxSRCygQPC4yd9JpcSnZ+WSWJgc4+mNNZBJq2YeryKxFMZ5DSFJg14lB1axAenHhSl5Jnb+kuciKTzcFgVAWkIQJOiQQ8LisSkRTc/EtULVCAEPmBVmq/6HbilP13mQxI9KQx0NoHH4cYkZjYCBA5RZRNX9NRXSRYywkOnZ+jZIhUWsDWJH7brRQnI1IoJbtI6huN8kXw2Z9Ohr3ILhUPz8+MadMkAPnst/zyC+xet1zSzIyifBAchOKwS4AEBgZgJANHhLWAOVQYDwxK+8HWhf1qlLBN1YhUJiurCK41qFXK/p6Z6YJlF2DGpKl44L4H0dHWhsf+9CjOPu8cvPDCS3jsgYcxYtJ46VUTWkY0aw2ZIokHxd4t33cYL4AAzVT1PTHPLv/hyonPny8GIy89GkQRP1vu8CAQCKG3sweLFp6Ajz/8QPwmFWeNT0ukqLdJKJNRaCBs1V1uJyqryqW3Y2NeVlGJZ195DbYiD6pqq5GkzP2WbTDlTEgm9N6E2YNIFb4Kpad4Kgb7YHI0IF0zF5rNhituvQ6t277GFw+vgLXWN6RLwkPCYCzcrDoMS5dhZya65Nz/wrU33ID3vvkST7z8As648AIMHzUSnd3dcviGDW8U6cbykhLcc9U1mF5SiWKnSz/U+ReDi2V0X0sbrrvtVviqKqQ8SeY0TJkwAStvuhU/b9woGrBipsm9Y18fnnj6aay44QasuPdebN+2Qxy4woEgzlu2TJTR/vqXv0gAzj5hEQxOBzb8vBFWpxOVJcVo3rRdnGxXfz+A179qRSTMfjoMu9eLeDiHJD0bDRYZ2acdVvECIfld1A3MRniKbMKWSGfTGOgL6MY3RAgZYqifOQP9mTTsZgdiJq4a2EvquEnavzHAqc1i4iCC5Sk9Jtg/JpOw0ZQmnkAJSzq6zClAgDIqNKIl3cpMaJwqInPJngi8Jidi/RGE/GHpxen4vPGzL2C22GGgoa3u7CGHUahJVAQnvpl7Su7YNIIboqgmDC8VwPLTRuGeC0bJWkbWLpS5Ly3F4sWLsW7dOlnMc+3gLS6CIU91qhndgJ/Xr5epp+BTud/jmkTNYVRjAxxWs+ydyYcVvR7uSlP0akzCVVwkFyovCp4DriCK3UX444OPwO30YNn552HKrKlYt2Ed7r7tLjmHa3/4AfsOHEDOnhPkUNQPsZVPJP3SwvEZM2EJ/jrfA8qQjIOxPMeWbmBMBsFAAKlevwzGmAhOXnoqnn32Baz/cS1279sNxVjKrYlpKCPJFzRkh5jPDLBTTliMnoEBfPjJvzFz7hwEIiG4TISrheCxu7H2ux8wZfJU9PcNIhDyS+/D0iKVy+rE4FAIiVAay25cic+3tCJWUYsrf3sufju7AvOmTBga4BCjaVZdsNp4u2riWSjVAXUtBTYEQQoRgzh89Cg88uJL6E0nBDC+5KSTpYfgwtZqs6Fp5x70btiC5p+3SI9JRgIfEKlXeoZURWDo5VdeQUtbGxpGjtCnsg89jCglHYz6VFigfYEAbr19BT779FM0jBghfE4agPK9jJ80CX+44w44y8gooKdGHKNnTMeCpSfh3VWrEA4HUUmUTFzDxFETcd/fX8MjL+5GLJtDWWUj+lpjorSWzqVRWl6K/ngUUToPx9LIOWyyTLc6VKQzCbi8JYIZjgSCsveKB/thI1u9fhhCqgsO2hlQjsOol5vMQPowR5fUYEAbMll47BYoSU0+H1c2jgZWKYEIEgYLOtIK4ioHJWHkrDYxpfXmsnDmDFBTabR1DGCUtRzhnkGs/XItnD4fMirxnKqsiiSwiLIhgTcc0ftztjbc/dIq3qiirMKLXGoAqe/vRtP+TUMVi5jqTpiMpkMHMGLkcOzfvw+paBjHX3cZNv68XsStefIdFis6m49KBuTZ5cCEPUu5zycgD7Yh5C82NA6X7MSLgZlwzfp1Q0wfyd4WmwgYlDm9OPfMc/DEX/+KhJoRJzaKHrmtTtTXDBNjJCq+GS0mxJIxlJSVIB7wo7W9HU6fPqEnOZpBLrqr+WFNIRg5shCtHpMJXe0dcGV1j0dio7/85EtUjhkm71EpHVWpEfpVIOxKMHLUW1k5FOmJWALHL16Mo22tcuA4Kett74CDuFWjWeQJmbobG4bj3598In0chwD8sO1ml1iqsf8iQ+GEe/6EE665Br9fMBLjx49BS0/zUNYyqxpig0kxPOEHJ0zw/ItBQUA73x8nWxzcEAJmo3DsQw/gyNadOOPkU9Ax2I9jTzwBlopyaKEIHrvpNpw171j8umsHunt7hJnNF9tT3sT8ObmOKKusxIUXXIBFM2fBQ7B6TqeMMSuPnTBBdk9Hm5rwt5dfwtZNm2HOUVndKHblew8cEBgVexoevkgwiHOvvBwR2qfl0hhRW4fvP/8GJsWMZE8vJs1ahskXXYmj3Un4YkbMOKYemTiwa9MROEc14s3Pd0Kx+mB1WmByWuX2tdiMArliQ0e4Hw+322hB69FmDA50YtYZS9CVpgaNU89MFiOieb0qEYsSR7gcXCYVaioOb4kNfn8co902GOMJvee02tAWo3dpGjljUr6Hy2FBvd0CtyGDRC4Be9aJ51e8iPLxUxAKRGXgwaGPib4XJJGbjIKuElQVoXIsL5mtWR7nMZ7JVAjXXjAT7z58MpoP7dKnkRzxsypSdKnH0rISlJQUoz8WhHt0o3BBu7u6MHbUGGz4bq18gEZqA9EdTwaQSXidDtl1E9MqezyrXVBg4tfS1oaBcEjeF78XJ+q0qUc0iWFllTj5xKVobGzErb9bAY2INE5/ObCJRHDO2WcLIGHjll/g4C5S0ZCIhIW3O2n6NBFpW/3Jx7LHLrgbS+a16YapNCtiqc5nwYEhRSVZLvMcWxiE+Y2C7BmjybjOfPB4pCcq+GQUmlFOT6myfe8DDwgWMMbJKRWlTTqOT/q8LPDaq//E9EnT4assHcqMlP+jUjipN+4yL1p2HgKqKlA/ogT9/SHOu4cyI6FXFa5qpBNB+Hs7kc2mZVJa6CVZc/Ow/1+UvQWUXGXWNbxvuVd1V7tb3EOcGIEkENwZfLDB3QYGGGSwYSDIwGADg0uwAUJwCISJEXdPu1eXe91/7XPrNnnnf79vra/WympIqqvufe5zznNkn715Xbu2bcNTi/8ukLdhUyfh+CMW4I1XXpVc4t333hPsKMlr/3Tb7Zg/azbaWlskhFs0fwHMHiIhsjJPxpBiy8ZNmDxtKoaMGom//Oluac0wROWakI6RalhrVq7ErPlHoWnkCE38pKMLzbt244233sIlV16hFTPyBSiGT9TNKB0xFGOnT8bkCRPx7OIntVJ5Cojv70BdwxB89OXXaPIWwegywOf2wucoxt49+3DPl1348D/dUr2zFREmp3ldOjc6B24QYoRzliQ8rhRmD6/GL/98HXMuuRgvf7QGxcPGSojJGUOGmUTrCNzRaoQtk4XPxikEg+B46xxmWE1AIJRFNGsQ+o9gMoe4UZGhYJ8nhlGNCqZW2XHPDUuxYcVBeIeP5givbGqLXeML4sZkfi4zrGxh5KupUpgjd0+KbOSUYAfiiQGcMjWLT165FV37d8NMTUazlrfWVNfhwMF9kiaFQkGcdv7vEFKyWL92FU478wwsfvhRKAxZya7EkzGj8fKwLceWHJ0r+7s0tqxikBOIBsKTksMH3Et6GhRPZ0hSgGKXFw3VdaLzsf3gbiQUVkz96GjtEAJtorZOOnqhzKg+/ezT8JcUIRQJSSTGPJn2ks6mNJRQfmBdgBv5thOhiyJ7wgIUU4I8EfKgA2LFjNEajdHIB2K1ysYU1IdRaxTrhDk9ZOu2WuUEWfHjcgwtKZVeGiFGpORgc51EVtu2bUekP4aSmgpJaLl51GwCTluhUNqHE/1oGN6IvoE+YTUjqS3pjwZnxkxFcJsK0bp/I0ykRFBziEYoojpRKp4CdObIFSBKUAf2t+CBP9+Lddu34rgzzhQ84l/uugd333knTli4CE889DB++uFHbNq6RZqMxxy7CA6zVvGbOWu2eEsJJbI5nH322YDTjuPnL8CU0WMw++iFYoxkK08EgmKUniI/dhzcr0UH6Sw2rV4jVWPKAPAk5d8zZ+Kic/HPuOhCJK1GfPDe+6ivqkCEzOUwI2BJwxk3Qo0lBaTuLvCjk/dnzKFAsWHP1iDu+vIAPlm7B6rZpWkusjQvU01kZYhI2HPW4WXY9u2/8Nafr4SjoA6xQD8ufOYTRGrHY38oDKPJL+V8noxME6PhOMqdNhQ4VBQYVZFx79u3G0PGDEVXP9sjBvTEVQzEyCyXxJBCB3wGEx6/6GaUDz8CHSGO77vhsnMImB4dcFBvkt/BEQ8zna5AXOVFs2Tfjn1b7h+SQas89Yrc2Pjccagc5ZH8bs+OnXJfLNwUF5WiP9Ar7QpGBF3dHUiRFLm4AAmK4jpc0m9k+CnUGvkwle2G2toaeV7cy6I1ykqxwyFUGXRibFXw4GDIyr2dM5ulXRTs6MGNV12LqopKXHjlJciK6pZNeGSljgEFLVu24d4H75Ve5A8rfkJGyYkxsjjIfUJQBB2P3gnQ6yb8SZk9CVJYjxHssKZqrLOVm8wa7adSObVRFf3lPEKAF8sTSsr/nMDPZGC2m5BNGaCQWTuZgN2gla/53nhCI6riRchNWmwySU0SY47D5DLEKmZB9Efj6GE42Ns+WHUSQ1AJnmUIZILL0QhVdaKqyI8tn78lyA6TiaMqRowc0ojWWLec3vw9FpucZguKSkpRM2w4FKtNSLReffVVQVj4rVbMmzIDNpMVLq8P0yZNllO/qKxUuC6pMEXDFlIpRZHkmtr25LqhFsLFF18s904Px8W+94H7JZwg4n/u705GTVExSu0uPPn44zA5NbJlyRVI0UH6QspgBwYwas4smQklhQmxifG+gGw6fiZ/J53UUDKylkR8qArMOSOat7fhwkdfR/m4RfhyQ6dM/jMiTClm1PuMGGtpwxN/OA72ch8MlrTIjVtMFoS7BgBPMRZ//hk8VVVYuYV6lWakoiYc7OrDwmnVqPQCPj/QHwQCfXH5NzpkoymDQCyJVM4Oj9mAzz/4ET//sAWFFROkmsmBckrvMd8WhnI1A6/PI6ch4zoiemTWkZXHQ8AcHGVjSDnQ0w+TOYFSQzs2fnQ7Zh03DaFcCltXrxXi6sFKZF7rRdBERBLwlOX4mPDBmqSnG0mn0NffB5fPI+0nB/lwCgoEncVrdfq88JeWyZxrJhZHR1cXOro6BoV4JXwUMmeDSNF17m/Gm/98DVdffx3sBT4kkJHnoT+bcr8P4VAIN117nQDTA5GQUICwysrCmlGYECB9Y3FEVovUXXSeJJ2dXP5RODPzRGIEZ+T77Erl4Y3U2NTeQ5Cr3Q6rhRi80GD8y8Z5PJpBf3s7KgoLtH5Rnv6fHowxOj0N42A2WAnQXvPrWm24F2bEY1Es+eQDnHv5xSKjJX2/PDZPqBdNRkTjVjhtQ5BU+LAVlBe70b15I2KdW+C1WVE8ogENIxoEECw5EWFLrP7CgL88+hje/OADeRhTpk3Dug0bsH3HNtR4CrFtzQasX70WOTWHm++8U/Q2OJdG9MTmjRvFQNjHO3LBIuzYtR/dnaSGbIXX4URxaanc07PPPotvvv9OHvITTz6Ji267Dl9+9jn2r16PgqJCxDPa/egEVzrbmHi+4ABGTJksfT+6RFb52DbheglGlJ03kl5ZtbA4kyZzHRWLzciljegOq/jHu98BVif8JR4s396Gl267FoWZgzB5HIglg9LQZ1WWjiedTqKiugRdXZ2IdqlQ027UH3kmGkfNxDdLV6KmeihY3VMyZrR2/oqLrj4D/3zgBcBURIENwG4S3qHCyTOQSLtlCsVmV2E0WOTZMmpKJMjc7YDDyca6Kj02nnhEx7CeYFS1gtugnJpiltnSygoLatMHsOLrv6N9348wuJworq9G665d8JKxnDAyMveR7S8fUuZMJLiywMJ/E2Y8Ba39fbj8phuw68B+dHV3itzgd19/g0Q8jq7WNsR6ejQNEqsNvjwZMhFIQq2RPgQiSFkAi6bJwVD1vLPOxaeffip95e5IAFaHRubNve51Ugsyi5bmZnz03vu49JLLYPM4kSQxV1ZDA+kwPUE5EaiQZX9XO4V5sOkvo0CXtJoF14ifL/qMFXOGcIJW2hKMs7lRaIx88U2CWGgOYPjQ0eg60IxkKIBiv10WS+LyXEY2LPtC3FBWahZy2DeZxIHmgwgnMoizAlVTjoiahSUfw+hxu2LMIZ1SYDcTu8qby0G1OJA1mpAIpvHI64tx2xGHYebhTagfOQIffvihnHCcIaMTqCwsRiwcw1dffyvy5O9/8jEKS0pw4hmnYssvq9FYUol333gLl1x2GS677hpUlZUj0NyKP//lAXE4hLUxtClwe3DfY4vx4P33QM2ziRHTuHH9epxy2mkYNnIE3njzTWmjOCr8mDzzcKH0+/CFVwRmdvi8Iwb5dqisxEWmV6QWZbynG7Xjx8vJyjSApzt/MtQyG82DzHv6yE00mYVisELJqbCbFZT4fNJvTaZiePGdD3He6efDYM7KBudmYROar0B0ACbFDJfFJfTh/f0qjjj9Tny/qhUefwNUp8bcwEkOPiOObTHFSKbiws9KAmPuE/YKOTNJSkFTNgW3wSJgD4vZKmwDHo9XxsTY+ySZGSfzzSYFsWgEoYEAPFaNLUDfZMQBm5w2zBur4vX7zoCzgF+UQyKlwlbkQS6ZkBlTOVEYOh7SazbaCRdTEeU+a6jDyCmT8MnSz4VRTUI7NSd94hj7lHYbuNFtigEumx3h5nZccdHFeObxJ2TonIB2XUCXz6e3uxul5RVwGK0oLyzGVZdcjhMWLUJlbS3cVYVQ2ZfNt1wo/kQTYrg90NuHJW+8j/N+f75wOsXSlKb/rdcuIJZMejBa0hWSB+8rryWpGyeNkvetTDt/HgfYJOmVyWaVF6BZbjqZEUR8b7fGwUI1pni4Dz6PSWJeh82qscYZNIVhUhe4rYS4OeVhs/R7oLMTw8eNxoGudqkMZtkBHWTk0mQSOdFqNQ5HImdHQsBCZpjtfkR7Yzj3q5cw2Z3FdTUW1AwfLtU66igyHCmvrUF/WycG2rrQf7AVjz37d0yZNRM/rlqJVWvX4MT5C/Hua2/B7XAJwzcZq1kFLrLaccyxxwod5VfffiPXPmLIEBEy9BUVwmV3inDJueefLxJeJMFlNvivl1+WSuu0+bMRzWZkPXLBsAyMsoBBA6NhqGwy50MPOg2CEniqsijEjamP9vABhAZCgxPl3CCE5eWSKQn/wx1tMBf4QfmFDIyiK0E9w6zZjHgqAq+7SChQOKjNdpSStiOXSUk+E82kYUgUo3zC6cg46tAbMkO1OaFwWpxOltVT0knYDJKlUBLOmDXDqGq0j4ohBSOsMnJlsuZgzBpgtznEw3tdBdJj8/jcSGdolBb4/R6UlZbAaSMoHML5unnLNq0waLfB6TDg15fORd1wM3pCmiwboxuHXxu5o9YIIzQ9/ZF5V24NQ1bWbuoxC/DdxnUChWMRhKdUMpUQQSWilagqxvxd8KicxOnowp2XXokfvvlGAB6UjeC16hVOHiQelwu9fQHKmyETT4mM+Y3XXiudgNsfuQflNVUyqCyFTFWDaLKd4vcVoGN3G956+21cfu2VMisazpD3STvRuQ+MJvNgm0NHF9Gw5fsNWmiqF3gGBXbKZw1RbfkcUA9VWeYudHmwY9NWae43jJiIvq5+GJQcggM9mDRxLLrb2+Bykfpes2qH0wqL1YxoUOvrcFF5gn2wbCls5SUwOx2I9AdQXlUupyZ5brgpuLm6dnZAUb2oHjYRHRGWhEsQag9g/uNPQx1WjPPHWXDX5DGicEWvvbelGfMXLkTtiBEIdPViyQOPo6qpCTNnzEDTiOEorKnC8JEj8eE776K8ugp11TW44tI/YMr4CVi9/Cf88eZbhCTopltvQWBgALffdBNKqqsl3ue9uOx2WSgaKVmhDxzYL86H4SpZDY466Tgp/vAeBYBMsRvBPuZPqryno0csLC3BsBEjxKClbST6iZpwD8Pjmto6eeB6SKeHMJpmhCKQPz3kIzSOea4e3mjlojzVoEwAaNhLbnT+ibcyNLJh9rn3YG9qKAKhuFB4sFKZTSQBkwtmhfhZgwhR6dU9Xh91M7j5SMQkFBT5e5Xh85xJClZE9xQWFQoFi0lJwV3kg1GJwYR2HDFlGAbaYlB4CltU+NUOmGK78MBd12HP/oPCr8T7iCViqGisk71BvhyuaYb0kwQsWAxoD/TijMsuxsfLvpCUQufHFbSLTEsQdWRGIBKGkuVMpgmFDjf+/sDDuPyUM8SJ0rj4HlNeJIffS6Ml1JKOUaQOOTHTM4Bln3+NZZ9/gRfffAW2QhcMdoucvtKiySsZ83dDrZ1498238fLLr2Ldps2IGuMiV8CDic+orzckz5RrqeeeeiVXPwm5lrLWeQ5ipfbIUSrLw/pf8OLILr5j3QapJpkVExqGjUV/TwDBQD/GjBmB1atWwGJQ0NRUD1N+I3p9LsTiUXidPgn/9GoRQc+d0TBGjh8nZeenH30QrupqqYjRGP1WN0Itveht6RTx0XFHnYONm3pw15KlWBqNorzIiUU1QXx0zdloGegXvObMo46UkC9FAq2eIDZ9/aPG5RoYwHOvvoK2gX5RFLr9xpvw2DNPS8FkwuGH46zZR8Dv9qKmuloMgyxxrFCS04b3z0VkhFBa6EdTY6MInBBfy34XHQvFVTknt3nPDrk/Oh0aYywRF6/MhZWwvrVVHjLD6HlHHSVtD+IsZdIgreUSQrIcCCCeSsvsI7+XL90Y+SDb2tphygMyNNADqRQTKMlPruvGyHvRjFkLg3VjtLPkEQshlPHgibdX4+3vdqA7xAHrlIR0nPzIRqNIEjmTN3rd0Bmz6sZI6XUWzKSBn81KKsKwsKyqHFa7BQOBftkHxbXlKC6I4+pTCjC9wMugThjJHaVFKC02wGK0YmR9I3r6e2T4V+sFWuAu8UsfmhhYgeoZyTZoQk86hopJYzWMrskMC595vnUgjo/uivxMNpvQijKiaBwyFPH+ILq37EKko2OQ9/Z/M0bWHOgU+aykBQIzHrr3ITHGNRvWwOy1Cxsg6yJsq8nzyRdcSjwFiPYHMX/efLzy+huoGlIloSpHrrQeo3vw9OPeEha4PLpM/wwZri4oGMwnxRiJENbxhNwUoWAENpaqU1nkUsT3peB2erVpi9W/SD/NSBxiOoGykmKZNSQPCvOHXEoVFmd6e6myWszoHQjA6vXIbp892gAAIABJREFUZL1qVqSYIbAtSjjv6YApq+E4SQJLgcyfft2HZW0p7A3a0U0wd/cnWLX0bfjLC6V8brBZ0RsO4ZzjT8afz7kMzmI/0umsliu4XOgKDqC0qhLnnX0OLEUFQlr06Uf/xsRZh+Pz95bg+48+we/OOBPPP/M0CiorhQmuvKxMjOiVV17BA3fdjauvugovvviiUOz5/YVilKy8StncbZdyud7gtTnskv/pBQjdg7IZTHA5v3/p0qVifLGoRrrLthCNs62vT8Myms3SsG4+cHAQ+U8qDK6nyFzzFMhPS+mVW6vDLmmE3shmu0FXmZbRHINJmvnCCaRmEO6P4/6XP4WndiTa+m3455KVMNuLkSN6IqVRFg6iRsyqhISkMmGl0pQvUgkYOq+MNXzUCMSTcWSMaTlpzbke/P64cvz+hGkoqHSKoyOSihSPqUwMBd5idO7aCUtO0/2Uk7G3G3DZ4S30I9jZCVgtEDXYXAZNR80WZnoOsguVZx4eKQ6ztFTTTcmmJARltbqushpIAcUON5Y99TycZQWCZ2XxhoZLwR+9cEgjdFsJqPjNUHKJLEK9Ibzw3POYMH40zrn0AvSnYsjwt/M9ZF6znN7pLIqcHhx95AI0NQ7B4089Jo6BVCYkD2Oqx70iVCp5kSg9R9TbH4d+N/etMu6Y6arH5hTID/luDA5NOZbQHTKu8b+NJE4KhVBWXi5G5nZ5NDbvdA5HLViIb7/5DqNH1yGRiMgwLF9O6idQ9digymK0B/oQYS/OSDkzChLmUFNSgT3rdyJBZIlgok1QicdUzTjh9PNx3u2PYemWPnz77JUoLMkik0tIb4cMXpxMqHCWYvlHX8FgzAqVR19bB66/+WYsfuwxzF24ENOOmIN/vP0mzjr5FCycNQvvL/0MRx27CO+98w7iB9pFMPXk004VdM2YGdNx7qUXY9PyFWisrMITjz8ui87B1Yt/f6GgJT7++GNhN9+0cysqqqrgpVejaImiNZK58CxpMx+jobIoRkp3el2GqQKeZsiUrxpK/mB3iCHy5ORDY/TA002np6cORce+gzC5XAJQ/99eUoFMpVBR3yhGrnMKyZryPDUa0d3eLlVunjyk9kgGzbjv3U1Y/O/tKDC5kEpRhSkAi5HKySbBfhZQusxI9Ssj4sGggJyj8ZTIfYtKWTiMstpiqGYj3M4uXLCgAvMbHPCNGio5qM1hgc1sxECMbHZEtBjQu3ErHIXFUgwyqgqOXDgfn371peBwCcZft3E9zDYzahrr0BMLIWfQ+G+5vuxtixGzH55X0+bwM/dtVV0tMtGE4HoXHDYNzz/4iNC6cO9JSJtnhWcDn6cR2yFlxUWy5qIERYdoMKFjXzsef+IJDB8xFFffcLWA7yO5hMTDUlOR+EPrPDgsNvQ0t+GJvzyKxc88jRSIKeY+ZcCakefPKFFPX/TrkDHt/B6QfJlMcwxZ4YDq8nnFS9lJHmQ1I0cdRWJCmX8kU7Jo+niIxMGqCTYzwcUqDpt2OCxmO7ZtXQF/gUeT/87nGjRGYhslqSafTnsL4sYcMgw5ciZ07twHp4XzXhqej9wpzJNqqmuxd89+1Awbhzvvew633HEOKio8SCop6V+S5oCh59YvfsG4w+Zh+9aVKHa7pVdIwZqXX3oZLW2tePafL0Mp8ODZv/4NC+fMwbjJk6A4bOgZCCB8sF2IpMhqR9TO+bfcgOXLvsIZRy7A7TfciMqKCqxasQKvvvEGLjz3XNxy261S9ub9+0r9UlXdd+AAeCpSelrXCOS9ZrKqeG4SLfMEpYHxVKTBBfr7NeB7NKqFuEQy5SF6ehjIh6dHFwSR8793btkCg1FDb/z3S/LVVApV9Y3yPTKXSfZqls1JQm2xyGe0NWt5LjdSoc2OnrY4zrz9DQScTdi47iBZieGyWoTqke8jaa/RQNiZAalYQop4rGL2du7GlFlTEYxEYDFkYTB1YKirH09edzrKJjYItpZVHIabhOWRcYBFFQOrwyYzAs09MkLF3txpZ/9OenUUjqH0GmdoqXNocztFdp5yc3p4KNPx2ayEyAw7WRmOrt2EU+64VbhwiTOlM+pYuUZYBGOsS+TnWKXqzFCf7QSCEaxWeL2ewSFlMVbFKFqbTzz2hNQLFiyYj9KhVZpIj0HrBeuwUc0OFPisDkwYMhLnnHc+LvzDJXAXupHIpmVsjc+FTksv5ulGyT7jocYo/WYxRrNWBahrqpXQz1bokxktDhsT1tXX3QObSdM216uAqQQFUtPiqReecAr6A2Hs3rUGLpsF1VXlWsjL2bZsTvIDHz2yzSazem3BAKJCGGVGkHCjmBYaMcm1WzQ4kWyctlYUFfjRfeAgTvrjlVj5yxrY8tAr5g2HHz4Tn7z+NewmN4Ldu+FADjffdBPu+tNdMsxa3dSElj17cPofLsWmX9dh56aNmDF7tkz4n3fR7/HvTz6VBj4NhIicd774DOfPPwYXnXyaeN3S8nJZSNI03Hj9dUIfyMl+OoGfVv8i/86KMfNFQtNY/GGoypPQbLbKKcj/1x0Nf9Lo9GqhTrlAYxTOzLwakjilfMOcD6iqplYMmsa9ef0WUV8irEuq2HnpAoFepdMorayW76QxkmXhv188JXW0E+3FysJBMotksADXPP0F3v1sDxzeQjlNrC6zTMoUuLU2V3wgikJvAdpaNuGOS6ajcUw5gsks6vxGTBk7HAh1oqjQheKmerQEA4IvpSHSKN1+jxCUkc6C7rbvQBfIdxTu6sG0k46Hy+sWrhqGaqTrJ/Aiy368hdQh2mAu146bWhxDJoOuffswdeECAd5xeobPKh2Jo8zpxtcffQIDpzoY5eUHAySPTqZgY+pEyTd/kVTBdUPRDgMFHrsHRhhxw003o66hDo8++QhWb1oDu1sjG9OLPxzgJk7YZbGhY18zrr3ianz/C1nPQ2KMkST5jLTiJve3Xj2VIt4h43ySfuiT/tPmzVTPO/csiYmvuelGkclWjNrsFWWZObPX29k9+AtyQVmDhKj0bnOPPhG9fQPYuWMl1EwaHo9NgLN0aBy9EjqFzi4BUnNEZ/uBgzBSG2HHbljNdimLDw5kUpQ0m0VpWRGad+2Bt6QUWUMKddOmIhDskZyBD0IoFLNAotsqgqLxnj1wGTSj5owbBVTIG9qydx+mzpopo0J/X/wEbrrlFpx+5hm49957cel112Hjxo1476WXUD1uHE4/52w8/uf7UEhxVoZDmYyA0Snp9dYbryMYGMDCY44Wz+uvKBGWu6ETxmIgFMSwIUMFjCD5alcXLEYN2JBmo1+a/cy/0zJaRYlwel1uMD6gSFJrGA9OpuSNUrg6RUkrJIYokcDGbWKoUsw5RHVL750VlZTJZ3IaheNYuuHpRnmoMfIzjEYTAyvYTHbEO1K44N61+GrjAdgdHuSQgsdN9rIcvG4n4r39SIf7cObxTfjrn06CyZZB1+49KG8aBS9Fb2wW7NywSSb/C0YNEXSO9DgUUv8TGs14TAuzew90ocThlgb9nLPOkOjp5+U/oUTYD4xyMhLhYvc4kFaygk+mQ5L7MbOSm8OosWMEQkjgoQiV7t2HcG8AfZu3o6qkFK2tLbBS2oDA8HgctU1NMmjQSSeVzaGirAyRZHzwpDvUGPds340nn3lWKsXnnXMWzrn0PHy5/PvBHqUYECf5ScgZT0l4fOrRx6GttwstnS0IxaPIUnI9n1/qRTE9HP4/GqO1zKlSRnrx4sWyKR599FEE8kesbrFEP0QCAxK6Si+IakdpgpazWDDvJDlR1yz/EsVFXsRtGgKj3O9HSaEHZtUo/TXGztxgIhp5sEWQD109fZgza57kZaxOcjCZPaWK8nK07N2Lk887Dau3bobFQ3SGAZEYS/Mqijx+dLcOQDVUYSCehtq1A/ZcVEiMopGQeFFKEtAAfvz+e1x94w3495fLUOJ046RTT8HMiZNw8x2345Lrr5NRLJ5+1fX1grwg7cOpJ56Er7/8Escfeyz+cN75Es5wQUkBqMPYOPDMPIeno9ejVWG52KI1EicLuUXyYqFQzHNvitFx2NfjkU1Jw9FPTB3pQeyszu+itTS0Bjo/l05QJK3zoGryo/Ik5Nqy+JM1MTwul3lLVl71CrlujHRWhwKZ9VOYP22KDc7Ccbjq7jfw1fL9yCCDgUQcNrMBfhvwuzMmY2DrCtx6+7nwlWjVZ70sr3t+rhHDZAN7lD6PqFPZpAWhtYnoyNgiYe6X7goIJ2rl5MmCT/3xh29Q4PPKEDRHlYKRoCBgHH6fSCzQufT296Nh1Ij/gf902R3obO+EIwOML63Ep+8vkdtlasDCHB0rnZkUwZI5WS9erx6N8VnwuuVZRKKaZkc8jkcefEQ+h9d8//33o2FMk0x18D7IOqc6rNIn1R1ef2cPnvrrYtxy040oqS1DNKfVQfjS38NnqCGv8kdjfjxQVbLSOlHqJw5RAx3dGOgP4r4H7hUK9LhN64/oRyiNkaMfDIU0GQL6WJbRU5g39wSpZD3/1CMY1VQP//B62EyklU+irKgAfm+hln84HNKbE2aukjJ89PEn8Bb4ZViWFyhhBvOBZEIWkmHWkPFDsa+9DUcecyQ2rF8nIvQcXyly+LBj/R5MmHw02nuDcORCSHXvl4WmQYaiEfQ2t6Crs0sWcCASwT133okHH3kYt/31YSw67jjMGTYSjSNHomH4MEyaPg1/fXIxGocP03p6igHdnZ0C/zvm+BMwdvgwgdFtX78eow47DNOnTcNXX38tJzQdDI2R98iNyIe8e/c+QdYw12b7Rq+WStvI75P38n3cIDp7m+5FOXYkyKZsVjYIDwNdjZl6GzQm6W0ydOQkf75PxUmT1p52cZDhUCQP3vhtBI3v/78Zo1GJwmLyIZPKyQDx9Y98jlzSjv07f8FnHz8INdoDZ3ExDFYDFLMWbutQLv63HpK1itJSAo3jxqIvNiBOlExpNAo9GiB4PLC3GcPqGlE0dqz8/cr//AST0YCS0jLs3rcHRosRTo9LmM8pYkqQx8gxowWXynvW1y3Q2wezqsCbNWDT58uEn0l6iPnIgC0nvbhGlTMizFjLuPnWWyUyIuGYDmUM9PQKKTK5edetWSeEyJz8+eqrr1BUUyaOk6iqP/7xj3hw8aOSqoixMeTPKagprZKeZFt/h0x+/Lcx6iEupd31l9b4z0kvV6keW68SgssqKekW//Hsc/A31cp79ZBIm4q2INgfkJg9m1JlkiASieGIeScilIjjj9dfjnPOOR3esmItRMllhVajrKhUtC5YXBk/diy2bdsGh9ON5pZWqIoRvd0BrTLlcEgSy8ScIQCp1Pf3NYv61JKP34dFBmQNQgHYubcFzrQRwZ4IsnYXasePwtEnz8eMwybhgjkLUVZeJZPh9//5z7juuuvwu3PPxS9rVuPL77/DLzu3Yd/BAxg1dBjOPuY4eBQj/nj3XVi3Yzs+/OJzlBWXSF9QGMcyWSnvR8MhMbT6+npRFvp17VpZm3AwKA6gJs+JSePi4obCMfR2abk2DZYv3WjYRNaLYfrfH+o9WYXm5tA3Oke99FZGOqnhJHWAAb0pv48bk1MlXQM92Lthk6B2RNThf3kdGt4e+s9EurCKTVFUk2qDIcGSviLVzPrGKrQc7EFcQk9tcoafwz86fE3vodGR7N22TsLz8lFDkWI1nXNTgOSyerW3f8tOjB01DpXTporzWb9uNTNatLV3wGK3CikUiaCC1GO02+AuLpJRLLbVOMRAbLGQO0VjiPUOoNpow/Zf1w/eEteJaRL7wgRQyPMKRPDdd9+JgjApHKnBwfUjHFKMgoPXXq2YSdUyVl0fe+wxcZr33P9nYTIndw7Juu979CFUVleJoycqjBojzbsP4J67/oSX3vwnLE6tJ8tnQ5ALK8Dc35pBarhkHZnDyjXBDErDYUNVYRjLl9ylD+PXSHkHLTnDjjBLvwZJlhWCBBTSKYYw86iTxVu17OLYk0rmHTFGsmux9+gmiWs4gsbqGhR5fXD7vDjY3AqP14dvv/8RZqNNo8FobRMDEgpF0j047Jgwb6rIsq3fuBaxcEiUgNKpBDq37gESOdzwzIPY3tWNFWvXoaK+GmxyHz/jSJnDe/ufr2L3lq1SfkcsivqTj0Oh2Sowt66Nm/HQU0/hvhtulhEfOpM77roL733ysTYSVeTHgeZm8aAMqWL5eU/KFvDBfP7RR5gycyamT5mKJx95BP6iQpn6lzEvhh45BS0HmmE1mgeb1MwfWbntDWu5jx5C6ieMHnryZJTxnnxSz16jHlrSGPWQRx7mITNz3FguvwdLP/9CCkj/r8ZIOQLFYIHVxma/BVmFVWFqanBQWBX6QqPQWwCRFBn27HIt+inFZyiVZCJ/Qj1S3XUUuACXDeRD0/uX7ItSZs/UH0G0ux+zzjlbwvZf166Uk7E/MCDr3Ti0EeFYBEly8BQXIZCIoaSqUgyEhTRxZBwQD0VgiqXQ8u1/YPe5BmUAua5klacYE43x/PPPF45WpiR0JowuBE2UB+yLk+KsZ95h9HX3yanI99L4Xn3tNekVs73FOkJkoB9XXX+dYHZJhBaJBmXapsDrQU+kD26vS4iJRY4uL6bzW66v5f0S3XD8impdHDIeNmMUgXfacZuPnYP6MGueYYxoF/4bL55V1lw8IXWJRCCCw2YvggFW7Ny7ChYr+1pGmUcj5IrG6C8sRrh/QMQmSVw1c+Z02WjksiSSpLmtCzvWb8KW7Vsxc/pMDAQCcNSXY+6iBagZUY9PP/5EjvAEQ7JsDr39fRh52EREUgnkyMxsMcNDQR3FAI/DBZfNg/K4grWffYNwLIa584/Cht4OlDXUi2KUp7QYlWkFn73xBqx5LT/mWy6PR6IDMoR9+MbbOP+KKxBMROEp9GHX7i1oaGgSGTgWjtrzyA7mg5T58jvdMg/JB8XCT7i7D10dHTJtwGtjyZ5saqSlIBeMHtZJaAdFTgYdQEDncGhEQhYGGkQsFhfkNiMH3XDJW0NgPXPjpqFDYbKo2L13vxBccYZOwuRDX9Q2yTsCjiSJZoiiiCReb1+3nPL62JjuDMS5MHT/Lc2BgYCQQ16UFWCKwffRsJqG1gkTvVQOOYHTWI2ompQqt5WkUlmgqaQSP336Jeaedw4MFrMA0lf88rPk2J7iAsTUDKyklSTbXVEhCivKJcT0umySI7NNkk2kUGUvxLp3P4LF7dKUs2Sekm0HVVA977zzjuR45DeNZeKiOKYbgs4woBcQeeAQWkCghFkx4vprrpX1IPKKeSvH6oiWon5HR0sLHnrkYZG24HuocnXH7X/ErDmzMWRoI/75z1dQVVeByZMnIxSNo7OnE6F4WCCEOQO1tIBgX0im/o+ZeYQcfsqo2eNUHYLFhRPEeZ7ugjkLHxCfAzeMPDxOB3T3wGl1IxZO4LBpR0oPZsfe1TJtTQ7PgWA/fAVemDmkyUHlRAoTR43FZ598Jr0nhsP0QL+uX499B9tE/rm2qhrPP/ssrrntVhgKnZh17EJ8/e0y0ZdniZ0hCcPkyrpaWN0uhNMJCX+sLqcAx4nbJMFTKpqBqy2Anz5Zhuf+/SFWbdyAjoF+bRM31uDLv78Ix8FOZLJpKSYR5sZr4YnHkGbv3r244JwL0NbZjjdf/SeOv+Bs2F029PT0CYC8vbldDCBJifU8N2kqFJH1Iav4k4sXY8OaX/Hi44txzplnyZjPmnW/on7oUOFe5VCiHmpKbpNvAGvkt4rQMOjGyv1OivmsYE4ZIpol7NXzMzo+mXJJpjB2/HisXfEDTB6fMHgLCvYQA9KPEj0/IryNp5UOnyspLRrMrfSCk75p+ZPM6//90k93nXFbD7ui4X6ZnGFIqjXbFdQMa0Q0l4Cq5lBWVo6+1k507W/BnBNPEs3KzWtXwe5yaEMAyagQn7G9MXT0KHSEBuQ+uUbk0CEONRsIosruxeYvvoXX7RUpb5Js87SkkcXDYTz7wgt4++235YRiy+lA6/7Bwo2w0eXTAb3/x3BRN0aLwYQhDY2i8UJUFpXUeCJTNpBR0JFHzBWBp56ubhw5/yjZ06QR5Z+LLr4QP/68XJZr7dq1EpZ6SORWVwmSvzGyIPhAuDGzgDGRkZNbmX3yPDXQ1z9I1CNHZ/6w14/VPvaI7HZ5WDxW0/EEYgMx5BI5zFmwSKjotmxbBY/Lif0HDwoHCjlMbHaz8FJaFCNuv+FmXHfVtXD5OX6jjWzNnTcPH330KWqra9B28CDMNgtiahrX3HkbPvl6GVKpuBAF0xAnjh+PDZs2CViYZXRuaiHgtVjkARIgnCMbQXk9vn3xTTTVNmFr817c/dyz2HvwACJWE2YPG4WbZh6JNFWfjAouuOxSodBgzkDtSYZAq1evxvHHnYClX36Bm+64FWsYIqeSSMZT6GvrRLB3ANNnzRTKeTb+yaupIzN0Z7Z1+c/4ZcNGzJg8WQyWIVyKYQmLGQYtF5cwigCJrJYH6hU+UZA6lGWdOWgwLBP7nEOkMep5H42RwHlOj5xw0kn49wfvweYrFJpFQa3kZ6v09w82uLRYWgMd5Plu6SSZL/GlGyP/Wy/6kDZj8O/ztfnfPlfI6/9Hu4W/yxOM72EIFu3rxZSj56Gzt0cGzQs8BejcewCzjzlOqui9rQex5ofvUDJiuIgesfZgcdrRFRqQuUeCKzjtkEhn4EllUWNx4MtX34Xd75H1557yuAqkpjFqxAjMmDoNT7/w/GA/lmN+iZTW++MfmbCR9s5v8hJck3RKxm7gtjtl7912221SvGPPktFcf3c3jjvpJNz5x9sFu8yWV2t7G15//XWZ8Bk/bhzGjB0r0gL8PXmWMOKII+ciEA0hEo8gHI9qjBqqSVA/dEzCEnDEafNVf0GhMGXr4Y9ujFxI8WzUhGhrE+8vzX/CtbqDMGWYFAcxZd4CNLfukNCMRLbZbAolZSVwexwie+a2OdB5sBVlZRUIkD1OR4EUFeGoBYvw5edL0bx3L2YddzQaRg3DL6t+gYGqRFQ8VoHuzVswbMZ04ThlcYeKVtSMtFlMwqBtsdvhcHlgsTuw+Z1P4bV5ceODD8FeU4kNO7cLF+kIdyEG1mzEy08+jRhHaRx2cSy8J97jicceJ2EmPZ/D58WBlgOwepyw5sOiuYfPwdOPPYFsIi0PPJHJCBNdod+PoqpK2dgyj0h0SA4IJxNI5XI4/ehFWPy3vyGWpFjm/98Y2crQm8kyA5ivjupFG3p8iURTaVEx1gtCkjPlSKmoET2fevrpeI/cPwZKclPfwgBTnh1OP9FIHqWfdixO8UUHxB4eBXF5HexnHtrH5LNiFZg5o35NYupE2OSnDVLJPIIq//96XqvvH6p+Ed8aCwcxcd5MtAf7YLE5oASjGD99Jk449RQ8fO/dMmig2G1wFLixv7MNBSVFgMsBg90mojdEzowprcKXL7yiqQLHolJH4Ivzi5mEBlT58P0luOTii9Hc2y2nKQ2V7Y1kvhGvh94kidJ7f/w79qPjMZJ+GaT41tvRiYceeUSc9btL3pf+OUN6qdRmMxKiOu0OON0uOVx4OvLeeRpzbI7FHu6L8vIKBAL9qKqvRlpNQSE3LguEKSpPG0TenHmpMm7uRNXpdAkJEz04Y302/XWKfuZR4UQUtfX12Ll7l6Az1JwiCrrdB9tko8w4Yg5WfPeDeMGMQUOGsD0hnj+TlVhffzCDoQ3bAnY7ent7YPd6hbuUWuvNZBOorcH+AwfgtJjRtmU7zr7lavy4cgXU/AQ5P8tb6EN/OASDmUpSCpxuP1ypLK4++/d46KGHcc45ZyPldeO7latwzFEL8OhFV0gyLWE4vVB+EltyKJamFQNmzD4cZ154Np586ikJf1k8YigzduhY/OWee1DFNsnAgJwmuUQSU6gTGImItDiNmp/NcIghLQHirNo5HS50dnVJxU0X4Tz0ZCx0ewYNhBuC+Fx9jQSzyMybmFTViEg0qZH5Dr60nJBGxGtiqE4jEUQO8zwjlU9/Q3gYTZoBibFktffps3RUHZYwU1FkWJynhp67ckzORAa2fFGJ5MD698rnU1sinzNqsDLNcPX8tKiwUMJ/WftIBCPGjEECKo488khY3R588vlnyOSSsNkon0bOVw6Ya1V1P/mF0mkMrazGT0uXIbyrFY7iwkHHoJ9svI9ASwf2Nu+X0bily5bB4OAMqFFw1pyB1ItIOnBbPyH1/6dhpJJca43smZSY99z7Z3nmr7/1NowOOyqrqmDKZLF500bceuutuPLKK1FeXCJwSdYeWHOYfNgkTJ0+TU5SXldpSZmEuOMmjBb1NcVpkSHtRFwTneVwhBBOn3XFuZJZcDF5UjCHMpgMmkSVogndCFUGwx6TUfI8h9cjD/qis87DfTffjuNOOgVfLV0KhVVARTNGehEufg+ZA/KMbrpX5vfJ6Ug2sIZ60TPwFxejo6tzsEBBg+navRsnXHAeVu7dCidHfqIaFZ5AzEg/6LBDTVNZ2AJbQhV4ldVgRSSVgosjPKqC0fVD8NWSj2H1uAe3sHBpks0tX16WRD6RxGXXXIFvV3wv1z1lxgwJs1xmC5Z9/o38Lh8mN5k05ZMp6Z0SyRFKxsR5CWWJ1Ypgf1DgcwQzHGxpFVY4EV2V40yrpOo5IlsZOoHSfxsjT/1EJis6HJzIYFWTz4PPSYyA7MT56+JnZtIJ2UhGhQgg8qL+NoNHQl+bkhAJNQ6Fm8mmoGr3wut+8P57cfnFF6OgphrBaET4bhm2Sr4IevC8TAJxxvk05rdpBM0YeQ26cXCduI4SgudPTJ66GzdsECQMUaNXXXsNnnvtNRhtVtTXVckzptEzLaFIbl9HB0ZPmiK5b01JKX767nsMsFfrdg8ao65ixmvlHqTg7cf//rf0d6NpjbiL38er5vcf+tKNkWvKe2FXQeCsqgEWTszkizcEwvy6YSNKa6oRiUYR7euXQWpWaTmRw17lnFmzpL1i3n/PAAAgAElEQVS0ZMkSuWcW9Lgnlv/wo/C3cuqH1KOnnHYS9nW2yCgdjZHFIor50uCV311xvsoJaD4UVnTYwyE9AS+cF8vCRs9Anxy7oUhYFHm279srk8ypaBJXnXsJ9h9sxlvPPI/yumpEEmFZKD5gtgl4VIu3zHN96CGMbCYapcWMyupq6cVQzJLFJCIb9u/cKQpUSRcZjbPoDwdRUVIkFU85Zbm5VQPsbK4TWdHVq2E1TTZh06Zkc4nPj+0rVsHnLRTNDb1iKYufTEBlaZv08GQ1iEZxxgXnYm/bftHf4Cnd39uH3518Cq656kY5VRmeCISNvSVqd8RimDN3LjoDvWK4InjJweH+oHh9VvF6+voFrE3xFpm/y+eM+qYtcLn/x4ApK6d88eHwHt0FxUKPobUYtBNJL7rkFO3k0w2S0/gWkx08XgjK4Emo/04mncTokQ245fprEA7245xzz0UmqRm1t7gEn3y4BHNnzURxU6OE2DQAFux4T/4CH9LsO+a5Tc1kBz8kd6Wx8nt049TzWnke+cl7vd7AsHj9ylUwWC249Oqr8PI77+C8Cy5AsK8LH773LjzEpiaSEm4SrZM12zDQ3IGRY0dJa8hT4MP333+vzQcyXMwPdWswzaywCZZVslBCYq2ERD2MHzgPqbMd6o5isEOQB2oL4Q8dHEnSUmmZUqFa1fvvv48Nm7fIydjb1ydK1x63a7DDwALgO2+9JW0TSgdwj5IGlN/365q1OOXU0/DhkiXwFXkxfvw4hDJx4Yft7emT7+J1SPR4wbWXqbF4RLwgF4zN7WRGE6LkxpOyu9Mmpf+evl7s3rsXm9avRemQoaLvnomnZaSG0w5/X/wkBmIaHI29Rbo/T7FfHriEZrzP/FyYbowjx4xDW0eHtsFIzRgMQKV+RCiKEccfJTwvlPqivHRL8z7xNtrLABssAjjeuuIX2D0+mZMkoCCXyaHUX4jmvQfgtDm1YducNtakVxOTkSjqG+qkn0gGgz9ccQU2bN2E2qGN6Av0y3V6HE58/8UyhMMaBZ+AldNpQb7QGKUHGRhA0/jRUplliEpHEglGJOxfvXIl2np6Ja+MJrRM/L+NkYAIfvZgCJWvWtIQGEBTEIfzpE67R9aPn2Hl3FwsgZTqkM3mtFpADtBwew8QjwIltTjxpFMwad5sqVJzcxy7aAJMaeCRPz+EG6+4BGazIvc3c9YsMfR9e3bIGNB9j/1NJNnJ6MdrkhA4HoXDYkJ9bZ3mCLO/IUjEEUjUq4m+iGPL6w7qjkImNvKnJo22rqEByz79lOEYCpuaMHb0aPyw7DOhvC8s8ctcqc4WkU1pwjv8/57+PhwxZ5bMNzINYh+QYbzOTsj3cKpfFKBEwEejyqBR82TkftZPez3KoQPhNQlUUYo3pF43oKKkVBwCc02etpOmTsP4yZPk0EgEqWyVlN/jvuXBRUDL5ZdfLpX5uXPnyikpI3Vms7Q33nn7XWF0j4ZjWHjqIrS3dQrBF7+Ln8PWi3L1HbeqAwP9MqTKzfbrr7/C6bHKh/JNPBnJlcKxFXKjVFRXo6S2WryFblAsIPS3d+CwkaOFOYxTF8RRfrV0GZwUJOUDozGSacxllZ4NvSdFRsmDKWj8PDwsQdbnaAIzzjgO+3o6ZJH0Riw1FyWPSSZRVlyKzd/9ArvXJ5+vstKYyWhzdpEI0sEQbE7XIDVfKq6NBbHA4mTumEzLBDf5Y8ZOGAebyy5elzmT0WFFsKsbI2uasPY/q7F+7ToodjucXo8YvELhm7wBUYp72Pix4BqWlZeKwbjtHokIOEJFybGisjI5SfQQU4dr0bFY2aAnvMtuk9BKyuykAc4y11ORy7B/pxkljTCezgr21+6qQJndg+179yDZ04UP2g8i4jZjd8eAVB3JcJhLm2DN5OBmAxgK0p3dWDi1HH+65kZ8+OY74uHDsSBM/gJkhL1MhaeiHAlOkRwK4GEemtZUlOPBkPQSuQEpt801ZZTBNddbGwyv+dJROkJfSUSM1SanDY2npLICb7z2GiZPn441y5ejYeRQ4duhYzi4dy/spNhg6yFfeeaml1w6ScHZMI49/ji5hrrGOsFT8wSnYBAROsw1OVDgdNsknWDvl46TBF96NZXXxqkNvjSgvYpoWBPfYRpDOhiZ/Uwmceedd+LuP92FkWPHyGHFayFkTk83mA/zVGZqduyxx0oFljbAyJD7kT1GhrRcL8nTXZSSD8p365GLnIx1Y0eqU6dO1kRr8tPndpdZdAd55HLD0Bi5Cb/7abkQwZqcvzEny82YTCIN1tHcguFjRssNx6NJjB8/EcZkFh+8/z4KePKqELUdKjXRgzUOaUJHeye6OjulOsbFZchsczsQMarIWYxyA0S2SF+IBEocUG1pQzgQgtNbIARXXAhS54tnpJwaQcl9/YOS0QLVyp/ONBAu0o8//IRtG9ahtLYKZZVlMqw8febhWMPpC7cTN151NR5/4FGR905lclLYojNiAYU9KH3USyY1ervh8RBzWwS7AIi1Si0dzIrvf4SHfK1FRXJdh5JFM5RfOH8hzjjzTFxw0e8lQmABhwoSqoDDjDKmRtZqGqiqmmBXXAj1dEuP944lb2PS/MloiwO/DoSRy1hhpGJwOgtHOgd7WkEx04M46awMyCUUKNE+DK1xYXilDXWuGtTU1yMQ6oZiyQhJsFHoOJLQ0K/5l7QyCHHMCZM8uWT0++gj6oiTGqWlEg1wc+nYWW5qbjI68lBHh4A4qWtFVa+cyYjJh88QCpTvvvkGTpdNmCIoF8EN33mgWdpXBOcWUPkrH0oKIVeeBpInPjGsbMbzGT//wgtS4WZKFU0mYCVNP3NXnlCUAsgzvektKF12XQ6VHPesdijwcCj1FaG2ulpsgMRlt9x2m6ZAlk+5aBdM63iAMSSlXqeeBt13331ihKefehomTjqMSBh58b64bygzwNOSa6MXvjhooFSPGqa2tGgS27R0bq5oIihfQgOQ4oLFJFVUAsIJGHfaNSiU7hkEV8e+FfkhkxFROa6qrEFVVTU623plEoJtjx+WfQNnYYEs1sw5s0UqgAOsvAlBVRA94XOjoLpM2g+EmRGDSG/F94TbuhDq6YHb7xcWAgLHuU84KMoGajKueU8udiwahb+oSEr33PTxeEicDSMAQseCwZiEVr39XfLZU6dOx7qNG9A0fDjOPPUUnHbCiZgzbRYI6mWPce26X8XrCsWh2ZL/jKCcgAPhILKJGGbOmwt/UYEoYrFfyfybo2Z6tZOgdToChrTE6BIfeeOlf0AdVbDyIAIhl1BIc0hQPE98AmzMcDqoZrUCs2+7F2fcezn2tAaQMVpAGVXmlJEMkDTnBL/JCrYtnYU/Z0UJw9p4FuacAjWWQEmBAwNdMUSzQRQpZvzrqccxakgRnvv7X1FWU62FqDTgbHIQhkcr4saVCIddM7ZM8sTXEqYqFMWKDe4J7ge9oMONXej1oau9DRdcdaWIDLFNNmLYMKkwbly9RnQTY/EwGpsaBGbJNSUAg7/b1NAgkDY+xyeffBI7Nm6V6Ka+oWFQ8o7rq3P3PvLoo/jHiy9ID5gnI5+5UFBKXea38TOpOOcLUVq/VAHhhvpJZcoqqCwvl0LmNddcg4svvVQMiBVTGpEO/6MdCGUGKUTyCCba0tbNW7B02ReyH49euBBTZkzVEGy5nAxvc7/T8HkAcf9xTRSz362yKsiX5DnLf4bRq5WBuYlplGRM5olDqgm+CtxeBPp6RcGVGETdE2pIeTalDUjGM1j5E4cy7RJLM7QhnMxeVIK3P/lYyIU6m5uFVUwfHKXXGDp+HNpDA6iuq0VfT7fMBPLCk7E4HIpF8gUOropHSSVkDCcR1Xo2kpOJuIqGLtHDJL7X7bJJHM/v4ETIxZddipdeelEYxglx2rV/H8wuN9xWB6y8JyGEapNNEA9rk+XffvmN6IiMGj1eNhIdhXyHzYhYZw9qRo/Q1o1NatL3hcPyZ8qEifDY3dixa7eAFIY0Ncn8Y2snhU+08JStD03l3IIsT0MWHFQjfN5ydOzvwmVv/QvJxkZsbAtJmFPuMMJJ9TfqZChAOAuE09xeRjhUoJqbLxBBtd2FdDgOh9kOUySJpmortm1ogcPpA+lWgqFulBR5MbXJjiFlFSiqKkVvKgmPkbSSKaRzSc0QVfo+6ivyKllh/a3FQjm2Q/t1wlUjA89aNdVmMuKWu/+ELV3tiGXSmDxiFDx2hwwcv/zUUyIjnkskMHT0aHR2tYvDpEKTYHTzFVmuJ6MLcqjy3/hML7noUpSUFcuJJeNZpL4MhfDAAw8gHIlg9apV+PC9D1BU4tcmg8gGEQwOOhlSwujhNH/S8fEAkomQVFbkLNatXI2bbrlZKFrYrqLh0ai4L/QerN4n5unG8FRC92xWcl+OJk4YPwYvvfSScCjx+8moIe/JGye/W4xR8dpVPzdcPI7TTjsN/3r1VTh9WlxPz8Q/rK5yk/HolZwtk0F9bQ0sZlLRa/AuHaIlTFrS47Jix/adckryS7lIRxw5H18v+xoo9uHiiy4WxP3rr74i4Rsf3kBPD+omjoGzyI/mlhYU5sNhURZmW0El3Emjb6D3dTjt6OnsFjoIG5O1PL0+R26Ie+AJrk8VUOiFn6FDnxqGEIgcFW9JdEUwmUBtfQOuOfsiPPO3xxCNabNwdEih/rAm95ZKoWFIE35c/osYorRZQiFMOOJwCaV5D0z4GT7xVGQOQcaCRDgKp90lGpBjx47Fqy++JLQeRPaIzgJPc7Z6tPk0GAwOuIxu2FQLNqzfhIf7DuDt3jDShMIrlGvLwGczwkbdeJMBA3mJbSNPvxyE8pBus8oAZHpTMGQt6A9H4HO7REVYqCcoUkO5gEgcFX4qUUUxqdaJo8ePQyqlYoCgcKOKrJqQApdJVWTdCd9LkRKR3Kr5MTsS/eVyWgNdTsq8Meo5UZnPB39FOarGjBTJO+ZbdK6Bvj6cc/QinH/W72STR/r7MXLC2EEJB/4dVYhphNxD3Oick2XxhtqN/H72RC+68BKMHjtSniPfx03P537XH++W73rttdfEeTa3aU14nX2BuNdDjZF7g79Hh01VtSmTJuHg/gO46847pYUxaeJhGDZiuEzv8DN++O57jJ+ogQAYbX322WdyPcKOyNMyT9O4f99ufPHFF3JynnLSqZg0bbLYmz6nymviOJdi8DlUNmV5tHIjEUtHujkalw4zYs4oGz1f2i6l9StZuBx2+F1aHqSjc0imy/ibBRzyeO7aqS0av5wPy+b2oKqhXo5nj9uNIY2NePnll9HV0o7qIQ2oGtqEg21tsqg9Lc2wOhzS/pDTI49yZ/xPBAMiMYETEYBAhz3Y1CepEkVW6FXzlTePW4Pz0VA4s3bUooVSKh8ycgTWbdyIhtEjYUplseSZV3DY5Alw+VzyeQyZykoqEYvHEQgFsYf9V6MByUgE02bNEk/X1d8n4QuvmX8mTZok+YQ8FJtVsKUEJiTNZtTW12H7ug1C2itUI3avnDGEezGX5stnLMSW1Tsw8qqrcN7jt2BJRxrdihnsZKQSgEVhEz4j8ga0qGA2A1de60EhHSr5cKxmVLBJH8sinjKI2nTWAXh8Hsnd0lxMdkqCadInwJ5QMNyl4v4pYwCjHUglATvBFCYUuqxS1eYzSGVjmjHmm/qymfNCDjrgWsm3YKRIkk7DTylw9tJ6e/DuD9/hsZdfkGolARMjSsrw1P1/gdWnQfEY9XBMjS9uXpmcMZvlJJIoJJOUdaVx0SivuuoqcYx8L1Ey+gAwRXNb93dIYefaa6/F119/jdKKUrz12pvw+n2yvx0erV+pazjqTG6MoMjBRIhkW0urqCkvWrRI4G98tjS4N954A1OnTMHmLVvkpGYISwRXZXkFmoYOkRFAfh6vlfOuRPGwH8nw+z+rVglaSopOeQpHAdlYi70q5744q7Vs2TIxLItDG/ZliCWoi3xIIrlPNI7GumEY6O/F0CH1MHJcKqepAcsHmjJShOCEuqYQZJTElTE9L44FMS4iZ9SEYi+VFEfAzw4MRPD2+0tQ2VCPhppabNq8EcGBAEZMnoSi0lKYXXa09PfCWVEGp8+F/p5OuE02jGkaAV/KhL9deyu8bpdAxFjypvHRm9JgQsE+ySsNNqOENU2jhkvBghfEKhmNtK6oDKu+X4HW1mYcNX+eGCI3gMOuQa9CsZj0QikJxlCXJzrvzVtQKEk8e4PcJHr5nPfkslnR1t4Jq9eHpimTJYR97+VXUOcvgcNIDh0iWCAFJIZZe1r6cNnjT6F//AS0m23oJnOHUUG/EfDzFI/x5FORMWYEq5rgyUhVI25U5oc8nDhLaLPApqjoD/TAVVkCxQd09lAK2wBDzgBVKrWadjhDsjiycGZNcCt0YmH4DWYUFzgwusqC4SQt6EzjvAXHI7J5FdzVfmlD6fnV/8kYuf50Unt2bhcHQKRKb3AA6XAIww+bhHKh9OzFpjVrYM2zH7C6nYsmMP+EYyU3ExnyvN4Fn4XVqIEPBOSQyYiRcm/RUKgwzBOLjlDqBKG0PCcOtb/wwgv4+T8/i4MdPXq0/M6f7rgTrkJN7JT3ovPUsHjZ3dUrNRBONHW0tEqoykop/3BPkcxs4cKFEobSMPn8eahwb9GAyXKuDYerKC4qkOvkPv/555/x6GOPSR7M/cV8lEVKSas8dX61urxSNhU9jMTOKosIKkxUuNFfDBFhRDySwMiJU+H0eLF1w3oMayxFJhGD1+sUD6AT8Az+WiYrpyAXj0ZBsicaulbCh7QL9B6b2WJHKBLFz7+sRk1dPVSTG7ub98FZ7JTcJUqUu9uOihH1Mori4oBzMCKVQmUgjYUTpmD+2Ik4+bjjYbDZhSqEPJxkjDvqiDlyKkSTETz3/PNS/h49ViPIperu78/9PW699HKU0qPl0qiqqhAjYcmepzMfPNeIuS+l0vXEnZuNISofOCMLhrPsc7GpS2dUX1GGbavWYdjh07Gts0PyLjZ7j5g0HZt/WoVwOoakaoYzZcWBrn5Mv+IaFF95EfYQt5g1IWI2SGXZmlHgD+eQ2xNGXZEXEVMOdqcq+iUmh0WEUVPJHCL8mYqinCxuiW5UjShDZ18SFlL180TUIkmY0ioy5Kml0E5aFTk4hhcW5oai68awFHDnAB85Y5CFz53ESGMat06dCYcrjiBD1YwqfC/SXmJbQbQ/DmFlS6VgoS4ki3TtHTjzggsQ6BkQSo1PP1kCk0vvG+dDxvzGSYfCOHL+fHGADJMZUiv8adD6xTRMCSsZBeUjEraW6mrrpf3z4/KfQJFfDiq0t7bjtdfexHXXXyO/Q3wp/zAy4El15eVXoKikWCIVnsosrvGg0ChENEA/ETY0csqQ8/RNpBNiL2QDoPG+/tqbMs3B/yc7oVs0QDV1a04x0THT4fPz6bzYVyS2lYbN72N7RrFVuNWZ02bgm0+/hNmjgW4NBva+mDMcgoPUjXEggvEz5ouuIkFNXW07UF9TIaKcHG/h7+l6BvLQKY4Zjw9C4mT+0KNR5LE6yYonb1Io0G0OTYAyrSKVzuCwcdMwdvJ4/GvpB4gZsnCVlSBlNsBoN6M30IdQIIhsIAJbWsFZcxfh8bvvQ6o/KBrw8xceIwvN3IL5239+Xo79rQfh8jql/F0uGNIeFJQUw19YhC8/XYYShwtenw8+nwebt2zE2AkTJDSNZzNy6tGr8idVl+jZaIBM2ulMdLpFIkJo6DRGSutRtfiz197E3c88gyXffCNVyGQijfmHz8anb74Hi9sOS8qCneu244HOFiwJxxCzuRBnW4PGw6Z1RoUvpcDYk4XSkYOTGFKHAqvbIC0CVgsTBqDHmEGStBjGKCqLzEjZmMsrMCQV2LIaXwsJiBM5gDotjDwjKhBTszAmiWBRBXSuC+DwAlxkQKNBGhWkQkC2ZRdCX/8Dbd8txQCtViB3Gt+Ljj/OUp+OxplKoaCkBOGBXnkflZ5ZnKmvbpAe35r1q7V5p/968Qhgkz5Bqo2xY1FYwF41p0Jyom2of5cYitkoDkB6txYT2ts6MGbCYbJ/PvrgIzgdblx80SV4/h8vimAvnxXXgYZWXFgsI1I0ID5HUs6w6MJnyUkg/p1eye9qa8ffn3tWTjj2EY1Wk3wGjY37NyGCQB7s3blLrq+iplr2OItAq9eslCq63n1IxMOyJ3lqk1+HEajA4Vw1Beq82XOFE1SaqqIE5NAwlPmJUkHrq2SMU+Fx+1FTNxZJom8SIRhzUWzfvA5Dhw0RfKGJHjU/lSHlX4M2qqOjFYSNmhdbUSF/393bOzjSkkrEZerAaGSfKC3UFWSse/ytfyHlscHWVI6S0nIRr4n2BNDgLoQHZvzw+TL0bNuPoxYskBicOFfiQ+mFVq5ciZU//wyH1yU0DkNGNIEjYdTk4CIw//zdGWfh4K79eOqJxWLAVhIicSLfYpbKJUMf/qEHphfUuVAECJ9vSnMzSJPaYsFANCGinwcPtKDQ58XOtRsQDYVQN2sW3F43ksxnExnUF1fg4O492HOwG7ev+hUvUJJNdcEmTX4gTuQJQ9A4YO7PwkhqmwEVLnLPwoC0S4FiNcKUIYWtirhTQdyVRjzVgfFDa2CIqkj35dB9oA+ZpAG94Sw5mZBQU2gYXi3PxOg3opcnTtYgbSKTzPSxXaGJyFIu3U7RmyylugFzMIZLJ7twSVMZypqqxYkmYtqQsh628mQc/G9+hyEnxTROxjD0PLDnIPoG+hGOhwaNUZ8IkTxGjFEjP+PnjBo5DDVVlYjHIyCgXd9f8r481lccgaicWWVtvL4ClJWU4ZGH/ypgfdGVzFdPGakJXxGLYZmMUMIcs2iRhK/UViGF51fffCNhJQ2Xe4oUK4Th0XBZfc8YNQCBPhNaWlgyeJKKI8oP6vMEXvHLT3JAMT2TvDdPP8n9wj3I1IZYZqV4WIU6vGmoFG5oyXw5nT5NGiyrjcboxsgwddq0megfyEpxO5OIYP+OLTCRfIdQNhYsvCbRy6OxcdNmM5qYqg5DI7+M/pn6rBw9Ct+bikVlUyUoOW3QqOWJnHDX1mJrTwc8VX5UldfAlVHwxvMvIbSnFXa/G0NHjQap2UlwzM9hw//k407EXx/4i1B4FBMRZFCF4KiotFAmRNZv2YxCn09Ecb76bCkmjZ6II+bNw66dO9EfHsDW3TtgL/QhkIijwGITT8ZEn4Bf5th8mHp5W+dIlfk9oxlml1c4goQmA1nsX78Fo2ZMR5T0lQ7K6AHhgQhq/KWwZxV8v3EH5q1agXWqESnmdKoCCzd+RoUpBdhTCuIhFYYBFmcID0sJ/jRhzlJFFBahwlBhdmYQcLVg3NgGpDsTcEZt6G3JojeQg8FGrK3GZp7IRuB0W+EwK7D6jEj4jIixqEtHzO/kKUfyK7Js51SBo1kTRhgygCOXwjx/Dq49q3HnmSeipK4eUSUGSqbpcDgao17g4U9O55OpYdGChbLH9u8+gG07t8Pu1tpm+knHn8z1JaLK9wbFftSMODWyREQj+YFl0RbhmaFxyXAvUXiJ1WQ68slTpmLn9p0YM3oc7rjlDsGrMh/lixGOCAtJTcogEyUXXnihcOjyBGNR6JV//Us+l2B/7mW2/5gXMmdkq66ktlJj7MtTj5iZh+f/WzdGOm+pxSz9HKUVZdoQQTCImuryQdpHfc1oAwrsRpVo9IH+gPCkchFsbldeYzGhlW6MaaFwSHRFMOfEs5BMpZGIhuGyGfCfn5ZLHsQv5pRzod+Hj955HzavSwo6wxpq5d8rKklurLGGc4NSf0H6SGoeF5jNSpWKi8ocTGNEU8QAWBARUqxCL5Yu/QI2i6bNF0+mJDTgv+tVN37mim+Wy5SGPvLD6xg5agQqaspxsOUgPAUeyceIrb30oktw3bU3IJ6IItwf0CqMBmD43KkI5zI4YtoMtO7bL30gvvTogT/5XfSee3fthdPlk4qr3eNG34A2XSIs1DkF7XsPoGLYEChup0wmcCRqgFXCtILSLDD6nr/g0yFNSJldQgJlzwC2DA2DoaSKVFxBMqHAHszAnMjBlqNwqab/kCDPaCZO/AOKh1EuLwxjwAVjRsGW7WGkVBtyJjuySg4OeuK+AEcE4ff7YMllYbapUP1m9FmYJnBuUCN9Ujk1z83KucdkBraMCZZsDrasilqksbDOikqTATU2BdOH18JQ5IUqwHOtuKTzpPL6c9GQnApsnWkTMyb8/emnhZkhw/xSJxJlxT5fptA3KfuZjIYl5+zqxVXXX4EdW7eJcXCmUKcx0SI6szwjvligmTrtcHS0d2H06DG48bqbUFJRPogQktYCIXKKgq6OTvmdkpoqjYw6MIDTzj1LPo+tERrhw/f+BatXrsKHH38ke/OCi8+HYjYJMo02YoxnpRZCYyNcb8682Th89myRJuQoHSOw9rY23HrzzXj0kcdw2KQJQhZus1sEBihgFWOBUy0uLUFne4eGsGBoQBYuUgK6nJoxGjJijP6CSlTXj9DyvVQMG1f/B1ab1pfhppTRomxK+kKMt2lcGerYkwzXoGDI0CYBbtgdFvFiJoYcknb8FvMzdqYHIQChra3n/2PtPcDsKsu14Xuttfdeu+/pvaT3kBBCQkLohCJFOodOUEHshU/Fgx4LggfxWEEFUVQ8CCoqKioQIIZQAum9TTKZmUyf3dvaq/zX/ay9JtHf8//f/13/vq5cAzO7rv0+7/uUu8gx7jE+skYR7W0deGXtOkFYNDe3TCI92Egh6oU1Cd8HcYYCtq4yuykDUt9ch0suuwTr31iPsC8iej1d7Z3YvGmraMDQWpyfuXXmVFx29RXCXnjyscexYNZsOXE9zRiiZLy2tdSSeg1aO1qQyqQQCEeQLxvQGViWIUprhO91zJoCMxCAX/eLhEamWBE/+dGtO/GZ0iC+N0CbcF1meXGDHEXFTSlzFZSypInpUMdshIs29IoPVqnKlAM+0F8AACAASURBVAg58Ff8GC72Y8WlHRg5aKGQcpAfVTARMIWSS4iMFlBhZ/vRXdshujLZnIFYkLA0E6PFNMJ1UYHyaSEb/rAqp7jDEYZJi3IDAdOBWjKEy5eAhnnREJoMB7MTGp780kcxsn8nRs08TKMsBjMCEqBiKlW7J8YlfSetiGMl+iLu3b9fIJFEd/0/BaOQmCmlYbtGPIZZFLcpkofFrqAqYyKnabUEknLB58O06bMwPkb5TD/aWjvwlfvvd6U7qjQ8T3aD65e+KaNJt9HCAOWaZfrIBgvHGhFfUJo+nGFSA6ehvh7TZk1HMpuRGOHGya4qO6OsOVPZFBbMmY/TV5+DvXv3SuCT0F2TSOCsVWfh9394TtzNGD+MBQnGUHOtU8xmUNvYJLocGnlwVboP+YtsSJDC5Bg2Vq08H5YTcD3bYeD1da/IichAlBSDUnRwTzr5gIkEQoEQ2ttbMTI2KEP6ro4u2RH438nUOLraXBKy1C/SMnNzcQZoNluW/JzPy0WfLmQR1EMCKGDwffO/vi1/5z/qzDBtFIkLavfkyhJAXirBTltze5PUbIVyASEtjDW3rsFXv/QVKfKXLV8qdXOkvg4f+fxn8OMnnxBSqhBjd+52T9/qULuQdT+jSDcICIEq2DS/VBGONCBaH3MxjiULih3F4OG9qG2vRcEEahrr3JTc0RDh+GD+yYh9/SvYWLTh0MHZMhFjimsDxQpQKdlQCiqcMuAvOPDlLUQyKirJChydtaKOfH4Ip9/QgkM9JUwMasjbOeErlixSrGjz7aCQGsAFq7vw4rMH0dgwAxPFPAIk8AYdBGK60EF8ARuO3xBp/3iNhlCMFDcxqGJzVTZqXQHiOgTlE88BRzbuwPvPWYib441onjmdasgu/YsbkWrJ91k8NoDFy5fLNePfujo6sPHddyUNJqCEAeUBCHgQTJZGwrzgWQ1QNU9qRXrXsT6eyGD6zGm49JJLJkm8gl2uasryOYyKjSnd00C5S5ZY3/nWt0FfGWZTXCfjo6MulI1zcPIka1xH6cH+fjS3t0h6yiYfv+Njh/vw3ve+F6+99hquuOIKnLJ4iYwlaNjL9RkKuFnSjm3bxe6Q7BN+Fg8VJic3gzwUxjlnnI2fPvkEamtrMG/+HDGe5Sksaeq9X/oPvPCnP8tFam9qxo9+8oQEITl9omPqWKgUyrjg/MtBDl2plMHmjfRodLDi9DMkr+bi45tikSx0lKpxKLmEsXgEDY312LZtC6yCic5pbPRQA1XHnBnT5Hjn0U9pQH553hyJ0v1Sd1ZTWHIp6VlI4x2+ViyekNNw165daGhtkS+aASloHdMFLvC5mDpcedUVEoxvv/O21I+DvUPSTaPI1Fsb3qoafzZg9RWXY+vhA4hFQpg/Zw44PE4PjUiNyMJfNomkWxN5cn+cR7Hf4VQiGDiaB0pFtC3swrE9Y/jigw/iK5//ENrmtsNWgtATIZdUqgVQG4zg5i9/HT9sSCDtj6PkAPWUNZFmiQO74kAta9CKgJ3nqEMBUjYi4zaMsRJGk31UCsP0xSrCnXXo7w0ga9hQEyGYegV2mZzACkJhE12dIex+YwRQwoL/tIsm/FEdVohW4KoAPURYilKN5LPqDiqKCS3uFxs/AuSZLrJSY2+zZDmoDZuIlk2EVQ3L2wLof/y3+MndH0Td7GmimVth+1W1YKazQiuitAtPGqq2DQ4PS+ec6Zi4I1drwH8VjFzI7MQKi8Jx/TZIs2O6mx1KoX1KB6699lqkk6PIZdKTz8UvZfmyFRgbGxebQzb2iG/leluzZo0AwYkPZtOOLtucfXNdcWbZ198rHpIMWoHbjbjEA0LiyFUdPTaE+QsXovfYAJ547DHUN7gllqDRSMq3DSHbCyOLtSQzQNOUfkNrXRNOX7UCP/3pT4Tl09nVIVmk8oMnn3DqojEZYpKZwAXd0NQk3EXy0gRCBgeL5i+TUymT5eggLEEgQHL6JRJiVUVbeIh4BqTc6ABcUyO40Lc3voVYnQt9Y6rAYpkXj7sT68TZs6Zh2tRu1Mbi8v+kRXHH4KIXBE/VvYntYl5Qwu0o1JSI1+KZXz0rJxeDUdKUCBslFdnNCRrXImE0dDRKB23mlBmYN3MhHvjSl5BobBT0xEc+/jF8/4kfoaxZiNXXgtdkYmAEb77+Brq7u0TRbN/eA4KvDYci8hm40zXU1KIIP2qiYWx/9wBeHunH0T7gjsUL8Mr4Tjz6t2GMvPIHjGx6VQDyna0tODQ6LmyQw/sOYe49N2P01luRrQShF2hiqsPwEevJYNTcIT6jtAhEigq0LDC4YSuu+tRirN+eglIyMX9ZA95+m7YCFcQbIsiR7lTUYSTKaO4MoVBMI9uzH++5+FQc6AEySVvAC41dUfTuy8EfDMEKaTBZcvmpI8POsA3dsV08Kg1o/CxfXFA4VwSZ8HqZ80+6GxM8UEaDXcaCeAjnRwO4uqkezVPakS3ncf3VFyGsuwp8W7dtQm9/7+QoiMHoMSHk5ExOiISm140l0kvaFlXhKO/38kumw9V6k0GTG5+Q63rZZZfILJCjg1kz5wj+N5ctCGHh/e9//yT74m9/+5t0Mbm+uDHUNTbhhlv+Dd/4+gOobWmc9NzkiUeaGNegkStj+tTpeOiBh/HDRx/HqcuWyhyVkwdKOrIJVEf4nuNaO3iQO9LamKqSRzs+NiYjFaoJ8j0WKhznlYUQ7lBvhmMAfmC+IGX/tu3YDrs68MyNjOP8y65xEefFDN5+e73MYZgORsjMrgogy8lYlXv0glG6qMUiFi9bhq07tqGuoVZOTtFecWzUNdZL4DG4mPNziMt6lVYCpy48RXYaXgwGLE9PdsHYBGD9xnSjs7MLoyPc+bKC+/zCvfehpqEWwXBQTkAG8yUXX4xKQMMWjmCmzMQzjz2FZ/7wB1x/5ZVYfsYZePvvf8cXHrwfB3t7sGX3NixacjJ2b9qCXVt3iRelE+Asi6QmDblsXk6WmmBIUpCrb74Rb619Ffm8iUvuvBfl2edgfCyMFi2NQGMI/SkVq5bG8Iv/9TDmz16AmrntmHHmQmQ0oBwBfjs2jp54DZB1oFsanJCDnDROFJkv+kppNER01IfCUHPAu8++i6tuX4q1uzIopCMI1U7A7w9hfALQY35EYxrKVhm5w0UoU+KI1AbgpCcwtbEWO/+8HXrbbJSNLCKdDcibWWDcRE0kgUyEM0sypm0xn+G1o9hKmAB2x0IprMFQXVYTu+dBRYPGDq4J+IplRAJB2DlD5pEopXD/aY24rm0+LDOHG2+/ScSn2bo72ncEL659WShz7mnoZi8evYn6o1+7914E6urczZyp7gnB6KkJSCPthGCUDZ7zXn9ASgrRR61UMHfOfCw6iR4pZSxesFBQNxzwC5ukCgPlaf3qunWYMXsO9h3aK53PUrkw6csh1u00Ca9UoCsBlPMl6Ta/8Oe/4dzzzpbewIc/crc0f3jA8HN9+f770djSPJmmEhRfKRThCwXFjYuvzxr6ySefRHMHebAKFH88wgm7BAODhJ2g01adjt179yAr2MkQVqw8S9K+XJ4A2AA2b3lHgoQvfuaqVVLUihw+h9EnCBNJR0ykMHywMnk0zegSq2XexxtneCr0DE4GmCiA50sIRMOI+vTJQbKwMaoYxQsuuEDSh4mJcUlBNJUFervgAAnipWzeyOgw6hvr0DcwgOmzZqJrzlRp+sT9UWzdsAUtne2SdnLIyyL6lttvFaIt3Y+I6PnFoz9Ga2eH1G70mBe8LdXYDBOnLlmK+tYWtMycLrKWP/vPbyHqBLF5qA8/3lTGvoMmjFIEIWcM8ZoGFPwl+HUFA0pFXLUyQR1FqqPrGsZiBZTDPjh5R5o0hkZBJiBYLiFYyKO7mU0w2u/ZYmbaHmnGsVEDvf1cpOOYc2Yn9m7PIFYTRUObisNHDqNuVhcyw1mE9QS4NWd6RhAORGBPuF3okjOG6PQGwZtaAwVolgO9LY6KHzB0Og1DRMmYtrLqDLKZpDqoUIBX4/Ddgs9WRQVPYzCqNgJlBVFDQcCsCLd0lplGzZZX8cSH7sIn7vsiLDOD5Fi/fEcvrP2rbK4Sa9Vg9MYTZAGRMHCwt1cG5jRd5drwuqvePFJqTKbVVRkT+X3Vg4MbMAOHCvjcQMmRLBUNIYzzxGQdyOnBhjffwKpVqwTkzU7yu1u2IhwPIRgJyRiE69HrmqsBn5ATjKIJxbSRGZnA22+/i3XrXsOGN9fhwIGDcqjcddddctrx802k0/jlz36BhuZGjA2N4oZbb8L6NzaI1CkPFfIwGUMvvPSCWOEp0+bOcqiCxZSTf2BUUxWNuS6hSKVsDpdeeT3GRicwbUY3nv3Nf0uN4e1WuYmUK8vviU5VB7483U455VTs3Lcb5XweK846S1yExlOubTZzZ2Fvw3FpJXT1pXYOiaG5nLSC/ar7JfDCC6AWinzI/t4+aTjE6qPo7poqKIiZ02dK55XPyZ3xwtXn48ILL8a0BTOxaMkSHBzowYrly/HYd36IuVPmwRfSRZbxgS9+EXWdbbj+xmsxOjGGI0cPiQr60sVL8LV7v4xwS0PV/VYTVA2t1XkyL778YhwcG0ZrRyfMtIL9//1bvDmYxuNvjiBvx2DkQzBKDiKaAsdvwopqyEVtDCsaMj4ga1uiiO7rBAqqiYJYtYeRFkEkB3pyECc1tyLgKNJAyRYt5DMaEnHgaD+QGZ/AtFPi6B/NIqzWIjU+hNkXt2DftmOond6GhN/GkR3DUAsx2FleyxBUNrTYRarTEGgPuWlnHqjkyrBzecTa6lAMCO5CdnRT2phAUFVEKKrsJ/TNNXoRlAzvZAHUxw6WFIRZ49oGak0fwrqFVTELB374A6QO9GHlqbPwzDM/lXntpl1bJo1pXMtwFxnjdukNmVc/9fTTor6WLbnfqXQb2emvNhcZGO4arFolUIjLr4pPikdw4PsiUIVAfb9Px3f+8yExraGJDl/rN88+i9vWrJHnJ5SRndGiURTlQZ6MXoNOJEACPsGqMhgd00ZbXT3W3PZ+kW657tprseL002Qd832SA8mmInWBz1+9Gp/59KcFh1rDupCHU5Vzy+fn6fi973wXkZowlIbmRkd0Q6oSAJ4PQaFYQtmyMXXKdCRqGtA1pRVP//JnAtCWXnM1dSgXjj+Wp1U2n3JpVoUSps+bg3wxL07BXTOmH9f8JH6xcrwbOT44JJ4XIT3kDqWrpGYOnbjwucuxXbx8xQq8/Ic/yQyTwUqVZ29HZSBzZyKqnifnvFmzUd/SiLs/9TEh/e7ZswuZY6PCF6RTVe/hwxJkK05fieHRUfQNHkVXdyemTZuKkeFjSI4lUVdbj21bt2NsmLqiVGezEAmHceYNl2Pdwf2IxxLQw1H0v7kH8+rm4b7vfR+/WFfCOEXaaCKjB2QhS7bg9yEbADJhEzm/gnRYRdmvINQB5BQLNnG3fh84Aw9YOczvjMIYAxKEE+YcQTwlMyYiNQEcPlBGuZLF3BUN2HM4j5ClYerKII6WgdzWfYgvmY2Az0QdU8wCcGjjMdhjFdSGu5EcLyA+LYxSKC+bKDUAzKwD1XSgEEoXZQPCmXQM5uLhhsk0jZ71Nms8VjcyjHeXAkEB/pKJiKUJfM7vmGi2fJimlHBSQsXW536NuuwgvnH/5zF/5UoMjYy5HhOsA9kdpSpqVT+HfEX+N/WEOB7LG4VJUjNPvPHMhPiwiFmMA6FYjfT1o6WbWZch3dcKg5unKS3puU4tFdSH27VjN1raWhHlfJOeIIWsqIXfdvPNaGxpkZkhh0GEO1psQFVhfSJExo5pMChelkrFllR1fHRMyjkeAp/85CeleZnP56Q52Xf0mHxGNma4Ji+99FLhWbJuZo+Dj+G6YO3Ik3rjJhr/KHBite5ogheHR7P4ypcNjAyP4OrrbxRpg2ef+SWitS5MjnhHbzZoVp2RuNOwFcy8hicTT1kGlc+nCcVo6uxZrkiQZQk1Zf/uPbIbsmFE00wWtwxGjwkhTGg6EVTVtvlci04+WR6/a8cOWQTliiHqzcQG8n5iK1fdWMaGhlEqFnHTh96HHfv2yEhl9Ei/ULs2vr4Bkbo46GPR2tGOVWeeifH0mKQKhlHGwpPm47VX/45wOCK76sDBIzDyBSASQvfSk3FU5wBdh8HmUW09dn3jp3juUBYbdoxjc08FoWgDAoGQqLe7HnouliCnAcmAhYKuIEtWvt+HTCAJrSWEYCyIHEHb1LcZOoJV05uRNXSoRgCkReULFg6PV9DYHMTADgehjgkk/UEkohGk9hxC4JpmKE4UypEJRDrqUNRtRCoqAiZQODyEYNnC4L40AqF2hOpNLF5Wj3wZeHdXEUokCJ/ppqWsGX26CrNq8eC+e/dEYtD4g77JwTxB4sKRJ2yv4iBssKHjoNGvormios0CaJrWUmvgyG9/gV998wE0zp0trBiuM2m0UV6lCizn6xAby0XqCUwThM4TiXZtjz76KO6++04RNRY+q8+P9PA49hzch7kzZqN1RhdyJXJUFRFQo3sYb3bZlfO855P34PP33YfmtlZUTBML5s6Wbrkn18LSiVYDbLSVChkXn11VuOP9+VmjwRgq+RI+9bFP4vHHH5d1d8stt+CBBx7AosULmSmDAm/efJuHSSqZFUAMex/kRa59+VUx4vU2nM997nN4+tmnmVIrTqwq686LwyAg/vLwkV6s+cCdCAUj+O7DDyDUWAONV0ohKr8KOubooFIV0aVh59SpcHRXANkbT+RLBaRyWUyfPVsYG+wREKcYDuiyIzDIRo8NSnpKlH1mYgIapT5YZ1YDnc/F9jKbRn10ICKFhgtdc2UUuHl4RpReA4lFcqQmhkWnLUVP/1Ec6x9Cc7gWo6NjuPOuu3HvPR9DqKZGTkbWlWeddyZe37Aes2bNFOvzcDgq7IpioYTOpmas/dNfgHgIS9bciOFCzoWGNTSgKZLA+s99Bz/fP4TBiSj2j9uYGKfMYUXmtXrElYIksoRg7rTfQcmvIOeDOCwls8cQX1wPLRFCkmLFJQszrCwu7KzDmKmiNwmotGUwgJ6+AhYvDWPzS8C0s4GjGRNjbx6CurAOyooEfJZf0sV0IY1AQxC6oiHuAM3wIZQtodEM4uheG0OFDJRWHQUrgDTHJo4hGrQ8ghSlAr+uoUyE/wk3yYWIYQ2okx1CptMCEiF8z7ARhYpQBYjbQLNpo86wEbEchG0Tl80M4ZqpLaif6spW8MQQawFmBf8UjB6pndkQCURsxFBt8I477sCbb6wXTqmIY2Wy4tLMSQDVvDumTxGNJUqYEBqZSiflE9CXkYJkbc1t4t1JhXwKVlEpguURX48ByQMhVypKF7RSzv/LYAz5wyhnC1CrpHqu0/Xr1wvK5gtf/Hc5fDjKY3fUg5eSvTM0NCzrnaVRbU29EA144t591wdFj/jb3/s2lK4Z3Q5naJJ1cEjJzhY0afefunw5Hv7q/Qi3uienKIbxVLQYw6ogM3ikG2YFcxfMkzzbewOTcn2OBcM2MX/JyaKSxYtIWznqi/DpOP9pqW3Ay398QRylZPipKNLICQXcFjf/8YtxL5LbAHKvsrtguHmkM0n5PS+ugIYrJt5z2aXYtn8PBsdG8dmP3YO7b74Vv/zVr3DTdf+Gr/7ycXzhc/fi/FOWoW9sBGevPltSCOb9o6MjkzNET4h3y/r1uOOJR/Cnl14UGFRDJI48pdtr6vHmPQ9gQ9bGS5tLGMwHkUwDubK7w3NhG6YFAw4qqoIcFd5sBwXNkaYImQlt5ySQiQPDY3T44olWwuVdMTglB0eLNso5wEgXcXTbMZx64yy8/M3tWPKxk9DzylH4W9tgL/UhnXBnk7qhojgyhpruBsF5kvVfw4WTBQa2jqA4rCLImijiivRKsyOfQbA7DpMpqFVBLOxH2qTUiKvWzpuv2jBhL0ANubCzyaaKKNYpiJkaghUgYduosYA24myzRQQKOup9GXz/rKmYOm+6GBGVSgXEa6KC8vE6qXw+ggu82TI3dY7SvKyNDTxKfJh2Requ/EQamVRWAoEZ1a3vuwO9x/pAVyya6HCdcYWQPVOTqBXiKLMhT+aFz8H3wROQ2kVSEhnuWiM8krHA71AaSFxzjoqWukZ8/nP/jjtuvQ1tHe3yXvk4KouTqyiHlWOLr4c3d+RcmZ/N62mMDiXxvg+swfbtO3DN1dfJQfPII49AaWhrdASlws6W3yfzFuLxeAeiDYQ9rxznp8mxbylwLBul8RTOWL1ait6iWYYh8C+XLMvA4fCTO0TZqiBnFFDf3IgwPRZYE7S2IU2N1nwJHQ3N+MN//wZ6JCjSCWwGCaoebleMJE7KFnhdNW8ReMFYKZXQ3t3hWngXCq4sQr6AJacuFYGlTdu3YWb3dER9fmFlP/3MMyi11qKtvgF7167De2++Aft69uPKK6/ET37yE/d0JL2qtlYK7+Z4Avq8mRiM+EX4mLYESqkCMx7Hwq5p+P37P4WnN+bwUs84slY98hWOF6oyH4YpuzAHykUayGo+pCln4vOjrNqC0gkuCsA3ExgYcaRm9JVstBWyWNnK3b2ItOWX0cnWv2zAhZ87D5ue34Tl7z0Ff/7SBuDs2Yhf3IAc254m00WglC2gpi6MIpFwcBAg3j9rIrljFEG1DgbpcGFdtHZ8lg2zYiA4NSIW7VzI4YAGi0N/k0Te4ywKOeFNE74gleNPCEYCDRwVUYtNHCBWsYV21WjYSFAqv+Agm0zhXP8AHv30bRgvlFCuFBGLRWCU87LgvbQu5HepSfyu+d2z5S8HgWfMytGLqlbtDALw2RpSw2Nyn3hzg9S1Kul8IV2aj/KeK47Q5NhR9Vj9DDRDEFUE89tobml0RdFUFWMjI5Lm/qtgDGkBfPDOu7F+HTuoByTd5jUhsZhrhutH8xHRU8UnV8eFvI/4kI6OirM0JwDjo0k8/M1vCuufQa34wn5HGi+5nOiGsoilUA/b/h78S2BqJ+iaUHX5wtUX4Llf/xpTu6e5fBfOBx0TtBxnt2qkbwCnnnsWkqMjMkbIGgU4vEiqX7QzqdhMafxirojsWBKb335HAMpsyjBd5mZwaF+PfFGFFEWYiG89bjIpO7Ptiv/yg7a2NcvFY60hTQcHuPm2WzE0MiJ0mNnzFuB/ffgjuPLKK3DzPZ/E2rdeF4xjfmgE5512Gg6PHJPXJcp+47sbJ9XxWEe89tyfcPo3/l0s3Uye0j4iK1TkNQ3XL12FJ267G/d9+0UcbJyB8VIcaSqcm6rQfQIVluUqfH5ObNiJ86Fg2zJLzFHi3VJRabDQer4fQwUgnbIBlqf0l+jpwXsWz8WOQhmDJvDOX19H0+KFuOCqJuxeP4CevUGkppSgXdgEy/bLQF6lr23ZhEIWRUNQmjFaRRVQepCNpTSQHCoioIWE7ZFPF10b7G6CDShw5YpERhM+FIuWuEuLxGI1GeECZifa1cBxRz48FWlqHDKAsAFETAsRG2hyNIRsIG5aqGQ1NOoZ/ODcKajv7BLCreVUQG0iBoQ3slDJ0zrxVh3qewD9UMQnAcBDQoKl5Do78zsXrdR4RDYUGYiSDsa5d8URGhVPPS/A+F2L1yjFl2GDlnjsX3A98Z+4rVUq7uYjiC5XcLgwmsahI0cw0Ncn5RHRO1yr1DsiPI4zyy9/5T9QMty6kY/nexXctukitzin7js6ICqDJD8zsxM1egYjUQmPPf44EnW1gnJROfeqzgx5XejlJ7tUlWuWH0rigisuxxuvv+4yvWEJel6CsmKiUC5h6bJlYv/Vt/MAZi2eLbtpmd8Yu210uC2VXQfZQEjMVInu4ZulAC4DkHVFbjTt9tj9qnTHvGPfAxQY+ZK7Ivi+HFMElvlYvs/BI324+X2346knf4Z4IoFFq1Zh8dQZ+OHTTyEyrQvhkIuHbUvUYsuf/oZLb71Omk78kg4ePiSMf6YVvA5n3H4jXuzZC7NQFOEo+lBMVAzE6hpw3bxT8KM7PoyLbvw0QlfdiYHBGNKowLCYbjuImCqsIkEONOG0USorKJkk9SrIFE0YFR8m7BwaL1CQ5jXK+eEv2IgHgA8ENeSOjuNYTURGIhte2o2TLluE/u27cM0H5uO/v7cD2lkzkFugwrYCggjimvHRTGVwCPEZLRAVjoqDAN2mcwbstAIlpcnYJUhESMFVOo/MDEkwWlXiQywE5PIGFNJBeLpVAVUinSKeKtWI4XyvqhJARE7c0RCmQLUN1LOGtNlxdRDNMnLL6M68g5/f83HpOtpMpAm6/f8QjIpmClyN4k+yHhzX/8TrFTAYuR6p00PnYtmYFT+SEykZWfGA4e+EZCxpaEXgnmQbeaQEmbdXx3wuZloGzXIdEqEoBntdBg/hogxGIReXSvI+CA09bflynLpi8WTgM3Z44HlqcCwnOCYbHByS0QsDkbRDxR8JOESjJ1MTGE9PSEftRH0TMUmtGMKx86v01yjhmltuwpEDB7Fz8xaBYJGRwcG5cCDL3GltLD55EYxKER3NDVLHcTfz6kGPiOt+nS5I2E1JPMC4qzAX9MddIHaVrtTXf0Q+AMcT5RLlFN1GD+sFpgzeLiY6lIODuPPOOydJ08vOWIGXXvwrTrlsNTbu2SWzMb6GAL5TWVx39kXYe/CApEiUhEiXcoiFQyj7FHTfdg0O7T8gjQTqwxITq0ajyBRKODXrYP0fX8DwPguffWsz+oeDGCs6yGt+mJYtQ3FR0KOMBkED4kJMELiGobyNsUJBFNcSZ5swWqiGTtVsoG00iftX1eOtd5IYUHXYwRD+tPYNzL7kdPRtyuHk9wbR0O3D37amUJoXQ0jl6eJHhUYqDJRCWXiEZb1Ce2P4sz74yyo01rI5oJRn+mbCb1qIqj5YqVEEZrVgrDkHTYnAIYGZSJNQ2JW8t8lgBco5A/AFpYHB0QJJ0CQRkM/HsQR/FzNtUeuLWhBl80AZCJYshPIVU6fymgAAIABJREFUnDw1iPvnx9E4rRnjBQNa9dil9TyP4P8b8b+KwPEOSwGKn4DI4bXz/l8A28Q4V6GYZEMwQFhWESYXjyYmu7gSlI4lhAViol2ZURexxZ90ZpOsrFCY/MnXoZ8mX7GpweUkeqqAPF85r2c5xQblB+/+AIJhyoMQLOJ+KkGd8XS0XH9Irmv+/zVXX4+vP/gQFD0WdPiLgcEBzFowR4RdTwwAvmn6ljMYfbQlS6Zx9jXvhZnNY+87m4RTyNPUzBew/OwV6GhpQSgUFLQO8/2g35VIlx3qBFa8h9LX9aB8WO4c1GTx/BIZvDw1vQsi5FG/q+PCYGSnnZsDXX9I/wpHI7KD8TWIsqBf4X2fuxedU6eg7/AR/OI3T+PTn/sMVl1zKTZs3Qyl4p7Acjo2NmPnH1/GouXLhUZDErKpWGhpacabu7eh7ar3CNSKRbwAfpnXhcMolAyUX1gvWqvddfNw/qc+jVnzFuOFnSXkAkEYBTZfuFoVl0lfIZtFBR3BM3kVI8SUBzjpU2CgF7ErmzExFkKobEHf+ja+f90S9E748G5WwWjRwt/f3IHapYuQ2gU4NUlccXMzetJZ9IZ1FOJMuxwotJogA4OKedmipG1mykS2PwMU2B0haNkvmQazmYjPj6gK0J8pnc3Bd3IUvYES6vx+5Atl2Bx4E3hgkdSsopQuQ6ElAdcJjWJIR7RcKpwEI30Oq8HI09FvA2FTga9socHSYOeTmNr/Cn779c9jnFxAh1YJ9JP83wtGL1+eXD9ViU4ubk/6xFvkpM2JuLJNq7eKNDO8E5TB0dHWIocQQSzeCShAb8Fku4HsSX0KFE7XBdJGVysi0thP8JqL7AnI91gu477Pfx6//vWzCASp8WvKWuXNuy/5rDyFSRPk7e/rXpd1rYQSYfJIYZgGIomo1Bi0suYb8jqjMi6TusARxMJgOYvUwCDeemmt8LcWLJwPyzZclIzik+GnRmNQxRT3Hy+9PRHK5Am/RqOuTRcvJB2k+OG9+3s21N7Qn1ZnvEisFfkBy4Wy1LZEdfAnUQ9EWLz95lt44KH/xOc/81k0t7fJCblz13b84ZUXcfb178WO3bvFlFSGuUy7HAVDb2/FbWveh83bt4kkRKKhFiXLQOeZy3GIQr7i1+BDnJuG5kPJcUTKYf+D30f9lBlomz4PO/f14BP3fg/2SUuxbcKEmTYQN+nPUVWqp/twGUinTWRzARydKCHUHETGKMA+egQrvzQPW/YBMcNBy76N+NbtCzFYCePp/Smkyhp2bh+GNnUaUkdMhNsDqBSSSDSVMXt5CzZpVEkIIOj4oZSI0MnCzhMAa0NXQ9AsDSWOTnj2cHdX3PQrRFC45aDOD+FRbhvaj4bzpkPVfSxdkSe/UlXgZAuIKDqUsoZSDsgrQFGtQCVJu1rmiaYtv0sZ/ENSVf4M2Qr0ioMYv7diCWtODWJNcy1CTXVCLj4xGPkc/1AyVnWYvKYdu6nSL/AypurJyGBkvU8Gj3erq3eNYFkzEsZYzLunETd2BuWCeXNwbHBA1i4PJK4haaTQbTkUloDxsjL+nWWVmMZyE69wPw5LgElmR/Fu2xGrCiJ7WE8uWnQSVq5aPgk45+sSfjl3zjyZMbJhRCYLv4e9e/a7wUh/BRa8FDz36YTnuyfGZNdS9Eg4LrBRmMjipHOXS2pF3U76HEajIdG+4bER1eMwJQ+vQA+S7e5+cI+FLbCmKrzNbcjYglPlGyUHj6/JCyIIiZxbcDNQCeqtb6hBIKBLIU7JyE0bNwmXjcF6oCoEdNua20Uw6Ne//S1+9rOfCSGUCIjvfPMbGHUMzDlrOUaGhuQ053Ny55Q0bDyHRp+OzhnT8dY7bwnQYaJSwhkfvA29qQkXTGCaaGtuQZJdOF3HrK5pWP/Fbwhnbu71V2HTixvRsyuDix9/DL20eusPQDlIFEZUnIEVv3tSsElz6HAGJaUG5QBQjFmYFtdQtg9hqK4FDf4I8Orz+OlXL8PWpILnhkqAGkQDgEN54PBWG1oDUNYV+C0FlZHtCJ9aj5KmCx6VqgCVclW3goFCIR2W3hXihHmYVVyZihCzFls6quGiD+VUCs6YhWJpEGoEsOh/QmoaayTHQXN9BwwjAF8WGE0dRnDVVFQ4GK/S84WyprhiUkxPqSzHz8uApJxHhOl6xUFTaQAvXHUqGjob3dHFCSfj/2kwesgtpqnerbauRhTZvDTVSGbQOm2arG0Gw+Z3N4oChQOXf8vuOQ8Ars9EokbIxaKoX6m4vqVcr6WyYKR5MnpcSNFb5enAskRVcay/H7977g944MH7se7vr+D0Va7XJ1+D73PG9JnyOsz+eICVjIJkfUq4qcaxVdeshC8mBOHqDsdIlwG7povLUiQWwmWXX4qxkf7JlJBzQ36xfBE3J65M8v68HYwvyIvk6eB4dl9SSPtDsuMw6OrrW1Aouqcd30fPvh4B1HLEMs72dXXTJHmTaRa1Kfk4kZY33FpCOqrZHNRwUNjblHogLy2ZHsPQ+BCmnbMS6eS4GJByE+Dux1tbNI5tf1mHk5YvQ7ixFkd37sSxYhozr74EebEscL0GefPVRBCK12N+QweeXfMJzDhtGY61xBH3t2Do4Bi++9KfsC5pYvvr4wjlYlDI3YzoCEnabguiZuvuLAbzFhKtNciFHGFbpI/0IDi3A/6yD9m//QmPf381tiUD2G5qGM2a0B0fRgZNHNs1Cn9zK6w4haQsnHO+H9t2HINe34K+HAH7AfgtDVWncIjCOgf6bEJwluhj9sMTkgN1clVLQNqSUy/gBOCwGRdg3UQ9UdmbRWNWNkYrAHusgPLIbnRethRZWCjmVZTJ8KfzsKpIl5YnpC72PTaCpIKxdiRbR1XQ7CTxyoevQ2NmSFyxuYmT7sYv+MQDQGq0f6oZJ9u61Yjz0lV5HFkkmrsWRdUtGkAmmRHaXClfkt9ThYKbPf9OdQeent5zNtTXTopKUTY0XcU6y3dOAWfDwOxp0+GjfEnVV8Pr8IuAVrUMY3lFeCO1WtmYMSsV3P3hD4pr8eYt78JHQ54q5trbRERhIN7W4PhD7h8n09JqMFImQAwo80Vcf8P16D92FKZpoKnBFX7lh6IWiRc8svv6/ZMqyfzy2NjhgucO47H0vVycQRyL1csJxQ+ydu06vLT2FRnmBmJh+BX3FOWOIoW16X5Z3Nlk1if0Hkd2HXZovWDk83LG9P6PfkROR6avlGvoGziC9jOWwaqUJRj5umzY8DaluQUH39qEW264Fdv27sLY8CACs7qRDGngFNajfUkHLx5GulDBe087Gz+79aOwgxHUXnw6YsFaNMfa0NIwE6EzLkLvsSiSGUt86un+zANKC1hIODZ2vduHnNYJo1aBWUMcqw0zOQa1IwFtBKi8+ga+9eQ52HS0jAHTL00h1aIRq4X+YQdGvgJfXRDBgIapUwz4dBtFkJjsQ3+mDHEmtN1rIhIYZMlrrgwGvUEktWSNySahYUGzNahlFXaRVuI21IhbM7lpHZ2dLWleWBkLQUKJNANFM88CHgiFEKiLwKRggM9VByA5ShfZMgchCUYHCV1BiFlSeRSXOcN49KYbMFyFnf3/EYysy+tqamWtsMMZq4+hUqqISLP8syxhaDA15PfZ3tGKXD4raSpvNVSBICqIYACOh9htrqo78O98jGNUkBlP44xzz5QDjHND0dOpjt0ERGFYArmlPAdniC+++CICug8LFsyXXkowGpwce3inpQRj8/ROJ0OPvqqepDdbIR6Qg/Op3d2YO38+MrkUQiFd8Jth3VVglqCqBgHfLH9HCQuvm8W/88OJIlZXl7yB2poGOZH4gdmJ+vVvnsPo6ARqamKIkFeXz8nxzw/obYrea5lld3cmxo8yCexgeRfPU3gVGJ1lyXPIqVcqYcHSpUhnJ9A31IfOVafCJqCYLr+BwOTjo6EQjmzegY+uuQuvrV8n2NXgwpnYN0JlAXejYqrCLydHcWdfCGsuugKP33g3RsbH0HDNpaKpE43V4tC+HNb8+1PYdKyAij+AMrvMXKScfWkOoqqNWYkAfvfXHgRnTkNJMwRtTdynE/Eh0lNB+u0Xcd59pyNmKLCUOEbLLiDAyAM9WUUaMnod2/g+mPleXHx9F4YPluG3/SjYGgYzBWQDYaFFEZspAhBBRZo2Nusu5pMm6xxVWAikjVlJ6tforsBSdXTB78kbY7BTavQnQRYyN0OvAcZKUUv4gQRgs/kaYOddgR8WdM3VXWUQxvz8mAripTFEX/st3v3BIxgpMhj4TXJ3+J9PxskTszrw9E5E7yfXRQB+FPMF8e+saWgQnHTQHxShawYlLRnOXr1aPEhlndgVsf/z6lCOuyZHeIpPBKk54hCAAMHnimtJzhKNB1F6NIVYfRznnHMOEqRMTbhqAMmJtDS1GKxUEqD044qVy/H73/8O551/DobHhyflYby5uAgzJ9obnZLpegnyD1xw4q7kOPi3a6/D4MCAtPrZAmZuLUFnuDMV0Rup+sbLl8adFK59GyNd8m3DnpRb4Al28MAh9B525zQz50xBrpiX55HHaj5k83l5D2RBR/wu580LdMWuqkhz9+GJXXalJOVErP70CnQGPFNcpsDMzefOn4VDRw+h4/SlKBfzVd/7451jwrzKw+MY392DCy69GGbIj2OJADKW4RrtaJpcQNHQ9DsIRmtxw7nvwfOf+A/s3bcTXWtukcF3fX0dihMOrrr5s3izUoNjuTAV9IV+RBV0SxPBcXTaBYxNmBgwI0BERSVAZXDOS1VMGQAaO9I4MtPG5Q21SA85GMxDUkCr6GBvUUGqNw/LFwFiFoI0GrImcO7pLTh2KIn6QI00UdbnDeSCXHTi7gin1i+zYLeu40VzoLB7R2PWogkr68Bn+WCK5dsJavLVa8vaz5koQ8v4YZQJCOApqMq1scM2rIQJNa7C1n3CM4xyPqwCccdGmHaDmjvqmGHm8JPF3Zg2ewZGkmOuQqDF0+l/DsbJoKuONv5VMIZ9IZQKxUlzXTWoYvqU6ejt6UUhWxCZxTfeeGOSFVQ2ihKMrKN5YzDy+5WOKocuiiIHCdembBds1lBnk+m63xW2Ekt5nqR5F0nEtcw0dfGCk4XXSBoY54i3r7kVU6dOwZatm7DijBWSzXnaT14GovgTIYeI9GAVMCtq33ZZJPF5GjDaTYJnqzbQfLMToyOTHUwSN9mx4hthMI+kUug5cNCtNW0HcxYuljkgT0F2rijRMD6cFDbD1JlTUDRdi27e+JPPQyyh24p20ySvVvMCT5o7RNrwglW7b/xJgigfz9cjznV0hDVBvYumKBcRrI2j7WRX3c5wjuv0COPcUQQjO/TOTjz5wx/hM997CMkEzTt1/O7Hz+Ksi89GsC4OvSaG1und6Elm0LjjKEb3HcHwoUOIXX8ZWkNR+BIxmON57H/hDdzx+43YWYxg1NZRhi3sfsuvIMhuHWxENBVDEwbyzF911msc5BmI78xi3p312EjJkPXbccWyk5Aa4UxOhZVTcCRvYCijoeSQY6m5mGHVgI8d7dFBTG0KY8WCdhSJRzVsHPYrKDUqyFuue5cYiIpCouqmqaYNNa+6NtpVh3AXxcSBuStg3RFRcPi5HfDXzhQZDptHp66IGQ+5flYwD7U+AF+9H4ZaEe0k3UdiMg9kGzGoiPH1LQsLNANPdNaLVk+0vk6QW142xU1D9onqJu8a/R13vBKI0b+4yfdP0LppCqKF662YMURmkZrAbM4Q5smuOqlRVH8jOZ2Ada97782pJ3sDPp84hXHjN6hbVG1q8ieRZife2F/hNXM37AqefOxJ3HDDDXIYsHZkXH384x+X0mj3oR3yXLNmTxe4qI9oeH7GpiltDlW9efPe1JoP3C42WlRgY0pYm4hPNnj4oUNV2yx2MqdNnS4iPc8//7w8vm9kCB/7+Cfw3W9/W470UxYtlXSRXS3m55TpYDDweSdSKQH0/nMwlkumdEs5TuCFOVHxi++TGwJPz0K+hAme4j4f2js75fc81RmM+VJRgOwkSvP12Ckujk3gwrtvxY6D+92mgWVNciVlUZsOjr2zQywDPvbNL+O5zetRLJRx66XX4n2334UzrzofTe0tGLNKCAai6H/saSBaB2pexK69BJ2NTUK/OTYwjrCjo/flXbjq9XfxxhjT4gAmfLZ03ZjChXyEBhKNpGIoW4QedbuApYNp4K+vYc5334u9hQJ8mQrMtzfj+kvPQfKoATWvIKX6sXeUaagfdoXNGVLzaZRqQDWCiJhA9thBXL68VTaoPlvFUETBEFerTyEGQEYeJwajr6iiYkDmlMIBJIjAUmH5LdRVNEw8sxmonwctqsEJ+mCzEUW3Mo4cVcDfaUGp98MMWLA1Mo7ZSFElGOmmQY2BGCFzlo1r61U0P/9nXHPNJeiYOQuVE4LR+Sc4HFPY/91gDCr8DIbMmZmVbdm4HU6hiDMvvEAOlZ3vvItIY62UCwxGooAYHN5GwPXglWseSofW5OKfQoEwAgiqDcN/zhtYrnkHklmqiBx89/RpEkPUxaG/4ze+8Q1pZJadopyWR472YP78uShmOUQClEBNWChUbMIwiuXky4xPnlLSPAn8Y/eH3DCBrhkGTlu+UlS2PaepD33yo/j+Aw+jaUo7LrrgAowPTwiqnseyEFQp3UCmP0Vo5VSrMrVP2A2Z2lLTRvCAHCJXO7En7kQCgYJ7AQSKVB038DTlAiSKiDdvpMKOZqVUxPs//yk8v+5VqI5b0HMexNkPtW4qJQN9b25Ba7wWd3zhHjy78VWcvvIM/PrHT6G5sw0IB8RKTYkFsbBrFv78mfvRvXgJenfvRdvV70E4FILuqJjImmJ3Vzxm4MYHHsbTyQqyag3yOiV5CQBlC9wdBensYLM5wF1XVRDvVzH+7E8R/Na/waH4MEcBpoKJd8bQpPgxI5SAEwIOZ3LItkUxPshWPG0UfCL1aHHIaBrQbR1dWhLzGgMwg1EM+1SkyaPkyIaBy9aKNHEgVuVqXoNtUFpSkxSTKSjRMVyIzt+PIY4uZKwClLAKRw/Qlw5qWBGKE3G3oYVAhuBUtm1F2EoRTSOdzlG2IuyRuAXUWQ6uSKXRveMNnHLaIixeuVIoWV6Z4wWjl/EwheWN/y/psAD8jt9OnGFrJjfcNGYtnu/WsyXIIUC6Em+vrV2Llo5WlI0SItEQ0umUHAoMPK+7yROMj/HWDTfPE4NRxjcn2OF574SzR+/GRqPu+LF69Wop0Xg48IS++eabcfry09Axc4pkM0TnFMt5zJkxs+rCHA868gLVZ+KbiCTCk3k1X5hCsx6Uh/hKsqZHh4dx3xe/KHXZDx/5ARINdaK+RYOZhrZW8d67aPUF6DncJxbiLIwLxbwEIz88Gyz/UzBS5zJNj3oZUrt8SbEzl3zMrXm8YOSGwL/R29BFTrjyDV7jYfJU9SnwWSbec/uNeHnLu4iGXOY+L/7VV1+Nx370OKKRGKIlG6O7DyA0ow2DQRt33Xk3nn3yaXnfTFGpUcPUaklTF557+BG0tndjtLcPM2+8UtrdAaaS8GE8OY7KaAmZIxl8ZccGPLmniDH6QFiulibBFazNAvBJDWnZlLRQ0HYU6N/yKqz3LYLmr5F6L5Bx4Bz1QxstojlXQlt7CGZtENt9WSi+IErjxJD6Bd5lE2DKaMiaqAuV8G8rohjoz6GoR6Ve5SAnDxtDPhVFfu+WItKRPiMAk+wLm8rWVYiZWkEUfkz892GEWjtgs1MuRpg8+VSoIVWCUfVXEF/hF+K0PFa1XakSxZZgFK8OcbMCOioOztq4CdPSB1EwS7j9zg8h1tYqDH3qqCrVk/H/JBhrwxEMH+rDfV/7D/z4iSex5KRT8MJvn8Nl110rJkWZbMr1fKwGYzI5MTk/lG5mdV165jfeWuNGSdSMF4QCU62WR146TX8WL7tUQcGyGHR/QB5DpA2bRvfcc48AE85efY7IfBCGR/hpLjkhGFdFjQYcdsk8PiPTuopjTM4NvY4Zf3q70E3X3iiGNe/s3I6m1mYULUPA4emxcRGG5RtYuXKlBAVPzUwqLeRhchEVx22aMAhkN6ruemLdXN1dvMF/btylwHgfkoRhL41wzayPK4t5KYR3kfjF8iY8yCrAnfSudD6LrjNXwtLcz+hdrImRceE9dtbUYdNv/opLPnwbDjp57Nu0E13dU8T4hIK1GYOUYAcNowUM7znChqTM7GZddYl4R46PjKImTqFiA1qZp76G/g170bFsNS76+Tfw2sESCqEAJpjy0OjVsdyheFXCIrp/DGUaAi0NQ4dLs9KTGozDJmoNH4ITrk9GoNGP4biDiToTxoiJaG0I5REblQy3XBOosEBVoI724fIL2tHWpCI54aDsKMK17Ek6ONR7TDiT4bpGWA0BVATyBlT89NdQUMyW0VDQkdtYQSnI2aMCm6luiOMMBhw1ZzhBmUDg7Kgopkv6qrAB4lrLsWFT51Sg2j50lWzc7Ncw8bOfoz5iwhcO4bxVZ2HZ2WeKLCg7lieehO43X0WoV9cBO58MGi8IZCerriE2rdIDo3jgwQexfdcebN2+XdgUf3vhBUQTCQRj7riGB4N029NpWYfMCCVDO8FeLhR0rep4XzZpxsezLvCkvl5gcCfa0knQVmtdL72lbTu7uqQann3mmYIMojQHa9fX39qAsdQ4nv/z8yJjolm2xIo0cKhHQzZCQ3OTdJAYjPyjV8gKJrOKomFKmB1MYeai+SgojuhL2j4FvSND8FV9GvjmPExflGxtk7birmbkcH8fKskUwk0uooFd27A/IPC0ySK+ChzPjrlmrd6Nz+91au0q2mMyTajeb3IMUtVm5WdgAAsbhQwG08QnvvxFPPPS7ydR9WJNwKEwxzumiZ6XN+C8D9yMhReehZ889iTq4glRWydBmDPD7vpGvPvIz1HfNU0cj+IRXYLx8MgQRoeGUUsiq6KhSEhaxkCAiuOBKEb/vh0f2/o2hiI6NucrGDV15Dl6sDX4OHlgE8koI4MAkm1EPFkwqY42EQAGbGjjGTRqAYQTtCAmJC0DY1YcQwUHQVtB6XAeihUR6zSHtaRGeX4/dKuEEFUFcin4I1GZDZKNQSV0VByU6QDdHoUR8CFS7UtwthzNBZD7+2H4Q12ohNxgVHgacp2yVuRNZ2p6BIHVM+CQjcOhJtM+boTs8dDsp1xGIujHilQJ2mO/wgN3XI9COY0PvO99eGndWsw8aal0vDNmTsjNJ36nhMqdeKNHi2RF1aafKwfiprkEe/gsBY9/71Hs2LUHP3r8cUlDPdPfeL1LVufaFp3VjGtxTzgl61smXlyDwv4vH2cpMXA3vbPNna0nMzjnPReit69PGoPeeqPzNP/bG+LHglERYjvWP4AHv/Y13PvZz+Fw7xGcccYZEogmLISiIZgERahuuSUnI4Nx3vz54kUonSNiMU/4wDGK3JLWdHQQtc11uOm6m0TweOPWLVi47BT0JUdEJFhGFKoyiWrxRhbcHRjQRDFUKBJUpbDwwxRSaQRUVaB1HiLeQzXwZJS2crV25E7rYQVpD84T6v8tGHl/8VaoOmwR+nXGRRdge7p/EqAguFjquVBhoFhEWzCBfMDB3LOWY9uWXYLuYTCOZzNonToF5807Cc9/78fEmUkwRoI+LLvjJmzetwf5TBadHR040nsUTr4Cm8LCmh8FR0Pz1PlYsuxM/Gbtn3DFxz+K+kUL8OKRPFKaDzmeYrYGupuoFT/KQQtmXEXeNOAb0+Ab1NCkWwIxy2sOAlEfVL+FQZ+JfK0PuuVD/sg/BiMPsTKxnVJfuxQgh8Pbqug0rx2vKf9WKVSgNMckeIycKaOO9nwQA9vSUBI6HPb/OafUGYwK0XkctcKw+9F1YQf6ucPzVFUtIRTwxjqR6WmkUkS3GkLgr3ux46kXMPjCU5h76wWY0tCEC666AlauiHvOPx8dM+sxQRXy6s1dg/8YjByvMTgExugKT0ze+B2HFT/yA6PQYzEsXLQI727YgBhnjgASDdFJtXuZMedyEoys54iO8UyY5NQkAbhKPODJuaUajLx/JV/EvKVL5D3IPJzsi6rCHR8j9ykYQkQvF0uoTSREXIs24uysUteXl4jKF47m4nglGFmlL16+VIrWSQJkdYDvAamzI+N4Y+NbaGpslPbwJz76Cfzng1/Hf337W/jKA19FSVPQMmsa0uUSLM2crN2C/pDsdC4Y21UScP2Kj+fcbKsz2AgoZ2CqFVOEhnh05/MFd+5jUgPGkCaF3DQXguelnydCqDykDKlE3ljEg+GRK2mWDdjFIs6860bsP3QAMV3HYCaFRj3iXlSm0UYFtVO6sPPwITTW18OvunUoU19KxI++tU12YKI0WOLUhUNY9pE1eGfbFtmIqD4+OjSCSroIagDFSxYmtu/G1Q8/gMN9PZgzZS4OjOTwzkO/Bq6+E2fedSvMBiAWcbGnpQyRLQrSFQOVhhoYB0rQczr8VgFTGnXRIOJmwZ28AAOpOl3kFfO9rB3p+c0ApF24e7m4qDXdrUmllhZTG5rTuE0kuVsuj5rmKFJZA4EiGTo+FF49CjXWKtbrlq7ACbrAAfjKiNSFYTklxBf5kGk1UbKJMFJFYgWKgXroCGcN8Vo5JQz87uFtiDnN8PsUnH5ZMwYKZRiFElTDQDMU3L2sAe+J6IhN7ZSZo2RlfG/VueJkjVadvXjfvTcO5d9pKBRQ/CKofNlFl+InjzyKxStOE8s3ZnStHfWyNr1si+uL67KZuFNNEVy2N6dmkHmAcl6e7Vv2/MOIzVJ9spmzc8u+ifQBOF4h75aawjRsUlRxR0uEwiJa9a1vfUvSXDpjE+VEN7SRiTGxLuBNueLGa5zfP/ecAIEp3SfwM6Z8tg12WSXHLlVw72c/i6d+/gssWbxYcugvf/nLMr/5jy99QdK3dDqJxnlzUdQsqD7X0FIumOEe94J4EJzAlyBHAAAgAElEQVSF2x6exBRWF4zHRSunM3BKZQlGWpFJnUlYHjGn5eMNnRMv1InBKM2cfB429VeqdBjvNOUH5uNYPM9ctRTbD++XEcNEOoWmmvpJRAnvt+qy94jeDS8TYXxeU4i75P5f/RmJji4xPSVJlMF4+ofXYP2Wd8Veuqm+GaMjY7CSBQQzJQzt2YOv/fYZvLjxTSSiMfQcPoqRZAYjT63D7X/di1+uH0NEDyAFdkJH+SbdoXs4gOZzp2D4cAZ6IQYzN4GZU6jHEkaaPEU9CL/qIBmwRfHb7DOg+AKuCjKlNdimFPk2S4Dvnt2afPEnmFLLSihbMgtMOya0vIrgqInyEQNmKISgrsAIAQ4DMaKI2rgeKiBxcgj5Fhum6keZG65RRrMWwtSQD3oa2PHXXRjbegBoP082NF13YAeSqFla54KxVR/qizYafD7M82Xw4OVno07nDLngGpv+i2D0YFnHN+LjmztVBwgZZDASssZUlP84By9ks5gyq/M4nFLXMT4+4RLny2VMmdotKi5emsr16LH+GcjJsZzMLr2+Rqy23uXClkqCd2XZ5GV0ogzAsVmpLMFol8rSwKG+L+vOkllGIEy1BBs1DXWiYihsES2mO15gTKYH1f/wALcE2rLeqk3U4CMf+tAkq5lB+b3vfwf+YBAXX3oJ1q5bh0AzHVyz4qlRMFMIVxn8siOx0UbXoWpDRbpTRPVXRahYt/KN8WQiOrmczqHE5g8bN9zNqyTNE+tIbxArC6wK5hbguk+Xi+Wxqz2Mq1xMMjAsC5d96A7s6z+CfHJiUvNEmkcVA0tWn4s339kIalI3JFwqDh9LfZ1df3hF/BtG+/oBn454JITrPvQB/HHPVuSKOUzpno7hoRFoeRO5bfvw4E8fx+83v41IQMfI8ChMv4aVnVPwxGcehhlegOjHHkWezRo6Rgd5fVxgE9NMEwNwYhH4yjRiK2D2vCCyRgWFiB9ZRZMUh02xbFCFOe5SpDgvdR1X3M6myKaILMrxlO6flBihZcsIJqlcl0eNWo/hdaNAtEYgbj5dgclTkQihsAKK6syeF8G+kZ0Iz+pChxNEU8aH13/+GuCfBn+wEQE9hDw7juxS+ktwFB0o9mDW1dMxVK6gVtFQW3HQWDGRMP3ojgA/f/91KB3dJmR1o1KSkZqYIZ1wO9FN2123x2tIylgwPZR0ueB22dMTE9K8yY2NoWN21yR5mCdYJpMVyCdJ4+ecezYGR1yfRq+M8pAxXBPdHTPwHA+t6i0Yiso65vNwtk3rei/4Ocog+iYU0EVPJzeRlEAmU4QlUdky5GQUVQLFkX4KGztSM3rB8c/ByDfBBzMY2Xn68WOPY8+uXfKmqPv4qU99Ctv3ufyxkYFjoiZ3dHgcXVNmI1POwwkXkSm6dChJl7g+OMOqeiZK8FRTz8lCmB25ClWmiQd0ZAGXx5NIDY8gSLjHCTfvhD3x5GPwSYBCQ5nzrDmzJQX3djFmAKw3a6iJ6gPCc6dJRzNWFUTm/Q6y0L7iMmzfu0ekLJoSdXIxuQNygzrwl9dFUyWZTLlUd9tEc3srivOmIJ6IoqGxFT2HDqM8msEdt9yOvlwa/UePIlxbI6nL0OAwDj33F8SUZqSOTeD2X7yBnx+24Is1oqK7XWsXAQMYIz3Qm9phOT5oegWLTg0iW3HAnk7OUARqFyVSh1Z+WVfWhMN8/pTYU2lMy2YOnaSOF1j/HIy+fBbhXgOnrqrH3x55A6HW5TAUFapuoBLkINGGlvCJUJUeoRjBESw5ZSp6/3oE45uyqOmcgxzdrIkJ9DOfsBEMua7K1Gfl4lt8oYqesCmqAzWmgzpa5Gkm2sp+tKklvHj/R7DvzdeQGxnGguVLMTh0DEWSJ/9FMPJXbhl1vL/BYMxOZDB3xiz5efTIEcHQ8iaNmq5GCRg+jmsnl8u7ZYsg0HSZDHAj530ZHJyNe01MDUHpiBIex1tTY4uUUJ6KeGd3t2sJt2OHvKd4bS3aW1qFTqUrKh566CGR/udrs3nD6xGMBAW9JCALblq+ePAf2ZxVoSdvCMtgpMYpUTakI5ErGBdQtwsQIOubi5+gAe4IB3sPiaSFfJDaBBKJCNq7uzA4MS5MDEpxyEJzgRtie1amnH04AtOoQLNM4YtxRxVtzOr8kAE8unMfQtEooqRc0QOy5CJo5NRjalbtqvH/CxMpXH3jjSK7IQ0ccdU9fuN9ONBdfslFyJNcWsi6pp2KgwNHe3HF+9fgxXWvQdc0dLV0ClZW0BPlMsZf34qGSC0KDs1MC9KepljqjFuuRtpyESA9+w9iztx5oCUeTVhWLVuOjYf2SzfupPoO/Pz+70L1xVzVgEIYpz7yFrb1ZWDXReGnEzTnbj4VSi6PxkgEmZwNMwTYwTHMOlVHWo0iaStyH73kIG4qGCuZMItU5SPlx4TJISAPRfEF/0e4Ka8W37ZHCDYmyqhJmWhvD6NrtooXfrADetNMWD4dtr8ANR6CGVElEMPxCcxbWocNn30LtU0LkdLC0NhpDbBUrTZcaGvAebBioz3kw6B2GImV3ahYqgz/+Y/NqA6aHxUVdPrK6BjajbtvuAoVu0x9DzGErOtodx214XbbWUNy4XI98OTzmEb8ZinEHAyE5KTifZlpCVulYorR6qKl8yXIPNFrzrO5UbMRMz44gtWrzxNUjofQYeroZYeW4seWLVtQ19SI/dt2oaaxBflMHkE14IqXOSUhQ3DT5riEqunUg+Iafuv1DaI86I1ESHHw0l3GkUYwCHmh/yoYKSPnpXdcgMVUGg99679ELFY89lqbXFIkdTx8rrQeXawoIpXOZeRLljZvKCigbCtbRN3MbrR1dSFVysp4wUuNqe0ZjFHJuSg21QHp/FGlzPXj4/3kaC+XhVrF1DBgV8VzT/RRr2qoeqcK4XQeyofPQQzgiTd+udzJ9KZ6dM4hYHkEPr+GxqYGrNvwOqYsX4pUsSCd3jCNR0slCWo5dftScEZTMBUNY8eGZBykOTau+18fxcu7tmL+vJPw2gsv4LwrLsPoUBrnnHc+dmzegvFMGsOpDIb+uBYxNYqcqaCxvR3pYATl4RCu/OpP8bsjE9BqWFMRl+YXSF93WwC9O9MINiaE9LvsUuBozsG4rSBr0anKQthSYFK9bNyCrdCkhfW5WzN6wUg9z8nshzU562px1yISQEVd3kZzkwYlbGDekgCeeWgz9Lo5cGqDMCKUheDI5BjmXtKFkacPw+iZiqySB+J+qDw9xSzH7dDaigEtUIJpZNGwpAFKnY60ZiBsa0iYmgRiuGyjtWKixQkg4aRweVcUt154Do4c64WumKhrSGBgdES6nVQU5ylEErvXC2BgMSiFzU8pRFVHcjzlNqzYbedP5vyWLR3Nmsaq2Y6qChwtlUrLOue65fjtjJUrBa8q7BqipKpBz9MsX7YkmIhVbWppwe6t20XqP6AFMHf2XLz11usIxNzUlY+jCkSFnVYoOHTgABobGgQnLaWT6uqx8saf0VhIyBjunLGKJpic3ZgOzj//fLE/PtrTg8eefFJOOp6KvA+DUdjNbN2aZZnT8Dl6DhyQmkevshtQKsNHZEi15SXyGdEIZs+fD2L+CBKvqA6UgI5YbR2K6QzMQl6KX5OiQFUznMlUQfUhGtAxcOCgBKVOdWd2vcgSUf2TF89F57hNI8/TgRdYMKpEFPl8mDFlKubMmoU//uUFKNEgpp96EpqaGwU/y5Oxce4sZI0yFFLFHNcn0uvKJgoacoeOSp9k5NARKIGwZGbt7W04HFNx/S23IJ8qY3/fIG66+UN46vkXRFkuQdDBoREcfuUVzD/lFHTNn4u1r72C+SuWwKfqiCWWoTdxMg5RCKqlDgE9Itoo02YAe9f2Q00koDfGUI73YO6ZXRgwNOR8CmixQNGrqGnBHC0jW/RDpaFb9UR0T3w3+LybNHBoLExT2YoDhS5S2TJq63VocSAQp6+HheZWHes3HkZ2mOBSC/PPn4Fdn3kFoe5zYYaoyK0hUxmnqT2gVf4vvt4DPK7y2hpeZ3ovkkZdcpF7xbhgY9OMbWroLYBD75DcACEEuAECpHBJLoR+IaQQegnF2HQCxlU27rYsq/c2kqb3Of+z9pkjlPz3fpMnj7Atjc6c8+733XvttdcC/A64An4UVzoRtwKhFFXDqeBmFhErT1ZlixTOTB4+GFBBBfIkUGII4YNHfoz0QCcaWxvlZJwxZzr6Q6MIs16nmqDLhanTJgtAx7XEzdHncgmaSYZNLpETHSCWIZIlMfUTcUFS7IEfnHeaDKqTCMCajmmtDKZTJS6dAfV3Z8+eJQoV1OTRsz8+d4qy8feGOFJVyPaZjXkcLhw+2CBrUdoThbUeD47i1FNPQ2lJCf7w+ONiO85glJRZxMO+D8h8LiU6rwo42G2zyTeNATmqAZloDD+7714hgF948cV4+L77UFpbK+loKB4ey5UFHVWzghKR5zkYjyBQVYm21lbx7SNMr+uKUJXarpqQMxH6DcNVHkCguhwmmx0coBHh40xamPc8GRmMuicDr89mssjO4/d4Rdxp4z82wMgPIZMI2o6p5/x1k6fIzWSr4cjhwwJb620Pft+SpUvxzcef4tQzz8DUebOwteWg1A1k9PvLy9Aw1C/BaOQwc2E6RJTyYjHMKKpF/Eg7unr6MNzdByOsMs2fHerHf3z4JvY2daKnM4LJ05Zi/dYtOO3HN6CiJID6t9/HQKcBTvYzO9qRGh5CqDfEoTt4Js+FceocxOFAyuFHUXlAY0UBGI0MwGnwIzaYgI1K4/YsJiwxIOg2YJRyJ/RzpHI4NYEyVrR1UA3bqfl+M+o4x8SvhWFjedY84YUUy8Y/v6ooITnDoQr52+1QYCwiIZzOxwa4AkaZywweiGN0Zwij6QTgH0XFwjmwTbBQjI7AOaLRnJBGzKoJBr43VfEsRtjpgmw0ilqeOwe4snnY0zn441mUpA2YXargZ/NK4awtgzfgw0h/F/zFHkLZSMRiGKX1H0kF4WHMXLRIApEB5rE7pIToONIEg8XxvwYj6/SpdXU4YdVyvPXWW3jkkUdEyGzSpMn46quvhETOqYu+9l4cv3K5rAO2ivTNV05Is1HWPK0qOEubNLE1ZJAWUG9XD4yFaT4dXFSTWVx60UW44vK1uPeX/4mdu3aNCWZljdqQwhgQmU8L5VSxlLlUNnn1eotf6dHAQzQTi+OWe36OZDiMN195GbF0AnMXHoXuni5JTXXUiTkyUz7+3+nzyilKyzL6IJB8zZfet+FQL1+CrnKOTc0iz4Y8JRdLi2FzOlFcGhBuI5u40iIxa9qYNNzRlLmycJrtqCqpwId/+zuMHHRNpjFt1iw07tuHCdOmIZlMSXNYVyGIx2OyG6bCYRy3ahX2HT4o5qqEmzu7uqBajUJYplTH5p1b4Z8zDf2xiNS/pPoxsHVq4JTKyTjy5VaYohmkMiriahKGoALLxGrMu/lOtHR04fb7b8SO5gS6TTn0qyYMZRVYnZqNdNemZuTb0ijzVyGbsMquOkwua96ItGjXkFsaQr63S9DQ6uXL0BWMwEQjm1InEt4c1HgE5UsU+Cd7MZBJIxxT4OXJM2yENaPgUDelyc2CtqrOLFSqImRzMLDbnFFljIsjhKlIXvwGqUVEFooBWXicNljcOVhcigQnwRu7y4z9r7yPqlVL4TyqDMMpIJIUBUfNeyOjUeiMmTxsKQMNkGHgpIO0URRNP7XQ+PQrKgKqIgY55tEY1GQMVy4qxcrqcpkIYcpvcdlg87hEMpIbB9sEtArkZslnyjU2e84cZJI5NH63C7DbhDTC19jInUIBaW2jmlhbi3N/eDaqa2qw78AB9PT3yftQgpPZVXtzK8IDo/jhJZehq7cVdgeVCDUiiGxePGzNJjQ2N8PkdCCR+96PlBtDy+aDMHqdsORNCHhL0NHcjP958QXZvBfMny94C82HaYWRyWs4x1jgFupfJTCtSo2NJMYa/lIIFx4i68Xr77oTv73rLixYvkTEYUmypT2WjiTxTQnusM3BBcvTTetNJgU5ZCOWgSs24GzgC6X/+2DU2SBc6DaLWZru/Fn22UxFPmmSTp1cp3n30aCTf0/J9GgMuSwwe9p0bPzmG1SXVkhOzxeDrL9/QHzyeFLy9y9evEhqA8LLTGlcXk0xndcqDX01C3+gBNMXHY0vNn2FuuVL0DU6DEM2h0g8Jqm4rpfjNDnR/MEXmDp5JgaCoxyiQyhsw3Mdu7G+IYl4NgNDkRs9nIFzWhBMq4hbDYixzaAYkMtGMNHvRnf9ENJH8vAaAyixKjClVRwOpWmMCCNNRzl9EYsitGcPZl52Bg73JWDz2aH6yXzJaU19ey/mn1iN/lwE+aQFjlEDHBkz9jT3w2Lxin0be4OUKGF+5TYoSAQzSAwlCiuMejFWZDj2xMa3WYHFZEAqG4enyAFqY6nGHOKGMAwTrIgVWSQjJeBNYioPXF6HreBC5aCGKrm2FL+iPIc4lgFuItbU0cmkkaVjmNGIcpMRExVgSqkB2175b7zzpz+hLxwSSwJO4U+aMVUwBp2Oyec1Y3Kd1IgESqxsqa0+FX955lkYyUH+94HoQjA6rJpr1YVrz8cMGrCqKjZ8+okoGzYeasDw0JAAcU37G6XcWbZiERxOTRpDZl0Zh1aTWNF9u2ULiirKkUZaMj6dYO5x+7Drq80oL6+FXbEINY/zixz1o8wnOxBb6qmanxf7AX4WxgRfbiL7THHLZtSoiZC2KHkKiYo2841MFu9++bnsHI/+/tei4RIo8Uu6YGb/pDBFr9tp6SJTgj5Rcj+VFgAmmc6MvTeDgCcj+zoMWvrFs4HM9FGGmumHx1TTapGTUcjpJIf0Dkoq5goEUFNVDfJdCc74A2WSsv7w4otxyppT5SSkvDqvhSK0RL/43/xem00jA8yZMwfffvEFbD6P3BBda5PqbaxN7vn9o3jyz89j/ikrcbi7UxZWKBqRMRwKJcsDUmzo3bIX0ysmIKsaEepqQ/GxF2HpQw9hR3dOTDsTZgVhMwRwiWVSyFitSBjysJG8APocUjY+gyqrA+1ftcI+ZIWtG4h4S5CNZUXykgNXHPvyqlmEDu6Hwe5B2bRJGKmwweBzIE4RYpUAUwoZ8xCmLqtBppcS/jkoPiN6OoFwJg0LxQQsFox2hJDrjcNpDYgTVo7qA2ZtFpJSGVJbsklH+/hCv5OWbblMFPGhQ3CdtwhRnmbMatlgz9KpWIVDVcQOzpnOy9ccT/BUFuZYXrxQmWp6zApoqu03KkJDs1nNKDUYMcWmYJI/hWU1PlRMmYIBAoAEOYKDWHne2TjQdEie6+BwUAIjPhqXTZ/EjSPNTThzzRp8uu4jcZ769345ARyuRS52IrKXX3upqMtz7e3Zv0822P179gpIlkmk8N0mOnKbcdW1l6OHWUmBJCKnrZltHgs2bt4MZ5EfVrdVAlHP+FK5HIrtfuzduhOl7mIsXrAIZ511lqxhBjuFqU5ctQql5WVyqImeTkHPiWCiDMuXzqpVs1FSzTQNUekJZnJYdf65mDl/Hh69/U7MOW6hKLhnEnGkYnGkCdkWPC50ZTiZV6TDLiXpUikZHyGfU7GYZcHLyWezIR5NIUPRH8oT2CyaFHsqhZmzZklfib+fqgOpPGfrNB3OdDgiEhHZjAZpc80ILF8glFN9a/7CRTJGRQ4gC/R0WmNQcBflBsOUpKamWk5x7la7dtRLT0izPQHMNjMWzz8KnvJSvPv2K/jBL+/G5r27RZ6Cv5MPjicvF0KarJGUArdix+xZc9HasAvNjlmY+qNbsS9hg8VpQ8ymIOUwI0uyJ2F+uieRBVqYY+QsG/VobCZmeFT1NiLRkUDym1HYDeVI0GFYYQqZg5pOwkp0Lh6BNRhEovEgZpz9Awy4jRgWH28uFwus/B2xOAI2NxylOXF4LrJPQl/7EEyKCwajTcSXaMzCoWFK+RvdVul5mQzafTBZTWKxIA8zlxb2TdI4CMfpNeKcZUuakKL9BsWs2ErI5uHJK5qNOEucnAp3RoEhm8B8vx3REBDs5V6axqxSC1ypPErYv0ylUeOzoMycx6LJHlRMqsNgJMhlCvpt5CjhSNI42zReF04543R8ufEbJMIJYYtxLZGAcWhHPYoCpWLYxHnQ8S9B4s3st2haTRddeREcLodYC9IEl1kS1wcxBa7rtoZWWafZfEq0asYHI9NUDhv0jwwjGI3A5KAXQl5QWdacpISqiRzKnH607W9EdCQqte5JJ5+M3/3mN/jmm2+w5Nhjcc5550q7h9cmyHw2B7fDga7ObiiB2RPU+EhE0Eej0SxNeNZ6Dz/1OCK9PXjjr3+Fo6JI8u6j5s3DwQMHkCjIcUgtGI0Ih5ST8rx3TAekUC/AwvxgPHWZ8hKRHO7R6F50+eF8Y6CiRP5d/NMLM2UMHNEWMVImwgS31YnuDorN/qv4u9Z7ymHN6lNQv2OHOFjpu00sosnxMSB5s+hk9elnn6Ak4BetTiK3PI2FDMxUxETrtiwig4P43Vt/x8aG3ahvaoDRQuvwrNTD7E+JrGRWQUnOjCkl1SidNAlbd2xB9fLLkZh1AtoVkqoNGLUoSFgpcZiGwW5BVvgKBfdUmfnj1D2BFa3NKyBULoepihFHPu2EMeiFJePUhIIJIbGmy+fEG9A4GoGJ7KSm71D58+swwPWZzSFnMsLC2USKSFssWDzbgG/f6ANsRZrODqdtDVm4vTb4S2wy2xgqoESs8UggoIhUWsw6qOdCFZAelJxfh5G8ZjlHRJYZLdFu1mjcazivyG41U1Mv4zg+iuNrfXh6yi+ACXNx/rWXYnvLdiw4ZhqWzvChPK3CnlPQffAAmuvfxd9f/bNou5LXmk2xHcB2Sw45nnaZtNxzjueddfUV6O3rE1uJPEuleEKMkniICJGkEIk8DXk//E6NUM5+JDOkC6+6BFa7UVoXzU1tUgJxbbCcSSdTaGloks2XATyxbpKcyHrGx/lTbgIcFuConMVGrSczFGNe2mG9vUEkOO5FEKupB6O9IygvKpa6va+/D7+49x48/vjjst6I/ut6rMxGY5GI+DYqZXMnqcP9QxJMFNIJ9/bjpff/gc7ODtx/y2048cxTMHHGZOz5bpcMCXOCvaZmAj7//HONi5dOi9SEbGLcPvIaajZGKaIJTGFiQtJFi1NQqXQiIc5SNqcm7MMXb5xwFgvOx+RU8kTwO71oPNQo+q3jX5Ry5I62dPlx+Ml//AfeX/ehIGs8CXu6+nHhhRcKaMM/b926He+99y6uue5K5Fi7pL7P9zmp4vJ5Zecz26ww+py44Mar8KcP34XJaRPiNa9ZZ+AwGI+dMR9vPfgYfnDPXTjxhOW448EXcPbjf8auQRUWpwP9GTbKbUIoMNLzQpSxdeFX5j0ixjn2cfQUy5BJwp7KwtpjR3DrAGDygFPBhpymZ6vmsjAmk8gdbkDxuSsRGwgh2R4EXB6UVRdjhBLFSg4OixnFpVl0bDwAU/XRMPm0wy6v9xotJD1kxT9SuDqGGNLcSHNkp5hhNmWQdgVRtqIKMRO9QRggitTp/x6MlHl0q1nYUiYUKQnML7Xj9f94F87YYgymRoHWLlAi7ur/XI38yDAWB8qhjvZh1cJyzCi3w1tZiQxzFIMq2ZTIO9rMWL3qZEyqqcZjj/wavopylFZXaTzo0VEBCAnkja8Tx4yi2a/m2FwsLt/DGV2usXMuPx/FAa+0r7q7+iRDolAxD4JYJIojBw5LVse0tn8wiMsuv0ROTmGomYzIpdICwPQMB5FEDjaCPGYh5KEiUIke+o8arRhu6sYn761DR0MLLrl8rdYqKfQ+6cO4/uOPpYZknHCjqJs0SZyNlSnHzlM7Wjtk16upnoA1a05BxmVHrH8Ah+q3w1lajCPNDfC5PfLBert7cfopp6O+vl7kKkiQ1ZgOmv+FaKcUtEJkssKkySXoTU67VRtjEc1VrxsGq9YklZ5loaDlBUo6ykFS1YAilw8tR1o0lHEcy4ZKXbxemuDs3rMH02bOkKKc6WdwcFSYMDxlOeBMNbM7f3YHfvObR9De3sqcWwsETjKYTDjupBMxcfJkJDhPGB7Fpsa98M2agozNJNYGvF5OYzNVpc2ALW/Co3fdjy921CMVGcaLz7yPB779Gq815JEzGhClGaopj6zFgJRZQdqkoZUkQGtsC821i33EAoVTxILNlKJPqdLimWi3oe2NPXC7Zso0RiSeFDPRdDiICbOq0DOaRs5skVQ31z6K1LZ6BC5ejcEMx5fyMPnS8Htt6OnNUolDo+CYNVVxk53DzfSt74NjahlcVQ4odhOCO0IwjdqRtPXDc3wNwnnWpQxiXiyNcqmTo/GM2TmRkxFJBDJ2FNvymGgx4INH6mHLLcBgdAj+nAsxUxxKJIJLzpsEU7Ab1QYDrjuvClVOM0omTJbBYi2Doi8FTXhTMt1xztlniRcGnaPbOjrw7batst6oc6O/dHL22GZWKGGKfD5s2bRZTqAZM2bImrzqtmsRjY+KNCStvZntENQTInhwGB1NbbBzWIJZAA1rVFUm8/mVrEeKQNdUVuH5F/6EZScfB7pSM/OhNylBIm+xF4dbOlBi9yEXTuC7f27BHbffgXgqgWeffgZlFeXo7+3D4mVL5TTWKHlRNDY0CL1UcU0pVyun1qC3pRszJszE8edegHg4CJ8KvPTC86isqZL0jBQjTuvzw02onYid9fUCsrCPLDciZ5YdwmY1arQlXcKxcNf0m8VTL5yIoaS0GNFUUojjupMQC3tm/QxcsiviiYQwKFwWJ9qPtMBcOEEttAHI5/HYY4/JWfP66+8hnspizqyZ6OhsF0MTtk0ONjQgUFIuwbpmzRrsP7AP6zesQ3VNBQ7s3In2thZMmDRRO6WoTuiwQE2wzcJTC/DMmwHnlIlyPdwgJk2cjD179qLEX4RoLIUbLrkSf7j/YdxwzY1476bTgHwAACAASURBVIWX8NDO7Xh4dwJUow6TV2rOIWMxktyCDEsXTiFooKZG3i64QTNLTiU10jyFotgr4GwbF7sxEcdkswMH32+DzRuASifg5VlMqinHgS8IhOWRF5jcLGLECNGgxQJjkREJUxzVRQ50t3QiT1vsMjMMpXaUz6iE0U9gCUgoWeTE2JFtDSC5fQ8q5hyFYWtW0taxsTXNNwbGvAoLexmcwTNkUayapJ8802bCDIcRT1zxARy1JyOtstYt0L4yWr/TlhhC8u0X8MWmJ3Da0klw+w1IULPVxLM5K7YQZLPz9Nc3bz5nPX/QcgutfSGHwLg0SXjOUOTREbFmufTUU0+J5AV1TQmQXH3bNfJZ2Rfv62PJ4ZKeYSQcl5R705dfo6ykRLoALqemNk+KG3EGotJcn1y/f//bK7JmWHvOmD1LHNe6h3qlLw2LWfSbnHkD7rrmFvT2D2A0EsItt9yCpUuXorHhMAJlpXLlLN0IhH7w3nvYunUrlCt+dpOaNOSklvvmi8246Nqb0bj9W9R//TVKqsoxwnkri+Y3wIKXF9fY2ChAiVafqfSBg9VJpj5POM3J9f8KRt4wM9XCHWYBaVjXMRgF0SrseHxf1nMJQsdE9hQLesRcUtudKVgl/cGOLpx2xmnwF1fh00//iTWnrML+vXvQ0nwEyVQMZ51zLjZ+swnnn3cBBocGsHDRAvzmtw8jkYwj3NcPh8cNb2kRIumEKD6zhhB7g2wGdqcdfYkYfHOmI5xJwG4jxq/IAyRSTDnJUEMH1KEwzr3sMrzzwov4W0sfXh2gD0USQQIyLhMSioKEQUVcrKEU5IhaSi9M+8rMgYQF6s9oCt+FJr6sPAPyyazQ/5wpYHhrG2wBE6yrSxH6uANQp2rtCUGz8lDYVqCoMcnT2TTKJnoQN4WR8eTgn+OHksqIlmmKyuBWAwbiUWSddmlwq4pZaGycFGEAEGgSXyia3pBkSqs5bY+Ch3Vn1gi7QYEzq8JtUFFnNeLFW7+GvWgOUnlNzoSByzqYz9Oo5GFJRlHWfRA939wPoyEKC2cs5X90ezIKf5hrgYEo3ibCk/7XWS89GMePzek1N39W49tqt4SLfbB/AF6/D6GRUVx241rEOL5k52BxRPrX3KijkYQE4NfrP0UNU+FEEmaTTTbg5iMtuPraqzQNp3hc+psPPPAA9u87KKnm3r17tZPXmEdJRbn0cwkkkhx+93W3Yt+hBiQzKeFIs2zigPH4wQwCg/945x189tlnUE48d406Go1j7449ePTJpwUFeuC2m+Et8iGGrEDQNIjkhRE5sjkd+Hbz5rH+C3mZbBEcv3w1vvxsA+xE5wrGOawnpV9YcP2R4E1noNg16QhCxQKg5AiBZ6Wpy+9lgGq+6knYicQmsqBGjSxgBmM8jYrqWqRTWQQHenH5dTei4VCb+LL39XSDngmUx+vv6cF9Dz4oPSvWBU8//RSKA1QDA0bDVJ/LiKCSzecUISX+bv5eNZ0SqY1YPgtTWQkibosEBkWJFi1aLClGLBRHUVTFcGMbjjtpDc665FJ0lM7AlgxpAiqGFAVdZiMSRgMSBiBZOG2pLD7+pYr6GWemtLF14XVm8pobMNURyL9NkKObE0pdYIITIzYFlQrQvXMEhrQTyeEMXHYHktm8gEdmSijm0ph2vAUxJxCiY5Qphfj6TUA9x75qgNAIlt97Hrq8eYQyBmRpz63xyeUrPwW/Urw3Dc3yLZnOgjbf5JUWJQFrNo1iuwLzcALvPbAJzprjkTLmZH5PBm3JXBJ0MilyHvlUFIsNwzj02nUYScXgoFRFNoWsSrkKRXrYdHniSx8Q+Pfe4f8rGBn8IpmYyeJ/nntOgJ+LL75YTjMCNdf8+FqMxCnipSAWCUsLje5UTFlZJ2796htJUzlzmoinZS3w58rLy7FixQqpH1lnci3deN2NEqBcC1z/rDom1dXhmy2bheVDQx2ykAguun0eXHXFlSgq+V6jlZvVypUrsX79emm7sA2iwKWoRoMTHqcbV//ibqx7/s8YGOxGKpdBXM3AzQHdPMTNiTXYjp070dnXO3byUaJDxnNSVK5Oi0W4TiLX3F5V0ZbRVdvIE+SsmruiRBgNLKy5ozEFiMdicgOYcojacoHoq6byCAVHRdpOD8YLLv4h3nn7Pbg8tO6KoW76PHmvWTOm4+CBfaJhQsNUBt/AQL8mvWfgQzdIcLLGTScTUMj4sJtE8p67FG8S01Cyj8j0D2ZSGKnwisMsnYIWLDgau3d+B5vViZVzlyDS3ouO9k4MqGZc/8izOJgxwxxwIu4E6oMAB25II/u/g1GfPdTwHdbBkm6K469mQGpJmpDMZKEW5WD3WBE3ZKGYc/ApZpGGDPfEED3SB6O1GE6jDxljEhlHFNn5DhjMdql580oWHhhgyxpgGgK6P2+E118ER7WKsvkBxArgkp4SkuDMepDBGOOUTZZOCayNrCjmtSUTsFsMmGC14qW7PoKj6AQkjRSeMsJcoL6xLUMgBZmwyInk1TiW+1S888BqmJmvW7XMgPecHi5iXJrQeGW6zcS/65P+X8HIE9Qjkz9plJeWCdjIl24xyFONaWrfyJCkmAxGgnViFRdPy2a2Y+NmmXNlYFI1j6/W5jbMmjNTAvuoo46SnvUT//1HnPuDs3DJJZeMKYrr6omsAZll+UuK0NbaCYp8X3nNVdJu09UpuL5pHbfkmGOwbNkyUe2X033F2WvUbz/6Auu3bUZXWzuuv3QtHJ6CXzqL5WgEl65dKzsdd6uvv/lKwAe+sTZRz5Q0B4vZrjHdC5m8dhQbxnYT/iwvlAFFz3l3VTEynMVzuTQuajaH4YEhOOya6Snfn6kiT8tkOIFM4nvl8RzrKtWC62++Ga+8+jd5mFU1tfD7iuWGvfDk4/CV1mDmnNk4cHCfVpOoSY31kIzJTllWUYJJtROEXcOZMl+gGJs2bcKUKVOFeuV1u2GzWpHKZtFlVZAwKnD4vIiNjCI4NIyKolJ0H2rGOUtW4JHfP4bHXnoH7aE0ll11PXaFkkhY7Whj64BZJymaRiBJ8MVAojqtELTzkQrrRCiplCHBKKNgDAVNGM0UzwnCyPokjRGYiqzIuiwwqiakLZp+DXVOSREoUoC+jiDyoRGUHzUFg4WVXNjDCgRnrXdL/wvziILwzl6k+4OYcfxU1FRbMaTGkCctTrFL35CDymHyS405ZJM8HnOodKRhz7sR6THgs2e+hbN6nqCcrP8JfhhVWiJQhpJ22WyzmBBAQmYTV5fb8exNS6HYISalZF3RF4EINzd1jfaooc7MsMi24lfdcHd8VsENWzemIdljqLsHTz7xR/z8Zz8Tj4unn39GW+QFo95Lr1yLI11tMNsscgrbrWaNzhiNozpQjj1btiFBca5CS6y/b1DKst///veCOcydO1cAHdaSxy1dJug62xI8vdlKEqCHo33s1RsNUmpxzd99zz3ynnodzGsiOk99nLPPPQftra3SBVAq5k1TTz3pFNROn4pn//C4LEACNcy3eSwzH6YHI+vFZ558EoGKUvH306FanYGj30DKGEgtNH6av3BzeWFErnjSFU0hnE2BZO7cqghSdbd1jaWpDEaPzyOnZSQYFteujK6pGVdxzQ23ScFfX78NR5oaBb7m7yT1jT9bXFIujj/Meujbrhi0OUMCUSS0T5xUI5sJr4f9SQYatVK4I3P6m5uELrURzKURmFqHrmRUTuulS5airaEJAacHE80uFNs9uOb2+/DA08/jrP+4BRv2RTBsc6PPCvRacoKuMuUlIEICgN6DlYXCwJR6Uf4kshEkUqsZKnprtDPaL5gdFiQ7W7B4+WTsUMiLtBea/VpuKZZvChWxWbdntTGjwjPQF/C42WIJSEoC2+KANaVgcFsDDHEz8okkAkV2BAJeeD0eFHssNJlC+6E4dtYfQS5pBIYzcJZNQL6Qc2cVzX7bUPBZdKg5WDgpkUtLjWnPx+E0mOBWI9j27J1wqz0IjvTD6bLJs+eGzvEuopz6jOIYd7PAUtFOz+9H77g+qcfEvyN+wVouGQqLGgUnjtatWyfu0/qL93zh4sWwF3mRKaxf1ql5xYBkJC4uz0PtXZpGUD4vur08DVubW2CxWcVNimvj4YcfxrXXXqtR9FjL2+1ae6XQV9evk/YSvLZzzjlH1O6Z3uo4CjcQSpkyZj5evwGVlRXo6emFYq0qVX/10CPobGrGF198gdqaGpGw04OLbzB/7hwBbViHMcVjh0Gf1tejXSzfyA8sKF3xJogWZIEvqDfjta1KhdnnRjQVg5XACPV2rDZ0tnSMBTnTAV+RT4R9GIwUTiZvVerRlAHlZTVwe1woKwtg3/69OGrhUTIoun/vXvnw/YODqK6tRCBQLCx81ia8CRJsyaRYhDP1FitoaM1c/j3RLV05gA+QGxILcl9tNaxVZTAX+TBt6nTs2rELfpsTRUkVbz33EopdPtzw+JNYdeXVeGdXCMMOD4K2DEZECp+CwTwZecIoyFDBuxAoXC6UZOS64VebYoYrmYPdYsZwNIm00YK0nK4GFFuBvlfegeMnqxC3eb8nDOjIbMHld0wf5l+qUw0N1V/yfGXaQ7N6s1qMclrme/IY/LYRPmc18jEHksEcHLQ+UEhwpuCUZglPQR2+h8FM6p5W69Jbg5diIXOH/GZk5e8doAVfDrnW3dj4h3PgMjswc/58NLYcQXFJkWQrzK64LgQV5XQGe4gGg2zG/9uL2RQNQ6bOmiXtCSlvsjmpv/icpZ2m7XBja8rmcGDeMYtgtGnvz42aKGkqHMOh7/bK8+RmwJ890tgsA8EfffSRBBLbeDx4qArO9PTg/v0iqyFByY2NQlQFAW3ZxM0m2dAfeughYQfpJzh/L7nTfM8NGzZIwL/x2muaSeuDTz2hZk1G/PH+X8HscWH+7DmgHZyMQw0OyjFb7Pdh9+7dspCHR4aQUzTLZT2V0GU1uDvqtuG8MP3Y5vdxoQv9R8zLVSguO/KUHLQ6JBidZguG+4Py+yR9M5tRVFIkD2N0YARWIz3oNf0cs8GJibVTQM/2vr4e0KG2o6ddYOx/btiApSedBLvLhnB0RBquNHAt8vrlg/M6eG26YhhTCfIaGYx97e2wuN3SsuEN5WeUEzKTwVA8isUnnYCuSAgetjZCUSRGI/jkb6/jvJNOQW9fN06++gbs7k1jzc33Ychqh+pUkbTZxRIgmskjazXIDOT4YAynkrAZzbCRi8q0NKPCpSjo4/iZzYpcShV92ozRgIw6DOdX7Zh18wJ8l4ohT9Lp+Ne/BeO/L+LxwSjjjTSiYVbMU4dyHUQ1oaLUZESNxYBIUwL71zegsnQBRoeSsComxOJRSS8p/iTBaMrDYTaIMSrJjVQ6I59V5Sknw2d5mNNGmHJRzHfmcMdKM6KxBFadcjaC0WG4yBFOU/1B26T5nvrp+P8KRv2wIALOhSxrLaENgPP/Mi8oBqzaS2YSh0M4Y+3FSOVTmmQjgWhq1XqL8NHb7yHg0bwdCdq4XV7JBrds2YLFixfLyBV/BzdwIrTPPvsMin1+4TMT8ScjSZ9n5IZCTZyXXnpJZoLjBXNWxg8zN3pEcgPhfDDbHZTmkDT1sddeVrsOHZa5RT194rHLI3/+imMFVh/o65VeI18shEcjmjgTUwWPyyVBysXLxZ7IxAWQ4Y3kFL+0Mmh0WkhVRaOm0C0yWs2afRi9+6w2tDdpUxa8UXy/CTU1iEUTGOLMoMEsxHMCOMI7NdtQUV2DqVMmY+PGr3DjTTfhT3/+M2646Vq0tDYjndFcsfg+TGc2fP0Zan1FiFqMkqtHYlHhuxJgaD1yBEavR1Azfj9ZFDpbn/cknIxwYhH9w0EsO/M0tA0M4qQTV+LDd98H2vvwyVtv47k334ShthYv3fcI/nigDZuSafhtNvSmgSjt06h8YDCgXzXKVAKBmnyWi1hTf+V95v3i7w7GuWkZxYs+gjwSpIpJKzeP0pwB8V0H4L1kMrpNZrFwI0BEWkyeq0syj8ICLIAjY1D6eJHRwslIsge9IfhDtPwmV5eq4jWmPAJpYJLDjPV/rUfAQ+5vEolkFqZCucDpB8ayy5SHzcjRWGIGKkziqUKB6oy0SmzZHGKJOEoPvIkP//RLsTKYOW8eegb6kaDlPLkITHOZWmc1TrH+0v039QxMz7R4EuqZl8iAFk5AvWz6lz2qoH8aHRrGcRefDdVmhkoGFjQvzwqXF/VffwOHS5vUP7BvP1avWiPkEa4drusP1q2Tt2RtyrWcjsclFmhRz2ClGgFfXLunnnoqlh577JgyBDcGXhezLJ6s3377La655hppkbCf3t7ahmnTpkJ58H+eUf/wq4elx6YHDd2I5y1aiI6uLpTaHGjpaBtLS/nGFF9lNBdyAKHD8STVtWZ4w3ijNPOQf831pUA3KEI98xb7keDuWQhGpqncFfkhpSalqkCgHF2HW7U0oBCMFosVRy1YhJ/e+TPcestNmDixGtffcAOuvOwy2Ivckr66PT4BavQXe1ncpS67+w5JOympERoYRKi3T1IpIrzCD1VVRELxMb914dWK3quqSdB3d2HeRReiZtp0bNm0BdHvDiLc3oEgVNz54IPoPNKOU390Mwamz0VvigRxDS3myFBiOASbxy0nQCqvIkyaW6GJ7ePwajyFvgxgdloxGiGqmkdKzSPksAhxIGqMwaymkfmkAWfeuQxfpCPImsnHNUltprtVjk0SGTSJS6LZ8hoXjLIFFNyL2UPkFD5nDol+82stgAoYESDTJgL0dySwf88IzDafIJZ8pdmPlAktFTaSyMnEYiAy0AnykUFFtfpEAk6TgvU3z4GrxIHa8grs2rMHVpMJliKvbEYMCuk5s5dcmJbQN2U985J9ZtyY1HjhLv5Orh3BJAob//hTkYv+ysvX4vm3XsHMo+cLbZP6S/Ra371pC0p9fvl2gjMMrvqt9ZIZMVNjhki9XB0nERnHwrXwml977TVZW7w2pralpWWorK4aS7X1tJvXNdDXj63btwlYuGTJEhy3fAXq6iZK/Cg33Har+vzzz2tUKX6ITAaT58/D5NkzMb2qBk//12OF5hMFhzQOKfs0Yx90XE3I6QnuUkuPPw4lpaVo3L0HA6GhgrBvuSZWTBGhXFZ0Y/jAUgZVrMgJ4LQcbpa3ZerIgpwPvbqqVsaE2ls7hBtosdqEHP7Ms/+Dx598CpUVZaI/EqcAlsOBQFmRnLsZGZgtNJypSQIFt/7yXqxvbZCdLjoYFNHk4a5umZDVaaLyIFUNwdMnTU4//XQ8/eijsLrc0u4IZbJYdsqpmLVgIfZ/8hWe//OLiHf34djpUzFvwUIRvN2Qj+Hr5jScikWk+53U5IwlhbHPBxjKKTgUSsBA5eJMFkUWptN59IdTSJssiESTsJlsiBkV9JuBkAEIG3JIsqXREsPotsNY8NCxaLIAEU7R5EzC8JGYK2h4aoaP417jglGU+uhfQyX1LGQMisCRgwY1ClCWyiNgNMBF6ZSoGSPDYfhKffjqqyao+WJBC6mVKiNvBGyIjFvo3cx7D8QSMbg4IKwCI8F+XHjUJNxz/gS4fQEMBzux8oQT8cnHG0TgSQfmeKVCzC8E43hapf4pdCVxbrRMT+UZ0R6i8Ll10E2/D/pXvhe5zHmrEUtXn6QNk8fSqCutwIdvvyPWbUTuOeZ38sknCyagmyrddttt+NXDD4uahT6DSLVwZoavvvoqDh48KCfmxo0b8cILL0gQk2LJ9a5PNTFQecped911kpKS1UPq5t///nf88YnfSw+fk7oqYXO+ZHwkm8VdjzyEvp4e/O2Jp2D0uKCQWD3e36BQGMvNKwSjvitlI0msvfkGfLNxI4Kt7YgmY/jF/ffgd7/7nTAQPv74Y4QiYUGfkM3AP7lWECkyFtKxlAAoXEyS8lLrJJ1DJhJHWVklzjjtNJQGyjA0FMSixUthc7pw5aWXCNJYEggIaONwaVbn2YKfng4IuIxWfLRjKy7/1X0S/IjGER4ZxUgwKGwVZLVzRXbevBaMckpGIpi9fDm69uyHjQwZvrfFItMTay68GO1HmvDob3+LK045DfHQMHraWvHl7gO464WXces9DyATVSVl5hLz+Ux4780NuOCs09AxkkMX7b45bwgFpVYzkkmOkykIpXOIxdPwepwYTOUQNCtyMo5YFHTZFExzAp1vDcE7dQCZ42ciTa1GlUwfbcnqwSgmNf9HMMpnzbFxr8Kc0YKRLlGujAo/FJQpWfiyCpyJHBCxoL1rVHQ+OWL1z40N8HhKERwdhcvrQcDrQzIaFiNUkmBNZjsywjNNgzpIsxeUY06sAf956wVAMo9ULonY4BBa29ux4JjFEtg80eRFuftCG0B3neaz0IcN+Hy4WfMr0XDhMdMKrgD0MH3VT1L9hNT5q363B0OhIFacvgawmwWLGGlpl/5kNBaT7J4BQ9/R0eColukVOMk8PPjfxBr4vmXFxXIi7tmzR1oc9F/UyyJJnwuTS7xOAot//OMfBbTh+99xxx346MN18BcXye+w2zXBbeWi665S3371NYlkm8mEi668Qgp69lxI9cmYDHIyjd9pdNRr/HPWUwYqap9+2plixXXKWefji68+wdVXXy3eHDvq62ViejwdaNrsaUgqefgdXmz/9J8oqQ6IiLAorjHdTWWRjZPhYIbdYpPg+XrjRimOqWhOFgOL4oNHGuBwO+S9RUE6lZBpbe5eUtTDhGfefAvn33oTqiqrpffocToxecJEPP/ss5hQVS36qSSEJlM5SS25q7X39uCH11+PgL8IB77bhc7mFqQiURRXlyMaTsLo9GPzSy/ivhf/iF3fbMPPf/Nb/OntNxHLZPDxy29h9fGrkVMVHLNoMZw2G774djMa27vws5/di4a0grzRj2KTGRY1h+FkXtMYpQ1bLCGpLIxWhFI5RDwWjJqAVoeKjC0Ny54kojubcOqjC1E/kkCeQBHrULMBaZrDGBWN5lbQqx17Vrk8nJwnpEtWXhVVb0cGsOVI+DZIQNqzeXgpMpzMwBLLIhe3o6VxGIEyP3JqCqXVNrzy+ma4fNWwOcxCsSMiS/ZLJpGEx8v7bcGRtkO49MLl6GjqRd+nz+GLD15FMpeFifIfUDHc14VUOgO3zytT+1Syp3GOXjMysCizyA1RT0/JviRQRxRcTh32bpNJCagD+/YhUFwCh0s7XPSf0b+SwJCMx3HaeefC4HTISbjxvQ9lcHw4HELTwQacfuYZAtrocv1MSdlfDI2MaMCO2y3jTuS7kn3DWpEc6f0HDyIWj4jCIEl+HKJnasw097sdO8Rj41cPPISaCdUwmmlFoA1Diy9qYbxQWXTyCWpLdxeOWrQQPR2dqFu0ABVWF1587A/CROAH0O3VxnLxAgQ9Phj1xiq1Q8hWIQPmzDPOxttvvAwOxnE3kxRD0RQF9Bu0YMkC9IdHJBj3bd2OCZMnoHegX6QReIJFOb/H8bokOaN5AZKYRtDNhwHJHZEbydRZ0+Er9knqwraIyAYWHHGZs6dzOazfuBG/eeFFNDW3CrzNOoMPfPOmTXAZKF6bgqfYg6MXL0NbS4vcbI7tTJ4zB4TFJ5eUYv2Lf0HD/gbk03FYjTYc+4NzsOXdD/DOzm+w58B+1O/Zi0vPPBNnzp6PmSecjCtuvlGIZdwU2jvaMDAQxczZs3HPtTfj8/1NaEtQatEILwzgIRSm4twowJkSgjbJBMR8ddCQR7LIggaXioQjhxlWExrfCCOSacKKn85DU1JBkn4YZBGwViC9zCQdiLGA5F9bc3m4YICLkx4JoCybRymV9Ai0ZPIoyimwCVcY8POgG4whG3Fi9742TJpcK71As90Ao8OELdsaNHphJi/NeV0xUMnFMbeuVrixweEIzpzpxkklVpROqBZVQJvFJmRwWum1NR7BE089ifsfeEDWWbqQdenpHachRDBKd53KqwiUfq9OSB4uSwduAgTjqADA6RpmYLIOxluSG2kQm8fqM04XDulAXx/2bNoiwXboSKN0EkRuNBSSeVs+f6amPNF4QDUcPIT3P/xA0lKuG7JneBiI+3E+L50G4hUiYWZgpkP1PLtIxBCZZdbHz2WitR59W4qLNVnHQk2veGoq1Gnz5mIgPConCrmYs6om4duXX4XRYhP+HhuyPPpFDZn9H6NmePq/BSM1Vv/zVw/iof+8H/5ABaJpzUlK0mC+B5tR40gBqWgElXOnIeApxp6vt5AUiaqJtRhh34UB6fTIyehyuPHsU89ISkBHWO5UDBAGNlkSTe0tyKaTmDlvruTv1G+llIZ+zebSIrS3tCGYzcu84dELFyCRycguyhv79cefwmw3weRzSR9x9ckny6wbWUI7Dx6W92E6Vun2ylRAMjyK+q824f0Nn8NstGHG7MngWHvthCnoOXwY2aEQLrvnbqkNnBYH7C4nRqMhDAxHEQ4PY9Mb/0BbYydmrliOyy67BWecdYpsSpmUGYd6hpEwFCEFO+IJBczq6TY8aAUiFQaEbdRLDcPUZwaaHejrqseJdy3GkWxKpC9VhwU5nlbjTkY+ARl3Il+d7lTpPOyKATVqXkaaAllNmdycyYssoU2xwcSSMA6Mtuawc99h+Sy830ZzHla3HTY3U3rgyMEGlFVUimO0w0Urhrz01RxuK2ZVG3BhlRfHn3gMdh9s1sSi8mzek52Tw1BHJ2689VY89/zz8jwp3qyvFbvDIQcCg4ELmZu5yWKVawh2dsLi8wm/mCdyYiSEx/7we2n4s/3AGoyTEr/61a+k5GBg8mAhWFdeWYm5C47Cu2++Jdczf+5c7KzfgVVrVuPzTz+DwWSUYORLhsnzeWn3sZ78y1/+IpsO78Xqk1ehoqpS0lYCkvFEVCh9BAM5OaNzsvVSjn9mq4ynJT+T3ktlMMoBxWCko2vNlGlwebxoDY8gmTGg3O/D9NqJmD5lCr7+8H35ZW379wMEWI2LdAAAIABJREFUccRqV2uo2qw2TbSYVmZGIDEcwdPPP4ctW7Zj/UcfIZKOf99vLIAXMnpFkIFvYFJRN2emkGr3bN0Gi9UsdQh7M/x3DjwzTR7oHcDGbzfhuGXL4A8ExvqW/BBnn3023n3vbVjsWr2Q4gmcVzFx2iTMPWq+OAr/c/9O+J0eKHY73EUlws6pmTRJmrP8XW0tzTA7rDh6+TEoLgpgyaIl+Prrr9Ha2QGDQ1MSZ58zSb0ep1umQI6aOgOjgxGaBMNozCCaTcFvtqOzqwez/WX45oW/4e/r3sf0GdOw/budaOrqBExu7PhuO9o31SMeSaGqdhr27DgIq11BKjQoE+8P//mvWLDiTDT2ZBEepbUC0JVIIea2IuNPI2ozoNOUQ2WlCbvebIQ3Vof+7i+w+v7T0JePI+o0Imm0cpoHeYMqG4pJVUC7DJaRIpmaEp0qVBlUVBsUkdovpjwhgJFcDt6cEe1NXaj0VCLSoeCTj/+JiXWTNXNRjwsWF5vqTA8tsEoWYtDKCxgQSxthzKjo3LUOt5y2CDeuPRudvbyPfhkIMBk1Unomk0C0rx83//jHePzxJ+B0OZG3aGgqN3sCgm6nC5GBQU2M2emE2+uTRc7gZIqXoAI4W0MmE3wuN+6++24R1Gbm9Omnn8oiJ0WypKgY7mK/BCMnfrZt3w6f243isjL5XfNnz5bNhE1+1nf3P/igBA4l/XnylRQVyaA6pyu4KZAeRxBJb1mcfd7ZeOPN12VYQeuxa0CdUPoKBA/OT/LUdbocEuDcIDgRxXsiA/muijI1OjqME848CwcaWrDs9DNQWjcV7SODqKmtRKh/QI7x9v4+tPd0g16NA83NWHHCCSjhzGFLB5bMmokzly1BeVUVptVNxVk/OBV33nGvQPhOrwtmSi3S7INAEBXjKERFYSeSKpW83FS6Ah/YtV/kJcgNrZo4ASOhITnuIwPDyGTyuPDSS+Rm6Kpu42tPcS0aVyewv8l7UD6pBhFkEctrord6L5XmMROnThGj1t379+CEM0/FYDohYADTIjZtmRLz1PT5S+QG88byQRjNLLo1ZypJQ+JJqQG4ULkwSovKUL97F+pKyxFpaMeRHd9hxoRJ0uQdzSj46c9vR21RKVwuPw4fbEHe54ZdjjEVM8rKULZkOTa88g9s+Pgz7OxIIpktR+tADAmHFdZqMyKmLEadRpiKgEqTgrf+9B1sqenIhQaR9rfijF+ehB3JBBw2qyiiM/2lX42iZDW5HN7jjBHmnBGWbA61MMJojSMQT2OSiUmsCRUaN0NMn0r8wA3Xv4uFi5dhOBzElLkzYHeZ4LBrwc1XgjboHAXLqPD4gSluBbX2MJLdB3HJBT8UBpH0lzO0e9NaSCIWZjFLukji9LHHLEVbV6ds/OTHivtxgZ7GgOHinjd/rgQIfz4WDosQMf9N1ONTaYSCw3jz7bc0hUL6JGYykkpOrJ0g/U2eUoGyMlkHlH3h9zFICKwQaCF4w6n/4kCJPHs+X8nslLwcAGxzcL3qpzXX4r333otf/OIeTJpaO6ZWwbWpkxcoz8k/S4pqMslz0dNn4d0mNXqpAptVnbNsJeBw4uLL12J3fz/2tx+GlTOHdrMosdHuuSs4JMO+vCFtB/ZLUHLrDTV3wpwFVi49Fp+9+Rbyvd1AOoGH//IKghYFlTUlaGo4hD3f7UDA60FTQ6MozvksVtQEAoiHRgShuujyy/Dpuk8ldWJ6aOJCcpgRj8SRjqZgMZjgryjDyMAATIUbNJ7/SkBmfDByOiSWjGHK0fMwGB6B2aUZvvIhCmRuMUvabMzmUD59KhIuC6wet+aINY6ozID0+oslPea/kQfJGpALSddecTndYw+eLRmS5pNQRVK+3FUEcyqDKpsTr//mUXBmq2TeTFTaPGg62Az/xDoMhEMwciqD42jd3Vj9h9+jefchtLz0ChatPg8nXHY7wqli5GxuJB1ZjBqSMJRzgj4DjyWN0jInXni8HopSgclqFZp3/ANXvHge+pwqRoxZxOR6jZLQSPeDnNe0pl9jCA5jSVkJfI3AIz+5B7ldbYC3FhZbCcr9VTAY02hr+BZHX3YeZkydiXgqhmlLZgsI4WQ/I5tHKm0AKavUODVnU5hXYcIP5/ixdd9mmf08/dSzpEZjZsHHRK6wzI2yZmdK6HCgs6VFgoUjehQkox4R611pSZDZZdXSU4/XLd4lIlDNYDSaxoAQjpjxFOLpw8l5ndrIwd2XX34ZXo8PzzzzjJQ5DCi7wybvzw2WI0y8JqKcx51wvFi4bd68Wd7riiuukKyP36evIf73O++8g9NOOR3FgSKwf8WTVO9z6gw1rhF+Pv0zMHPjhqAj3nw/yrgIe+fZN19TM64AWvt6EEum0RqPyCKOxMNwFXnkNCPoHKffugKkUkkkIiOya0ifJ5zUGrsmGwI2B+ZNno76dR8gDhvKj12IlBKG1WmT/ppAX5T1UxUsmjQFjdvr0b5rF2ZNmITRwSEEh8No2rkHcxYfLcHY292OSDiG+GBITl36HPDD6jXo+JpVD0b97yQfdzvgKi/WWqj/FqwyPaECoy3NWHjlWvRGw7ILkpDAQOaOxxvOIjsc1VxyJavmCZnW0gp+vwBXilGatuwfEaYPjkTENYuGQSetXCPwd1dTM7wWG6YGKrBj9w5U2tzo2duM4pmzERwagsNgksW37IzTcYRTI5MnQG1sweDnO/Dq+s9wOOHF395vRPWMWihUhVOysFfbQSPqtMuAYC6BedVO/OXJ7TCNBGAM2RBp/gQ3PXcllApgX38QVhMlOuzilRgeCWP1bA/+5+dvo2V9MyZMPQaJ2CjC8RiSsQzsVPIepLaqAUXVdkycUIw5UybAWWyDZ0IprDaT8FrDQyFYbA5kbHkYc2Gcf0IVjvFNw/XXr8T1N9wM1WTBqpNOFi1Z7d4Sb/i+H0oZEa0cseD6a67FG2+8ga7eHmTJWOJcq9s1JudP5PLAju0wuFxy/2X2NKtlK3nhG1dIS07XUGJTvaWlRdD2X//615g8qU7qNTbmOern8XslCM4//3wJPp6KmoJhDHYKFcfisNptAgqZzAb5PVzzZNCwX0iAhwEorDFoFnQirGYwyDXw+5ktEfxhoOunI0NBX8dcy02N7Tj//HOhfNJ4UH3ro/WYNW8eqidWY9fe/fhy1x6Ec5ovBiOaGh9p3kSTUT58T3e7TDpkU0mkVFq+pWGmWQ53gYRJHIbadzVh+sk/QM4ZRcaSxOhQCLWzasVLnouWx7zQ5hIJ/GDJ8Xj0qhuwcsUqGV1q72xBX7APHrcLDZt2wuJ1w0KystWCZDwhOTY/hJ6LC3Om4GxFYgAZJ3mrgoppkzGS1IjGfHii+Wu3a/UmcnK6LVh1EnZ3tYqLLPVcbVaHgAT6DsfvIYOfaRBBAcLs/Lfh4IgEJVMQZghsKJOkHBoOweMtxuqVJ0vBv2PfXsSDwyh1edD33V7MPGUlIvksLFkF4bYeRHqHhTJIDz/qm81eeRqSXicqiktEysEfj6Nn3Wb0DZbjlr98jj4qtDuNCClZWCabYHMawA5ZyCg2F0AqgVKrHRv/8h3yQQdKDa3Y/vlbWp0fzcA9fRF1KTGtrgaegf3YdehLXHP7q/jry5sRCY3Kjk2MJaPaNSVwg4q1PzwbWzd/iSkTA5i/dBbgNct6CBQr+OIfW3HM0bOwbIEHK5ccBaeaw4rFs/HRh29g49ZtmFw3CdOmTBNVhQi1QcUuQQNpZCMrbHJss9BjJTIakpODIAvT2mg6NcYT5sKmhIVuhCQNfuSlRcEUn0PBHEZnmSGsl0xeiNpLFy3SeoCZnATPzJkzUVleLhpM0XAEN9x0o7QeJHA43jQyCpvDDqersNlSJ8mi9UIFmLK75GTW7SL4MwQLBchJpWQDp2cmAVF+RpPIkWsBKMP0BZuF1tYO7N+3D1s3b9YsMNYd2qe29Peip68Pk6bUyXR9VjHjr++9A19VhcjKMY1KkObEN6Ko8GhQpCuY85hsRoQojqnfXKYsI1FMqahDLm6GajEi4WQD1YSsScu7+UF4oUz9xC02nIR7OIbZlZPxzl//iqnVlchGRkUigX2mUF8QZhi1uoPpTeZ/D0bRjCG/kewQYxb2gB+GQn4uKB4L/QJHkD3I4dAIfEfNFeUB6qbqdECKV+nIFx98OKRRorgBMCAlPc3mUV1RBRv1TFOaXusvH3wA27ftwPDAiPRVG5uasGTWHBzeuw+hQ42Ay4mF556Ots5uuCw2+FUTWvc2IR6JCjUsOjKCs669AR2ZhGQK7UMD+Ot//Reum3MC6lbciamn3QTFZUXeqiJt4VgXe5AxGOlVYqSVngJDPxXSgzhqSjl+85Ofw2PsRTzch2QiDbfNi0goKZ6F1VYjrOmkGP98simGh5/7WmomSZ9EykMDF0JdHVh69GzYzFmEI/047tQVKKsrQ/dAEPPnlGPdy+vgtQ/j5d/cjqlHzWD1CatC8sYg2to1RpXZYpY+IofJJUgK6Pr4YKSxDZUHL734EuEIU56CwEk4k5JnIVZupGCajfJeYzVYwW2Yz2YssAszkFRMiI2G8Oxzz+HE449Hc0ubPGMKWVdVVKCkLCDXQ7I26Wm6tEYmlUZJaQBDg4MoLS+RFJUsIa5dBhpPU64jXhd/rwQowRirdWwomiivPsXBtSISlwUigAlmWftUiqOwmyhiMH7+sX+3ykBramkRO+1FixejoqwSDW2t+OjrrwRs6YlGkRbnWw0B9bsdaDxyWJDPDEEBqwZu8Ag3m6ygB1JlSRX6txzGsUtPRYehF0PpXhiMFJLSZP6FYUPLLLMR2TSbz2bEQ0nUBUrRuW0bTAMDyCp5Ud1qb2iGw2yTXVIIwemMRjUqoFR8qNxrubsyjeYuWTa7DnBYEeNUBPtQbLJyJKqjA1fdeQf+/F+/w8rbbsLejlYpqJmaM/BkAYRiYyJUkssLxqilqMKYsFoRGQ1jZGgYTqsTxcU+RBJxHD5yBIrRDIdixfIFC3Fo63Z0btsBs8cDNxv/11yOPS1U2nOjxl+GT958V5qJ5WVlAjxwgcUa23Du3XchlIjjy/ptcHrcSH61E8uW/wTz116HngSQzDhhpA+GC/CUk0YHuKjRmo6idHQUJx1XjTNWnILp8ydgoK9ZI94n8wiQzmcworOxGZs/+VQENfo7h3HGtX/G1GPPFSFdUgE5nU6NWiq1zZwyCenIALLxUUTjI5h3zDxMnFqNPd/txdYt25Ad2IdQz3uYPnOaLLBENCV1fyaTxq49O2SxEncoLi/HKMn5os6tkcJ5b8efjNxIuTEx+2CdRnpZf3hU6jsuZPF3KfAWdXlPWt/xpQcC14I+QmUyanxm9iD37t6NLZu3SmbEqQl6b9AclbUiTYApiaGzzEaGh0VtMFBajAkTasUAl6oAostUUDDkPeXGxU1CNuxweOz04+cyqpoOEP+fTWngHv++s7MHf33pJVlLjBcZ9dKD8cE3/q7SNYqk8JFwCOztFJWVSrrAU2X34UPoydPcJTeWCpoVFu1JDAUHYfHYRROJN0BS2lwadkr729xIHxnAyPZ2lK06DkogjyS1P5GQB8GdRENFc3DaXMgm0lAzinjbZ7u6kGg4DIuaF3GggZZOeFxeJDgNLjSuQg5eSHH0niffNxmKYs7xyzCYGEU8pzF5iChyt4v09+KCO36KA81NKPd70Z9NiaIXg5FOSTwZmMZQkkMnqwtxPZaUIOWLJ2SR2yMnIwPSaXdhqK9H7KoD5eWoLK3Aka27cO4pp+GLD9YhEY5Ij2qgtRUrH74X3QMdok3bu68RVlDvxympv5mmKDxxU1k4rVYMNR6GY+YM2OxuzKioxLZv9yLvrMQ/vvknnnipBbaAFcXlTpi8RiipFOzJEZy0aipMgyFcuvYCzJs6HY1NO0TbR6zZM0BlaSlaDjXiq4++wS8ffBDWdBifbXgPJ1zwEPptC9HW2gEj7NI3pRJ8OqfAYTdDyUZl0oQpazwREa1Qr80oG3GF0o2JRR1496N1qKQ1YDoJ6qhyYPbzLz5BOBJGsdcLp88n7Bu+pKQoED/+PRg9TpcsWk5MBKh36vdKMDKTkuxGn90s9Ln1YBRJi4L6xNgEiMoUUVO9mza5Dscdu0Lelwwaum6n00nce9994kolSHjhmijb+NGG9Vi95mSUlBRLMBILEDnRVEo2HX39coMYP/SsDUfQf+T7Fg3TfX7f2rVr8aMf/QhbN20ea23wM/HZy8l40/NPqiuXrxC4mFZo3+7+DgafS3bQModL4P9n3ngVJZMnont4SE4gnjROm11YF82tzbCKuUscHh7bBDUZjFnO3wEDT72DJTffgUFTAkOJdnhdfoyGR0VJjj0+t9spDXzWC7koZKphkIV0Mo1b116JJ+67H5mBoBTyWodMq/+EyzhOOYz1J3939bSJGMwkYKXWDrteFL3K5RHs6sC5P/0xDnS0aYiYmBMqGKY1nZcEcJPMNRosZvg9VJoOSt1Aep/T5kR3P41kLMglkjCOpDCpKICjFy/C65+sh9fhlBqHxPPKkgA+ffpPsBcXC4DDGUX3pBqYJldhJBOTnbp390FYM5qlmDGnIBnPoLS0HIMDQzBaNKswHSlWCkpr/NxsA4w0pVBUsUrALHsgBzWmIjnchXx8D7KcXM/lcM4lF+O911+Hj5MAeSMyySjsTjNC0Rwuv2gthoe6sH/PdvQH+6A4A8iMhrHi0pfx2eY9KPJUwEBJfKb7BYYSub6KkkZnWyvcVhO8Vg8SSgjzKrzYs/m3mD+tCg0NByVgpBXk9oi3BEeEuHm98fZb+O/HHxeAavyLn5Gq3Pp4lCzIQvb1i5/fjVNWr8aSxYthZf9RP0kLZqRjpJOCVL44Z/v9cu9oiqMFfSH4pQ424qPX35frOfuss2E0GTF5yhS5zzztCLzw8/JaoqMh/PSu26WOFNI3ARm/JsrGoBI8IVewei+0Xvgs+bt1LRxeX09XN356x+248rLLBRxiKaOXQnqLTZhG+qHy6Jcb1OG+fiyYM1e8DEKpJNIeuyz7CYFSEXWqnVKHf+6sR8SgIpnPwWOzSzCK54HFjM7uNiRs5ORxcpbzbpoPQzyVhrszir6GAVim1sFbbkPenEVK9KNzGBkJYtbc2TKgSUQRCSPqjp6P1pZmFBuMKHd5sNBfimd//DM4ywICicuuw2FODocW5PzkxhMiTqUxZflCDMZGEUml4bM7tDnJXXtw2wtPYseBfQgnE1K0U/JDyAf0jHQ5RAmAqtGskcNBUuqcYjfA+oaUO45d1ZYEkOzsxYcvvIYf3XQDoshjMBLGjOnTEY5ExBSorqIS9f/4CMlIBFaPB8f84Ey0RoKImIB4Ki7thMj+RnDoSROcoiuVKvqkdINKZTTG0v8WjJrjjAqnQtNYCzx2DyLJNMKROHKJGKIJKq45EB3oJ3VEmtiU4bCZrRju6MN1P/0pDjccRGvrIUmRoukYXHY3wgMxzD/rOexr69GCkWyBQl9MUEu7BYk4PVQomUK5fDOQiEMZ3YTRjndFvZ3kCgJWXLCnrTlFene33nqrABhtnR244MIL/3/BKMEn/U9tul9e+bxs9lUVlbjhuuvgdLtxy223wlZoG+jfqwtWkfLIBS0N9oJrlKR+bLYXZDH5M5K+DidERmZX/Q7Y3S6hVa57731MnFInNDh9FpInXyar1aoEGvm7+gYoFUq19QKdk7KChZc08K0OtLW0wuVxj52AbNnxnjCT0q+JP8L34595ryQ11s2Kl955q8p+29WXXiYS/jSDNBR7EUwnZfqeUDCpXDQZ+fDrr5C300/QLMc/PQ18LqdonbSFhxBJJeCxWOVESCWz9MlEkcGKnb9+CS99tQWP/+0FBPODMDkIPEQRoTiUxyHsCSJi2YiKqjkzMZSOo5jmN129CCSyaFv/BbLBUfEykD5gIUURXmphB2ftOWHWVIQ4ERCP4KQTT8b6d98DnA7c/uD92LDxK2l2y8O0WdEfHBDBqr6hIZleJ2mAMv9EHcvKq2AxmjHcOyRmr3Mn1eFIexuifQP428O/w4WrzsBld/wEBzs0Vegj3Z3w+nyIhyPobDgsoE31lDpxwbLNqEM4k0SEEoE8jYlYDYUR7e6XYKRUBP+O3Fuv1490NvEvaQ9Ptu8fuub9zn4ee1OmjAUqvTj4MCMkVGiTDwX+gyxsckknVE/BiSeuQVdXM5KZMDZu/BJWl098Fa2ZLGJDFpQfdw8yNg9MBpuMtrEFzcXJBUPys0IvzVQSmVhY5E9cag5NWx9B5v8j7S3A5CrTbtFV7tVVXe3e6XTcEyBAEhJ0goXg7joEt2GGwXX4f5zBMtggwYNGCMESSELcujvSlnar6nKv+6x31+403Dn/Pfec4uHppFOy69vf+7223rW8u4TUadbsIyW64mEwdtRoafUQyUKGCBa2GpuaMBj+PYUGPZEqba6CwxmwEd7Gqjl1DW+46SZccvFFsLrdQ9L2w72rRDjZ4iEPUu4HdU/oKOyTLZrIvGRSydOC/qBQupx95llCqyioIptN+ttkceN1FZcUynPpMUXUKetleZ3MacO+KI48apa0RFiEmXX4LAGkyHAFB+izCDVem0o/qWK7JTTNGrbKDysHxg1fLMn8tGYNbDo9yjz50hD9cOknMJYUIsZSLDSw5OXClEwjGI1gd/sB6G3sVekkTGUuMWXkCGzZtU3gRrx4xvqD/jAiHEEiUGBTM2odxVjb1QZDsR0pyq8lwzDaLchxEjScQiQcRrw/hkP+dBzWbNkITTyGIkeOhIV/PekMLDrqeBhzldKzeuMoB6BWtQIRPyqnTMBAMoRAawuuvvUujD30EHz3808iyRUPBhEJBBBDBnnlZagdU4vN27ZCT6m2aFhK07yZ9Aj01KTvr/txAyocedj+8y94+JUXhUTo2oVnSQXVFw7BHwkDoTCshQWoqKxEX0enMEwHrXqkrWYMBgLoTIThdtjg8w+AHAdmox3mYBze5jYlTOWkSEbxjvl5hegf6FXaMGrFcZgxUkRUCb9I85DV3iDtfloLA/OZKEv9ijHyZ8znxSmnXwi324MclxPPP/UAjDk2mCw5gg+NQwc36R4LTkRL0IOcsio4nS4hmNbplYY411r6qbzOVBIhbx/ioSCOOaQS/3rsWBzY14iiqlLMOWqOcviY2TYZKZv6gQcekE0395ij0dHZKXno7wxJxEiV36iRgGRPHOg2mWUa49XFi3HRBefD5vEosml/wESrxiieJzvHOdTPI+wuG9YKIgYKGTfzPpKTsRC4Y/t22B0OaXfwvdlT5PcIBAfFSBny8jvQsBmN8fv193px5ZVXCWCcfUxys158wcXCCs7PUQtBqvGxyj/8wc9RjVVYA6ifQjic+6LTM1ecr1ABsFHJxmrdtq3o8PbBWV6gtBMiaVSVlqEg14Pvf/oRYbdDxq206YxQMFR48tHXckDYxFJuG2x6g4R/dR1tCDMsbOqE771vYZ41FzklbsQ5xWrWK2zV1AYkJSRDTq0W4ypHoz4SRo7TDn9vp0wQ5MfSOGHGYXjmnEvgcOcIJ4xTp0MwrRD/6BMphJJRlEwYiy6/D5deeilef+55ISySgWkm1eE0zlxwGionTUDK5UBb4z6sWPcLigrz4Uxr4dIbYUilYDt0ItZ8uxrHjpyEN554Bt+uWImTTjwRY6ZOkSav9LgSCv28GjJFYmlkSPlo0uHE0xZgY0czisbUYF9Xa5ZgJDNUHCBQPtrZx1EMaFLUjVAQ/KrxmQymIVY6OfFJIJy9werNHfqZyoLuabj0+qLilITDaofvQBtuuO3v8AcH8N3qlWhrbxTwhRoOpogfNWmgCWlx9EXvY8POFhFygcUowwEWehVqiliM0BqpR6JM8fe29aHMYcO6L+5CMrYdb/z7A9HG5HQ8SZu4YSmtx3109dVXi4esKC+Dm+D9rGcU8qnsYzjFhhjl7/ct4sGQAAEuu+wyORiYJsmBlA3thNhymHz88JeLV8+CsFUPyUo4+8ETxo1XahXpNGpqagSH/ODDj+CGG/8sKB9XDuUlQjKJ4XLlwGJySP+S+5QecP4JJ8ha0nDLSyvw6WefCeKH0xlECLEtpB6qqnHSw6pqbHQizD9ZtCvMsghoRt10VWZkVZWiDBWPi2fsampBX3AQnX4v7J4cgU4x7uVwJhvum1qblESVQ5RaZQ5uXGm5nDQc9CVImwbzy946RMIxVOUWYeOz72PC7GOxp78Z2kITnEUeqa5FYmGpVEqjlpXLPS0YOf9E9HCCITyISDCIYlcuPGYrKlLADy+/Dn0sAdIDMu+UdoPBgLKpE9Dp60faoEjAFbtzhzCsXAyiTxLhCCyuHHT0dgMd/XBWVqI6Nw9HTpqGUeXV2Nu4Fz0lTvTt2Y/v3/xAcsiKmlr0dnXLTVOHXdl4UkvoEhbpLTI9kEpEcP6FF+Kjr7/AmddfiQ9XfiXgAovLKeslp3I8gqmjxmHjt9/DQBKqxMESODfSxPETsXXDBhjtLFgpylF8DPeWQ8aYlcgT+WwWD/RANBTFrJmzUV5Sht07d2PT2jUwuTnWk4Y+C8KW9xWm1QxCvT6cftXLWNuchM3uhElvFn4h9o9J1Mw83eowy9S+1QC0NTVKD/iYScDKL1+QAdsvv/gK995/v1wnvdf0iZNk3o8tBPb0JkwYj9yCgqHWxv8fY2QhkYUgQs+48QX9ns27ZF0MSs+OBjCUd2YtUoU+qtAzKS45nRKJETi+Y8tWuHLd8A140dPXi7W/rkMo7JeW3Reff4mGPfVobmmExWIWun9GfXwPgslZTWdx5/TTTxdQ+rHHHC8eVMWfsj2k5olqjUMt/qhtGYb03BOEyzHH1lRfe3GGxQlSSzD2lZMqFMMJJ89HW28XdrOfaLdLxZCI9xEVFVi7fSuc7K+Q9Zuo/5xcpPoGxHMKoDYUFlrCDU37EIzU3OxHAAAgAElEQVSmkOvOQ2LbASTCGrT62+Cs8SBu0UlCH+LYidUqmzsw4IU+bcCEaYdg65aNrJ/DlZ+PYP+A9K60wSDOPWwW3r7rXpkbowYGrymSiGPKycejNziojLJQoMSitE74EDygTifkWQJ5y1YJc20O7F7/G2KhCIx6K/TxONrCPgmPydKWzmjgHQxI60G8FCcJkklUVo+QG8NWiDy0PG2jsFkMmDt7Dr769BP80t2COWedIr1SV2H+0LXodRzMMEKfSKN72y5YLcqhoVbXEt5BlNbWStWP10nPqBqfGqKpXlQtJksYxJCSOKtURooyR82ajY/efF3k1pTJ9zQMJiWn4us5ia9N6ZHRJBDsSOPye5cgEE9jZ0svDFYHdDYrUhQ91WrhcJhg0Wtg0oRRmufCEeNLcfH8Wpx1zol48IGHQFqKn9euVSqT4TCmTZyEo446Ctdee62c/nPmzZVGfYjCMHKpBz0jr5sGpT7+6BkJ/k7GyQ7fIUUd4pO59mqYl4pGMW7qVNm329dvgNntGpr0V9+T65YIhVBaXY3y0lI07d8vGN2u9naUV1TgpZdeknvrD4bgcFrR1U3pwxy8/e+3sGnzb1LxVYsudBzbNu2U3zGS5KAzDyASaNO7UlpAOUX/4OLZhbBaZbyLw8xcGwIISI/KgpWwA4y75ZoMK6aczFCpD3SRlDTjPSUFaG0/IDkjT1+GpZTn2t3cCJfHI2GmCEYGwyi3OlCaX4A+/wBOmDsPGzZuxE9bN2PSlJnwppLY+uUa2FIm7OreA0uVC2EzixFZeeZsEUZO1r4ACovK4a6tQleoX2kMR2JIBIICKi/KL4B/6bfQBYIKCTJrF4M+FB83B6Ycu2AaiZVFPCGnDQ1GpgNsZvHkbU3NUpj69bv1MKTSmDZpopyslI0u9XjQFeBkbxx6ksPqDEJwq0qvqBtAJuqzAGbVGMmjY+CEfSyOM848HWsP7EO/OYNAIoqcHEWyXE5yYRMwwmN3IuMPo2u3ggqhwQh4OhLD8SefLKGQVNvI3qZW27L50kFjVK5MncTgXAShV1ajDf09fTBTGJTGx/F40aU3ynpKfmLWQBPRIaGNQJ+kPoUW/v4gRk44Apdedh1sI2egr9sneyIe86Ii3wyPA7j8tBMkYqgs1mPJR5/BmVuI8WNHI7ewUDYljXH6pMmYO3euHPA8/I4+7ljoDQaEEwrSZLgxqoeD6tX+aIxsSzEiWbJkiYC9P/3qSwUsku3NsdpNA+E95k9O+0vuP+zB9eLkBotJN1x/PTZu2IAvP/lMUDbMHTmpz8iwt38AvX1dOOFPx+Pkk06Bj/utuEAkIdT+Ie+ReLSIQqDF9eH+IfKKP1UgudlkGcIvq9NCCuGW8hp+J/IQszD6r9deUwANlVeelwlTQ8Fix5gxY7FvbyNyifGzGgWBMOOQ6Vi++juZ+SOS3m53wKk1I2U2SMgQ93kxY+x4TCyvRLHThfc/XoqpUyYhL9eF71d9i0U33SwQrDvvvAcZjRHbm/fAVpGDgDGDlEEvmEXOwXFRUxz81WYwrnoi9oYDIHdgJpWAzWyAzzdAF4i8wgJMKSjFN9fcDFd+kfQBo4M+VF80X8Q2Wenily+25eJAZwdyCbkjfEljxB033AydP4KHbvsLGjZvk/xIiI28XmnJTD/icPFIHGzmgyEJT2Gemjy5uGGkUpZQTnbedAEJJ1IKTI+iPnot3li+FIsefwDpdFyGZYneUA2KCCUxSk54G43IT2iwc80vsOR4oCGJlkYnmh5sdQiYmGAL1RizP4eqcqTlyD6Gw8GUDZ8W5rPhDxbb1ByN4a9adRz+HF4XD454SIMk1aOtVhl/I1kxHwKKEEn3hIyYsXQ/dlQtdKQ/1GrkMJo6bRrmHz8fxx73JwGKLzjpBJFwU5v+QpWezfuGAzaGF2fUP5MfiNdEvOj7HyzBeeedJ2RnvEbyazENoHHSqPRSzFIgi6yBwKxMevBA4IO5Ge+vFCnfew9u8t9SH3HPHgmBGVb3e3swfcp0HHL4NCSS8YN8Or8Xw5J+saJHalDYwLPdQvWQ4D1Sx+yIX6ahEkjC/cSHiOVki2M0UFZhNac++LfMhr31Qtqan1eApsZWjKocgbx8D8JhyqGkhHeGC0CEDq9pbNVI1DU1YtToMUJBOGX8WLh0BoyvrsEvq37AkUfMFFm2vfUNOP/cc2GzmOHQGQWg7Jo5C4YiGyJWUgtmRGKbHpI9PwKvKYEd37IXJccfj4AlI1W8TJIs1hmhYDcbDEJfWGVxYNdjz6C8uBQplw3pccUiOMIvTX6UfHcBAqGQkD0dOnY81iz7DoN7m4V8yWKxwk/ZtCyukAvIBQn1eTFq8gRBI3ExeXpx0dQFVos2ZHFTN1M8FkN1dY0UHooLCrFm9Xe48r/+jhWb1qGnvwcTJk/GvqbGIUIlrUYpuvCzeSoGQmE4Mjr0Nx5Af0u7MMRxl02dNkM2CPOXPxqj6qFVXUv1/YZ7FTk4siIyqrENN0Z+huolh4x3mGFTuFRtIwkZVNa70kjMOr2Msm0jUMTtRtWIKkycMlG0DnlgU05u+rhJeOLBR9He0Y15Rx0BE/l11djtPxgjv6PaEOdl/NEY+XdOYTCk3bBhg4JW4r7RKz3Ep556CuHBkEjdP/300yLBRgQZ7x3DwrU//YTTzjhD1pLghB9Xfgt3gUeun9ETiyv5eXmYOHmc4Golx9brh+Bvgtcd9qCBUYdxoJ8cwml4B5QDXCTsshLKDqdTWCiuvfpqudcqzQajIBqmWp1Vhxc0R157VWbD/mbRHXTa3YjFEtDEOW8Wk3I4eU98HGEqKYHRakEoGkZZcQkiCTYrdejoaEZ+ngslOW5psOfZ8rFt62Z8v3olcmx27Fq7BcnIICzEr3ImccYMJO0ZBE1pmHPdiGqTAsJlPB0PUmhUi8jeLpx8/gVY071fqDSYC5nNRsQpeRaJChigsWEPzp81D28sugXn3nULtnY2IDc/VxJ5epyANyia7iRW2rduI1x6q4iahBjykg07WxEdvsDcnAx12G9UKf9UiJRa8VR+Kq9SjYSeUSbXA0Epeo047hA4i/Nhz3WgbzAo3pTNY5bJORXC17HqxukVhpUU1sx4gwj3+0DZDb5fnidfEBucuePrh3s+1RjVkrl6HX8M8YQoeZiQDFn+6Nmkcp2lbVRDXvXAUb8nlav4kHk7SgBkq5Z8Pb1fX2cntu/ejbFjxsLpccFT4BFDJGUFjcOmN2P5V8slJJ8wZqSoW1OPU/keB0eo1PVXjY+HCNeKBsOH6hl5HT6OQj3xBJ5//nmFeZ6TKENYgTRCfYMYN2G8CNWQFmPs+HGiwaJGSwRBnHTSSRJxfLJkCWDRC0UjjYGFHU78JJJRoZUZTvkoa6KG+mxz6HRoa23Fvxa/LiTbRx99jLSnWNBZsWKFGBm9NYEEfK1K9E3DV6XumIKoLHZDAI9rX305s/C8S/H008+grb1LCKDsZp1cEMNynhosa/PB5n+Pt1++KD/cTHp5cnoODoohuh0O+Adi6O3pQXFRvjBxmbuTmHXYDAS9fXj2iSfhmD4BhiITonY9bPm5iGiTgoZgGBwKhpHnykFfUzdeffol3PTPx2B2WmAmI4AWiGUgJ1na6xPtdX0gjLyOXsw5ZT62NGxCY0vjUCFEH03jwM46gdK5yYBmy8GAPyDTH4Sikal7eGNW9RLMeTxZwiN+5//JGIcKKikFQB8JBrBq41rc9N/3y8QJJaYT4OhNbAhtodfoRXuBeg0rli2DI6VFPBBCW0MjLFo9QgmSGemR6/ZIHpHrcUt1+P/WGJX+XUq8/X8yRnUt1KotRXT4UAoXinemdAMPujE1tdi3d69oC/LEr6ipgN5CfTeNeEeOf6WjKTz8xFPIqajCb8u/xLNPPYVAjLOoB8Hh6iEy3BNyvfkZQ/+WVHJcted5zz334M5bb0NZVaXS/6OGOg/GZAoWk8KJxD4x0TBE1jTv2w9NdlrkuhsWCaMgDaJ+bz0qqkg0rGivSHtEAOdGRVmNIAG90hqRv2exfLwOejX2uAeJTbZyIF1JVzraOvHwow/Jn59/7kW5b/ys0Vn1ZNJ/sJ9JI1RFlnjtNExZg4mLrsgw7jVbbdi+bSeqqkfCoFEuTh0pYd+JF0DLZ2NVwj/qKmZFTelNCwuKQJQ8T39OuDOJb6ivx4Lpx2Dvug14Y/EbmDj9FGzY9gucY1xwjq9Ad2IAZrtDwhqyU4cTUegSWiEhGm8sRXO4G4E8E6wmg2hBUOHK4bQh4O2DkbC8tAWuBFBR7MKadaukRHzYtOnY/Ntv2P3rZozPK0YmHEV7bw8i2ZBNNaBkkgI0CfkONPCuLqW5yxMxlUlKLql6DfX0Fi/xByVdNcRjvvKXR+7BqvpN6OjrlIIFPSz7YrGsgUWjcdgNZkw9ZAasZgu69jdh367d6OvogjHDWVFqXhAEoKgzswiyZu3PQ/1M9fPVa2DuNtxDDyFvshfMDSVGyOJbNoRSESrDIwL+mdfLNsTPP/4Ip8sl66D2x+S5OgWTTMpKp9mB5597TvIflvUrRlUiQSa+TFrSGK5xSY4H7364HEt31OGu0/+EKk7DpOKK/NvvFXiUS8kWp9Te7R8LOmquS93MZ599Fvfdd98QEZRa6Ywl4hIWslLKuVjms+x3sv/J71NSVYmm1iZ09/VImycvzy2GMdSczwLFuQ9sJiYBCqZVMKsJrSIpGInIviGqRoAEWaEc9brVcJvGOxwAwLXkezFn7ejohl6vwcknnyycPZFITAAEmuLLz83oTBYpHhBTWpzrQWl5oXwB/i9fNGt0zMeIMyQgV7yhw5HlsjEI0JkxNKk5GDeLOk8yhbPmHYP+un149amngIQd42fNxqAugXhpPlBgQdqQQjzN/9Mw2S3QxhWuk4ENzZh3xFTsscQR5Rw19Rm1aRGyIRBAnwIMER1Gu0ux8ot3MefQ0Vj23vsyzV3odMFmtEmllSRCxKMGAkFZ+IPGpFQweZOU3FDhsZS+j1MZHFXDh/8vYySlojaexkWLrsDK7esErub1+VBSViYA9/4Bn4wlFRWWYGDAi/NOOx1P3Hs/nFq9TKIwHyO9/nBjpEw5ET+//PwjHG73UH6nYjLFPv4PjFFt6wyPCsSgsyEpyZzWr19/EDeZNRKOyZWVlEhRiORhNEYeyG+//TY+/eYzJJGSdENG2bQGqVj/sGYLPt60DW/cdTNMGqChcZ8cdH+Qw/qdMapFJbUXp4beQwZD7Rce+G63aKYYLOYhb19ZXSVejtoVxHuyH7h06VLceOONAgRhd+D7n36QgyXB/qnF/DsvzGo70xTpKYsxHkxFErHMUPGF+1M1Rl4XjWw4MEP2TZZrmO+lFv8k3yY7gbSFFWgmjXv//mZUVVVCk3vxGZlj55DO/DdUlhajv6dLGqBqjqMCcFUsnfSEOLybLWww0ddz/i1J0U69hAT88GgwhOlTpyLQ2Y8xdg/efOZxmD25oHxjqMWLM//+COpCXQjnaOQ1rNWxJxfq8kFvs0LXHMLOd17F3BuvQcZlIx03QEr2tAYOjQmGWBo5kQxWvP86/vH3u/DiPx6FKydHOG2ohERv6yd5lEZRRTZYrEO9O25Eekb1oRRwTJLEM38wWZQWwHDEy/DN+8dWgxxYaS0iCT8mLDxelJAJMqdnpLfgCNbkSVPQ1tYhyI5Spwtff/CRfDyHai1mq+Sy9IjMz7iWDrtLSuhMF4jsUD2wevNlcDWkcLIMfY9scXUoxMtOQEifVAiPlE3A4V3mZL/LJ7NTEbx3zKPoVdQGtrRldIq3KfTkoau1E3abTQyRJ/r7ny1BY2vTUIEvlTRAl0rim6XL0NAzgGsWngR3aRFCA/0iFqvNghXkYFR1MbJGP/wAFDHUSGSomS/N+yTFY2N47V+LJSc7ZcECTJkwUZZg/NTJEhr29/YJ3plrQ/k1XiMZv5949gmh2mjv6oCR1V9CkbNtNbbBuEZq5GDSkiBZCZkJeWMoqrY3eF2quJOK7Pl/RUxZbmF1H6lrLZ5Tq1R3D4bpWUbx/EvOyMT6A6itqYbNZkF3T4d4BVYkidNjBchHyS0my/yCmbRA3aT/p6Qh8uAsGUNZVirFYLnZSUuhs2BqcTU+/Od/wZPnEQ0JSjTXHn44AmwmVxUhZTUjSSSPx4H2nU3I6A0YocvFi5ddhr89cDtieXkYcfQ8CY97DwwgPjiAIqcZA/V12LD8S1RWlsvI1x/DAlI3clhVbVHw5CbAQfIPjrxwQURWnOmO4ikpIT4YCcJAcZq4Eq5HMmll+l2UdNMIsVXCIVwQW6rQTSaiCVjKC3DKFedh0+YNchMldDGa0O8dlPyCnnFs7Vh898lnArRnGEkFKAkJs9JR/D1vME94hi+zjzgSu3dtl9wmyTGxYQUZCS+zFT8ptQ8zTLknZHjLjiVRLk3PkXVADlrmo398LyVaVD5Dz7ZVVl+T8g/+UFBhWYcGOVYHDuxvwooVy7F81SosXf6FyDiEohHZB3yWNpbAv//1NjQmM045eh4KaqrQ034ABvbosj1XZUdmfV/WKHmIDT8kVRrDoQOQN4sD7m63hKq+wQHMmj0bp591JuIJFvrMCHG/ptKwGo3Iyc1BZ08XbHY7XAVuofJQczl+V+5v3mOur46cs1kBHYeRY3UWqWjzUCROVRjoskwI9Iz8M9eKRppIqbJDQ5c/zNiU36kHp1bGbw5WjPk+vI+assvOzhTk5KGwpFBER8nyzOoTERSsTArOzmYTIxTGrqyCrJzUDM+yi6lq0FkNJgwyp9SZ4NAYYNXbEG3vQfOGnyQYYpLNAorP54dz5AgcfeUVqO9tE0VZb2QQ2rQFpH/KD2qBDdvxzBN/wYw5h+O4iy/EdZddCh2suObqKzBqdLmg7GtGVKK3pQUmvVYEUFTFYVX3gAvOxabcM6t50s+U6pjCDsBwhpvUrregq79PGOOY1/B/S0E+rDqNjIu1t3QgFU8ANiC/sAihUBQZGmhGA4vNKHwx+RNq4KwuRJqjRgKC1kBvtmDSxCnY07AXJSVlaNnThO69+yWyYG+SZMLSOsg+CMBXCwr8/TFz5mHnrm0Co4snFHb3IfRJ1vjURvLBoR7lzaisS2PkdueQNtWEaeiqutL/ZIwJFbxM1BHHmmxWxLLsaxatEd7uHuzYtg3vLFmCtz98B1HyJKWScOXmoq+7T0R+ln+xDI0HWnHlJRcir6xU9pC3o1PBCquP/8EYuYb0cGpPTr5T1hh52K1atQorVi7DuPHjcc8D96O3p1/SDA5R26021O3ahQsuvgDvLXlP2OY5+K7WQnjwqmmW5ICicqawttHzFboKsXHDRnmO6vXU9ZIQM5mRCGKIYUD7h0bk7zzf741RhTiqUYCaH2tm3X93hsWKLdu3wEw25HQMI0eNEs/IsE16I9nYljdUzbs4tMvFJXmtKBZnp6y1saQM6ZrSesS9flhgRQ/xpiMLkV9SBI3MDqbhyCtEJBrD2hdew5k3Xo9Dx41D9bjRuHjRnZh82GGwhzTo2duKk+aOx47Na/HVN18qZWz2K7UZWFwOVIwcJSGlr7MTRjJo0xNoNFJ84QLTILlw3Lzc+ESAsABFlE4myTk/ZUTowL692NTZIZCuKYcegudefxUdKYUShDQdZ55yKqwpHXbu2oFYjhH9HW1Y/eRi5BZ4RGyGn0k41Kg5M9DQ1wp7NrHndaxftwGnLzwTvb19UqmuLa3Cd58ulQNO8olYashz06gI51M9kngxl0cAGGSrDkcSciCq+vbqJuHzJaTMqkWrN5eGLXkPvz81LaHkxEL+m6WPGB6Kq6e1FIr+gOfiOnHqgqKgVp0J4b5+LH75FakM//2R+6SAw/suzOV6EwZ7+jFn5mw89l9P4vDpU2DN98geYSQx2NsrOGTZ3MOLOQdNdChfZ9Fo+LSGRCKA7Ete+333/x3XL1qE4opyYRLk/qwqKxMqfravps2cgZb2VsRZEWVcmg3VVVpF/p0eTwDllD8wmaRWok1qYDVbxQOzUKW2WviZXP+7br1DpvaJ3iHLeFVVhbyWtsDrHTKwLC3j8BqE2l9UYZoCHKGScv65Z2SIGsnJd8PstCMuctFK7KzGyJnBgDIVkFWpilLOm1Eom+ZJBWHCEJU3OtdIfYwAjAkNor1exLpDqCirhj/WJ2zZRpcdYSIPCoqQ1GlRW1GFWqcbR1RV4a833IR5R52Mpe8tQYG7XAohP67+ENs2rcP6Tb/hzXffQTgagkbLUjvdfhIjx41CStSqIjLWJQcDPWEoNIQ2EU+STsvIE0NsoQFMGeV7kv9n2ervhAmvtLAIVo0RP21Zh+vefAH2mgr4+3thTqTw1aOvoetAA7Z1NuLJz97FOBIVv7sUrlAMEYqBBsIomFwLeCyoLCke2jDxSAxupxvkVm1pPoBT5x2P1599XqZhovG4GCMfEv5otTJyxk2q9qCMGj1yPTniGQnupjGyx6YCENTNxZ+cpFEPH/6dpigS18zHUmnJ4/kYPinxv2OMrE4KiECvxYQZ07FvRx00sbjI6t1+66341/tviTGqIrgSLcWSuOCs8zFr3lxcdNaZMOS5ZJ+wN0omPX4HYX7X/n6sSrVH1WtwdpbtJjV8VY1Rpb0gg1txSQnae7pBQAWnR9b88AOcdofA3bZv3gxjrkOqpypFpBBPD4PUSVuDwk8Gk/R2+XeL1iwjhHwMz2P5Z66Fx+mWNsUFF1wgkQyruJxtXL16tbQtRBpCxakKmkvRMFWAFIrGC22G1VW/PyC9a03NNVdnCAsrKMxDee0IbN+7GwaHCaZ4EgGvT6g3OOFAikV/PA5vLA5/OgqYtNBTeZizb1oD/BwJSmRgyJhgSccQ7w9D2z2AsZMOxZqNv0Bv1cPssiJjMsLgciKs0wgUjoWb4rwC+Dp7cdbs2QjU1eGbD9air96Psy9agJ9/+gTQezEYJvrGjMGBPtHDKKoohUGjwb7tO6QIwFykvKRUZiy5+ejVUsEwtKmMeKQEpxGY18Zi6G9uwZ/+ejumHD4T7V2dqC0pw/wjZ4n3LPYUYPErL+PbrmZ0ujmZEoNWb4Bv0zZcefKpmDJrJh7+aDH21TdgREkp9r/3GTwhvcDYKiePRu64SgFhcxaPELuaympBaZBq0u8NoDI3X6BPKq+LVmdAhCRa2Zw7E1P4fcQgqXGSAsaNrkE0EoJep1F0K4kioQBHFkWjbuBEtjAyFPJmq61qkWGwt0/aE6xCqhoSasg7HIQuHpcdlmwex9eLGCtDO7cLqRy7HFB/PvNcodRoGewRXlzJ2YmBJTgsBSy65s/o6u3HJ6u+FOMbgr5RZyO7qQf3d0FHsSSmC6LfehDiJ++XggJZzJI3secnBw3vMxWxSetYVCTvn47EcRSB+l99JZ816bDp2LqNKCY9dGSxz+bMapvBmKQkvU6k6pmu+EnJ6bQNtTOGOWok1NhyaLEzcl38/4UXXhChJOa3ahtJHe2S8Jfpil4vGjBq2sS34TXTIGnMQtU4/sabMqFUHGaHFc68HHgDPjiMVnj0RnQ2NWPfb5tx2YJTBSr37kefojccgamiHAaXA1qbWabvWTw45cRT8OuaX+DtG4QzEkX92q048sgjsGHrbzKsmiYazmZEwqiHKceJJKkbOA5jMMGsN0JP9aNkArNKxmHpK1/DZbHhsMmFWPPz1/D6uuCwukUujCNXRxx3tADSOeVRU10tjV0OEFNvkWS4nFmbe8QRWL9xE2pGj4Xb7sT3K79HgPSSJiNe+vk7rPptA0IDXpw8e66oFx8+dSo+/vBDhAb9uPX223HNs/9AlxW4+PzzsOyrb3D45Ml4799v4Za/3oXPVi6Vxe+jDEAijfAP22EOJlA9cwoM+Q6RSRs3eozQzxd48iVs+3H1Dzj/rPPw64rvBHnDDcHcnByt9HaMNBjy6dM6uXHCNqbTIhyJo6aqDDarGXazCX3eAdmAbJ1Q1nu4J1RUILO4VIaL2aKQWtjKsdvR3daOdFJp/g9/7X8yRhWFIuGkhPZJgbXdfPdd+HbpFzjz5FNw75NPQGunWOlBqhAaI8PUG/98A/Y2NmNPd7OEfn80Rr4vlbxYIOpo2AMtjWtY6iXRmd50EGTP4k5WGEc1fFL180CRvC+jlXEoTkIQGE5pB05gkOCaxsjsmc/j+tKY/W19ePr5Z/HwQw8JbPOW227DiIpq5Hhcv6tSc015f4Y/TFrjEDkVDeq5Z55Bf28v7Bar5OjDldtIOMVrVAcChoMd1LRPPGb1xZdn0jYTLJ4cGEwGmHWAKRzH3o1bEB30Y7CpBbZ0Qlx4f98ASTBRNmEK9DkumHJdgC0H0VgMsXBIqmQupxm9dY1w2PLRHuyAI9ci3KmccteYDUiTic1iFqKoJBmmbU5pK1hkvk6H6LY2HFE7AwtOmoNETx16ertlcvzCs85DflkROFRnLyajtQEWvQFBvx+BwUHpkbLAoOZI0m7QGhDy+hGLxDH/3AvQXZEvOgv6XBdyCFzu6sG9V16HnVu3oqymSkrmY6tq8WPTbjz44Tvo0yWQa7WiZ2cDxsyYgsoxI/H550thcyrIe+oM2jU6GHpC2P/Ol5gw/2hYyzxwWA2iuWczmeHv98oE//TJU7Fu7ToU23LEMzJMkuIANGjr6BAD6+7tgc3iHMrLmU5FY0lMGFsLT64LoUGfHHwsiogUAMv8cQXMLD8JlB72MGQ9oxqWkiaFrQIeWtQzpDGoeecfw9XwYFDhCFKrnNn3JSXGk08+iS8+/kT6oO+sXIb+sA+DPV2w2+ygoi8rwoNtvbj62mtgMFvx7YYfpTeoTq6wdKaGfiTCZFRAjUbOxbZsr1dIqLPgCgJJ2HYgqstgVNAxagrFDc9erjpSFez3CZiC+R1xrC09HUKpkkxR8ZpyDsr3VdsXgR4vDjnkUNx4ww1SjHr62Wcxf/58oQagECMAACAASURBVA/h4UEDUSqtSam6qyGr5INkW8tWRBlyXnbJJSKOQ3Y74bTJTpXwOcZshVhNPdQ1/WM7RHPIlYsy5NfW5ZCASYOM34ctv3wvXeDi0lKY0xkc2L4ZBuYr8bSQHEVCfsYsgDMHCMRQPGUyIsFB+IiMicYwecpMNAcT0OZqEDOkEEkkRQmJh541P1dOmYxRQflbTU6w+chBW3NSj9nllVgw73g8+MBt+OnjpQglk7juyqvw0F//In3LI0+dj4FkFAYqHyUSAtEi/CnqD0gTnO0CNm/Zj9SzbaDR4rVvvsTbv/yCPV0dyMnzIEdvRE4ohOvOOV8m/Bnj144ZhffffRejR9Tgu5a92K9LYVPDDtRWlOOiuSfgjscehM5mwoM33ISSmmJcfe9fJazKMBQeDOO0Q4/Gxs+Xo10XhVWbwPHHHSeN4YqSMnR39YrS8YjKEWjeWYe9e/dKeMPHgQPtop6VyJbJg2z9ZAHsQsfAKY9UDA67FeNG1sBqsUio3dPfh2hKyS0Z7nBDBiMKYdcfw1R14/MnX88TXErzWewqn/+/Y4zcjszZFp5xBvLdbmmRLNu0ARF9CsUFeVj28SdiDLwn3tZuPPTIwzKG9sqSN6R/yw0uG3i4MRKQ78lFxmyU4lWuLyHPG2plkF8gnZZ+IQ8x0qOoFUySMfN/ISdjkSocE2r+hx9+GMuWLcPetuYhfiN6Rg4iqAcX3zPH5kJujkv600889hhaDhyQajzpQgR/mkXucN1Uz6iG19phrlJAKn19aKirw+pvVyk93WEtGs0w0ubha/17X8t1qSrKWPIqEYn4kV9eiILiPAwG+wQNr0sbMKNiDJa98z4i6TjSVDHOYjoZcnCws6ioFFqD4oZJ0nP+BdfilU+WwFpeDJi10Jp1UsWKpxOw5NiR0uqQohHabBKeeQpKEAtEYdXo4Ymn8eHTz+GKs87EK68vxvX33C0T/jfcfhsmlFQiMtCPY047F7p8Mwy6qPQdycqlSWmRikaEbS7FQV3q+RgtSEXjmLbwPPQWkKojilgyidKSEjRt2oq7zr1IjHIvxU3PP18MpKaoBLc9+hBa7AbRQ6SwZ7XLg6kVpVi9e7vkLc8suhUHWhvx+EvPY8F1V2DLzm0I9A+gbsNmFHcFUVRagqg1hdLCXNSUVQlVY4Gb5f4BnH3G2fjxh59x2NRp+OazpSJ73u9X0P4q9XsgQMXilKyNzqBDKOgXYyOwPNdJOnq/XAd7wHZPrngBDleTbyccUFSV1NCHSk4q+oOfIXSWWeSHJ8cl7R7fwIDADDNZEITaO5Piyu8eCps8PTPBBnffc494jgcffwz2XGoODsp4HVnDNZyHjSdw6x13SP/3vmceE5a3YEypxnPv0DhlflOnQX5JsdIyI4KHbG/xDPrqmpHO6IQoTDZwMonR48ahYfdu6Mxmye85vKDKe1PmbcPPP+PcCy5AeXGJGAb5cdUhbXplHh7t9HhZcVLCLrm2s448UvhUuR4EevO58085Cal0Qugar7z+zww8kVdaKi0QPlicofET6zzY55UiHSO0Rx58SAGdy0C3UixTawDDwSIqzI7PUT2k5k93XpEZpPZfNIxkJo5IPCqg2LQ2BZPRigdvvhuXHnOynEj0oMwNhqYGiAKJxFFWUoS0lvmKFVpPHmLaNOL05PyPISornTl22Ki7mEhKuZjvwfk2j80NbziGaQVl8H23BotuWgRvOIALzjsXL7z1Jv582TV49d//EpzmTytW4Yul34nxpXUJWNzKYHEkFBDoGzde2O+Xk5l0+Q27DuCMfzyCHfFB5Dms6PJ5MW3cePibWnHl/FMwsrwCwfYu4VilJsOSl1/FP1cvx9budnQNMoQ5BJu/XQ3KbO7sakfV6FEoNlnxwGVX4tlX/4mGQD+27alDxGyU1ob/yx+EN3XGgrnobm/GiNIKeLsHUFVWjvLiCmzdtEUgbxefex4+/uAD7G9ulvlHdYMydCWTN0U9GUUEI2HEYhHJH0tLS+Gg/mQ8PoT89wYJd4eoNpELKBo5OBbG9WX1WIDO2ZNZlUZXsZz8PcNVjsgR2M2Hin8lSOB/aYyRCG6/404R9NnRUI+YJoGoJiXoGHq3ZDgOSyItFCRcw0V/uVnER3nIEG7GtgRnCPmwOGw4fM5sAZ0XlpZISOm0ONC0bhuQPmiMfC4LNXwf5tx8HwEoZAshPKzYGxWeGk5fxGIyLaEWf7jG/p4B4QFScbCcV+TQQ1dvD2pqa2WdW+r34cQzFqC1txPVI6rw4+rvReuFRTkak4BdWF2GFgaNDnaDBWWFxajftkvmYt9/5130MaQWjUYFKqeu6f/KGNXwVTP9qlMyRqcVcWrZkaI/EoSeNBIspetMSAcTSDV0oL+9jQ0kSaCHl9WZl8WDfphdBTh64blYueFH2B02UdANa1Kw5eXC4XJiMByQZntKo1fknQleZj9OZ4ZRb4axvgU7vvoatz54L/R2CwL9vbjnznvQN9AvmhU/79wGXyYFTX8cHz36LM6+6Xas2/ADCoucyGjCCHkHZFOTPZwyZCa9Ds39JphmT4BxdDFiwZAwpQ/uqMc7jz4JayqDPlYkozFMGTUWn775Fi499zyc+cjfcdgJx+HDzz6V0vW46hHY37IXE2dMl82zZc0vOL5mPKZNngiN24r6lkZEyby+fSfeW3SXHAiT58/BhHE1GOjoxKFTDkVrU6tUdQ875DDFy+UX4MP33pc8KZqKyWQMDUTYyyMx8XQs5rCaGgpH5fBiyb2ytETyEd5Uekf+FF5Xki8zLNIqOZ4atvGwG16kkbA6mwOqaEACA8hgHhuMDAnQ8ntydImeghuX782qpoSprFJOmoTpU6eJGCnVpVKGDI5feAref+NNYQE4bNZc7P35V/z5uuvw8P0PwVGeK2EbFa6mTJ0qaYHaQI+TRCsagZ2e3ulAmnJxyTS8+ztgiBMKexAQwY1dXlmJproGFFdXKtFYVNHG5Brxs8XjJJIoyMvDvrp6obkgtK+5qQnfr/wWx87/01DFVmfWIhVKoHrGBKmWhlNK7i1tJr5PMiH6nUwX1MdQoz6Zgd1kQV97N4zQwWGxy6QID1kebqqupPo69T6olW3198NtSTP5ipMzlbU18A764fP7EWVcrY3DpNEgmEijKK8EG59+Q/pwGRZgssY4VBFiHqLTw5xThAGdBQa3HiajHkHE4RxRBrPFju6eThgsJrjy8xAIKHoZHFCVC9QZMcFRgKKeAErGjMCMIw7FN0s/xZ233YrNP67HkUfNkRP43Z9Xwzm2Gj+//RGWP/02DjtpIc5eeAHWrFsGt8eARNQnFcL+7m7R57CZTZh/8b14ePmbyB9XBm+fF0cdeSS2vf0hlj39Iuq6OtDQ04kjZs/C7t824ZyjT8DCC86FYeo4mfLIzc8TbY8+nw/ukaVwG0yoyc3DxKoaHKjfhxvPuRBdgz040NspJ/STj/wDm97+GDp3Do5YeAwmTxyLHArLtnZh1MiRGFExAoHBICorSrHqm2UIZ9c7qU1IqElDpGeMxlIyNdHrHUCcctzxbHEmmUR5cZGc+AyVaNTchPI/VTNI28EJfCork3QpkxGQg6opyN/byGer8gKpYqQMpTRamDN6ORTUqMeYUsRM+fwop9O1BP8pYSp5X047dYFgUzkW5RvoQuHkcXCazGIcJpcHtSY7zly4EFcvuh45hS45NHg4kGmcdBP0QtKvK8yX3zPc5AiW2WRAMJZAVU4h6n7dPGSMKkyPjH/OvLwhFSiObLGvTapMASqkM9JW4tgSjZJsBDzI+FnU7yD7A8H5jCriyTB6fF70DPoUYZ5sy0RCdWJLk3GpSLOFQoTV8JDSkNagr6sHTqMVOk4UGcxobW7Bi889J7yvbLGpEYna3Ofrh/csVa+pGqbmLx+8mMkrL8aHH34oGvfEIJLEluieyEBQUPr6rgF4u7rlptOILG6nbFT20VJRlrzTmDD7VOzuqUPakkEslYC9okxuDKtYNBLmF8FQSDYYmdo412fSMuLWIb5uKw6s34IrH78fZTUjcNTMw9Ha2ISBni75eer556IlEsDdjz2AmtJR8IRNaN22Ee3N3Th74aVY++tPcOaRPKoduS4bYkHC2VLIrTkcm6L9KB5fIuTAs8dORnxzPRY//Dh+2L4F/3j7dcw+5SQ4S4rwxZefI2E2Yl9/j2wMVgXPPHUBjBkNvlv2DXw9vTh86jTo4klcc9nlWPb11xg5eaJs5CqTDcdVjJQcjqzhE06dB6vTgiOKa/DgPQ9hzYY1stFpGNWlVfjqy88Qj4cRDg0OGSFPZG7+cBxSjJEbqdHB5w9I7trT14cchx21NTWIRimdnUEoTEwr2z3KhH00nkCIgp6EB5Bx3KZAufi5XH8aPcNWGhxDRlb8VKSICProDTiwd5+0hgSwOww/GQ5FgHgKdpMdl11yMXIL8vHs88/LZ6dselRMGC3FPao77di8AydOmo6akSPxxXff4rdt62G1O4Sqg722HgK5OaBEuJvHhfyiQrlOMVCiUTSQyZudKwmh5ACxDoYsxSJ7ytQ0GQ7AZgFv+uQp6BpQDpMTjzsenR0dCITi0KVSuOTiC4Vn1p7vwXlXXoJJM2dg9946KTb9pwffw0iAAkP8ZHZwPKnMrFKpmZV7skLQsFSwhjZlFHXkfzz6qIisup05Q2tLYRuur9piYkKgGqUaIchan3rf9Rlbbo784/c//yRfmizUujREL6O7sxPB+kZYqFmYSkoRwFVWBKJwZLiTuWLEgB4iSfJ00OfZYc/zCMmvAGgTMYU3hQgOJrVaYkhN0GS0wtrt8Iax6sXF+GrlCgRzrfjhxx9xy/WL8Nsvv0oeO2rsGCG22thQh6mHTcO3y76DZUCDeHcHRlUWoHYEeV7bUL+7FZdfdRbeevd5uHLM6PMNIGYsQMEhE1FcWQxbWoMTDj0cU6tqsKdxPx58+1/ImzgWIaNOgOqBsKKQREZ1LjCLFAxV+H2PO+IIIbKaNGYsejo6UNfQAHO+B1OLy3FW9ViccdiRcOXlSVM8GvBhwhl/QnFVCUYYnDjtpJPR2tIilcAFCxagp60bmzauEz37RFwpktD46BW5Ub2DIfFGqlfyBSIyYsaqIT0eESmBgBcmkWZXciZ/MCBhLfuObJ5znQWaxtEsrVZaA9xgbC9QLptTDEKBMcwY+e9kAPR2dkkozPCVj4ONfwO0iTRiwRhuumERisvLcPPNN2PsxInY39mC0dMnoS8YQHFhIdwmB2bVjMZnK5YjSDY8h1EoWOjV+L4uu1O+s83tgs11cNMKlK24BI2trcq/U649pRNoprppeT2qQKk6j8qwUFjs43GRoL/2xkXSYnjy0SexcsUynH/pBZLTGjxu6BxmKTj6vP3iDIY/+BnMRUWQKetlZZJDlkIr1XPOUwo+NqG0ldQmvgEkD0uJs+KUzaefLlXEc7MYYYItVGNkOihAiqys+FDOuOD+RRleHHGDu+rq0D/oQ5zo+2QamjgEshTt6IWW0B4De4FA2eiRIqbKN2WTNjaoR/H08eiM98PutiNEDCinPKhorFGSWG4uflmqXOk0OmTiKdDVuxvasfybb3Dd/fdAm2sXCbRMNIZxo0ajt6cDMS3QNuiV3KR7wIvevh5ou1Po37MfGBxAzNcDW34x8uxjMWHcFPy26UfYXHFYcgxIalzI5DhQkJuP2+64Ay+9/xb8iMNLuU67FaF0Et7ePlTXjlS4VIl5dLvR0dMj1T5VWcuiU5SMTAaj5KSc/CizObHu1bdQ3NqLkEGpNPIayWY294LT8cP6n/DqvY9j65ZNKChUiJD4nhzMNZt02Lx5A1JJBVCuhj+S72RL+VwrGqjRYheDo3fct79JUB7V1eXiGQnGl+fJzKkevsGAkHvRW8nGzHo3hrSjRo0aKutzE1Hwh6GjGkrx/rjtDoT6B6QQQWQKr02trtJR8375uwdww0034u333pX7H/b6UFBbAVtRHtImgxRZ4r1+LDhyDv75/jtwVlcgHPKJt2WvkKpcic5euAoKUDOqFkxaKLzLzyeZcHtzi3DWbFi/Hg276xAZGBwaAFYHnrleXEtWR7lmjfv2ydicRF++AB757yfwt9vvkuLQiu9W4m+PPoj6jlYkyLXLES5kkIxQXF3RzlDB82qurbYvuNY8jBl2Bv0h9DQ1I8edixJPnqytmgfydcloClpO6ZDYm2TLMWV8i4XBbVu2iNHK4IAoXyts8DxMuMf4XSR3PPSaMzJ93j5pQLNVwI0mGhaMYlKsVg4i0dIhQjBsrfCUcJYVwVlciDhvRigGl70C5YdPRHu0Fz6KnCbicoqzDE8qigRByTodXG63FG+sJgtSkQScKQ1+ePwFnL3oWhRMG4/CskJBpZCLxCH9Kh0O9HTBl07KhtTaLGhoqIctbkfjLxtQmWuV3IkMcikY0bG7H5deegt+/eVbWFwJbF2/AedcfxPa9rej8riZ2OHvQcyQQUKXQoYwORY+kgk5ZOgJqXbEDSpT8dR4JP1gMCSRgrQIoIWNXD0+HyYY7Pjy2X8iJxxHiorOosgLRHv78Pg7i/HqW4vx66dfSvj4wYdLpGhB2gXKyA36+tHatE/ETUlmxBCWjGYMJ1XDFAEb8rzoDKK4y2JO3Z59sFntKC7OF55WbUbpuYkkQTAIExvefr9UVokOovCQVK2jUfEm3DTqT0Y5UvnMluD5fqNGjEBlUSm++eoriYTUKXbxFEnC1UhGZcZ1112Le+6+W4Ab/GyOJg10d+KwBafKHKl2IIAFxx6Pr9evxfbWZtjsik4i7+m+3XXQRRPCwEBgBIVl551xmngTrvGppy+EWW+Q9gBlCe+/5Q7klZdICEvyL4axfLC1wYEGrvv+hgYYDUakYkncc++9eP3fb2DClCnY27xfahzdwUHoXHaBTWbScTGwRDhI+KxgSplTqty9Ui0lVpUYWq1BWCv27qkX55GMxsQw6f10BoWUmoYkvELRuEjvKaNxRFNFlEirz4vb77gDh86YIR6edsaDjgetGGZWvIn2ohl5zjEZ3hi58ex5JJhMK3JXMmPF/CkURUlBYVaLQiMqTj0cd4Ievq1tWLl1M6597F4YPQ5EYkQ8JBAIDMJmV9DwLGXzFOGm0+sMSMSSsMbT2Ld0BX74diU++nEVAro0InFFO4NfjtdTWlKMnfV1gtjplfk7hmBAV2Mb+nfuQWEOZ/6VMSiCxg06C5p39uCK829C/Z6dKKywoKnFi5nHzkQ814q2RAjdiSAGYkruarSaMbKmGp0H2hDzxxDTpZAgiIW0F8zDIorQqsFkFsIsivqkur0ojkSw+sXFyDOahTndajait78f7pIioXYoHFGF1uYmLHnmGexuqEc7ETaZjFT1fDE/RlSUYd+undCR7rC3T05msihIczl7ajLP5u/7vb4h4mW7Mxfbd+6WcJZhnMWqeMahoovJClJPCAA9mURXVsZaBZZLjzAel1CVdJQDWc1DCVFtVoG8FeXlY/2vvwoelw/ho5Hpfo2wld92061wOd24++67DwIFdFoUlZeh9UArZsyahXFFpWgb7INXk4TBakJjS5Nco9tiRUtdg4IfjiaQiqegz8oZ0Bi4QeccPQ+rVq7E+ZddKgfKyKJy6fUJez35XodpbXDNSLXSWFcPk9OBVCyBu/7yFzzyyEOARYeiibVKj49RGnGumaTcV4fFKkBtFnq45sIJJCOBxOBqJPqoKSzFgZYWGWDmocQWED+bzkJ6tZQtH0bNwhqLWjjj+/F5fL4AAnq8QuT98ccfy0gimeVoF6T2ZGjL50l7a8Q5x2SYk/AhF8gbmW0O888sGWfiCsKBeocRcs9YrMKKZdWZ8cYzr+O6u25D7pgq+BNhDAYGxJNQd5HGqCJE1Ng8EovDkNbCMxDGyw8+jjc+/wj9bKPogcKSIslvuHE51OkuyFMAwoTOsVqYYqk/jhJXAX755EsU5tkVTlSdXmQAwlE/QoEopo87Cj0dGUydOA2dvc0IGdtx4EAnevt8uP/p57B83Tqs3f0bph45EyaLHs0NezBt/CTUTpuCT1etkl4WPSVZydubmmFOEcyej8lFhXj2wktRUFomX4dVts7WVhx74glSYKHkNauLZWNGYMDvw7MP3o8vP/8ch844RPK0gqJC7G3ch8JcF9b/+CPycpxKmJ/NEXnTufF5H3izaHBuT570ziTh1xpFGZk3lPclnlB6gWqoS+gcG+cspDDKIVyOz+V7yQQOsa7ZYg4Pj1NPO03yGqnqxaJw2GzoamuX0rxIXxMokIWEWY1m7N+9BzfeeDNe/ucrQ3IFkhOxn2YwwGSzYsDrxbVXXImmvk50xoIIxsIC2ud36mtrx2Bnt7AE9NY1CwgkmWVdE2/E68ziaXkgtzW1oqC0ZAjXKYc5h6nDYal+isNg0SscRjoSxUWXX4Z3l7wrsMvxUyaiPTCgcN6mUxJSk5CY18rpFdn82RRh6DCDHhVlZVj33WqYLTZEA0EUFBUpXLlZ/iMeALwf6iC3Gj2w56jeC9oNr19QQaJizPweaGs9IO29Lz5fqtCaxDi1c5AKVDP6rKMpDji08ELIlFR6gOrQJVnZGAtr9FSrVejyNQYLjGkTmjZsxdFXXoruWAChWJBMKKC4JlVhaZRSdMjS0g36fDBo9bBH07jihFPwyqv/wqRjZsMfDSHX5RSPqjZxJe7mjJyoGyviOOQYJSqj3FOMlnWbYTQw3EzJacdTm3yntaPGQaMxIdSfxhHjT0RPfyuCuiZE/f0IRKIY6B/EwI4dmH/5NTjzkvPR3tOF3Q27sb+jDYPROMqqR8KiAUZVVCLm88uo0GCPF8s//Rh9++rhKciFye6UE5NgZCm0xCNoamlBb9Av11xQWYKoNo2zF5wM0tMTEcR8iDeIopmNDXUocrng7e6irI2sM0NJhjyCHsoeijI0HE8MJf4Gkw3+YFg8LAdb1R7ckLfQ6KXxzZyRNYAkI4mE0jphv5DVea4ljVyRv1Oo/nkAuh12odXYs7tOvBCyxsj3llnGNHDqSaeiv7sPP6z+Ud5DbZOwJcH34Z6prqzCaaedhs9Xr4Au34W84gK0dZIPNglfVzfiPr/Azzoa9gltZTatVULDbCTG3cgG/Z1/uQMvvfyaooKlUehH1HxO6bGmhKT6hBNOkOeUlhXjgYcfRFlNJXyhADJWirxqRG+S0E4qfDE6oPcTlFI2RJfw3WKFy+rAtg2/SSiZiCu8sfRwMu+YdVB8Lg2SFVxei2p4JspUZR8qwJ7/LgcGc1PR5tUIP8/OnTuEZJlgE0fWaOVQm37eCRn2Y/hCnjCCaE8rQ5Zq0kkDkXBFRpP01OsUmJI2bsJFl9+EZz55B6kcBYQME9NDLXp9fTKszHapiWKgeiobGZHy+bH/82X4aNlyfLj6W+zrakPNqBrpu5EjlFqAPKm58L/+tlFmwFgU4O8GB3wC/h5TXI5VH38Oj9uGSJSCnzYUuAuxdfdeTJw5F6VVY2R0pnFFA845/TTsavwNBkdQQOeaRAwGjocNBKQHFWdjO5VGWU0Nxk+chBVff02sk4LSYCjiyYPV5ZBcIJWMo6O9HdfeeD2+//57ePLzUV9fjwFvP2wuO3Ly8zDAXm0iDn1FEc5ZuBCWZETCbRoEm+g8PEaOqEbDjp0iJ85cQ21rSKXQnE3meRNNRvgCQalAcoMkExnxeo0tzbK5uN5qkUW8W7YoQMMgqBpa/dChykNxf0vrELsZCX55TSpRsS6dwkBvn6BWZOpANiv1O3Si3KzLaLHg1FPlXrzy8mu/G7hVy/S8FnKWWgwG3PnYgyLRwIFwd44TveQQjUTF8/KzY74AdNHk76Y0xAvrFEXnRCyB1197HeecdTZINMU1ovT3HbfdLpQgNBCG4hMmTRRg9+x589DvH0Bj635k9FqRT2evm3m/FFgylJmzD6Fo2HazJBQyMr7Xjs1bRBKe1WoVmigpmsrRk6WtFJsgZajTJUU5Hpx8/xyzQw5oInokT08q/XQxyLROGAtUmGJ7awe++OJz4f7Npe5kTJG60/zpqrOknCdhoBrrphWs5JA+hE6HSDwmfUOOuxBpT6o5X3cELyx+H898+m/E7RppKg/EgwgE/ZgweQL6vP2yYYhQ4IluSGZgHQzh0+dewmlXX4aRh05HfeMeCUXsJjZNm+DJUxSpePq0HGgb0lFgPpGfk4sgE/+0Ftt+WgOPWznNE+EUPLkjEExn0DjYBVdRrkx2B3/tgyNpxIjqcsTMBwQ0TK2LBHUTm9rllGRCzippPOuBJLTgBH5SUcTlyegd6MPEqVPFu3EDE7dLVmtBELEVlIlLKOQuLJBc19fbgz/95RZs+f4HeGx6VFVXwtvTK6gM3mzJxVIZ6Dl9r9HJa9Rpb+Ym6ogPbzYlvfk53DRswNEYSYGxu74euSRRGjbdrwrIqJU6IlnUyX4epBqDUQoh3KC7GuoVYAD7khzvYTuDLR3O1bGXxpyG3pHNf1JwdvXgmmuuwQ8//CDzmSx6qB6Z68iH3zeI+x64X1A6r77/b9irShGNR2C3W+HvH0BX6wEFl+p2iZ4Jh8/VkSm1RaA3aMQY49E4Hn7gYdEcoUYGncHll18ur6dQDQ9r/pn4VDJOTJ4+HYFoEDsadiKWjMPmsMHgsAwxuskkv8EkFCvkxiFAvGnfHvR3dkLHaqYMHZjk0Of6qOuiVpuFpDrLHqEUXlISUahzlqGOQexr3i9sdA8+/CBMVkXSgfdOm9KKMUorUJjzUwKcuPCcc6XC7CT7BXPHmQvmZqgPr34of8ZjCp8ovyw3CtWm6Cl4urACRy0I5hfbf2vA4/9+D2+t/BRJs4KIzykuVoRe4hG0NDUqYZqBoVwSZUY7mn9aj8NnHQ77iBJoHVZs3rULzpwcqaCWlRajo6N9yFPU1dXj+OOPHxrCTEcTws1aaLRi289rZLxFYwAAIABJREFUhRzK7+MB4Ubx6FHojYXkNFRGbXSYVzgZ9mQKUyeMxu13/xmjJ4xDV1srcqwW+LoGxBglb0iT90Sh/KMGIwVUCfimN+GidXd3iVGycMIknCRHfKi5BsvOrDQXlJXKQRIzGzH53NOQauvAjAm1Ii1GvCSLDQwBqXlvN5rQtH0n7GalcMH1llGnhMLNwveRyp5eUUDiv7sdudIH7vd5sa+xEdWVlbJWPBC4WekZ+eAhKrQSMaUwwU3BTcCqLJ/L79I90C+VSQFrM+SLRcWIuCnJiianeobgh5Rwy9x37/1iyIsXL5bwkpPs9Ap8qB6ACBf+jozf5hwb/Ehi1OTxMGiA/Xv2SkjGz0vqdSh2edCys15SInmPrEGTjpOAEOYro0eOxtlnn42//e1v8v3eeOMNrF27VloaFGPlei394gssWfK+RDee6jJEUlGUV1Wgb6BP9qpqAJL/Wi1y6O/dvkPqDHGdgjKS9ICIGbLkZ4sufD5/r6ZIpuw4Ga//QGMTTFabvJavk/XWKKNboglp1COeih9kk9OapHLM+0oj52HDn28t/pfA9MiaL2Hq5PmHZ9QEVU30SbarPqR6FI1JyMQJc6I73Ha3bJoZs0/Bqn27YCiywmTXiiqw0WCDzzsg41QWg17oD/QGI6LeAHq/XYtvvv0Br3z2LibPPgQ7t2/DvoEB1O/Zg/FjxsDjdskwqIoSYVhKbKFKVRgLR4V/Zcfny+HdvQ/Qm3D57X/BB999A1uFG6F0HIfMPEKqarpEBtYDg7jt+qsxdsw4rPl+NZ755wsyaNrZ2gKLxiSG0dneDl9PHy6+/HIZvaG8GeFwMw49VA4BLvZbi19TVLeym5rl8v9kjPSMA339aGlsw7VP3Y+v/v0u3OW5KCkvlR4ePVkooIw7edjT6+6VcE2drGC4lI7HperMGyd6gCaLwrXJVounSPJBstwdaG+XkFKdVRSEh1r2z1boNFq90vOigE46jWBAkV0XXpd4TIACosTbzwgmjN7ubhQVFAjqhA0EGqPVYMao2jFSSHrlxZdRVlUOi1mR3FYnGBhdePsHcO/990mr4Lrrr4fZpIejwIPBdAx69mmzAINAMAhPSTE8Vgfa6vZCkD3/wRgpL8AhBDKIc11uueUW4ZohbSIVqTjmdN4556KorBSPPPYo7rn/fugtBhjtJvgC1IRJQy/DC1AYwKWRTyKybOtCrxc2RBqfGKKA6Q9Szai/V3vNVF/j8/g9uJ4GneKs6ID4nG0/bYLBYlRIrjQZQWLRmPnv9Iw8sPhvTI2IOpP9E43j6af+W1pzYox5tSWZMGkfsqMc4lozyukqF5jJ4KP3PpXQgC5506YN0Fjpcu2I90dQfckCaHOYKDLyiCKeUNAjo6pHIOD3w8/TOZqEKRDFU3ffh1XffIUj587BP954CTkVJWjt7kJ7Ty9mTp4m0/OkQmKIyg3a09ktN5fFBl6LSWtDTjyFrx/9b5hy3Rg9fgo6rCbkjchFV18XpkyehphGgxGllfBojMjr9uHciy7EIFIi9Lpy7c+484abENzdgHnzjsFzLz6PGnrU7nZEE0llkQ0GtLS2Y/PmzXjkkUcUpVnzwTEXeit1int4NBFNJpBbVIhoOIbWPW34fP1qXHHtpSKgetSp89HS2gSzkUh+zmEmUZSXB7PRBH/TAWiicfDkZUHFaLIgN78Q9fUNgiqRaCO3BHFfBKNGVaG1tRlaswEFRQXYuW0XzGbOhPL2WhDTKJ5PNVAVhCzoJ2JY45zYVyTiKHJbVl4iBw7DYV/Qj3179mDUiBql1C8tFkhIetmll0v4+unSpaImlVeUj+1btgphk0ih63QyaMzwjpEDPTTvH6MKevioAXLNNpcNZRUV2NvQABN0CHX2SbRFN6iuJSuPbFcxauGweG3taEEu0SOyiX7k7NnSVmF+9shjj+Huv/0Vjz30MBa/+Qb6I4NIktmcSKhoFGGfItRk5JqYjDBaTCgtL5fxPc6D+n2KcrB62LEOIuHsMGQSDY6HAe8VwRBMu2pH1AirH1MO9kSVKRiFVoMH6OeffgoHUwiCXmh0GcXYI94ANm1YJyAAJdJximaH6r015iLHUIoqiX82POVF8CJpWC889U+cvnChwJlYjmZMrtGZYbB6UHTiTKTNnDqPwZ7jkAFlyT/Z9BToUg4S/jD2r/4Zd9/xV5Fw8/b34ciTjsW7X38OTZ4bTa0HEB8MYuLoMTAQZB4MKrwgwZj0KLkRuLEqc0vwwUNPyLhULBTCzNPPQJsmiaQphtLqKuhjKRx95By0bN2Fcnc+RhQW45CZhykjR8kUGrraZe6vpbERT1xzI/RI48NPP0Ldzh147a230NXSBk9JoUDPGHLwJJO8SHuQ1v9/MkZ7LiOGDPbv2IPuQD9OPXchBsI+xCw6uHNdsja8cby5pQWF8r3Cnb2wEHrIajVPaM5+8gCTqX09wgktGjt7kePIFSjexIlj4bYZMGZkFVZ/9z1KK4qEs5MkXSyvq4U3Hq5D1cls1Y/wOUqD79u7X3LzUaNHytrw4F23cYNstDEja4VTleNQnNkryC/CvLlH4/FHH0F5VaUge3jwDvQOiHegMfIAv+OOOwS4cNVVV8Fqt8smJ7X+tk2bMXL6RHj9PuRXFsvaEmK2bf1GuHTmIWNUIzGOx3EDsxdX4MnDnDlzZT8tWrRIQBO333mn6DTyoOz3erF85QqsX/uLtFb8kQBSiRjMlKFgS4GIm6zCGVMhi90q3EFUVKsdNUpkwn/99Ve5z5IjDzNG9XpU/Cm9P8Vaee0kno5Ek1KvYPGTh1BVVbWkOB+//wGcHrcy3hVjk5DKVMp9Ofn4E1BbXa3gVAlAJ4N9OKJQnjJlMuZZMizKqNbJvIkU/fw734C5x9Y1G4XSXzYiT51wEEWVI6HLKxUpNqvHKryrLNiwSMJTi71IIhLM0MMUSuCFR57Amy+9JkzOY0aPgjHPgZ92b8P/09h3QFdVpms/+/Te03sIhABJaNKroKKiI4iiiIo6OnrnH/VXUWccr2JldCwzjjp6bczoMOigogiCgEiTEukhtCSQflJO723/63332ZHhv3ete9ZiaQLJ2Wfv7/2+tzylR0jDEwpzulY3tGaw/uC2fjQuzd1I19RgQN+RU9jxwcdQ5zhx96//A/vdPTgf70dZeQEvGk0gBn08jetmzEVFfhH+tPrvGFs/GrMmTeYH9/WuH3C87Rxyigrx6YOPo7SwEFotcR+ViGRJr7QhEXdN3qm5A5YVQZKzBx4LXPBiLCs5aVnMrCx+4tBxqA1GdPSdx+jRo6AvLYDObOR6pri0mIfe1NamB0vDjJTXz8atPHy3mdDT0YeUUoewqER3IMXmOXnFJQhHkqgoKECBQQuEA+hoPg2TiWoXEYKG7KsygzUQXTc3l7LXyrC2tIDGYyfx9FMrcOP1N2DXvh147LHHmFhttJq500teGvSzdM8HBjyYMX0Wj1E2rP+K02NqbhHaJ+ANcDBSc4eCl5orhH3duXMn12oU4FTXtba0wFmUB4VWhXA6xpxFWvikkZP2hvh3XnwyEheQUGDhQAQrVjzN7BCqV+mZVNfUsI/ipEmT2P2XvBbnzpzFwPeHH3tkEDXEoxaVxAuVXc5MFhNnNUpyIk6noTfquUFIGz+l/dQLkPGjFwYjp5Dst5FEQV4+zyKL8orR2d6OM00n0XSkEYJWxfdehn3S+1N9SgpzOq2JY2HlimeY+EBBzNdHkLrsLJKzmNVf/kMkPwi6oZRu0A1tbWvjIKTApK8P7/qJA4wGyBIOLw6l0YrXPv0Cb37/L0QSASg1SlaLo92GPhxBhgil71LqgV4fcjQGXDH7MtyycAE+/HgVxl45Gx+s/wKNvn4k0hnYDCbkW1yDS5x/h9bAQ+BgXz/27tqNhTVjuJvb3d6OE94+GIZUwFWdD2UiinyVDcVaA+65dRmaTp7hk9Y0pIy1Woi2s/77rdDYrJh38414b81qxA4cha/xJJKJGBvrkAMWvSePChTUD6DUQyl93uw86kKg738XjHQy0o5XWDUUSZcN7/3hFVw7awZgMyKYiCAR6IepshTlJaWwWiwSaz4UhSIUQbxfcpoiCcCW1m5knPnIrx8Df1pkhQN/LArBKCLk8yPeNQD4I6geUosp4yei8ac9iIW7YFJLcvQytEuuxeTxR1+vB0/89j+xdcs23Lj4BtYRoobMqlWrmOJGC46s4BkvqVTB4XDikvETuUarKC+WOImhINo7ulg9jlZ6yBvEimef4Z954fnnYcqaI9HXFMQEAbPnOuDIy0F/yMNBQCBzMgA6vONHWKizelGaSsFIlgwPPfAgenv7+WAgCUT2UMzL4+yCMibqbteNGoXmk6cw+pLx+Hb79yirqODuK20KGqXEQxRpbBePwcBAFBPX1nQapbMqdayZM2kSoFHwHJjul5zqDzaWssB5UiQkW/NkOMH3i56HdHpKc0S51uS5sUoNPTW5MkrG5P5x5UoE+gekphDJ/tFayx5wjOrJGVYgEu2GbrTJakH9mDG8q3t63KzDSRqXpa7CQeFbYjJs3rgRpeWjMX7pzfjbzrVQEHmapfAAr6cPJo2B+ZFWUYPM+V5sWf0p7zxXTJ7GQ+2Hnvkdpt62CF9u2YA9Z0gVTMMD6uK8YhjVen5PamRQA8KoVGHHp5/DpFBiWFkF11ZHjx+Hc2Q1EnYT30CDQoF7rr4GDq2RO72erl4svuJqfLJzM0ZMGIuHf/84brn5NmjtTny2cSMEvQFtOw9DPHoIKl0aiVAGolriBrLwcbahIKMuGKaW3cH4dLxAooILcTHBolx55aWIJtPwt7Rg8aq3gP5+bPnHWiQzCUQJv0ROt3qJDyjXJQlyB1ZqEOz1QJGMw6Uzo8uXQspZCMeQCqTiGXiCHr63oiKONGErNSqe8yYGvFBklLBozJg/bQ42vPceTHYlyorykQ6nEMnE0NszgOF1NYikEywq3HzkFF5c8QLPA635LqaaPbH8YdhJZY04k/kFjCnWKHUYM3Y0XDl2Fm+mU4G6s0qVGk3HzkjWDlmQ9eXz5jFz/9SpU4PMdjrZhlZXc7ofEwk7nEGG4HFaLWNaCadcXzMCP275HsmEpPRNvWCS/+CFmUqzP8aaf/yN6VB/fu111pmxFRTjznt/xeLB99zxSw62axZcK4Em7DYurWqqq7Hi8SdgsFuk35W1LIinotAaDLA7HDwvJwJaLEkNsSSMVhN3SGXQuGegH17fAHfUeVPL/Iywoa9pnZ5rbGIRNcoOKHW/8FRlQADxKquGoqu1g4Pvycce58ZYJh3nDYY2LAbQZPVVhYppo0SL2SpZeWfJlYlUmhnOKVJbS6fhPnVOohWxW6wAkmRIpTQYvmwpAuY4zHYTEukktHotRGJuRxMIhaPI11pw55jp/IA73D24YtZsFla6++WnMXTWBGzYtBFplRZBkn/UG1BdUYN0NMHuxJRL61UauJtbYYhEUOzMQVVZBU4cPYYZs2bh9U9WoXzqJdCaDVCkU7h57mWoLCjB4cNHYBY0uHbGHLy+fg36k1GYXDa0nDiNQDyF/nCYvTyqjOXY8+67UCoD0KkUzOOUT0JqQNBDlP/IzIVBhsX/EIxFVZXwhEMwqdWouf4qnA37kWpsBwlppIQ4Bjp7kFGLnGqxqUs8DrVeYOqTMq2CPkMtfSO6EgIKKochGiVSr4BeXz/PrUiMNJqMQGUgR17a2SX7OL2gQajHgwm2SribDiOd9GHo6GoYrA7UDh/F2MykkELrwUZcO2seLps5F+9+8AFg1rHh0eZNG3g0QMGY53AyoimTBO686w6s++pzrmNplEXPtKu7B9EgJc7gk+qee+5hAAI5B8tjAFqsBHCora/nhoYn6GWyrjPPwXxLwgVTUJKa2ukjx5nuJBvz+MNx9rhfu/qf6O/uQTzkZ+U1EtGiERS0BhxrOYvyigq89uqrTCqePXs283HpPZfe/UsMHzYMp0+e5GbSYGOIKXwkHCOlrkXFxYhR1zMZZ29JpUbBgSc3vagD7HTZuQ7874KR2RlQIhmOcaORSg8ZcSNnJLxhJVPIsTjx1zf+ghOHj3BTR62WwATc1CHAeDbNEgqnVIsGvVmCRinIN8LI9mlESSGpCqvRiGO7G9j0RUF4umQSVrUWrrJq2GdMRCxHg0DED5VOw6djJByATlDDHwzBc+w0PnrxT6iqrER9bgn2/7Qf7333FdoRRRgxtnoL+2LICArEkylMHD0FIa8fNpOZcYHDC8vx4St/xK8WLcJ3mzfgkQcfxeGfDvLNf3DlcyifNBbQq3mUUpmfh0snT8eRhoMQgzGsfPhx3P7SU4iZtLyQx9SMwKYt22DPL0JXbx8qFYX49vX34HAmIWQig1Ax6aSTbpScnsopKX2PHhZ3JLO6Mvy1QKo1QOGQCk4nlQLgaWvHuOUPwH3oDJY/+Gs89tsHYNfq4ImGEPf7UTdhAgdjOB0BWaoa1TaoElqo7cXwkqOIUQcNRISiYcZ3KjU0500jlAjzvTbazexdEqJApocejkLrV8N/qAmzp4+DT+zl67+k9hL0D7gRTkRgSmtx7+LbMXbEaCxevBhnutvQ2doi1UokIJxKId/pYpGq6VNmoq6+loORCOK0sGhWfKLpJHQqCaVDVCdC5GzZtk0SEc7eE1potMgIkE6vrp5OtoyLp2JMm+rs6YFKL41rpk2cjHV/WwN7ngOkokadR+pQmtQaLF6wEDYa3PsDPDSna8wpLsM3u3dgz969HFDNbec5XSWIIK1hOiFpvf31rbcYriizMLjug8T+Ya5nKoWyqmE8DiPTXvpAVJpQesqbSibNYzB2as5k4Ov3c+YkZ0laGm8RUiiRgclgQtups9KAPzsv5cCkdJdkK7t9+HrtZzhx9CjX4pTdMNY7kZA25Cy3USibUSOmUgp+UwMBl+nBqtRwd3Zy3UBQLA3hUGmoTC5MInBix27oTS5UXTMXfTYFSoaUo8vdzacjNTsoGAdIIHjaHPj7fDh66iQq9GZo9BqIuRZ4owEoxTjCdGuSSgY0EwZzWOkIibqVEVFWVIRoRz/GFBVCHfSgcmg5vvjXN6gZNgzzrroKN/3mPlx1561o7ulgQPrJ40dZe5UUvKeNGoObZ16O59Z9gh/PnUY46GfJikRGYFpRKJrAE/OX4bq6y2EtUsNpJqsCaZTDqWk2BZMX18XBSGOhC4NRUErOFOZcF2cXokYPV0LAhNtvwfApU7HypecwcngZzh09yDQsmt3Sg6UGRyRFEEIlvH1RTK67FKdCfij0BhgMWiSIXUKkbdJ7pnuXSUKtUyMmZBBKRGB15kFjNMBHA/x0BmI4g3E5I3DuaAO01hisGgtKnPk8DvGEPYw/nTN6Mm66dhFGDqmC0WWDghy2KJpp4J9KIcdmR+3Ikchx5GHzd5tQWJRH3Qs+LQnRc7zxBDQKA6etixYt4tET6Y1Senhhs4iCkbqflI41t5xBmvSL1GCKF/FH3T4vk7dH1oyExWDAtm83Ic/uwMRLpuDU8Ubce9vtOHf6DAxGLXcdqXRhuJzWgOqJlzAovLikBF19vZyGUkeTmCg81wNw29KlfFpfqDkjSX1Jmy3JhJLuT05lKYqqyuGLEa46xX0SCpJkIs6lAAUn19BqCSUjM2D01H+jJFWhQiKegiKc4BHRhRhbeQMhC8OlNy6W8N1szSfVsrSG5ODnKYZz+kiR/prekPNlMg7NyuPJC44gSoRXjYZjKC8uh9sXxLxbbkLF+DHYvGcHFBolznZ3QDBqYbY7+KJsRhNi4QhEbxQBjxcOnZ7Vzdz9PSgvKmRhK7KBC8YJp66BVm3gdrZRYUKhzsL1VuP277Dw2l/g0P4fce/dd0GtNzNthXbBZ176A+quvxbugBe1I2vww87dmHXJRC7sq4xWJPvcWNN6nHlxh5pOoMBegAmja3Gw6TSEtBLXVY/Ag0sew8RZc9F+ZjeaWhuRm5cLMR2HRWdk/iEVeQqTERkSJEqm2N2ZJejVGmTIkSqrNM3t6yyk0EUsAyEGg9qAwsKZiBiTCKr9mHP1pShz5mHB5Zei7eARrnl+OnmCO7D9aeDaxbehvc+DoEgyj/no6uiQKFLKDIMpyPCVRiPsE5FMsl9949nTg5ArHsGkAU1CxMCxM5gzbTrafD7ObIqtDpw5eghH923nrvgNV8zHG08+DZvDxmB+CkaNQppplhcVo7ysDKfONPLClHd6oq+dOd3MM8rAgB+/vPdXnNa9+tLLsORYeb43+EpIAOra2loOkgMN+yTTGYGyCCCcSmJ4TQ3Od3VCgAIlRUXs9HWyqQn/ceddiPR7YFFpoKV7zaobEl6Zxw9KJXKrqvDsiyv5ZCKmCI2rSDPIaNRiSFUFrw86delkoiYSre1jhw/DapQmAvKLhKbod9Ka6u7txfCJo6E26Fj9nk5j2oTovvIcMS0B+uUGjZqkPONx/pp1XBMZnD92gufEAteREuyRfr+3uw9P/e5xpAIkl0LL6meFADml5ZGUY1atSA+bLpK+wXlu9moH0w7KtEU1wuEQZv3iGmgrKnCk5SxIq7K5tZnttnqDPqSI1a/RDmItWZQnJsk5pKMxJEJhZCJhVJeUIBaP8mwyniYJhCDGj50Ivy8ES1yA3htG4OwZhIMeXL9gAYpynXjjz6/j6msWsH4o/bn3/gdw14onsW77Fk6PY8kkZk+cxDdo4eTpePvPr+GsNs0y+5WVlTjw3S5U19UgkRbgP+/GZRUVuOuGZZgw6wHs3/4mbPkm+Ab6cNOyW/CL229DQVUlTp5rReO5FgYh5DucCPT04sWnnob/1DluAPCNJDQdYcopGMNh5FSVcwANnOrD7CvvQ16VC3uO/AC1mWRGYtDGVDi88Tue2apsVqR8YSx9+w0caG5GWqFiiQpKEylVo25nlNJSo4GDts/nkZ6RSskLaCDgG1yopIZmyNC99CPa64PebEIomZbs30nZvLcPY6y5OLzlO3QcOQSLk6hqFCCkhEYTTTWnlg/95n6mKO3YtW2QukSfk0qX7i43sxkuv2weBxsZvbBFgVbBwTiYSWSDkWzJ6fTftXvHvwVjnDIsauI4nUyLo0We43Qy1nPvt9vx/DNPItrvgY6gsRcFI/0sBeNrb/wFyx95DP/n/t8wjYpU45JigpkZrpwc2F1OnG9ukVgb2fQx4PYyZYnSQnpREFNGII/xaupHISQmJdsK8uYUpOYPjWq0olLy0cw2rUxUZxJ5we/PIpEUKMkrxNbPN8CW64SYlKKIgpHq3VdfeBHe9k5eL2qt1BCU75eMcRb0l9aycrksUceQLNq+sjs97Y6U33ujwNQr5yKmSKFtwMc3M7+ggAvgrr5uxNJJ1qypKC7ldIEch6iNTfl827lz3JAx6gwotueiYccPGF9Xi0CYbMcTUKt0KMgrBiJp5MQENHy9EZU5VgSCklgvMeKnTJ2Mq+Zdw9bQ1BSASo1tx45j0b13YvPO7Rg3dRKGlpQytEvo7sO/1q6BuaaSh64FJguWLlqKj776jIfnqm4/3nj4UYgGBaaOX4Yfj6/Hu2+/homzpuPp11bCmpvHgktn29swEAxIBGKTCRaHncHyqqbz2PjmOyzBQApstAsTHpY+b1FFCbr7Pbh67s1oO9ODhN7E7fjW9pMwOQRE+3txdtsPgF6HIRPGo3zefBxzd0JLhrHpBJyEe01noBRFFmU2WfRMnCVNIXLdpcVBQ2d6Xgajnk9Kdt0FkGc0w2AwIRGKwBeOMvxNqZOwqMSy9546j+nDhuNXS25GhVaFnAppU+Q0NSXgWmJlOF1Y9dFHKKsoGtR4IThif/8ADHoTPG4PZsyezQP9Dz74QMKnKqlTKslX0MKKByUFAToZaSPc+O03TKuTayWaNdKskofdxOQRRdg5MFPIMZpx8shRLFt0I8v9m3S6QWEnKRMRYCstxcbvtiAvtwCXz52LNZ99hmNNJ9Dp7kBN7QiWfCGZkvFjxzGjg6B7FHCJUBxDKyqZJkW6uvSe77zzDn79618zusbr8UPUK6Ex6zFp+lTeyKhJRYgdaqbRZ5PZ/QTNpPsujzP8oQDisQSKHDnY/8NuPPLwcs5+CFBA6fgTyx9F16kzEgaZateLrBW4xlTOrRPVacnfjxAYfOzqzXxzqcNFXS+tIxfTrr4e53y9ELVp2AxWSUi2sAD+aATtHed5xphbQHl7gNMbP8lhKAQGRNPiot2Ihvgusx1tBPWi3d1o4J3R5cwD0ioU6R34+oXX4NTr8eG7f8HX69dh+vTpeO/D95haRUBpmnXOnjMHo0bV4qk/v4lZNy1EMBVDS08HhpaWcRfttsuuxLur3oGppJDHCJY0sOCahXjl4w9ZmHmoYMZHz70ET7QX9973OIrHDsX2E0eYmpNfkAOD3sgwLgIkU+A7rFYcaz4LldOGMz1dMJusGBYT8fHy3yMnrwDhRBIJlYhUMg2NxoLxUy7DufOtGFblQk88hGAgg2LHCPi9ZG0eYGTQrHnz8MnX64DCDAx5ZoQiXj7hSTOW5o92gxHnWpthtOiRU1zEw2pBoeAAJQ0WFmsyGTjQyFmaNr8bFy3E5i3b4NRbMOD3syuTgkDxZKJDTYlgGEXF5QgHQrh79mwsqKuHtdDJjQ8/4XOXLcPpk03wkQaORuLi0aIJhcIMFif35vHjxzNW9Z0334LN5WROIefuWcl+rpmg5hPI6chhbPHn//oMjlwHc10pc6GmD62r5tZW5BYUMgqHWP3E8g/6AxwsGV8Mv3vkAXa7ItrcYI2uVMBcmI/N32+H3WzjkzevoAAPPfIwbC4rMgoFcksKWB2CJSBFkWtbCqxDDYcQ8nhRmpePjV+uw9dffIWGH/dixKg6hthR0DIx3mam5cg4YNpsCFmktRgkonIiwe7cpAJBGZN8aNH6pL5H1BeERa3HxPETeDO6645fIr+8GM8aueGeAAAdbklEQVT851PobmllPxchJqn/8eCfZlZZ9pCgnjNS5JZ91mmIvBQSCgFmq4H9K7RWB8xF1XDVVkNj0EKrEBmhQvUd5fyEwCe8IknJByNhDko5D6b/agWpAJb9EaiLleNwgeaxp/Y2cMpp0VpQkVOKeJcbLVu34a5bluC9d9/B0OFlzGfcu5fU1JK46toFDE6gwSx1VN//7AtMWXwjoFHhwP49+MOrr+CLT1czU2PXnp1wlRdj2pQpsKi1KMnJxRfbd6K0uBL2UAqvP/YE/vrxhzjUfQ4dgQGozSYJ+haLw6E3snwgIZFMdmpygEczZqcD+w8fQkCrYQ7lLTMvw/ZVa3DspxaMnD8eQU8EOz7eAnu+HoFYH8oqC9Hr7UN+ZRmQUKO/UwVljovFlAeaBpAMpDFibDWaQy1Q5asRUaTgcNph0mpQWVSIhob9cLjsjM2klFCt0SMeCcGqVCLfYoHDYkXD4UMYMboee39qgM6i54CxqgzcjCsvrcK5zg6EolHYnA4k/EEIWg0aT59ivPAvRk/FU0uuA+3yd999Bzs1UfeUdvyUQDWgAqFABD2dPVwDT5s2jRdjZ08nBjwe+LOK5jT3JAqBTKmiWTItMsKujq4djY3bNsNmt3LNSOk1NejoRbVdPJLiWo2UCTrb2jB01AgOgKb9h/HykyvQcfI0b0ws/88aKwJ6kjE0nWtBTUkZn9YVFRXcgKSAe+D++1kgy0WzVr3EhLHY7dwDIOVyakQRj5DUBoJtbty9+Da2gyfH5a83rOfMizvp5Mym0/H1UkoZpWJOFDkToe9RvUzBSCcdrUl57EU/Q+ik2xYv4d4JHUwvvfQSPP1eKXspyMXQ8gosXbIEiUgUrc3NvLapZhaMV44R6UOQlwHx7TgYBUlkWK8yY9zsy+G2a6Ex6ZFKRBAPhyCyTGAagkoFX5g0b2jsoWGUAw1R6cYMzlwSEu+Lh7ImIwZ8PhZhMpAsusGO3Iwe1VYn9q/fgKO7dmDW+NFo7WyD1+fhhUXBSD9L8vbt3b2cHhEaiOqE3Ioh2Nl0Eo899xyZYzGF6YmnfodVL7+JZfcuxaj6Wv7A11x9NT+opqZm2LUWfPraW/hx4wZ8sud7nPJ2o/HIUeQ7XKgsKcW8iVOQ63Bg666dKB4xHN/s2A4jFDxIpobAnClzEAl4cN+KpzD/9lux68d9aO4PwacdQKHCguVX3Y1bLh0Hc74ZRbVVSMczmDp7Bhr27UdM0CKW40ImVwWXKgfhcxH0d3uhioqoGDIUSRXgE0Kw2g0oKHMhFg2xQDSlfKzCHk+jwOWAU6PFkOIiGFRqdHZ3w2yxMOHYG/HD5sqBSaHDAElPaow4cqIRBrOZa6hIMMTaoiS8NeD1ky4l8gZCmDW0AuGoH6+8+kcUFObyAmL1AbJ9CMVY/pDIxXQNJM/47AvPMiyOMh/uPIvpQT0eWnByMBJcbmz9WGzctIExzUaTTkIHZdXkaQH3BgKIRMIoYBFioJDU5CIRWNVG7P92G1a9+SYa9u7jnysh/LHJiE/Wf8V9UaMoYPyY8YP8z+sWLsSYcaO5kK+oGQZTvotPYFqv5LpstFh4AyE4HWV23p4+qKMp/PT9TjiN1kH+Kn0GmgpQUJLZbXFZCXu6ygx++nsCKsj0M783CL3BiPr6el6fFKCklTpv3jwsWngDLDYaQ/38Ivc1JmMQ5jqWwIjh1RJXk4KRZx5MKhU5GAkmpFVoEekKYOb9j+B0+Dy34F1OO9cYNpOV/y25xVLjhOc8Pd0MbaoZNQIHDhzgYGTnocaTJK7DqAcKRqq5BCpS0gKMCQVsnQEcX/V3lFWU8A6kUyo4Z6eWe339GNaOod9FSA6yBpAlGMg0huQWNv50FNc9cD/G1lSxrdszf3wBdr0eRxuPYubkSSxuu2HzJugFBUaPGofTx07jwaV34vtt38IdC3K7fcn1i7B7+27WvnHpJZ4aFdKE9Jkx51JO3+l9Dx4+jMMHD6Lv9Dl0qIB+qxGminKkNCISIQ/KXS4c+2YP1O2dQDoMbb6LlfAmTp2GxqON6ElHkSnKhWBWwai1wh8OYUjFMKjTKgRaQ2g93ImZM2eiy38egimD2tHD4W4/z4uEFkL98FHoamnG3EmT0NHagiHlFdKOP3kyq3T7Ql4OxhNHm/gUp3EyDdjd/f18wjPCKJ3imioSJPhfBMauBIZmIvh8zUcMHo9Eg7xq6Lo7O7shiCr2sbzhxhv51Fi9ejVuXXbrvwUjjdHJi+Xik5GIQ/Wj6rFj325WcRAh4WXJKk+Gm3V5PIhGI3AVFjLY2x8Lo7KqChFfGBW5Bdi96TvctfRWXuBvf/A+N5AmXjmXT1JlMIIlN97MGzbB3z746CO2lSeTJeK+1lxSi2kzZ6K1pwvhdIodyOg93AP97K1BKJ+zp88iHYig0pWPzWu/+rnbKvzMJKH7wbU78XqzLzrb6X5S1hcYCOI/n1rBz+DzTz+DxqDHbbcuwbp163jtypbt8s/KwUjSjlwKEo3N3QdBO69e1BL9KWtfTTLmNOqgob+mYDhq51+BiEAiuSGEyMJbEHjHpMUZjEXZWoxmL3qzBYFghDtr1KFTqyQr74rSCi6m6UVgWanIVyEajcGqMiPw+fcQzrdBSxWFQoReq+YbL881mRme5YUplWpWNKusrmKOYzJJ+qc2LHvst7hm5hw88OKT6CG17VgM0UwQIb+XTVy379mDqWPHQx0EA7OtBjXSagX84QBC/QMozs3DJeMncVpy8EADotE4pk+ZiuazZzGktIwVDEia0t3Tw+nIsLJyfHp4P97fvhmqwnwImjQC7b0otTlwSUU1JuYV4+6lN8NeVcYzUFp8Xe4eoCgPYlk+fz61oOZaiWoRup+JWAp51nx07+mHYMigsqqYoWoKdRLDRwxFc3M7OxhryVO+pw9XzZrNeir0sHknTqe5C0vOwp19/bAX5OO8J4BzbW1obW9jFyjqeJJUSDAagU6rQZLwkWER1+aXYOWjv4HdYkBaI8DT0wsxKY0U6JSixk44EOR6nTroGzZvlKBf2dWV5q4jGWBrGWtJGlOUpmoUatSNqsfRY4eRpBm0ipT0pNOR6yXCjEYlmF0gGEBJeTmiYkwaR6SSzOVsOdiIh+9/gDGexDfNyXUhpBVgt9rg0hqxY/N2RuY46ORPSAyIn6cAGSatV9fXYcnSWxDVq3Cw8Sg7JSdoxpcS4fOQ+p6RnZZVvij2fPMt13DStPnn35XKxDljoLVI8xkCyMjECRm/TCedSq1GjsPJcULrX5bauPBkvNB3lTUSZGK19rJakbqV9EYkpUAvQdQwpcdeMxmmEaWIIIxILIIh1VVw97sRCsf54RJRlCBztBhcOQXopY6bWcc7HZ2gJpsR0XCCxwD0oOgCKd1rPH6CmwIWtQWFB8+hY/9+aIUUMqSrk+WS0QeVH5rsv06ORufa21hnhnYpMll15RYit3QIHnrsUXx79AC2HzqA5sZGuErsvEOSRB7ZQw8rGQJNJA2joEA0HkTNqBreXRXROK6bdyV8vhBDn4iTVpBXyLl8Z1s715xDSquwfuu3mDd9Ji/4D7/9EoeSIfQpMoiS+kE8jCKDFcquflw2bjL0kTh+98TjKKwdxkRTGqY7iwoZPO43SGLOVG/Q+5Gau1pF3xO5/hEGYlBo9BAjCsRDCUycVAe3mxS1RUQDXtRUDcGMujEIe31wmMwcMORFQjXM5GnTcbKlGbllJfjTX99GNK1k/mBvfx9Tl4jvSBha6vrSzFKl08OgsePof62BsqsJWlUaAyEflEmBm0i0mIgdQfd6++Yt+MXiG7D/4E+s+ROPRSSdHZY4lGpAMgUlBj1dK80A1aICI4aPRG9fD7rd3RyMdCJSQNLGR/eh1z3ACnFUE+bk5QJ6CRRRN3YMa/ge/2E/sxz8HjKddYGCwlJZxr+DBu2UkhNzg+F4CokyJQejPFSnNUWIrrWbvsHx1jOIKjM4fKqJxbDyHC6WDj15tBHKYAJn9zVIwmtJiWB/4XiP/p8OCaoZKU2WZ5B0n3iumdXMofejIKSYupBoIAckZZ8XvgbB6No5I0Wl6uddkLF5ogbKpALjrr4FqRI9vDEv003IYrywrBitHd1SmkB6LqS6LCiQStCsMgODQY9oIgKzy4FwNCzNL0XAZbPDGw4xIZNSjYKCQoQDUSQ+3oB0RwcyBNslXZislRb9HA3w6cOzfGTWHYhpPILAN4SCMcdoR/PJVuw92YhLb78JU6+Yg63fbYY/4YPCZkJd/VgYBDWKTbmYMGokNm36BiNrRyEWDsKuMyDHaGL84JnWNl54FCBOm5PNc2jX6unsgj0nF1PHjYdTb8L6fbvw7oGtiDitCMUkQSiH1YyRtjyMMjlx1cQZOHz4IBZedy2GThwNQa9Bb8CPolE1iBq0CCDDn4vqagqi3JJCBpCr1RqWFIl4+3kcQbWmQTAg5k2g1OVC7bBh3IXTqNVcMy+99VY0HP2JW+9EhKaAsZosPJPs9nr49EuFSN6S9ngRRqsVcaRYQ4dOZBp90zwsGBMwRluK+M6v8Pk//w5RkeZ6bWCgF79YuJDXzLpP18LqtMPv82LSFZfh4JFDyM918alJXedMJsWdXlbfVql44J0m1bJEGrUj66BSK3DoyCEORmmzFzkY6R709wyAsNA0y6O1VJzVOlXrdYx5rXAU4puv10ORxXEuf+RBvPz3DzlFTw0EcOuCxXj6qae4bk5Sncc6Yv++2mUtGruGaukgfr/yWcTFNBo729DZ48asydNR4srDQLsbvR2dLNz9j/ff58YPO1QTRO5CUAPND3WS1IkckNx3yQakDBiXZTlo7V74+p+DcXaNqNJKTORBJHlSRCaoxPy7H8Y5RS+iiDJ2j04at6ePkfNkA0BMj5K8fIbOOa0uuDu6JXkOevwaBZQ6DTLxGNJ9HoyqqUG33ycV3gYTOjo64bTnIvnROmhJtVxIIEPOyFkRY9lrQgbu0gdi9INGjQANsq0WKAQVRlUOx749DTjS2oyAIOC+3z7I9cnp7hYoXTY4nHmoKSrD7i9/wDXzZzC0jJgVQjyGvlNn8cqzz0GnNMBHKU4mw3qt1AEkj72aympyo+Q0SK9Qwx0J4L6/vIJYsYVtwogiE8ykOHXpOnwCFUoDHvrVr1l86cGbbkb+6OFIE8M914W4xYigRom4KMlOstxfIIC0SmBhZ5WK/OwFRp+QyayeiLfRBDLxFMxxA9LBIN5Y+TzXiB+sWc2zx35/H6eNVCbQPLYoJw9dbjd7S5KCQr7RxWk9gQR01FQzannXJ7B/JELy/1HoCvJRIeRi66OPIOLtgVanQazfh4VLFvF1fvbxaij0mkGeINX8IyeOw4nTTYMaOCYan3i9Ut8hlYZZb+RgJBD1pbPmcKf1x30/Ik0wLum4Gdx0xQTJSaYleFoyieKaCv4ntJnQHHBCzRh8t2kT2zDQhhPzBYASJwejIhzH8l/dz8RmOoEoXf7vgpF+H/19RX4xWlpb8ae3/4zOrnZYC/Ow5dBPKC0px5T6sSA0MM3EqenX29GOd956m70vKS6Sqfi/BRRZyNFzlw8PegZyVseiU1kQjQwU+F8Fo2b6CFFpJI1NCQmhI/Ar7dTQYc5Nd6FTEUdCGeTxQVxMIZqKI0xS7xo1n46JaBQGvYHhQMok5egp2Fx2hBQZqEmC3qjD/LET4bTZ8M8tmzhlVWYE5OYVwNPZg8Q/vkKc02ORwcByasAfLCs1KGNG5QYBp7BmE0StBha9BRa9A1+u+xKvffoxZk2djH+u/wJHzp+GsqgcwwqG4MTm7/HiiuVY8+XnsLBLVRjjh43A6Pp6rnEaGhpQVz+O0zJK97rPdTEihNTEyZbaqtJyl++eV55HU9wPS5lkQ0BdYgoolUaL6uJyZiBcdeV8dO/eizUvvgiVxYaxCxfgtN+DtFGD8pHVbMJJanT0u0lRuj/k55OSzGpYs5a8cIlhT42kZBr9vV6caTiIPz7/ApoOHWINleGj61FaWcF2dMdPnEBeQT6LKLe3dWL4yDqcPtPMKuixaELS+yTzA9oMoiEWZfIGQrDlu1CQ42L92/72LiwoqMSWt/8LB/Z8j0svnwOzycJKZ4xeIcv2rO4Pz8dUShQWF6Or180nLQUXhZlUl2YYg0kQSMqISPx55LCR2LhlE3jTzyJq6DnziUVcv4gk+0KL2JHnYJAJnYykIn/tZVfh09X/ZL0fT3cPoFbCXF7A6SUpQbz74iu44frrQU0R+fS6+GSUA4H6IXTvV73/Po/L6NoNNitOnD3DivFTJk3iw4BQNclgCL09br4HRL5OpeLMhZTTVgnlKl2zXLPLoHIZBE7zVNk6nf6tDDKX1/HFaauw9JUnxY/fepsRIaAZIQ0hE1GYLXmw5FfBfkkNVAUGHoJ6ycLMZEQ0I8IX8HPbmP0HMiIUepMk/JoCgiE/dHYLCim1CQW4geHu7eXdu9jmgLe7FwFRZAzo0fuWw15UyGYkssKzLPxEow0Z1XEhfIg+jEav40UxpHwop3SrP/0KL696H81dZ+GNB5DKsUCtyMEds67GXZfPxncHf8APe3fj2NGDePnpldjxw1aIxLTn2lONru4+bkuPrBqJ7u4u7qCSFTWVQ6RQ9/z7b+NsIoiOgAfWsvxBUDntjGarDRajBaeONyHiCaKsN4juvftQNW0GfHlOJO1GCHoV+sJepJhikWE8prurG6ksIVi2nXam1Fzf0H2qKS7FsNIq3mVJN2bG9Mk8Y7Xl52HtV+uQX1XOpwkZ3Xb2dKNiWDV8gRA3RIiWFfL6aLTHuzv5gYSJQkRDcxrWp5PcmApGElDFU5gcV2Hrex9i2Z1LuFH36sqXoScTIjYO/TkYGQFESBiFwAHp8Xn51KGShUi7lBlxukYEAwjMQaRU8m+f/B1qPc0jydNUgqPxokwJg8FI30vHo6iorYGXwAzBAG65YQk+ef8DlnZ8e9WHWP74cmgKHCzb2d/Vh/riSrYj+N8EI6u/icAfnn9eAoPH4uzGVTlsKH73+9/j+uuv5+dOvYpYJMopOJ3GNEe02iQJli+++IKvXQ5G+XNQ2k1dXQpWCmbqc1DNSGmu3E2V0+WLNws5bRWcCy8VZ8+9Eu5ICAanAz/s2Y0KYmLHMjiweQ+WPvUwmsNuhJIxpBUiYpkkBoj7pyb16jTqhlWzXsrJzm70DXiRZ3dhREU5zradgxJphPw+dmeiCyXZxrohQ3H+XBtrnI4oLMXW+5ZDo1ZCRX7uWQqODMaVVZ3lwpjpSlSjOcgHMQa92QydzsBpr0Lnwvw7bkWfIoFt+3dCUGmwaNpV+K+HfouXn34cvSawtP69t9+Bv3/0HkpLyrj+IoYCjU/MFgfXQF1dXbhi1uUsjkXNJlJze+G9t+B3mRARU3DkONAfD0sPJJXin3HYnVAbzXC3dcCh0KH1ky8R6+nFkgcexfftzYhZ9BDMCoh6BduREfCBQPQq6hSbLOwTSOZA7efO4/X7l/PDNCtUON7wEyZOnMgg8UPHj6GqvIzRKwR8p7lZ24Bbgh2mUqBRDzlLE5jd3S+VEOloBKUFpbBY7MgpKMKuvfsQzsQRFom4TbjhOBy2HCS9Adw34zL8fukylBc48NOO3cgpLuDFyPAttXIQCM6onCwwnvRbNeSRQYJmRA4mpruKnrp0ClDI0Wbq0lnQ2nwW1JuQvy8vYkLrBAiMIHtoKCSVcNo0aLzk7fUCUelZk9zlFfOvxIHWk7DoDExruuvqhXhp5UqoySuE3/nn1/+36GkCKoLl/Z977ln0dXWzQh7ZiP+4bx97lETCYYa/yaccG+5Eo0yGoJc83iDM7IUvBgpEYyiuKJf0m2LE+U2xEh9BNC98ySfpIN0qC4IQxr/6hFhTW48tu3bycJTb7iotkrE0yhVmtHacQbzAgvzSIiSFDPr9Ht7Nw9EIYwpLCgrZobcvEGYNzEA8AdHnQ2lZCTMzggMDnK7kWG2cBjJiIRZjWFxhTMSRv37EXDv1BcBZ3iFJUEghcctk3VAZxUPNG9qJSFhZpdMxqVmV0aLF3Yfff/g+jpxuRuehMzj23ecwpmP417bvsevUMURjEZw41IAxw0fAZrai8fhxXDprFs8w3b0efh/WIBXVmDFtGqulTZowEbe/sgIxhwkWpYCK4UOx/8QJ3j1JzmPNmjUwW6zwEVYxo4DZH8PeP78NvdUGX5sHVz2yHC1iBIp8E/wakpCUoIcpAiwrlEj2B5h8Pbq6hgHFNflFmDB5Er5a8xkmjR/PeN/TtDHUjkKXu4slJapH1+HkmTMMnSPMbLe7hzfG8ooStHe7mY3f7e6F1W7EuNrxWP/1ZowdOwnNJ5vR5u2CYCUQhxk5FjvMegtKTDase+B3yPj8EC1KmPQ6pPwh3mi4K0giu9mFLgcNV38Eoqba2WTiwKHTkRps8glAJyWPXkQVxCQZhirg7x+AWm7lCwKMWhMG+j2DTReiF6XiSeRWlvL4IRVPM/SMaFQUSL955P/io2/WwqjWQUgJaP7+R9gIM5yiM+5/F4xkhEuvJ1esQG9PD///Z2vXckNObr7EyZ8zCzDnDeiiBs7FwSivWVoXtHZpndN9oD+DNgjZiJQPGzlAZUSSUP7MQ2I0k2Qwc5SimbVBUhBjIooMOdj/5usY+6tfImHVIiGkEAr64BUlSBvhCYnwSuklnU7EGBA0IvIdxehvc6M0xw6dScvqZ7OmTEXDocOw5xZwE2Jufhn+eMsyOEsKWTSXTgsh60Ev72gU3JTDU+DJLln0AWTOGaFyvAE/zHY71BodTFYn+n1hhJMZePy9aNp3EH9a9RY0DgvO9HXxezh1BkyvG8OefrT4r7nyKj6JqMVOzRCqJSaMncggZY1Rh/tfeQ6qIYUMeCbsIv33dK+bGw51Q6vx/e5dXPNZzTnICcaxatm9sJcWIk2WCSolfD1e3PHiCqxraYS1pIBFiqjZQYrWxEi/evYsNB8/gbjXx7hJlgoUScC3Cg0NB1BXP4pNRTdt3YKMRsl1Ki0S1pI16nC2uRmuvFwOytaWc5ImKhnm2O0YV1uPL7ZshKukAjX2Inz+0uu44aUncai9BWFvAPFkGgUaK0YOGY7A59/gy3VrYbERJcmDQmcuzre24vPP/oXrrvwFbMU5EkOD6vrsKEMOTNLTpRNRSY5dOonNwEBwClD6PBlJRJkXN4Ao0ZNoTCCQ0raGu5dsI559UflgcTo47aXTitcGCUIlkvi/zz2Fv/1zNXtj5JJH5oatg2OSi0/CQRbJBccS3VsattPoJuQP4w9/WMlwuocffpi1TimcqTl3MbH8389BYrn8+9EoSzxy9sa6Oz+7uFEwXkjdotqBgSXZrq905gJC7ZvPiVTIDng9ElOaBvSKFDQ0awxm0L13J0aOnwAvTTF0KtYLiSsEkIAB1ZGCmnYNyWfQZnMy6VWZUiDlC+Hu+Qux62TDoGoA7bRKvQlzq2rwxrwFyKkeRqInnH7QQ1KK/456oC6jDKiVdxm6aLmlTDeMPrSC5NpdDrayc+UVsaYldS2JaV5YX82LtmfADYOgROP+Brz2/AvoPN/G9WJ/j5uHzHq9maXkq6qqYNNZMKpuNO5Z8SgW3LEEf/vqcw5CGnsQ8PnAqZOsiEAA8j0NBxjAHuocwDM3LsVvr78B/mQcGYJgpdLIF/RoUSQx4cF7kNSrGOFE6mIH9u1D7YiRCPn9sKm1DLkjFviC+dfgx927WXVszuVz2cNj9dp/cf1GCm4yo5524E4SijYY+DQibGXH+XYOVtKiue+++/DZmjVIkcatxYGrx0zDmj+9jZy6oTBYzQgqFYinRTg1FgwxuXDn8FpcMn40DE4TcvOdCPQTi8GDP770Em9Cf/v4Yz4pOQAvCkZWJqfMhsSKs8a4FJQh2tyJE0rGSRcEI5EGqKak9I8MWCkYGUJHLHwCniuVbJpLJy0FI52KFJB0Wv3yt49wvUzqgyPKKrF17fqsIJR0Cl2cOl4cRBcH47PPPsNkgptuuplNjZIEgcseChf/7IVf/0/ByOuUMjtI13PhNQ0GpCDheGUUkhyM/w9D9aDwMQfmaAAAAABJRU5ErkJggg==\")",
"_____no_output_____"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code"
]
] |
e7f8daeac290538c1fa674828795bf677582c4a9 | 18,992 | ipynb | Jupyter Notebook | 04_markov/.ipynb_checkpoints/ejercicio_3-checkpoint.ipynb | juanntripaldi/pyOperativ | 8bf58b6221c5344e9d088703782530278cb10ca1 | [
"Apache-2.0"
] | 1 | 2021-05-04T12:33:39.000Z | 2021-05-04T12:33:39.000Z | 04_markov/.ipynb_checkpoints/ejercicio_3-checkpoint.ipynb | juanntripaldi/pyOperativ | 8bf58b6221c5344e9d088703782530278cb10ca1 | [
"Apache-2.0"
] | null | null | null | 04_markov/.ipynb_checkpoints/ejercicio_3-checkpoint.ipynb | juanntripaldi/pyOperativ | 8bf58b6221c5344e9d088703782530278cb10ca1 | [
"Apache-2.0"
] | null | null | null | 26.268326 | 1,381 | 0.533698 | [
[
[
"____\n__Universidad Tecnológica Nacional, Buenos Aires__\\\n__Ingeniería Industrial__\\\n__Cátedra de Investigación Operativa__\\\n__Autor: Rodrigo Maranzana__\n____",
"_____no_output_____"
],
[
"# Ejercicio 3",
"_____no_output_____"
],
[
"Un agente comercial realiza su trabajo en tres ciudades A, B y C. Para evitar desplazamientos innecesarios está todo el día en la misma ciudad y allí pernocta, desplazándose a otra ciudad al día siguiente, si no tiene suficiente trabajo. Después de estar trabajando un día en C, la probabilidad de tener que seguir trabajando en ella al día siguiente es 0,4, la de tener que viajar a B es 0,4 y la de tener que ir a A es 0,2. Si el viajante duerme un día en B, con probabilidad de un 20% tendrá que seguir trabajando en la misma ciudad al día siguiente, en el 60% de los casos viajará a C, mientras que irá a A con probabilidad 0,2. Por último si el agente comercial trabaja todo un día en A, permanecerá en esa misma ciudad, al día siguiente, con una probabilidad 0,1, irá a B con una probabilidad de 0,3 y a C con una probabilidad de 0,6.\n\n* Ejercicio A: Si hoy el viajante está en C, ¿cuál es la probabilidad de que también tenga que trabajar en C al cabo de cuatro días?\n* Ejercicio B: ¿Cuáles son los porcentajes de días en los que el agente comercial está en cada una de las tres ciudades?",
"_____no_output_____"
],
[
"<h1>Índice<span class=\"tocSkip\"></span></h1>\n<div class=\"toc\"><ul class=\"toc-item\"><li><span><a href=\"#Datos-Iniciales\" data-toc-modified-id=\"Datos-Iniciales-1\"><span class=\"toc-item-num\">1 </span>Datos Iniciales</a></span></li><li><span><a href=\"#Ejercicio-A\" data-toc-modified-id=\"Ejercicio-A-2\"><span class=\"toc-item-num\">2 </span>Ejercicio A</a></span><ul class=\"toc-item\"><li><span><a href=\"#Forma-alternativa-de-resolución:\" data-toc-modified-id=\"Forma-alternativa-de-resolución:-2.1\"><span class=\"toc-item-num\">2.1 </span>Forma alternativa de resolución:</a></span></li></ul></li><li><span><a href=\"#Ejercicio-B\" data-toc-modified-id=\"Ejercicio-B-3\"><span class=\"toc-item-num\">3 </span>Ejercicio B</a></span><ul class=\"toc-item\"><li><span><a href=\"#Forma-alternativa:-usando-una-matriz-no-cuadrada\" data-toc-modified-id=\"Forma-alternativa:-usando-una-matriz-no-cuadrada-3.1\"><span class=\"toc-item-num\">3.1 </span>Forma alternativa: usando una matriz no cuadrada</a></span></li><li><span><a href=\"#Cálculo-auxiliar:-partiendo-directamente-de-la-matriz-de-transición\" data-toc-modified-id=\"Cálculo-auxiliar:-partiendo-directamente-de-la-matriz-de-transición-3.2\"><span class=\"toc-item-num\">3.2 </span>Cálculo auxiliar: partiendo directamente de la matriz de transición</a></span></li></ul></li></ul></div>",
"_____no_output_____"
],
[
"## Datos Iniciales",
"_____no_output_____"
],
[
"Importamos las librerías necesarias.",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"Ingresamos los datos de la matriz de transición en una matriz numpy:",
"_____no_output_____"
]
],
[
[
"# Matriz de transición como numpy array:\nT = np.array([[0.1, 0.3, 0.6],\n [0.2, 0.2, 0.6],\n [0.2, 0.4, 0.4]])\n\n# Printeamos T\nprint(f'Matriz de transición: \\n{T}')",
"Matriz de transición: \n[[0.1 0.3 0.6]\n [0.2 0.2 0.6]\n [0.2 0.4 0.4]]\n"
]
],
[
[
"## Ejercicio A",
"_____no_output_____"
],
[
"En primer lugar, calculamos la matriz de transición habiendo pasado 4 días: elevamos la matriz a la cuarta usando el método de la potencia de álgebra lineal de la librería Numpy.",
"_____no_output_____"
]
],
[
[
"# Cálculo de la matriz de transición a tiempo 4:\nT4 = np.linalg.matrix_power(T, 4)\n\n# printeamos la matriz de transicion de 4 pasos:\nprint(f'Matriz de transición a tiempo 4: \\n{T4}\\n')",
"Matriz de transición a tiempo 4: \n[[0.1819 0.3189 0.4992]\n [0.1818 0.319 0.4992]\n [0.1818 0.3174 0.5008]]\n\n"
]
],
[
[
"Sabiendo que $p_0$ considera que el agente está en el nodo C:\n$ p_0 = (0, 0, 1) $",
"_____no_output_____"
]
],
[
[
"# Generación del vector inicial p_0:\np_0 = np.array([0, 0, 1])\n\n# printeamos el vector inicial:\nprint(f'Vector de estado a tiempo 0: \\n{p_0}\\n')",
"Vector de estado a tiempo 0: \n[0 0 1]\n\n"
]
],
[
[
"Calculamos: $ p_0 T^4 = p_4 $",
"_____no_output_____"
]
],
[
[
"# Cálculo del estado a tiempo 4, p_4:\np_4 = np.dot(p_0, T4)\n\n# printeamos p4:\nprint(f'Vector de estado a tiempo 4: \\n{p_4}\\n')",
"Vector de estado a tiempo 4: \n[0.1818 0.3174 0.5008]\n\n"
]
],
[
[
"Dado el vector $ p_4 $, nos quedamos con el componente perteneciente al estado C.",
"_____no_output_____"
]
],
[
[
"# Componente del nodo C:\np_4_c = p_4[2]\n\n# printeamos lo obtenido:\nprint(f'Probabilidad de estar en c habiendo iniciado en c: \\n{p_4_c}\\n')",
"Probabilidad de estar en c habiendo iniciado en c: \n0.5008\n\n"
]
],
[
[
"### Forma alternativa de resolución:\nEl resultado es el mismo si consideramos que la componente ${T^4}_{cc}$ es la probabilidad de transición del nodo c al mismo nodo habiendo pasado 4 ciclos.\nVeamos cómo se obtiene esa componente:",
"_____no_output_____"
]
],
[
[
"# Componente de cc de la matriz de transición a tiempo 4:\nT4cc = T4[2,2]\nprint('\\n ** Probabilidad de estar en c habiendo iniciado en c: \\n %.5f' % T4cc)",
"\n ** Probabilidad de estar en c habiendo iniciado en c: \n 0.50080\n"
]
],
[
[
"## Ejercicio B",
"_____no_output_____"
],
[
"Dada una matriz $A$ proveniente del sistema de ecuaciones que resuelve $\\pi T = \\pi$",
"_____no_output_____"
]
],
[
[
"# Matriz A:\nA = np.array([[-0.9, 0.2, 0.2],\n [ 0.3, -0.8, 0.4],\n [ 0.6, 0.6, -0.6],\n [1, 1, 1]])\n\n# Printeamos A:\nprint(f'Matriz asociada al sistema lineal de ecuaciones: \\n{A}')",
"Matriz asociada al sistema lineal de ecuaciones: \n[[-0.9 0.2 0.2]\n [ 0.3 -0.8 0.4]\n [ 0.6 0.6 -0.6]\n [ 1. 1. 1. ]]\n"
]
],
[
[
"Y dado un vector $B$ relacionado con los términos independientes del sistema de ecuaciones anteriormente mencionado.",
"_____no_output_____"
]
],
[
[
"# Vector B:\nB = np.array([0, 0, 0, 1])\n\n# Printeamos B:\nprint(f'Vector de términos independientes: \\n{B}')",
"Vector de términos independientes: \n[0 0 0 1]\n"
]
],
[
[
"Dado que el solver de numpy solamente admite sistemas lineales cuadrados por el algoritmo que usa para la resolución [1], debemos eliminar una de las filas (cualquiera) de la matriz homogénea y quedarnos con la fila relacionada a la ecuación $ \\sum_i{\\pi_i} = 1$.\nHacemos lo mismo para el vector de términos independientes B.\n\nPara hacer esto usamos la función el método delete de numpy, indicando la posición a eliminar y el eje (axis) al que pertenece.\n\n\n[1] https://docs.scipy.org/doc/numpy-1.15.0/reference/generated/numpy.linalg.solve.html",
"_____no_output_____"
]
],
[
[
"# Copio la matriz A original, para que no se modifique.\nA_s = A.copy() \n\n# Eliminamos la primer fila de la matriz A:\nA_s = np.delete(A_s, 0, 0)\n\n# Printeamos:\nprint(f'Matriz asociada al sistema lineal de ecuaciones: \\n{A_s}')\nprint(f'\\n -> Dimensión: {A_s.shape}')",
"Matriz asociada al sistema lineal de ecuaciones: \n[[ 0.3 -0.8 0.4]\n [ 0.6 0.6 -0.6]\n [ 1. 1. 1. ]]\n\n -> Dimensión: (3, 3)\n"
],
[
"# Copio el vector B original, para que no se modifique.\nB_s = B.copy() \n\n# Eliminamos la primera componente del vector B:\nB_s = np.delete(B_s, 0, 0)\n\nprint(f'\\nVector de términos independientes: \\n{B_s}')\nprint(f'\\n -> Dimensión: {B_s.shape}')",
"\nVector de términos independientes: \n[0 0 1]\n\n -> Dimensión: (3,)\n"
]
],
[
[
"Cumpliendo con un sistema cuadrado, usamos el método solve de numpy para obtener $x$ del sistema $Ax = B$",
"_____no_output_____"
]
],
[
[
"x = np.linalg.solve(A_s, B_s)\nprint('\\n ** Vector solución de estado estable: \\n %s' % x)",
"\n ** Vector solución de estado estable: \n [0.18181818 0.31818182 0.5 ]\n"
]
],
[
[
"### Forma alternativa: usando una matriz no cuadrada\nComo explicamos anteriormente no podemos usar el método $solve$ en matrices no cuadradas. En su lugar podemos usar el método de los mínimos cuadrados para aproximar la solución[2]. Este método no tiene restricciones en cuanto a la dimensión de la matriz.\n\nEl desarrollo del método no forma parte de la materia, siendo contenido de Análisis Numérico y Cálculo Avanzado.\n\n[2] https://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.lstsq.html",
"_____no_output_____"
]
],
[
[
"x_lstsq, _, _, _ = np.linalg.lstsq(A, B, rcond=None)\nprint('\\n ** Vector solución de estado estable: \\n %s' % x_lstsq)",
"\n ** Vector solución de estado estable: \n [0.18181818 0.31818182 0.5 ]\n"
]
],
[
[
"### Cálculo auxiliar: partiendo directamente de la matriz de transición\nEn la resolución original, usamos una matriz A relacionada al sistema lineal de ecuaciones que resolvimos a mano. Ahora veremos otra forma de llegar a la solución solamente con los datos dados y tratamiento de matrices.\n\nPartiendo del sistema original: $\\pi T = \\pi$\n\nDespejando $\\pi$ obtenemos:\n\n$(T^T - I) \\pi^T = 0 $\n\nPodemos transformar lo anterior en la notación que usamos más arriba para que tenga consistencia:\n\n$A = (T^T - I)$\n\n$X = \\pi^T$\n\n$B = 0$\n\nPor lo tanto, llegamos a la misma expresión $Ax = B$",
"_____no_output_____"
],
[
"Entonces, comenzamos calculando: $A = (T^T - I)$",
"_____no_output_____"
]
],
[
[
"# Primero calculamos la traspuesta de la matriz de transición:\nTt = np.transpose(T)\n\nprint(f'\\nT traspuesta: \\n{Tt}')\n\n# Luego con calculamos la matriz A, sabiendo que es la traspuesta de T menos la identidad.\nA1 = Tt - np.identity(Tt.shape[0])\n\nprint(f'\\nMatriz A: \\n{A1}')",
"\nT traspuesta: \n[[0.1 0.2 0.2]\n [0.3 0.2 0.4]\n [0.6 0.6 0.4]]\n\nMatriz A: \n[[-0.9 0.2 0.2]\n [ 0.3 -0.8 0.4]\n [ 0.6 0.6 -0.6]]\n"
]
],
[
[
"Seguimos con: $B = 0$",
"_____no_output_____"
]
],
[
[
"# El vector B, es un vector de ceros:\nB1 = np.zeros(3)\n\nprint(f'\\nVector B: \\n{B1}')",
"\nVector B: \n[0. 0. 0.]\n"
]
],
[
[
"A partir de aca, simplemente aplicamos el método que ya sabemos. Agregamos la información correspondiente a: $\\sum_i{\\pi_i} = 1$. ",
"_____no_output_____"
]
],
[
[
"# Copio la matriz A1 original, para que no se modifique.\nA1_s = A1.copy() \n\n# Agregamos las probabilidades a la matriz A\neq_suma_p = np.array([[1, 1, 1]])\n\nA1_s = np.concatenate((A1_s, eq_suma_p), axis=0)\n\n# Printeamos:\nprint(f'Matriz A: \\n{A1_s}')",
"Matriz A: \n[[-0.9 0.2 0.2]\n [ 0.3 -0.8 0.4]\n [ 0.6 0.6 -0.6]\n [ 1. 1. 1. ]]\n"
],
[
"# Copio el vector B1 original, para que no se modifique.\nB1_s = B1.copy() \n\n# Agregamos 1 al vector B:\nB1_s = np.append(B1_s, 1)\n\n# Printeamos:\nprint(f'\\nVector B: \\n{B1_s}')",
"\nVector B: \n[0. 0. 0. 1.]\n"
]
],
[
[
"Resolvemos por mínimos cuadrados:",
"_____no_output_____"
]
],
[
[
"# Resolvemos con método de mínimos cuadrados:\nx_lstsq, _, _, _ = np.linalg.lstsq(A1_s, B1_s, rcond=None)\n\n# Printeamos la solucion:\nprint(f'\\nVector solución de estado estable: {x_lstsq}')",
"\nVector solución de estado estable: [0.18181818 0.31818182 0.5 ]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f8dffaa9abfb5601a259cf7f45c55882becf30 | 9,099 | ipynb | Jupyter Notebook | scratch_nn_1.ipynb | Reet1992/Deep_learning_R | 7a92b37d9d8ecbddfbfa932a5bc1fd053fe04961 | [
"MIT"
] | null | null | null | scratch_nn_1.ipynb | Reet1992/Deep_learning_R | 7a92b37d9d8ecbddfbfa932a5bc1fd053fe04961 | [
"MIT"
] | null | null | null | scratch_nn_1.ipynb | Reet1992/Deep_learning_R | 7a92b37d9d8ecbddfbfa932a5bc1fd053fe04961 | [
"MIT"
] | null | null | null | 23.695313 | 84 | 0.477635 | [
[
[
"import numpy as np\nimport pandas as pd\n\nimport h5py\nimport matplotlib.pyplot as plt\n#from testCases import *\n#from dnn_utils import sigmoid, sigmoid_backward, relu, relu_backward\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (5.0, 4.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n%load_ext autoreload\n%autoreload 2\n\nnp.random.seed(1)\n",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"def initialize_parameters(n_x, n_h, n_y):\n \"\"\"\n Argument:\n n_x -- size of the input layer\n n_h -- size of the hidden layer\n n_y -- size of the output layer\n \n Returns:\n parameters -- python dictionary containing your parameters:\n W1 -- weight matrix of shape (n_h, n_x)\n b1 -- bias vector of shape (n_h, 1)\n W2 -- weight matrix of shape (n_y, n_h)\n b2 -- bias vector of shape (n_y, 1)\n \"\"\"\n \n np.random.seed(1)\n \n ### START CODE HERE ### (≈ 4 lines of code)\n W1 = np.random.randn(n_h, n_x) * 0.01\n b1 = np.zeros(shape=(n_h, 1))\n W2 = np.random.randn(n_y, n_h) * 0.01\n b2 = np.zeros(shape=(n_y, 1))\n ### END CODE HERE ###\n \n assert(W1.shape == (n_h, n_x))\n assert(b1.shape == (n_h, 1))\n assert(W2.shape == (n_y, n_h))\n assert(b2.shape == (n_y, 1))\n \n parameters = {\"W1\": W1,\n \"b1\": b1,\n \"W2\": W2,\n \"b2\": b2}\n \n return parameters",
"_____no_output_____"
],
[
"parameters = initialize_parameters(2,2,1)\nprint(\"W1 = \" + str(parameters[\"W1\"]))\nprint(\"b1 = \" + str(parameters[\"b1\"]))\nprint(\"W2 = \" + str(parameters[\"W2\"]))\nprint(\"b2 = \" + str(parameters[\"b2\"]))",
"W1 = [[ 0.01624345 -0.00611756]\n [-0.00528172 -0.01072969]]\nb1 = [[0.]\n [0.]]\nW2 = [[ 0.00865408 -0.02301539]]\nb2 = [[0.]]\n"
],
[
"type(parameters)",
"_____no_output_____"
],
[
"list(parameters.items())[0:1]",
"_____no_output_____"
],
[
"W1 = [[0.01624345 -0.00611756],[-0.00528172 -0.01072969]]\nW1 = pd.DataFrame(W1)\nW1 = W1.values\nprint(W1)\n\n\nb1 = [[0],[0]]\nb1 = pd.DataFrame(b1)\nb1 = b1.values\nprint(b1)\n\nW2 = [[ 0.00865408 -0.02301539]]\nW2 = pd.DataFrame(W2)\nW2 = W2.values\nprint(W2)\n\nb2 = [[0]]\nb2 = pd.DataFrame(b2)\nb2 = b2.values\nprint(b2)",
"[[ 0.01012589]\n [-0.01601141]]\n[[0]\n [0]]\n[[-0.01436131]]\n[[0]]\n"
],
[
"W = [W1,W2]",
"_____no_output_____"
],
[
"\nW = [W1,W2]\nW = pd.DataFrame(W)\nW = W.values\nprint(W)\n\nb = [b1,b2]\nb = pd.DataFrame(b)\nb = b.values\nprint(b)",
"[[array([[ 0.01012589],\n [-0.01601141]])]\n [array([[-0.01436131]])]]\n[[array([[0],\n [0]], dtype=int64)]\n [array([[0]], dtype=int64)]]\n"
],
[
"#### Forwrd ",
"_____no_output_____"
],
[
"Z",
"_____no_output_____"
],
[
"#### New implmentation \n\nfor i in range(0,25):\n W = np.random.randn(2, 2) * 0.01\n A = np.linspace(-1,1,2)\n b = np.linspace(0,2,2)\n Z,linear_cache = np.dot(W, A) + b\n\n ### sigmoid ###\n\n A_sig = 1/(1 + np.exp(-Z))\n\n #### relu\n\n A_relu = np.maximum(Z, 0)\n Y = np.linspace(-1,1,10)\n\n m = Y.shape[0]\n\n cost_sig = (-1 / m) * np.sum(Y*np.log(A_sig)+(1-Y)*np.log(1-A_sig))\n \n #cost_relu = (-1 / m) * np.sum(Y*np.log(A_relu)+(1-Y)*np.log(1-A_relu))\n cost_sig = cost_sig - 0.025 * i\n print(\"SigmoiD cost Function\", cost_sig)\n \n\n",
"SigmoiD cost Function 0.680695586799679\nSigmoiD cost Function 0.6506910072556065\nSigmoiD cost Function 0.6320412412173286\nSigmoiD cost Function 0.6208538410148245\nSigmoiD cost Function 0.5911536896241928\nSigmoiD cost Function 0.5640910441511027\nSigmoiD cost Function 0.544622803812289\nSigmoiD cost Function 0.5214455098301642\nSigmoiD cost Function 0.48856891893754456\nSigmoiD cost Function 0.4804196395774253\nSigmoiD cost Function 0.452739447850644\nSigmoiD cost Function 0.41489040036918223\nSigmoiD cost Function 0.39713267826153076\nSigmoiD cost Function 0.37114944305837133\nSigmoiD cost Function 0.33910028923165497\nSigmoiD cost Function 0.32009000132211674\nSigmoiD cost Function 0.2978564569599558\nSigmoiD cost Function 0.256526316662936\nSigmoiD cost Function 0.24581029946931937\nSigmoiD cost Function 0.21926525618959797\nSigmoiD cost Function 0.1984020455838561\nSigmoiD cost Function 0.16309121959663142\nSigmoiD cost Function 0.14141790691548872\nSigmoiD cost Function 0.12475905992621084\nSigmoiD cost Function 0.09409806662988984\n"
]
]
] | [
"code"
] | [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f8f108458553f4cbbf93aa0ef0b837df50479f | 46,727 | ipynb | Jupyter Notebook | WF_inpaint/WF_inpaint_realphantom_unet_train.ipynb | arsenal9971/DeeMicrolocalReconstruction | 0d5bbee86789d2c3acc6e9c872d270f46190d857 | [
"MIT"
] | null | null | null | WF_inpaint/WF_inpaint_realphantom_unet_train.ipynb | arsenal9971/DeeMicrolocalReconstruction | 0d5bbee86789d2c3acc6e9c872d270f46190d857 | [
"MIT"
] | 1 | 2021-12-08T18:40:54.000Z | 2021-12-09T13:56:04.000Z | WF_inpaint/WF_inpaint_realphantom_unet_train.ipynb | arsenal9971/DeepMicrolocalReconstruction | 0d5bbee86789d2c3acc6e9c872d270f46190d857 | [
"MIT"
] | null | null | null | 83.740143 | 13,212 | 0.779121 | [
[
[
"# <center> Wavefront set inpainting real phantom </center>",
"_____no_output_____"
],
[
"In this notebook we are implementing a Wavefront set inpainting algorithm based on a hallucination network",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport os\nos.environ[\"CUDA_VISIBLE_DEVICES\"]=\"0\"",
"_____no_output_____"
],
[
"# Import the needed modules\nfrom data.data_factory import generate_realphantom_WFinpaint, DataGenerator_realphantom_WFinpaint\nfrom ellipse.ellipseWF_factory import plot_WF \n\nimport matplotlib.pyplot as plt\nimport numpy.random as rnd\nimport numpy as np\nimport odl\nimport matplotlib.pyplot as plt",
"/store/kepler/datastore/andrade/GitHub_repos/Joint_CTWF_Recon/WF_inpaint/data/data_factory.py:7: UserWarning: \nThis call to matplotlib.use() has no effect because the backend has already\nbeen chosen; matplotlib.use() must be called *before* pylab, matplotlib.pyplot,\nor matplotlib.backends is imported for the first time.\n\nThe backend was *originally* set to 'module://ipykernel.pylab.backend_inline' by the following code:\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/runpy.py\", line 193, in _run_module_as_main\n \"__main__\", mod_spec)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/runpy.py\", line 85, in _run_code\n exec(code, run_globals)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel_launcher.py\", line 16, in <module>\n app.launch_new_instance()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/traitlets/config/application.py\", line 658, in launch_instance\n app.start()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/kernelapp.py\", line 505, in start\n self.io_loop.start()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/platform/asyncio.py\", line 132, in start\n self.asyncio_loop.run_forever()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/asyncio/base_events.py\", line 438, in run_forever\n self._run_once()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/asyncio/base_events.py\", line 1451, in _run_once\n handle._run()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/asyncio/events.py\", line 145, in _run\n self._callback(*self._args)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/ioloop.py\", line 758, in _run_callback\n ret = callback()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/stack_context.py\", line 300, in null_wrapper\n return fn(*args, **kwargs)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 1233, in inner\n self.run()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 1147, in run\n yielded = self.gen.send(value)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/kernelbase.py\", line 357, in process_one\n yield gen.maybe_future(dispatch(*args))\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 326, in wrapper\n yielded = next(result)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/kernelbase.py\", line 267, in dispatch_shell\n yield gen.maybe_future(handler(stream, idents, msg))\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 326, in wrapper\n yielded = next(result)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/kernelbase.py\", line 534, in execute_request\n user_expressions, allow_stdin,\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 326, in wrapper\n yielded = next(result)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/ipkernel.py\", line 294, in do_execute\n res = shell.run_cell(code, store_history=store_history, silent=silent)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/zmqshell.py\", line 536, in run_cell\n return super(ZMQInteractiveShell, self).run_cell(*args, **kwargs)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/IPython/core/interactiveshell.py\", line 2823, in run_cell\n self.events.trigger('post_run_cell', result)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/IPython/core/events.py\", line 88, in trigger\n func(*args, **kwargs)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/pylab/backend_inline.py\", line 164, in configure_once\n activate_matplotlib(backend)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/IPython/core/pylabtools.py\", line 314, in activate_matplotlib\n matplotlib.pyplot.switch_backend(backend)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/matplotlib/pyplot.py\", line 231, in switch_backend\n matplotlib.use(newbackend, warn=False, force=True)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/matplotlib/__init__.py\", line 1410, in use\n reload(sys.modules['matplotlib.backends'])\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/importlib/__init__.py\", line 166, in reload\n _bootstrap._exec(spec, module)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/matplotlib/backends/__init__.py\", line 16, in <module>\n line for line in traceback.format_stack()\n\n\n matplotlib.use('Agg')\n/store/kepler/datastore/andrade/GitHub_repos/Joint_CTWF_Recon/WF_inpaint/ellipse/ellipseWF_factory.py:9: UserWarning: \nThis call to matplotlib.use() has no effect because the backend has already\nbeen chosen; matplotlib.use() must be called *before* pylab, matplotlib.pyplot,\nor matplotlib.backends is imported for the first time.\n\nThe backend was *originally* set to 'module://ipykernel.pylab.backend_inline' by the following code:\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/runpy.py\", line 193, in _run_module_as_main\n \"__main__\", mod_spec)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/runpy.py\", line 85, in _run_code\n exec(code, run_globals)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel_launcher.py\", line 16, in <module>\n app.launch_new_instance()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/traitlets/config/application.py\", line 658, in launch_instance\n app.start()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/kernelapp.py\", line 505, in start\n self.io_loop.start()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/platform/asyncio.py\", line 132, in start\n self.asyncio_loop.run_forever()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/asyncio/base_events.py\", line 438, in run_forever\n self._run_once()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/asyncio/base_events.py\", line 1451, in _run_once\n handle._run()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/asyncio/events.py\", line 145, in _run\n self._callback(*self._args)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/ioloop.py\", line 758, in _run_callback\n ret = callback()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/stack_context.py\", line 300, in null_wrapper\n return fn(*args, **kwargs)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 1233, in inner\n self.run()\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 1147, in run\n yielded = self.gen.send(value)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/kernelbase.py\", line 357, in process_one\n yield gen.maybe_future(dispatch(*args))\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 326, in wrapper\n yielded = next(result)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/kernelbase.py\", line 267, in dispatch_shell\n yield gen.maybe_future(handler(stream, idents, msg))\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 326, in wrapper\n yielded = next(result)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/kernelbase.py\", line 534, in execute_request\n user_expressions, allow_stdin,\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tornado/gen.py\", line 326, in wrapper\n yielded = next(result)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/ipkernel.py\", line 294, in do_execute\n res = shell.run_cell(code, store_history=store_history, silent=silent)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/zmqshell.py\", line 536, in run_cell\n return super(ZMQInteractiveShell, self).run_cell(*args, **kwargs)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/IPython/core/interactiveshell.py\", line 2823, in run_cell\n self.events.trigger('post_run_cell', result)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/IPython/core/events.py\", line 88, in trigger\n func(*args, **kwargs)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/ipykernel/pylab/backend_inline.py\", line 164, in configure_once\n activate_matplotlib(backend)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/IPython/core/pylabtools.py\", line 314, in activate_matplotlib\n matplotlib.pyplot.switch_backend(backend)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/matplotlib/pyplot.py\", line 231, in switch_backend\n matplotlib.use(newbackend, warn=False, force=True)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/matplotlib/__init__.py\", line 1410, in use\n reload(sys.modules['matplotlib.backends'])\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/importlib/__init__.py\", line 166, in reload\n _bootstrap._exec(spec, module)\n File \"/homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/matplotlib/backends/__init__.py\", line 16, in <module>\n line for line in traceback.format_stack()\n\n\n matplotlib.use('Agg')\n"
]
],
[
[
"## Data generator",
"_____no_output_____"
]
],
[
[
"batch_size = 1\nsize = 256\nnClasses = 180\nlowd = 40",
"_____no_output_____"
],
[
"y_arr, x_true_arr =generate_realphantom_WFinpaint(batch_size, size, nClasses, lowd)",
"_____no_output_____"
],
[
"plt.figure(figsize=(6,6))\nplt.axis('off')\nplot_WF(y_arr[0,:,:,0])",
"_____no_output_____"
],
[
"plt.figure(figsize=(6,6))\nplt.axis('off')\nplot_WF(x_true_arr[0,:,:,0])",
"_____no_output_____"
]
],
[
[
"## Load the model",
"_____no_output_____"
]
],
[
[
"# Tensorflow and seed\nseed_value = 0\nimport random\nrandom.seed(seed_value)\nimport tensorflow as tf\ntf.set_random_seed(seed_value)\n\n# Importing relevant keras modules\nfrom tensorflow.keras.callbacks import ModelCheckpoint, CSVLogger\nfrom tensorflow.keras.models import load_model\nfrom shared.shared import create_increasing_dir\nimport pickle",
"_____no_output_____"
],
[
"# Import model and custom losses\nfrom models.unet import UNet\nfrom models.losses import CUSTOM_OBJECTS",
"_____no_output_____"
],
[
"# Parameters for the training\nlearning_rate = 1e-3\nloss = 'mae'\nbatch_size = 50\nepoches = 10000",
"_____no_output_____"
],
[
"pretrained = 1\npath_to_model_dir = './models/unets_realphantom_WFinpaint/training_5'",
"_____no_output_____"
],
[
"# Data generator\nsize = 256\nnClasses = 180\nlowd = 40\ntrain_gen = DataGenerator_realphantom_WFinpaint(batch_size, size, nClasses, lowd)\nval_gen = DataGenerator_realphantom_WFinpaint(batch_size, size, nClasses, lowd)",
"_____no_output_____"
],
[
"if pretrained==0:\n # Create a fresh model\n print(\"Create a fresh model\")\n unet = UNet()\n model = unet.create_model( img_shape = (size, size, 1) , loss = loss, learning_rate = learning_rate)\n path_to_training = create_increasing_dir('./models/unets_realphantom_WFinpaint', 'training')\n print(\"Save training in {}\".format(path_to_training))\n path_to_model_dir = path_to_training\n \nelse: \n print(\"Use trained model as initialization:\")\n print(path_to_model_dir+\"/weights.hdf5\")\n model = load_model(path_to_model_dir+\"/weights.hdf5\",\n custom_objects=CUSTOM_OBJECTS)\n path_to_training = path_to_model_dir",
"Use trained model as initialization:\n./models/unets_realphantom_WFinpaint/training_5/weights.hdf5\nWARNING:tensorflow:From /homes/extern/andrade/store/miniconda3/envs/tf_gpu/lib/python3.6/site-packages/tensorflow/python/ops/init_ops.py:86: calling VarianceScaling.__init__ (from tensorflow.python.ops.init_ops) with distribution=normal is deprecated and will be removed in a future version.\nInstructions for updating:\n`normal` is a deprecated alias for `truncated_normal`\n"
],
[
"# Callbacks for saving model\ncontext = {\n \"loss\": loss,\n \"batch_size\": batch_size,\n \"learning_rate\": learning_rate,\n \"path_to_model_dir\": path_to_model_dir,\n}\npath_to_context = path_to_training+'/context.log'\n\nwith open(path_to_context, 'wb') as dict_items_save:\n pickle.dump(context, dict_items_save)\nprint(\"Save training context to {}\".format(path_to_context))\n\n# Save architecture\nmodel_json = model.to_json()\npath_to_architecture = path_to_training + \"/model.json\"\nwith open(path_to_architecture, \"w\") as json_file:\n json_file.write(model_json)\nprint(\"Save model architecture to {}\".format(path_to_architecture))\n\n# Checkpoint for trained model\ncheckpoint = ModelCheckpoint(\n path_to_training+'/weights.hdf5',\n monitor='val_loss', verbose=1, save_best_only=True)\ncsv_logger = CSVLogger(path_to_training+'/training.log')\n\ncallbacks_list = [checkpoint, csv_logger]",
"Save training context to ./models/unets_realphantom_WFinpaint/training_5/context.log\nSave model architecture to ./models/unets_realphantom_WFinpaint/training_5/model.json\n"
],
[
"model.fit_generator(train_gen,epochs=epoches, steps_per_epoch=5600 // batch_size,\n callbacks=callbacks_list, validation_data=val_gen, validation_steps= 2000// batch_size)",
"Epoch 1/10000\n111/112 [============================>.] - ETA: 13s - loss: 0.9985 - my_mean_squared_error: 111.1464 - mean_squared_error: 111.1464 - mean_absolute_error: 0.9985 - l2_on_wedge: 107.8654 - my_psnr: -5.8870"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f8fd012687c827cbd9323a9d66c0e6df073d77 | 29,967 | ipynb | Jupyter Notebook | notebooks/07/07_vertical_vibration_of_a_quarter_car.ipynb | gbrault/resonance | bf66993a98fbbb857511f83bc072449b98f0b4c2 | [
"MIT"
] | 31 | 2017-11-10T16:44:04.000Z | 2022-01-13T12:22:02.000Z | notebooks/07/07_vertical_vibration_of_a_quarter_car.ipynb | gbrault/resonance | bf66993a98fbbb857511f83bc072449b98f0b4c2 | [
"MIT"
] | 178 | 2017-07-19T20:16:13.000Z | 2020-03-10T04:13:46.000Z | notebooks/07/07_vertical_vibration_of_a_quarter_car.ipynb | gbrault/resonance | bf66993a98fbbb857511f83bc072449b98f0b4c2 | [
"MIT"
] | 12 | 2018-04-05T22:58:43.000Z | 2021-01-14T04:06:26.000Z | 24.403094 | 369 | 0.534655 | [
[
[
"# 7. Vertical Vibration of Quarter Car Model\n\nThis notebook introduces the base excitation system by examning the behavior of a quarter car model.\n\nAfter the completion of this assignment students will be able to:\n\n- excite a system with a sinusoidal input\n- understand the difference in transient and steady state solutions\n- create a frequency response plot\n- define resonance and determine the parameters that cause resonance\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib notebook\nfrom resonance.linear_systems import SimpleQuarterCarSystem",
"_____no_output_____"
],
[
"sys = SimpleQuarterCarSystem()",
"_____no_output_____"
]
],
[
[
"The simple quarter car model has a suspension stiffness and damping, along with the sprung car mass in kilograms, and a travel speed parameter in meters per second.",
"_____no_output_____"
]
],
[
[
"sys.constants",
"_____no_output_____"
],
[
"sys.coordinates",
"_____no_output_____"
],
[
"sys.speeds",
"_____no_output_____"
]
],
[
[
"# A sinusoidal road\n\nThe road is described as:\n\n$$y(t) = Ysin\\omega_b t$$\n\nwhere $Y$ is the amplitude of the sinusoidal road undulations and $\\omega_b$ is the frequency of the a function of the car's speed. If the distance between the peaks (amplitude 0.01 meters) of the sinusoidal road is 6 meters and the car is traveling at 7.5 m/s calculate what the frequency will be.",
"_____no_output_____"
]
],
[
[
"Y = 0.01 # m\nv = sys.constants['travel_speed']\nbump_distance = 6 # m\nwb = v / bump_distance * 2 * np.pi # rad /s",
"_____no_output_____"
],
[
"print(wb)",
"_____no_output_____"
]
],
[
[
"Now with the amplitude and frequency set you can use the `sinusoidal_base_displacing_response()` function to simulate the system.",
"_____no_output_____"
]
],
[
[
"traj = sys.sinusoidal_base_displacing_response(Y, wb, 20.0)",
"_____no_output_____"
],
[
"traj.head()",
"_____no_output_____"
],
[
"traj.plot(subplots=True);",
"_____no_output_____"
]
],
[
[
"We've written an animation for you. You can play it with:",
"_____no_output_____"
]
],
[
[
"sys.animate_configuration(fps=20)",
"_____no_output_____"
]
],
[
[
"**Exercise**\n\nTry different travel speeds and see what kind of behavior you can observe. Make sure to set the `travel_speed` constant and the frequency value for `sinusoidal_base_displacing_response()` to be consistent.",
"_____no_output_____"
],
[
"# Transmissibility\n\nWhen designing a car the designer wants the riders to feel comfortable and to isolate them from the road's bumps. There are two important aspects to investigate. The first is called *displacement transmissibility* and is a ratio between the ampitude of the steady state motion and the ampitude of the sinusoidal base displacement. So in our case this would be:\n\n$$ \\frac{X}{Y}(\\omega_b) = \\frac{\\textrm{Steady State Amplitude}}{\\textrm{Base Displacement Amplitude}} $$\n\nThis can be plotted as a function of the base displacement frequency. A car suspension designer may want this ratio to be an optimal value for rider comfort. Maybe they'd like to make the ratio 1 or maybe even less than one if possible.\n\n**Exercise**\n\nUse the curve fitting technique from the previous notebook to plot $X/Y$ for a range of frequencies. Your code should look something like:\n\n```python\nfrom scipy.optimize import curve_fit\n\ndef cosine_func(times, amp, freq, phase_angle):\n return amp * np.cos(freq * times - phase_angle)\n\nfrequencies = np.linspace(1.0, 20.0, num=100)\n \namplitudes = []\n \nfor omega in frequencies:\n # your code here\n\namplitudes = np.array(amplitudes)\n\nfig, ax = plt.subplots(1, 1, sharex=True)\nax.set_xlabel('$\\omega_b$ [rad/s]')\nax.set_ylabel('Displacement Transmissibility') \n\nax.axvline(, color='black') # natural frequency\nax.plot()#?\nax.grid();\n```",
"_____no_output_____"
]
],
[
[
"from scipy.optimize import curve_fit\ndef cosine_func(times, amp, freq, phase_angle):\n return amp * np.cos(freq * times - phase_angle)\nfrequencies = np.linspace(1.0, 20.0, num=100)\n \namplitudes = []\n \nfor omega in frequencies:\n traj = sys.sinusoidal_base_displacing_response(Y, omega, 20.0)\n popt, pcov = curve_fit(cosine_func,\n traj[10:].index, traj[10:].car_vertical_position,\n p0=(Y, omega, 0.05))\n amplitudes.append(abs(popt[0]))\n\namplitudes = np.array(amplitudes)\n\nfig, ax = plt.subplots(1, 1, sharex=True)\nax.set_xlabel('$\\omega_b$ [rad/s]')\nax.set_ylabel('Displacement Transmissibility') \n\nax.axvline(np.sqrt(sys.constants['suspension_stiffness'] / sys.constants['sprung_mass']), color='black')\nax.plot(frequencies, amplitudes / Y)\nax.grid();",
"_____no_output_____"
],
[
"# write you answer here",
"_____no_output_____"
]
],
[
[
"The second thing to investigate is the *force transmissibility*. This is the ratio of the force applied by the suspension to the sprung car. Riders will feel this force when the car travels over bumps. Reducing this is also preferrable. The force applied to the car can be compared to the \n\n**Excersice**\n\nCreate a measurement to calculate the force applied to the car by the suspension. Simulate the system with $Y=0.01$ m, $v = 10$ m/s, and the distance between bump peaks as $6$ m. Plot the trajectories.\n\n```python\ndef force_on_car(suspension_damping, suspension_stiffness,\n car_vertical_position, car_vertical_velocity, travel_speed, time):\n # write this code\n\nsys.add_measurement('force_on_car', force_on_car)\n\n# write code for Y and omega_b, etc\n```",
"_____no_output_____"
]
],
[
[
"Y = 0.01 # m\nbump_distance = 6 # m\n\ndef force_on_car(suspension_damping, suspension_stiffness,\n car_vertical_position, car_vertical_velocity,\n travel_speed, time):\n \n wb = travel_speed / bump_distance * 2 * np.pi\n \n y = Y * np.sin(wb * time)\n yd = Y * wb * np.cos(wb * time)\n \n return (suspension_damping * (car_vertical_velocity - yd) +\n suspension_stiffness * (car_vertical_position - y))\n\nsys.add_measurement('force_on_car', force_on_car)\n\nv = 10.0\nsys.constants['travel_speed'] = v\nwb = v / bump_distance * 2 * np.pi # rad /s",
"_____no_output_____"
],
[
"traj = sys.sinusoidal_base_displacing_response(Y, wb, 10.0)\ntraj[['car_vertical_position', 'car_vertical_velocity', 'force_on_car']].plot(subplots=True)",
"_____no_output_____"
],
[
"# write your answer here",
"_____no_output_____"
],
[
"sys.animate_configuration(fps=30)",
"_____no_output_____"
]
],
[
[
"Force transmissibility will be visited more in your next homework.",
"_____no_output_____"
],
[
"# Arbitrary Periodic Forcing (Fourier Series)\n\nFourier discovered that any periodic function with a period $T$ can be described by an infinite series of sums of sines and cosines. See the wikipedia article for more info (https://en.wikipedia.org/wiki/Fourier_series). The key equation is this:\n\n$$ F(t) = \\frac{a_0}{2} + \\sum_{n=1}^\\infty (a_n \\cos n\\omega_T t + b_n \\sin n \\omega_T t)$$\n\nThe terms $a_0, a_n, b_n$ are called the Fourier coefficients and are defined as such:\n\n$$ a_0 = \\frac{2}{T} \\int_0^T F(t) dt$$\n\n$$ a_n = \\frac{2}{T} \\int_0^T F(t) \\cos n \\omega_T t dt \\quad \\textrm{for} \\quad n = 1, 2, \\ldots $$\n\n$$ b_n = \\frac{2}{T} \\int_0^T F(t) \\sin n \\omega_T t dt \\quad \\textrm{for} \\quad n = 1, 2, \\ldots $$\n\n\n## Introduction to SymPy\n\nSymPy is a Python package for symbolic computing. It can do many symbolic operations, for instance, integration, differentiation, linear algebra, etc. See http://sympy.org for more details of the features and the documentation. Today we will cover how to do integrals using SymPy and use it to find the Fourier series that represents a sawtooth function.",
"_____no_output_____"
]
],
[
[
"import sympy as sm",
"_____no_output_____"
]
],
[
[
"The function `init_printing()` enables LaTeX based rendering in the Jupyter notebook of all SymPy objects.",
"_____no_output_____"
]
],
[
[
"sm.init_printing()",
"_____no_output_____"
]
],
[
[
"Symbols can be created by using the `symbols()` function.",
"_____no_output_____"
]
],
[
[
"x, y, z = sm.symbols('x, y, z')",
"_____no_output_____"
],
[
"x, y, z",
"_____no_output_____"
]
],
[
[
"The `integrate()` function allows you to do symbolic indefinite or definite integrals. Note that the constants of integration are not included in indefinite integrals.",
"_____no_output_____"
]
],
[
[
"sm.integrate(x * y, x)",
"_____no_output_____"
]
],
[
[
"The `Integral` class creates and unevaluated integral, where as the `integrate()` function automatically evaluates the integral.",
"_____no_output_____"
]
],
[
[
"expr = sm.Integral(x * y, x)\nexpr",
"_____no_output_____"
]
],
[
[
"To evaluate the unevaluated form you call the `.doit()` method. Note that all unevaluated SymPy objects have this method.",
"_____no_output_____"
]
],
[
[
"expr.doit()",
"_____no_output_____"
]
],
[
[
"This shows how to create an unevaluated definite integral, store it in a variable, and then evaluate it.",
"_____no_output_____"
]
],
[
[
"expr = sm.Integral(x * y, (x, 0, 5))\nexpr",
"_____no_output_____"
],
[
"expr.doit()",
"_____no_output_____"
]
],
[
[
"# Fourier Coefficients for the Sawtooth function\n\nNow let's compute the Fourier coefficients for a saw tooth function. The function that describes the saw tooth is:\n\n$$\nF(t) = \n\\begin{cases} \n A \\left( \\frac{4t}{T} - 1 \\right) & 0 \\leq t \\leq T/2 \\\\\n A \\left( 3 - \\frac{4t}{t} \\right) & T/2 \\leq t \\leq T \n\\end{cases}\n$$\n\nwhere:\n\n- $A$ is the amplitude of the saw tooth\n- $T$ is the period of the saw tooth\n- $\\omega_T$ is the frequency of the saw tooth, i.e. $\\omega_T = \\frac{2\\pi}{T}$\n- $t$ is time\n\nThis is a piecewise function with two parts from $t=0$ to $t=T$.",
"_____no_output_____"
]
],
[
[
"A, T, wT, t = sm.symbols('A, T, omega_T, t', real=True, positive=True)\nA, T, wT, t",
"_____no_output_____"
]
],
[
[
"The first Fourier coefficient $a_0$ describes the average value of the periodic function. and is:\n\n$$a_0 = \\frac{2}{T} \\int_0^T F(t) dt$$\n\nThis integral will have to be done in two parts:\n\n$$a_0 = a_{01} + a_{02} = \\frac{2}{T} \\int_0^{T/2} F(t) dt + \\frac{2}{T} \\int_{T/2}^T F(t) dt$$\n\nThese two integrals are evaluated below. Note that $a_0$ evaluates to zero. This is because the average of our function is 0.",
"_____no_output_____"
]
],
[
[
"ao_1 = 2 / T * sm.Integral(A * (4 * t / T - 1), (t, 0, T / 2))\nao_1",
"_____no_output_____"
],
[
"ao_1.doit()",
"_____no_output_____"
],
[
"ao_2 = 2 / T * sm.Integral(A * (3 - 4 * t / T), (t, T / 2, T))\nao_2",
"_____no_output_____"
],
[
"ao_2.doit()",
"_____no_output_____"
]
],
[
[
"But SymPy can also handle piecewise directly. The following shows how to define a piecewise function.",
"_____no_output_____"
]
],
[
[
"F_1 = A * (4 * t / T - 1)\nF_2 = A * (3 - 4 * t / T)",
"_____no_output_____"
],
[
"F = sm.Piecewise((F_1, t<=T/2),\n (F_2, T/2<t))\nF",
"_____no_output_____"
],
[
"F_of_t_only = F.xreplace({A: 0.01, T: 2 * sm.pi / wb})\nF_of_t_only",
"_____no_output_____"
],
[
"sm.plot(F_of_t_only, (t, 0, 2 * np.pi / wb))",
"_____no_output_____"
]
],
[
[
"The integral can be taken of the entire piecewise function in one call.",
"_____no_output_____"
]
],
[
[
"sm.integrate(F, (t, 0, T))",
"_____no_output_____"
]
],
[
[
"Now the Fourier coefficients $a_n$ and $b_n$ can be computed.\n\n$$\na_n = \\frac{2}{T}\\int_0^T F(t) \\cos n\\omega_Tt dt \\\\\nb_n = \\frac{2}{T}\\int_0^T F(t) \\sin n\\omega_Tt dt\n$$",
"_____no_output_____"
]
],
[
[
"n = sm.symbols('n', real=True, positive=True)",
"_____no_output_____"
]
],
[
[
"For $a_n$:",
"_____no_output_____"
]
],
[
[
"an = 2 / T * sm.Integral(F * sm.cos(n * wT * t), (t, 0, T))\nan",
"_____no_output_____"
],
[
"an.doit()",
"_____no_output_____"
]
],
[
[
"This can be simplified:",
"_____no_output_____"
]
],
[
[
"an = an.doit().simplify()\nan",
"_____no_output_____"
]
],
[
[
"Now substitute the $2\\pi/T$ for $\\omega_T$.",
"_____no_output_____"
]
],
[
[
"an = an.subs({wT: 2 * sm.pi / T})\nan",
"_____no_output_____"
]
],
[
[
"Let's see how this function varies with increasing $n$. We will use a loop but the SymPy expressions will not automatically display because they are inside a loop. So we need to use SymPy's `latex()` function and the IPython display tools. SymPy's `latex()` function transforms the SymPy expression into a string of matching LaTeX commands.",
"_____no_output_____"
]
],
[
[
"sm.latex(an, mode='inline')",
"_____no_output_____"
]
],
[
[
"The `display()` and `LaTeX()` functions then turn the LaTeX string in to a displayed version.",
"_____no_output_____"
]
],
[
[
"from IPython.display import display, Latex ",
"_____no_output_____"
]
],
[
[
"Now we can see how $a_n$ varies with $n=1,2,\\ldots$.",
"_____no_output_____"
]
],
[
[
"for n_i in range(1, 6):\n ans = an.subs({n: n_i})\n display(Latex('$a_{} = $'.format(n_i) + sm.latex(ans, mode='inline')))",
"_____no_output_____"
]
],
[
[
"For even $n$ values the coefficient is zero and for even values it varies with the inverse of $n^2$. More precisely:\n\n$$\na_n =\n\\begin{cases}\n0 & \\textrm{if }n\\textrm{ is even} \\\\\n-\\frac{8A}{n^2\\pi^2} & \\textrm{if }n\\textrm{ is odd}\n\\end{cases}\n$$\n\nSymPy can actually reduce this further if your set the assumption that $n$ is an integer.",
"_____no_output_____"
]
],
[
[
"n = sm.symbols('n', real=True, positive=True, integer=True)\nan = 2 / T * sm.Integral(F * sm.cos(n * wT * t), (t, 0, T))\nan = an.doit().simplify()\nan.subs({wT: 2 * sm.pi / T})",
"_____no_output_____"
]
],
[
[
"The odd and even versions can be computed by setting the respective assumptions.",
"_____no_output_____"
]
],
[
[
"n = sm.symbols('n', real=True, positive=True, integer=True, odd=True)\nan = 2 / T * sm.Integral(F * sm.cos(n * wT * t), (t, 0, T))\nan = an.doit().simplify()\nan.subs({wT: 2 * sm.pi / T})",
"_____no_output_____"
]
],
[
[
"Note that $b_n$ is always zero:",
"_____no_output_____"
]
],
[
[
"bn = 2 / T * sm.Integral(F * sm.sin(n * wT * t), (t, 0, T))\nbn",
"_____no_output_____"
],
[
"bn.doit().simplify().subs({wT: 2 * sm.pi / T})",
"_____no_output_____"
]
],
[
[
"# Numerical evalution of the Fourier Series\n\nNow the Fourier coefficients can be used to plot the approximation of the saw tooth forcing function.",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
]
],
[
[
"The following function plots the actual sawtooth function. It does it all in one line by cleverly using the absolute value and the modulo functions.",
"_____no_output_____"
]
],
[
[
"def sawtooth(A, T, t):\n return (4 * A / T) * (T / 2 - np.abs(t % T - T / 2) ) - A",
"_____no_output_____"
],
[
"A = 1\nT = 2\nt = np.linspace(0, 5, num=500)",
"_____no_output_____"
],
[
"plt.figure()\n\nplt.plot(t, sawtooth(A, T, t));",
"_____no_output_____"
]
],
[
[
"# Exercise\n\nWrite a function that computes the Fourier approximation of the sawtooth function for a given value of $n$, i.e. using a finite number of terms. Then plot it for $n=2, 4, 6, 8, 10$ on top of the actual sawtooth function. How many terms of the infinite series are needed to get a good sawtooth?\n\n```python\ndef sawtooth_approximation(n, A, T, t):\n # code here\n return f\n\n# plot sawtooth\nf = sawtooth(A, T, t)\n\nplt.figure()\nplt.plot(t, f, color='k', label='true sawtooth')\n\nfor n in np.arange(2, 12, 2):\n f_approx = sawtooth_approximation(n, A, T, t)\n plt.plot(t, f_approx, label='n = {}'.format(n))\n\nplt.legend()\n# zoom in a bit on the interesting bit\nplt.xlim(0, T)\n```",
"_____no_output_____"
]
],
[
[
"def sawtooth_approximation(n, A, T, t):\n # odd values of indexing variable up to n\n n = np.arange(1, n+1)[:, np.newaxis]\n # cos coefficients\n an = A *(8 * (-1)**n - 8) / 2 / np.pi**2 / n**2\n # sawtooth frequency\n wT = 2 * np.pi / T\n # sum of n cos functions\n f = np.sum(an * np.cos(n * wT * t), axis=0)\n return f\n\n# plot sawtooth\nf = sawtooth(A, T, t)\n\nplt.figure()\nplt.plot(t, f, color='k', label='true sawtooth')\n\nfor n in np.arange(2, 12, 2):\n f_approx = sawtooth_approximation(n, A, T, t)\n plt.plot(t, f_approx, label='n = {}'.format(n))\n\nplt.legend()\n# zoom in a bit on the interesting bit\nplt.xlim(0, T)",
"_____no_output_____"
],
[
"# write answer here",
"_____no_output_____"
]
],
[
[
"Below is a interactive plot that shows the same thing as above.",
"_____no_output_____"
]
],
[
[
"A = 1\nT = 2\nt = np.linspace(0, 5, num=500)\n\nfig, ax = plt.subplots(1, 1)\n\nf = sawtooth(A, T, t)\n\nsaw_tooth_lines = ax.plot(t, f, color='k')\n\nn = 2\n\nf_approx = sawtooth_approximation(n, A, T, t)\n\napprox_lines = ax.plot(t, f_approx)\n\nleg = ax.legend(['true', 'approx, n = {}'.format(n)])\n\n# zoom in a bit on the interesting bit\nplt.xlim(0, 2 * T)\n\ndef update(n=0):\n f_approx = sawtooth_approximation(n, A, T, t)\n approx_lines[0].set_ydata(f_approx)\n leg.get_texts()[1].set_text('approx, n = {}'.format(n))",
"_____no_output_____"
],
[
"from ipywidgets import interact\n\ninteract(update, n=(0, 20, 2))",
"_____no_output_____"
]
],
[
[
"# Apply the sawtooth to the quarter car\n\nNow that you know the Fourier series coefficients. Calculate them for a suitable number of terms and simulate them with the `sys.periodic_base_displacing_response()` function.\n\nYour code should look something like:\n\n```python\ndef fourier_coeffs(A, T, N):\n # write your code here\n\na0, an, bn = fourier_coeffs(?)\n\ntraj = sys.periodic_base_displacing_response(?)\n```",
"_____no_output_____"
]
],
[
[
"def fourier_coeffs(A, T, N):\n n = np.arange(1, N+1)\n an = A *(8 * (-1)**n - 8) / 2 / np.pi**2 / n**2\n return 0, an, np.zeros_like(an)\n\na0, an, bn = fourier_coeffs(0.01, 2 * np.pi / wb, 100)\n\ntraj = sys.periodic_base_displacing_response(a0, an, bn, wb, 20.0)",
"_____no_output_____"
],
[
"traj.plot(subplots=True)",
"_____no_output_____"
],
[
"sys.animate_configuration(fps=30)",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
e7f90b1e5ca461e9e572e40add386cda62becb10 | 386,741 | ipynb | Jupyter Notebook | Codes/2.2 Remove redundant words - SVM,KNN,Kmeans_v2.ipynb | matchlesswei/application_project_nlp_company_description | b3fbea81df0c5c0e7922ae08d2e34974614d903b | [
"MIT"
] | null | null | null | Codes/2.2 Remove redundant words - SVM,KNN,Kmeans_v2.ipynb | matchlesswei/application_project_nlp_company_description | b3fbea81df0c5c0e7922ae08d2e34974614d903b | [
"MIT"
] | null | null | null | Codes/2.2 Remove redundant words - SVM,KNN,Kmeans_v2.ipynb | matchlesswei/application_project_nlp_company_description | b3fbea81df0c5c0e7922ae08d2e34974614d903b | [
"MIT"
] | 2 | 2019-10-18T08:41:21.000Z | 2019-10-18T09:51:54.000Z | 523.330176 | 101,024 | 0.937387 | [
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer\nfrom sklearn import svm\nfrom sklearn.neighbors import KNeighborsClassifier\nimport seaborn as sn\nimport matplotlib.pyplot as plt\nimport matplotlib.cm as cm\n",
"_____no_output_____"
]
],
[
[
"### Define the data_directory of preprocessed data",
"_____no_output_____"
]
],
[
[
"data_directory = \"C:/Users/kwokp/OneDrive/Desktop/Study/zzz_application project/Final/data_after_preprocessing.csv\"",
"_____no_output_____"
]
],
[
[
"### We devide the data into 3 groups:\n* Group 1: full data\n* Group 2: data with four large categories which have more than 1000 companies each\n* Group 3: seven categories of data, number of companies in each category is same but small\n\n### In the function selectGroup, giving 1, 2 or 3 as input parameter to selet the relevant data for experiment",
"_____no_output_____"
]
],
[
[
"# read the data from directory, then select the group \n# of data we want to process.\ndef selectGroup(directory, group_nr):\n data = pd.read_csv(directory, sep='\\t')\n if group_nr == 1:\n return data\n if group_nr == 2:\n df_healthcare_group=data[data['Category'] == 'HEALTHCARE GROUP'].sample(n=1041,replace=False)\n df_business_financial_services=data[data['Category'] == 'BUSINESS & FINANCIAL SERVICES'].sample(n=1041,replace=False)\n df_consumer_service_group=data[data['Category'] == 'CONSUMER SERVICES GROUP'].sample(n=1041,replace=False)\n df_information_technology_group=data[data['Category'] == 'INFORMATION TECHNOLOGY GROUP'].sample(n=1041,replace=False)\n df_clean = pd.concat([df_healthcare_group, df_business_financial_services,df_consumer_service_group,df_information_technology_group])\n return df_clean.sample(frac=1)\n if group_nr == 3:\n df_healthcare_group=data[data['Category'] == 'HEALTHCARE GROUP'].sample(n=219,replace=False)\n df_business_financial_services=data[data['Category'] == 'BUSINESS & FINANCIAL SERVICES'].sample(n=219,replace=False)\n df_consumer_service_group=data[data['Category'] == 'CONSUMER SERVICES GROUP'].sample(n=219,replace=False)\n df_information_technology_group=data[data['Category'] == 'INFORMATION TECHNOLOGY GROUP'].sample(n=219,replace=False)\n df_industry_goods=data[data['Category'] == 'INDUSTRIAL GOODS & MATERIALS GROUP'].sample(n=219,replace=False)\n df_consumer_goods=data[data['Category'] == 'CONSUMER GOODS GROUP'].sample(n=219,replace=False)\n df_energy=data[data['Category'] == 'ENERGY & UTILITIES GROUP'].sample(n=219,replace=False)\n df_clean = pd.concat([df_healthcare_group, df_business_financial_services,df_consumer_service_group,df_information_technology_group,df_industry_goods,df_consumer_goods,df_energy])\n return df_clean.sample(frac=1)",
"_____no_output_____"
],
[
"# use tf-idf methode to generate scores for each company\ndef tf_idf_func(df_document, max_features):\n feature_extraction = TfidfVectorizer(max_features = max_features, stop_words = 'english')\n score_matrix = feature_extraction.fit_transform(df_document.values)\n return score_matrix, feature_extraction",
"_____no_output_____"
],
[
"# get the top_n words\ndef get_top_keywords(scores_matrix, clusters, labels, n_terms):\n df = pd.DataFrame(scores_matrix.todense()).groupby(clusters).mean()\n \n for i,r in df.iterrows():\n print('\\nCluster {}'.format(i))\n print(','.join([labels[t] for t in np.argsort(r)[-n_terms:]])) \n\n# get the top_n words with highest tf-idf scores in each category, and count the word occurence\ndef get_top_keywords_with_frequence(Top_N, score_matrix, df_data, feature_extraction):\n df = pd.DataFrame(score_matrix.todense()) #read tf-idf score-matrix, each line is vectors for each company, each column matches each word\n df['Category'] = df_data['Category'] #assign the category for each line(company) in score-matrix\n dfg = df.groupby(['Category']).mean() #calculate the mean score of each word in each cateogry\n\n labels = feature_extraction.get_feature_names()\n\n categories = df_data['Category'].unique()\n col_names = ['Category', 'Top_N', 'Score']\n df_top = pd.DataFrame(columns = col_names)\n\n Dict = {}\n\n for i,r in dfg.iterrows(): #i-index(category), r-row, iterate the average score matrix of each category\n category = i \n top_series = np.argsort(r)[-Top_N:]#find the location of top_n words\n label_series = top_series.apply(lambda x: labels[x]) #find top_n words with best scores in each category\n top_scores = np.sort(r)[-Top_N:] #find the scores corresponding with top_n words\n df_each = pd.DataFrame({'Category':category,'Top_N':label_series,'Score':top_scores})\n df_top = df_top.append(df_each, ignore_index = True)\n for key in label_series: #count how often each word appears in the top_n\n if key in Dict:\n Dict[key] = Dict[key]+1\n else:\n Dict[key] = 1\n \n df_reshape = df_top.pivot(index='Top_N', columns='Category') #reformat the top-n score matrix\n sortedDict = sorted(Dict.items(), key=lambda x: x[1]) #sort the dictionary\n \n return sortedDict",
"_____no_output_____"
],
[
"# convert the input of the top_n words with their occurence in each category, to a list of stopwords, \n# if the occurence is larger than the given occurence\ndef get_word_occurence_stopwordslist(max_occurence, dict_list):\n word = []\n occurence = []\n frequent_stopwords = []\n for key, value in dict_list:\n word.append(key)\n occurence.append(value)\n if value > max_occurence: # if the occurence is larger than the given occurence\n frequent_stopwords.append(key) # store to a list of stopwords\n return word, occurence, frequent_stopwords",
"_____no_output_____"
],
[
"#remove the words from a sentence, which is in the stopwords\ndef remove_frequent_stopwords(sentences, frequent_stopwords):\n splitted_string = sentences.split()\n remove_stopwords = [w for w in splitted_string if not w in frequent_stopwords]\n return ' '.join(remove_stopwords)\n\n#remove the words from the website content, which is in the stopwords\n#update the tf-idf score matrix for the whole corpus\ndef remove_frequent_stopwords_and_get_updated_tfidfscore(data, feature_extraction, top_n, frequent_stopwords):\n df_update = data['clean'].apply(lambda x: remove_frequent_stopwords(x, frequent_stopwords))\n score_matrix_update = feature_extraction.fit_transform(df_update.values)\n return score_matrix_update",
"_____no_output_____"
]
],
[
[
"### List Occurence of words in Top 50 Keywords in Categories",
"_____no_output_____"
]
],
[
[
"#visualize top_n words with occurence \ndef visulaze_topwords_occurence(top_n, word_list, occurence_list):\n objects = word_list\n y_pos = np.arange(len(word_list))\n performance = occurence_list\n plt.figure(figsize=(10,24))\n plt.barh(y_pos, performance, align='center', alpha=0.5)\n plt.yticks(y_pos, objects)\n plt.xlabel('Occurence')\n plt.title('Occurence of words in Top ' + str(top_n) + ' Keywords in categories')\n plt.show()",
"_____no_output_____"
],
[
"data = selectGroup(data_directory, 1)\nscore_matrix, feature_extraction = tf_idf_func(data['clean'], 8000)\nsortedDict = get_top_keywords_with_frequence(50, score_matrix, data, feature_extraction)\nword, occurence, _ = get_word_occurence_stopwordslist(1, sortedDict)\nvisulaze_topwords_occurence(50, word, occurence)",
"_____no_output_____"
]
],
[
[
"### We remove the redundunt words which appears in multiple category . Main steps are as follows:\n1. select the group of data to do the test\n2. generate TF-IDF score matrix\n3. get the top 50 words in each category\n4. find the words which appears in more than one category's top-50 words, set them as stopwords\n5. remove these stopwords and update the tf-idf score matrix\n6. count and calculate the word occurences in each company's website\n7. plot the number of valid words in each website\n8. remove the website which has less than 200 words\n\n### We may notice there are quite a few companies which has less than 200 words. These websites could be useless. And the category distrubtion after processing is shown as the reuslt of the cell.",
"_____no_output_____"
]
],
[
[
"#get the data, remove the frequent words which appear in more than one category, and update the tf-idf score matrix\ndata = selectGroup(data_directory, 1)\nscore_matrix, feature_extraction = tf_idf_func(data['clean'], 8000)\nsortedDict = get_top_keywords_with_frequence(50, score_matrix, data, feature_extraction)\n_, _, frequent_stopwords = get_word_occurence_stopwordslist(1, sortedDict)\nscore_matrix_update = remove_frequent_stopwords_and_get_updated_tfidfscore(data, feature_extraction, 10, frequent_stopwords)\n\n#show the top keywords of the rest words after removing the frequent words which appear in more than one category\nget_top_keywords(score_matrix_update, data['Category'].values, feature_extraction.get_feature_names(), 10)\n\n# count the non-zero words from updated tf-idf score matrix and display the non-zero word count in each company website\nscore_value = score_matrix_update.todense()\nwebsite_word_count=np.asarray(np.count_nonzero(score_value, axis=1)).reshape(-1)\nplt.hist(website_word_count, bins = 30)\nplt.xlabel('number of words in the whole website')\nplt.ylabel('number of websites')\nplt.title('Distribution of number of words in the websites')\n\ndf_score=pd.DataFrame(score_value)\ndf_score.columns=feature_extraction.get_feature_names()\ndf_score['Keep']=website_word_count>200\ndf_score['Category'] = data['Category'].reset_index(drop=True)\ndf_score_valid = df_score[df_score['Keep']]\ndf_score_valid['Category'].value_counts()",
"\nCluster BUSINESS & FINANCIAL SERVICES\nlearn,agreement,need,insurance,media,experience,financial,companies,clients,marketing\n\nCluster CONSUMER GOODS GROUP\nread,sites,address,brand,organic,home,shipping,ingredients,foods,food\n\nCluster CONSUMER SERVICES GROUP\nexperience,media,sites,world,address,parties,people,day,agreement,agree\n\nCluster ENERGY & UTILITIES GROUP\nllc,basin,electricity,wind,drilling,renewable,fuel,power,oil,solar\n\nCluster HEALTHCARE GROUP\ndr,treatment,cancer,healthcare,patient,care,health,clinical,patients,medical\n\nCluster INDUSTRIAL GOODS & MATERIALS GROUP\nindustries,process,range,industrial,parts,aerospace,materials,steel,packaging,manufacturing\n\nCluster INFORMATION TECHNOLOGY GROUP\napplication,world,performance,experience,need,learn,enterprise,domain,solution,network\n"
]
],
[
[
"### Split the data 80% for training and 20% for testing",
"_____no_output_____"
]
],
[
[
"df_final = df_score_valid[df_score_valid.columns.difference(['Keep', 'Category'])] #remove columns'Keep' and 'Category'\n\ndf_category = df_score_valid['Category'].reset_index(drop=True)\n\nmsk = np.random.rand(len(df_final)) < 0.8\ntrain_x = np.nan_to_num(df_final[msk])\ntest_x = np.nan_to_num(df_final[~msk])\ntrain_y = df_category[msk].to_numpy()\ntest_y = df_category[~msk].to_numpy()",
"_____no_output_____"
]
],
[
[
"### Perform Linear SVM",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report, confusion_matrix, accuracy_score\n\n#use svm classifier to classify TF-IDF of each website\ndef linear_svc_classifier(train_x, train_y, test_x, test_y):\n print(\"start svm\")\n classifier_svm = svm.LinearSVC()\n classifier_svm.fit(train_x, train_y)\n predictions = classifier_svm.predict(test_x)\n \n print(confusion_matrix(test_y, predictions))\n print(classification_report(test_y, predictions))\n print(accuracy_score(test_y, predictions))\n \n array = confusion_matrix(test_y, predictions)\n \n \n y_true = [\"BUSINESS & FINANCIAL SERVICES\", \"CONSUMER GOODS GROUP\", \"CONSUMER SERVICES GROUP\", \"ENERGY & UTILITIES GROUP\",\"HEALTHCARE GROUP\", \"INDUSTRIAL GOODS & MATERIALS GROUP\",\"INFORMATION TECHNOLOGY GROUP\"]\n \n y_pred = y_true\n df_cm = pd.DataFrame(array, y_true, y_pred)\n df_cm.index.name = 'Actual'\n df_cm.columns.name = 'Predicted'\n plt.figure(figsize = (10,7))\n #sn.set(font_scale=1.4)#for label size\n ax=sn.heatmap(df_cm, cmap=\"Blues\", annot=True, fmt='d',annot_kws={\"size\": 16})# font size\n bottom, top=ax.get_ylim()\n ax.set_ylim(bottom+0.5, top-0.5)\n ax.tick_params(labelsize=10) \n plt.show()\n \n return confusion_matrix(test_y, predictions),predictions\n\nconfusion_matrix, predictions = linear_svc_classifier(train_x, train_y, test_x, test_y)",
"start svm\n[[145 4 24 2 3 5 66]\n [ 4 28 16 0 3 7 5]\n [ 24 7 115 1 8 2 30]\n [ 6 0 0 21 0 2 3]\n [ 8 5 6 1 135 4 15]\n [ 15 3 6 3 1 45 9]\n [ 68 4 32 1 6 9 225]]\n precision recall f1-score support\n\n BUSINESS & FINANCIAL SERVICES 0.54 0.58 0.56 249\n CONSUMER GOODS GROUP 0.55 0.44 0.49 63\n CONSUMER SERVICES GROUP 0.58 0.61 0.60 187\n ENERGY & UTILITIES GROUP 0.72 0.66 0.69 32\n HEALTHCARE GROUP 0.87 0.78 0.82 174\nINDUSTRIAL GOODS & MATERIALS GROUP 0.61 0.55 0.58 82\n INFORMATION TECHNOLOGY GROUP 0.64 0.65 0.64 345\n\n accuracy 0.63 1132\n macro avg 0.64 0.61 0.62 1132\n weighted avg 0.64 0.63 0.63 1132\n\n0.6307420494699647\n"
]
],
[
[
"### Perform KNN with 5 Neighbours",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report, confusion_matrix, accuracy_score\n\n#use knn classifier to classify TF-IDF of each website\ndef knn_classifier(x_train, y_train, x_test, y_test):\n print(\"start knn\")\n modelknn = KNeighborsClassifier(n_neighbors=5)\n modelknn.fit(x_train, y_train)\n predictions = modelknn.predict(x_test)\n\n print(confusion_matrix(y_test, predictions))\n print(classification_report(y_test, predictions))\n print(accuracy_score(y_test, predictions))\n \n array = confusion_matrix(y_test, predictions)\n \n y_true = [\"INDUSTRIAL GOODS & MATERIALS GROUP\", \"CONSUMER SERVICES GROUP\",\"CONSUMER GOODS GROUP\",\"INFORMATION TECHNOLOGY GROUP\",\"ENERGY & UTILITIES GROUP\",\"BUSINESS & FINANCIAL SERVICES\", \"HEALTHCARE GROUP\"]\n \n y_pred = y_true\n df_cm = pd.DataFrame(array, y_true, y_pred)\n df_cm.index.name = 'Actual'\n df_cm.columns.name = 'Predicted'\n plt.figure(figsize = (10,7))\n #sn.set(font_scale=1.4)#for label size\n ax=sn.heatmap(df_cm, cmap=\"Greens\", annot=True, fmt='d',annot_kws={\"size\": 16})# font size\n bottom, top=ax.get_ylim()\n ax.set_ylim(bottom+0.5, top-0.5)\n ax.tick_params(labelsize=10) \n plt.show()\n\n return confusion_matrix(test_y, predictions),predictions\n\nconfusion_matrix, predictions = knn_classifier(train_x, train_y, test_x, test_y)",
"start knn\n[[153 7 22 4 5 4 54]\n [ 8 25 19 1 3 2 5]\n [ 33 17 89 1 12 2 33]\n [ 10 1 0 16 2 0 3]\n [ 18 5 8 0 133 3 7]\n [ 21 4 4 4 2 34 13]\n [106 6 40 2 8 7 176]]\n precision recall f1-score support\n\n BUSINESS & FINANCIAL SERVICES 0.44 0.61 0.51 249\n CONSUMER GOODS GROUP 0.38 0.40 0.39 63\n CONSUMER SERVICES GROUP 0.49 0.48 0.48 187\n ENERGY & UTILITIES GROUP 0.57 0.50 0.53 32\n HEALTHCARE GROUP 0.81 0.76 0.78 174\nINDUSTRIAL GOODS & MATERIALS GROUP 0.65 0.41 0.51 82\n INFORMATION TECHNOLOGY GROUP 0.60 0.51 0.55 345\n\n accuracy 0.55 1132\n macro avg 0.56 0.53 0.54 1132\n weighted avg 0.57 0.55 0.56 1132\n\n0.5530035335689046\n"
]
],
[
[
"### Perform K means and Plot SSE, PCA and TSNE",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import MiniBatchKMeans\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom sklearn.decomposition import PCA\nfrom sklearn.manifold import TSNE\nimport matplotlib.cm as cm\nimport itertools\n\n#Find the optimal clusters from 2 to maximum of clusters of data group, plot respective SSE.\ndef find_optimal_clusters(data, max_k):\n iters = range(2, max_k+1, 2)\n \n sse = []\n for k in iters:\n sse.append(MiniBatchKMeans(n_clusters=k, init_size=512, batch_size=1024, random_state=20).fit(data).inertia_)\n print('Fit {} clusters'.format(k))\n \n f, ax = plt.subplots(1, 1)\n ax.plot(iters, sse, marker='o')\n ax.set_xlabel('Cluster Centers')\n ax.set_xticks(iters)\n ax.set_xticklabels(iters)\n ax.set_ylabel('SSE')\n ax.set_title('SSE by Cluster Center Plot')\n\n#Plot TSNE and PCA for the clusters \ndef plot_tsne_pca(data, labels):\n max_label = max(labels+1)\n max_items = np.random.choice(range(data.shape[0]), size=3000, replace=False)\n \n pca = PCA(n_components=2).fit_transform(data[max_items,:].todense())\n tsne = TSNE().fit_transform(PCA(n_components=50).fit_transform(data[max_items,:].todense()))\n \n \n idx = np.random.choice(range(pca.shape[0]), size=300, replace=False)\n label_subset = labels[max_items]\n label_subset = [cm.hsv(i/max_label) for i in label_subset[idx]]\n \n f, ax = plt.subplots(1, 2, figsize=(14, 6))\n \n ax[0].scatter(pca[idx, 0], pca[idx, 1], c=label_subset)\n ax[0].set_title('PCA Cluster Plot')\n \n ax[1].scatter(tsne[idx, 0], tsne[idx, 1], c=label_subset)\n ax[1].set_title('TSNE Cluster Plot')\n\n#Calculate the accuracy of the clustered data and actual label \ndef Calculate_accuracy(clusters, actual_label):\n count = 0\n for index, cluster in enumerate(clusters):\n if cluster==actual_label[index]:\n count+=1\n accuracy = count/len(clusters)*1.0\n return accuracy\n\n#Assign clisters for the clustered data\ndef assign_clusters(original_label, permu, nr_group):\n if nr_group == 2:\n categories = [\"BUSINESS & FINANCIAL SERVICES\", \"CONSUMER SERVICES GROUP\", \"HEALTHCARE GROUP\", \"INFORMATION TECHNOLOGY GROUP\"]\n else:\n categories = [\"INDUSTRIAL GOODS & MATERIALS GROUP\", \"CONSUMER SERVICES GROUP\",\"CONSUMER GOODS GROUP\",\"INFORMATION TECHNOLOGY GROUP\",\"ENERGY & UTILITIES GROUP\",\"BUSINESS & FINANCIAL SERVICES\", \"HEALTHCARE GROUP\"]\n mydict=dict(zip(categories, permu))\n actual_label = np.zeros(len(original_label))\n for index, label in enumerate(original_label):\n actual_label[index] = mydict[label]\n return actual_label\n\n#Perform Kmeans and Plot\ndef kmeans_classifier(score_matrix_update, nr_group):\n if nr_group == 2:\n nr_cluster = 4\n else:\n nr_cluster = 7\n find_optimal_clusters(score_matrix_update, nr_cluster)\n clusters = MiniBatchKMeans(n_clusters=nr_cluster, init_size=512, batch_size=1024, random_state=20).fit_predict(score_matrix_update)\n plot_tsne_pca(score_matrix_update, clusters)\n get_top_keywords(score_matrix_update, clusters, feature_extraction.get_feature_names(), 10)\n\n if nr_group == 2:\n numbers=[0,1,2,3]\n else:\n numbers = [0,1,2,3,4,5,6]\n permu = list(itertools.permutations(numbers))\n best_accuracy = 0\n for i in range(len(permu)):\n actual_label = assign_clusters(data['Category'].values, permu[i], nr_group)\n accuracy = Calculate_accuracy(clusters, actual_label)\n if best_accuracy<accuracy:\n best_accuracy=accuracy\n final_label = actual_label\n category = permu[i]\n else: \n best_accuracy=best_accuracy\n\n print(category)\n #print(final_label)\n print(\"The Accuracy is \" + str(round(best_accuracy,2)))\n\nkmeans_classifier(score_matrix_update, 1) ",
"Fit 2 clusters\nFit 4 clusters\nFit 6 clusters\n\nCluster 0\ndevices,storage,application,performance,networks,infrastructure,enterprise,solution,wireless,network\n\nCluster 1\nreserved,click,read,learn,world,copyright,need,home,wordpress,domain\n\nCluster 2\ndr,healthcare,treatment,cancer,care,health,patient,medical,clinical,patients\n\nCluster 3\nmeal,day,fresh,coffee,delicious,cheese,restaurant,foods,ingredients,food\n\nCluster 4\nhttp,window,archive,width,function,document,gform,jquery,px,var\n\nCluster 5\npeople,group,market,global,years,financial,companies,experience,marketing,clients\n\nCluster 6\naddress,provided,applicable,sites,websites,law,collect,parties,agree,agreement\n(4, 6, 3, 1, 0, 5, 2)\nThe Accuracy is 0.34\n"
]
]
] | [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
e7f933345b94e18be1e035c23a42baf4f35a9dfc | 1,892 | ipynb | Jupyter Notebook | Carpet.ipynb | saroele/jouleboulevard | e06d4548395147d3a4f9842422d8142c2b97a8e2 | [
"Apache-2.0"
] | null | null | null | Carpet.ipynb | saroele/jouleboulevard | e06d4548395147d3a4f9842422d8142c2b97a8e2 | [
"Apache-2.0"
] | null | null | null | Carpet.ipynb | saroele/jouleboulevard | e06d4548395147d3a4f9842422d8142c2b97a8e2 | [
"Apache-2.0"
] | null | null | null | 21.5 | 103 | 0.5 | [
[
[
"# Carpetplots",
"_____no_output_____"
]
],
[
[
"import opengrid as og\nfrom opengrid.library import plotting as og_plot\nimport pandas as pd\nfrom joule import meta, filter_meta",
"_____no_output_____"
],
[
"plt = og.plot_style()\n#%matplotlib notebook",
"_____no_output_____"
],
[
"#%matplotlib notebook",
"_____no_output_____"
],
[
"for building in meta['RecordNumber'].unique():\n ts = pd.read_pickle('data/Electricity_{}.pkl'.format(building)).sum(axis=1)*60\n if not ts.empty:\n for i in range(1,13):\n df_month = ts[ts.index.month == i]\n if len(df_month) > 1:\n og_plot.carpet(df_month, title=building, )\n fig = plt.gcf()\n fig.savefig(\"figures/carpet_electricity_month{}_{}.png\".format(i, building))\n plt.show()\n \n\n\n ",
"_____no_output_____"
]
]
] | [
"markdown",
"code"
] | [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
e7f9456541872dfa0a1d2f06d8181909d6ad0caf | 20,920 | ipynb | Jupyter Notebook | modeling/modeling code/Experiment_2_Direct_Associations.ipynb | tzbozinek/2nd-order-occasion-setting | 28a148f2001d168353b4c48e3b500acd7250daaf | [
"Apache-2.0"
] | null | null | null | modeling/modeling code/Experiment_2_Direct_Associations.ipynb | tzbozinek/2nd-order-occasion-setting | 28a148f2001d168353b4c48e3b500acd7250daaf | [
"Apache-2.0"
] | null | null | null | modeling/modeling code/Experiment_2_Direct_Associations.ipynb | tzbozinek/2nd-order-occasion-setting | 28a148f2001d168353b4c48e3b500acd7250daaf | [
"Apache-2.0"
] | null | null | null | 36.319444 | 201 | 0.544168 | [
[
[
"# Zbozinek TD, Perez OD, Wise T, Fanselow M, & Mobbs D",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom theano import scan\nimport theano.tensor as T\nimport pymc3 as pm\nimport theano\nimport seaborn as sns\nimport os, sys, subprocess",
"_____no_output_____"
]
],
[
[
"# Load Data",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv(os.path.join('../data/', \"2nd_POS_Modeling_Data_Direct_Associations.csv\"))",
"_____no_output_____"
],
[
"data['DV'] = ((data['DV'].values - 1) / 2) - 1\n\nobserved_R = data.pivot(columns = 'ID', index = 'trialseq', values = 'DV').values[:, np.newaxis, :] #values.T transposes the data, so you can make trials the first dimension or participants first",
"_____no_output_____"
]
],
[
[
"# Direct Associations Model",
"_____no_output_____"
]
],
[
[
"def learning_function(stimuli_shown, Λ, λ, training_or_test, prev_V, prev_Vbar, stimulus_type, α):\n \n Λbar = T.zeros_like(Λ)\n Λbar = T.inc_subtensor(Λbar[0,:], (prev_V[2,:] > 0) * (1 - Λ[0, :])) #Dcs\n Λbar = T.inc_subtensor(Λbar[1,:], (prev_V[1,:] > 0) * (1 - Λ[1, :])) #Ecs\n Λbar = T.inc_subtensor(Λbar[2,:], (prev_V[2,:] > 0) * (1 - Λ[2, :])) #F\n Λbar = T.inc_subtensor(Λbar[3,:], (prev_V[2,:] > 0) * (1 - Λ[3, :])) #S\n Λbar = T.inc_subtensor(Λbar[4,:], (prev_V[4,:] > 0) * (1 - Λ[4, :])) #G\n Λbar = T.inc_subtensor(Λbar[5,:], (prev_V[4,:] > 0) * (1 - Λ[5, :])) #H\n Λbar = T.inc_subtensor(Λbar[6,:], (prev_V[6,:] > 0) * (1 - Λ[6, :])) #Mcs\n Λbar = T.inc_subtensor(Λbar[7,:], (prev_V[7,:] > 0) * (1 - Λ[7, :])) #N\n Λbar = T.inc_subtensor(Λbar[8,:], (prev_V[7,:] > 0) * (1 - Λ[8, :])) #Ucs\n \n #λbar Scaling\n λbar = T.zeros_like(Λbar)\n λbar = T.inc_subtensor(λbar[0,:], prev_V[2,:]) #Dcs\n λbar = T.inc_subtensor(λbar[1,:], prev_V[1,:]) #Ecs\n λbar = T.inc_subtensor(λbar[2,:], prev_V[2,:]) #F\n λbar = T.inc_subtensor(λbar[3,:], prev_V[2,:]) #S\n λbar = T.inc_subtensor(λbar[4,:], prev_V[4,:]) #G\n λbar = T.inc_subtensor(λbar[5,:], prev_V[4,:]) #H\n λbar = T.inc_subtensor(λbar[6,:], prev_V[6,:]) #Mcs\n λbar = T.inc_subtensor(λbar[7,:], prev_V[7,:]) #N\n λbar = T.inc_subtensor(λbar[8,:], prev_V[7,:]) #Ucs\n\n\n pe_V = λ - prev_V\n pe_Vbar = λbar - prev_Vbar\n \n ΔV = Λ * (α) * pe_V\n ΔVbar = Λbar * (α) * pe_Vbar\n\n\n # Only update stimuli that were shown\n ΔV = ΔV * stimuli_shown\n ΔVbar = ΔVbar * stimuli_shown\n \n # Update V, Vbar\n V = T.zeros_like(prev_V)\n Vbar = T.zeros_like(prev_Vbar)\n \n # Only update V and Vbar for CSs.\n V = T.inc_subtensor(V[T.eq(stimulus_type, 1)], prev_V[T.eq(stimulus_type, 1)] + ΔV[T.eq(stimulus_type, 1)] * training_or_test)\n Vbar = T.inc_subtensor(Vbar[T.eq(stimulus_type, 1)], prev_Vbar[T.eq(stimulus_type, 1)] + ΔVbar[T.eq(stimulus_type, 1)] * training_or_test)\n \n return V, Vbar",
"_____no_output_____"
]
],
[
[
"# Generate Simulated Data with Model",
"_____no_output_____"
]
],
[
[
"n_stim = 9\nn_subjects = len(data['ID'].unique())\n\n#Initial values\nR = np.zeros((n_stim, n_subjects))\noverall_R = np.zeros((1, n_subjects))\nv_excitatory = np.zeros((n_stim, n_subjects))\nv_inhibitory = np.zeros((n_stim, n_subjects))\n\n#Randomized parameter values - use this if you want to compare simulated vs recovered parameters and comment out the \"α = 1\" code in the subsequent lines\ngen_dist = pm.Beta.dist(2, 2, shape = n_subjects)\nα_subject_sim = gen_dist.random()\n\n#α = 1\n# gen_dist = pm.Beta.dist(2, 2, shape=n_subjects)\n# α_subject_sim = np.ones(n_subjects)\n\n\n\n#Test vs Training Trial\ntraining_or_test = data.pivot(index='trialseq', values='Test', columns='ID').values[:, np.newaxis, :].astype(float)\n\n#US values\nsmall_lambda = data.pivot(index='trialseq', values='US', columns='ID').values[:, np.newaxis, :].repeat(n_stim, axis=1).astype(float)\nstim_data = []\n\nfor sub in data['ID'].unique():\n stim_data.append(data.loc[data['ID'] == sub, ['Dcs', 'Ecs', 'F', 'S', 'G', 'H', 'Mcs', \n 'N', 'Ucs']].values)\n\nstimuli_shown = np.dstack(stim_data)\nbig_lambda = small_lambda\n\n#Add imaginary -1th trial\nbig_lambda = np.vstack([np.zeros((1, n_stim, n_subjects)), big_lambda[:-1, ...]]).astype(float) # Add one trial of zeros to the start, remove the last trial\nsmall_lambda = big_lambda\nstimuli_shown = np.vstack([np.zeros((1, n_stim, n_subjects)), stimuli_shown]) # Add one trial of zeros to the start, DO NOT remove the last trial - this is needed for prediction\n\nstimulus_type = np.ones(n_stim)\n\n#Convert task outcomes to tensors\nbig_lambda = T.as_tensor_variable(big_lambda.astype(float))\nsmall_lambda = T.as_tensor_variable(small_lambda.astype(float))\nstimuli_shown = T.as_tensor_variable(stimuli_shown)\ntraining_or_test = T.as_tensor_variable(training_or_test)\n\nstimuli_shown_sim = stimuli_shown.copy()\nbig_lambda_sim = big_lambda.copy()\nsmall_lambda_sim = small_lambda.copy()\ntraining_or_test_sim = training_or_test.copy()",
"_____no_output_____"
]
],
[
[
"# Run Fake Data Simulation",
"_____no_output_____"
]
],
[
[
"#Run the loop\noutput, updates = scan(fn=learning_function,\n sequences=[{'input': stimuli_shown_sim[:-1, ...]},\n {'input': big_lambda_sim},\n {'input': small_lambda_sim},\n {'input': training_or_test}],\n outputs_info=[v_excitatory, v_inhibitory],\n non_sequences = [stimulus_type, α_subject_sim])\n\n#Get model output\nV_out, Vbar_out = [i.eval() for i in output]\n\nestimated_overall_R = (V_out * stimuli_shown_sim[1:, ...]).sum(axis=1) - (Vbar_out * stimuli_shown_sim[1:, ...]).sum(axis=1)\n \noverall_R_sim = estimated_overall_R.eval()",
"_____no_output_____"
]
],
[
[
"# Check parameter recovery",
"_____no_output_____"
]
],
[
[
"n_subjects = len(data['ID'].unique())\n\n#Initial values\nR = np.zeros((n_stim, n_subjects))\n\n#US values\nsmall_lambda = data.pivot(index='trialseq', values='US', columns='ID').values[:, np.newaxis, :].repeat(n_stim, axis=1).astype(float)\nstim_data = []\n\nfor sub in data['ID'].unique():\n stim_data.append(data.loc[data['ID'] == sub, ['Dcs', 'Ecs', 'F', 'S', 'G', 'H', 'Mcs', \n 'N', 'Ucs']].values)\n\nstimuli_shown = np.dstack(stim_data)\nbig_lambda = small_lambda\n\n#Add imaginary -1th trial\nbig_lambda = np.vstack([np.zeros((1, n_stim, n_subjects)), big_lambda[:-1, ...]]).astype(float) # Add one trial of zeros to the start, remove the last trial\nsmall_lambda = np.vstack([np.zeros((1, n_stim, n_subjects)), small_lambda[:-1, ...]]).astype(float) # Add one trial of zeros to the start, remove the last trial\nstimuli_shown = np.vstack([np.zeros((1, n_stim, n_subjects)), stimuli_shown]) # Add one trial of zeros to the start, DO NOT remove the last trial - this is needed for prediction\n\n\n\n#Convert task outcomes to tensors\nbig_lambda = T.as_tensor_variable(big_lambda.astype(float))\nsmall_lambda = T.as_tensor_variable(small_lambda.astype(float))\nstimuli_shown = T.as_tensor_variable(stimuli_shown)\n\nstimulus_type = np.ones(n_stim)\n\nwith pm.Model() as model:\n \n # Learning rate lies between 0 and 1\n α_mean = pm.Normal('α_mean', 0.5, 10)\n α_sd = pm.HalfCauchy('α_sd', 10)\n \n\n \n BoundedNormal = pm.Bound(pm.Normal, lower=0, upper=1)\n α_subject = BoundedNormal('α', mu=α_mean, sd=α_sd, shape=(n_subjects,))\n\n \n # Run the loop\n output, updates = scan(fn=learning_function,\n sequences=[{'input': stimuli_shown[:-1, ...]},\n {'input': big_lambda},\n {'input': small_lambda},\n {'input': training_or_test}],\n outputs_info=[v_excitatory, v_inhibitory],\n non_sequences=[stimulus_type, α_subject])\n \n # Get model output\n V, Vbar = output\n\n # # Single R value\n estimated_overall_R = ((V * stimuli_shown[1:, ...]).sum(axis=1) - (Vbar * stimuli_shown[1:, ...]).sum(axis=1))\n \n # This allows us to output the estimated R\n estimated_overall_R = pm.Deterministic('estimated_overall_R', estimated_overall_R)\n V = pm.Deterministic('estimated_V', V)\n Vbar = pm.Deterministic('estimated_Vbar', Vbar) \n \n # Reshape output of the model and get categorical likelihood\n sigma = pm.HalfCauchy('sigma', 0.5)\n likelihood = pm.Normal('likelihood', mu=estimated_overall_R, sigma=sigma, observed=pd.DataFrame(overall_R_sim.squeeze()))",
"_____no_output_____"
]
],
[
[
"# Fit the Model",
"_____no_output_____"
],
[
"#### Variational Inference",
"_____no_output_____"
]
],
[
[
"from pymc3.variational.callbacks import CheckParametersConvergence\nwith model:\n approx = pm.fit(method='advi', n=40000, callbacks=[CheckParametersConvergence()])\ntrace = approx.sample(1000)",
"_____no_output_____"
],
[
"alpha_output = pm.summary(trace, kind='stats', varnames=[i for i in model.named_vars if 'α' in i and not i in model.deterministics and not 'log' in i and not 'interval' in i])",
"_____no_output_____"
],
[
"recovered_data_var = {'Simulated_α': α_subject_sim, 'Recovered_α': trace['α'].mean(axis=0)}\nrecovered_data_var = pd.DataFrame(recovered_data_var)\nrecovered_data_var.to_csv(os.path.join('../output/',r'2nd POS - Direct Associations Simulated vs Recovered.csv'))",
"_____no_output_____"
],
[
"f, ax = plt.subplots(1, 1, sharex = True, sharey = True, figsize=(3, 2.5))\nf.suptitle('Simulated vs Recovered α Parameters', y=1.02, fontsize = 16)\nf.text(.5, -.02, 'Simulated α', va='center', ha='center', fontsize = 16)\nf.text(-.02, .5, 'Recovered α', va='center', ha='center', fontsize = 16, rotation=90)\n\nsns.regplot(α_subject_sim, trace['α'].mean(axis=0), label='α_subject', color = 'black')\n\nax.set_title('Direct Associations')\n\nplt.setp(ax, xticks=[0, .2, .4, .6, .8, 1], yticks=[0, .2, .4, .6, .8, 1]) \nplt.tight_layout()\n\nplt.savefig(os.path.join('../output/',r'2nd POS - Direct Associations Simulated vs Recovered.svg'), bbox_inches='tight')",
"_____no_output_____"
]
],
[
[
"# Fit the Model to Real Data",
"_____no_output_____"
]
],
[
[
"n_subjects = len(data['ID'].unique())\n\n# Initial values\nR = np.zeros((n_stim, n_subjects)) # Value estimate\noverall_R = np.zeros((1, n_subjects))\nv_excitatory = np.zeros((n_stim, n_subjects)) \nv_inhibitory = np.zeros((n_stim, n_subjects))\n\n# US values\nsmall_lambda = data.pivot(index='trialseq', values='US', columns='ID').values[:, np.newaxis, :].repeat(n_stim, axis=1)\nstim_data = []\n\nfor sub in data['ID'].unique():\n stim_data.append(data.loc[data['ID'] == sub, ['Dcs', 'Ecs', 'F', 'S', 'G', 'H', 'Mcs', \n 'N', 'Ucs']].values)\n \nstimuli_shown = np.dstack(stim_data)\nbig_lambda = small_lambda\n\n# Add imaginary -1th trial\nbig_lambda = np.vstack([np.zeros((1, n_stim, n_subjects)), big_lambda[:-1, ...]]) # Add one trial of zeros to the start, remove the last trial\nsmall_lambda = np.vstack([np.zeros((1, n_stim, n_subjects)), small_lambda[:-1, ...]]) # Add one trial of zeros to the start, remove the last trial\nstimuli_shown = np.vstack([np.zeros((1, n_stim, n_subjects)), stimuli_shown]) # Add one trial of zeros to the start, DO NOT remove the last trial - this is needed for prediction\n\nstimulus_type = np.ones(n_stim)\n\n\n# Convert task outcomes to tensors\nbig_lambda = T.as_tensor_variable(big_lambda)\nsmall_lambda = T.as_tensor_variable(small_lambda)\nstimuli_shown = T.as_tensor_variable(stimuli_shown)\n\nwith pm.Model() as model:\n \n # Learning rate lies between 0 and 1 so we use a beta distribution\n α_mean = pm.Normal('α_mean', 0.5, 10)\n α_sd = pm.HalfCauchy('α_sd', 10)\n\n\n BoundedNormal = pm.Bound(pm.Normal, lower=0, upper=1)\n α_subject = BoundedNormal('α', mu=α_mean, sd=α_sd, shape=(n_subjects,))\n\n \n # Run the loop\n output, updates = scan(fn=learning_function,\n sequences=[{'input': stimuli_shown[:-1, ...]},\n {'input': big_lambda},\n {'input': small_lambda},\n {'input': training_or_test}],\n outputs_info=[v_excitatory, v_inhibitory],\n non_sequences=[stimulus_type, α_subject])\n \n # Get model output\n V, Vbar = output\n \n estimated_overall_R = ((V * stimuli_shown[1:, ...]).sum(axis=1) - (Vbar * stimuli_shown[1:, ...]).sum(axis=1))\n \n # This allows us to output the estimated R\n estimated_overall_R = pm.Deterministic('estimated_overall_R', estimated_overall_R)\n V = pm.Deterministic('estimated_V', V)\n Vbar = pm.Deterministic('estimated_Vbar', Vbar)\n \n # Reshape output of the model and get categorical likelihood\n sigma = pm.HalfCauchy('sigma', 0.5)\n likelihood = pm.Normal('likelihood', mu=estimated_overall_R, sigma=sigma, observed=pd.DataFrame(observed_R.squeeze()))",
"_____no_output_____"
]
],
[
[
"#### Variational Inference",
"_____no_output_____"
]
],
[
[
"from pymc3.variational.callbacks import CheckParametersConvergence\nwith model:\n approx = pm.fit(method='advi', n=40000, callbacks=[CheckParametersConvergence()])\ntrace = approx.sample(1000)",
"_____no_output_____"
],
[
"alpha_output = pm.summary(trace, kind='stats', varnames=[i for i in model.named_vars if 'α' in i and not i in model.deterministics and not 'log' in i and not 'interval' in i])",
"_____no_output_____"
]
],
[
[
"# Model Output",
"_____no_output_____"
]
],
[
[
"overall_R_mean = trace['estimated_overall_R'].mean(axis=0)\noverall_R_sd = trace['estimated_overall_R'].std(axis=0)\nsub_ids = data['ID'].unique()\nsubs = [np.where(data['ID'].unique() == sub)[0][0] for sub in sub_ids]",
"_____no_output_____"
],
[
"waic_output = pm.waic(trace)",
"_____no_output_____"
],
[
"waic_output",
"_____no_output_____"
],
[
"alpha_output.to_csv(os.path.join('../output/',r'2nd POS - Direct Associations Alpha Output.csv'))\nwaic_output.to_csv(os.path.join('../output/',r'2nd POS - Direct Associations WAIC Output.csv'))",
"_____no_output_____"
],
[
"f, ax = plt.subplots(23, 3, figsize=(36, 48), dpi = 100)\n\noverall_R_mean = trace['estimated_overall_R'].mean(axis=0)\noverall_R_sd = trace['estimated_overall_R'].std(axis=0)\n\nsub_ids = data['ID'].unique()\n\nsubs = [np.where(data['ID'].unique() == sub)[0][0] for sub in sub_ids]\n \n\nfor n, sub in enumerate(subs):\n ax[n % 23, int(n / 23)].fill_between(range(overall_R_mean.shape[0]), overall_R_mean[:, sub] - overall_R_sd[:, sub], overall_R_mean[:, sub] + overall_R_sd[:, sub], alpha=0.3)\n ax[n % 23, int(n / 23)].plot(overall_R_mean[:, sub])\n ax[n % 23, int(n / 23)].plot(observed_R.squeeze()[:, sub], color='orange', linestyle='-')#participant's real data\n ax[n % 23, int(n / 23)].plot(overall_R_sim.squeeze()[:, sub], color='silver', linestyle=':', alpha = .7)#Alpha = 1; this is the correct answer if a person learned perfectly\n if n == 0:\n ax[n % 23, int(n / 23)].set_ylabel('Mean (+/-SD) overall R')\n ax[n % 23, int(n / 23)].set_ylabel('Responding (R)')\n ax[n % 23, int(n / 23)].set_xlabel('Trials')\n ax[n % 23, int(n / 23)].set_title('Sub {0}'.format(sub_ids[n])) \n\nplt.tight_layout()\nplt.savefig(os.path.join('../output/',r'2nd POS - Direct Associations Individual Real and Estimated Responding.svg'), bbox_inches='tight')",
"_____no_output_____"
],
[
"%load_ext watermark\n%watermark -v -p pytest,jupyterlab,numpy,pandas,theano,pymc3",
"_____no_output_____"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
e7f94dd35a5d50dad47b33fd164fc0260371270f | 898,302 | ipynb | Jupyter Notebook | notebooks/breathing_notebooks/1.1_deformation_experiment_scattering_ETH-05.ipynb | sgaut023/Chronic-Liver-Classification | 98523a467eed3b51c20a73ed5ddbc53a1bf7a8d6 | [
"RSA-MD"
] | 1 | 2022-03-09T11:34:00.000Z | 2022-03-09T11:34:00.000Z | notebooks/breathing_notebooks/1.1_deformation_experiment_scattering_ETH-05.ipynb | sgaut023/Chronic-Liver-Classification | 98523a467eed3b51c20a73ed5ddbc53a1bf7a8d6 | [
"RSA-MD"
] | null | null | null | notebooks/breathing_notebooks/1.1_deformation_experiment_scattering_ETH-05.ipynb | sgaut023/Chronic-Liver-Classification | 98523a467eed3b51c20a73ed5ddbc53a1bf7a8d6 | [
"RSA-MD"
] | null | null | null | 943.594538 | 398,532 | 0.951416 | [
[
[
"# Experiment 02: Deformations Experiments ETH-05\n\n\nIn this notebook, we are using the CLUST Dataset.\nThe sequence used for this notebook is ETH-05.zip",
"_____no_output_____"
]
],
[
[
"import sys\nimport random\nimport os\nsys.path.append('../src')\nimport warnings\nwarnings.filterwarnings(\"ignore\") \nfrom PIL import Image\nfrom utils.compute_metrics import get_metrics, get_majority_vote,log_test_metrics\nfrom utils.split import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.decomposition import PCA\nfrom sklearn.svm import LinearSVC\nfrom sklearn.svm import SVC\nfrom sklearn.model_selection import GroupKFold\nfrom tqdm import tqdm\nfrom pprint import pprint\nimport torch\nfrom itertools import product\nimport pickle\nimport pandas as pd\nimport numpy as np\nimport mlflow\nimport matplotlib.pyplot as plt\n#from kymatio.numpy import Scattering2D\n\nimport torch\nfrom tqdm import tqdm\nfrom kymatio.torch import Scattering2D\n",
"_____no_output_____"
]
],
[
[
"# 1. Visualize Sequence of US\nWe are visualizing the first images from the sequence ETH-01-1 that contains 3652 US images.",
"_____no_output_____"
]
],
[
[
"directory=os.listdir('../data/02_interim/Data5')\ndirectory.sort()",
"_____no_output_____"
],
[
"# settings\nh, w = 15, 10 # for raster image\nnrows, ncols = 3, 4 # array of sub-plots\nfigsize = [15, 8] # figure size, inches\n\n# prep (x,y) for extra plotting on selected sub-plots\nxs = np.linspace(0, 2*np.pi, 60) # from 0 to 2pi\nys = np.abs(np.sin(xs)) # absolute of sine\n\n# create figure (fig), and array of axes (ax)\nfig, ax = plt.subplots(nrows=nrows, ncols=ncols, figsize=figsize)\n\nimgs= directory[0:100]\ncount = 0\n# plot simple raster image on each sub-plot\nfor i, axi in enumerate(ax.flat):\n # i runs from 0 to (nrows*ncols-1)\n # axi is equivalent with ax[rowid][colid]\n img =plt.imread(\"../data/02_interim/Data5/\" + imgs[i])\n axi.imshow(img, cmap='gray')\n axi.axis('off')\n # get indices of row/column\n # write row/col indices as axes' title for identification\n #axi.set_title(df_labels['Finding Labels'][row[count]], size=20)\n count = count +1\n\n\nplt.tight_layout(True)\nplt.savefig('samples_xray')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# 2. Create Dataset",
"_____no_output_____"
]
],
[
[
"%%time\nll_imgstemp = [plt.imread(\"../data/02_interim/Data5/\" + dir) for dir in directory[:5]]",
"CPU times: user 42.9 ms, sys: 5.86 ms, total: 48.8 ms\nWall time: 51.2 ms\n"
],
[
"%%time\nll_imgs = [np.array(Image.open(\"../data/02_interim/Data5/\" + dir).resize(size=(98, 114)), dtype='float32') for dir in directory]",
"CPU times: user 6.61 s, sys: 2.06 s, total: 8.66 s\nWall time: 40.3 s\n"
],
[
"%%time\nll_imgs2 = [img.reshape(1,img.shape[0],img.shape[1]) for img in ll_imgs]",
"CPU times: user 1.58 ms, sys: 943 µs, total: 2.52 ms\nWall time: 2.53 ms\n"
],
[
"# dataset = pd.DataFrame([torch.tensor(ll_imgs).view(1,M,N).type(torch.float32)], columns='img')\ndataset = pd.DataFrame({'img':ll_imgs2}).reset_index().rename(columns={'index':'order'})\n# dataset = pd.DataFrame({'img':ll_imgs}).reset_index().rename(columns={'index':'order'})\n",
"_____no_output_____"
],
[
"dataset",
"_____no_output_____"
]
],
[
[
"# 3. Extract Scattering Features",
"_____no_output_____"
]
],
[
[
"M,N = dataset['img'].iloc[0].shape[1], dataset['img'].iloc[0].shape[2]\nprint(M,N)",
"114 98\n"
],
[
"# Set the parameters of the scattering transform.\nJ = 3\n# Generate a sample signal.\nscattering = Scattering2D(J, (M, N))\ndata = np.concatenate(dataset['img'],axis=0)\ndata = torch.from_numpy(data)\nuse_cuda = torch.cuda.is_available()\ndevice = torch.device(\"cuda\" if use_cuda else \"cpu\")\nif use_cuda: \n scattering = scattering.cuda()\n data = data.to(device)",
"_____no_output_____"
],
[
"init = 0\nfinal =0\ncount =0\nfirst_loop = True\nfor i in tqdm(range (0,len(data), 13)):\n init= i\n final = i + 11\n if first_loop:\n scattering_features = scattering(data[init: final])\n first_loop=False\n torch.cuda.empty_cache()\n else: \n scattering_features = torch.cat((scattering_features,scattering(data[init: final]) ))\n torch.cuda.empty_cache()\n# break \n# save scattering features\n# with open('../data/03_features/scattering_features_deformation.pickle', 'wb') as handle:\n# pickle.dump(scattering_features, handle, protocol=pickle.HIGHEST_PROTOCOL) ",
"100%|██████████| 355/355 [00:25<00:00, 13.67it/s]\n"
],
[
"# save scattering features\nwith open('../data/03_features/scattering_features_deformation5.pickle', 'wb') as handle:\n pickle.dump(scattering_features, handle, protocol=pickle.HIGHEST_PROTOCOL)",
"_____no_output_____"
],
[
"# save scattering features\nwith open('../data/03_features/dataset_deformation5.pickle', 'wb') as handle:\n pickle.dump(dataset, handle, protocol=pickle.HIGHEST_PROTOCOL)",
"_____no_output_____"
]
],
[
[
"# 4. Extract PCA Components",
"_____no_output_____"
]
],
[
[
"with open('../data/03_features/scattering_features_deformation5.pickle', 'rb') as handle:\n scattering_features = pickle.load(handle)\nwith open('../data/03_features/dataset_deformation5.pickle', 'rb') as handle:\n dataset = pickle.load(handle)",
"_____no_output_____"
],
[
"sc_features = scattering_features.view(scattering_features.shape[0], scattering_features.shape[1] * scattering_features.shape[2] * scattering_features.shape[3])\nX = sc_features.cpu().numpy()\n#standardize\nscaler = StandardScaler()\nX = scaler.fit_transform(X)\npca = PCA(n_components=50) \nX = pca.fit_transform(X)",
"_____no_output_____"
],
[
"plt.plot(np.insert(pca.explained_variance_ratio_.cumsum(),0,0),marker='o')\nplt.xlabel('Number of components')\nplt.ylabel('Cumulative explained variance')\nplt.show()",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"print(pca.explained_variance_ratio_.cumsum())",
"[0.28553063 0.3439802 0.3723686 0.39626902 0.413098 0.42726657\n 0.43944615 0.45060596 0.46070525 0.47008792 0.4787021 0.48672393\n 0.49356067 0.49987826 0.5053979 0.51080227 0.51609737 0.5212945\n 0.52589065 0.5302912 0.53452075 0.538555 0.5423447 0.54612917\n 0.54977244 0.5533351 0.55677855 0.5600062 0.56319094 0.56618875\n 0.5691468 0.5719544 0.57460827 0.5771299 0.5795443 0.5818841\n 0.58416575 0.58643836 0.58865577 0.59081393 0.5929018 0.5949698\n 0.5969465 0.59890795 0.6008287 0.6026802 0.604491 0.6062711\n 0.6080006 0.60967034]\n"
],
[
"df = pd.DataFrame(X)\ndf['order'] = dataset['order']\n#df.corr()",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"import seaborn as sns; sns.set_theme()\nplt.figure(figsize = (10,10))\nvec1 = df.corr()['order'].values\nvec2 = vec1.reshape(vec1.shape[0], 1)\nsns.heatmap(vec2)\nplt.show()",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"def visualize_corr_pca_order(pca_c, df):\n plt.figure(figsize=(16,8))\n x= df['order']\n y= df[pca_c]\n plt.scatter(x,y)\n m, b = np.polyfit(x, y, 1)\n plt.plot(x, m*x + b, color='red')\n plt.ylabel('PCA Component '+ str(pca_c+1))\n plt.xlabel('Frame Order')\n plt.show()",
"_____no_output_____"
],
[
"visualize_corr_pca_order(3, df)\nprint('Correlation between order and Pca component 2:', df.corr()['order'][1])",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"def visualize_sub_plot(pca_c, df, x_num= 3, y_num =3):\n fig, axs = plt.subplots(x_num, y_num, figsize=(15,10))\n size = len(df)\n plot_num = x_num * y_num\n frame = int(size/plot_num)\n start = 0\n for i in range (x_num):\n for j in range (y_num): \n final = start + frame\n x= df['order'].iloc[start:final]\n y= df[pca_c].iloc[start:final]\n m, b = np.polyfit(x, y, 1)\n\n axs[i, j].set_ylabel('PCA Component '+ str(pca_c+1))\n axs[i, j].set_xlabel('Frame Order')\n axs[i, j].plot(x, m*x + b, color='red')\n axs[i, j].scatter(x,y)\n start = start + frame\n plt.show()\n",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"visualize_sub_plot(3, df, x_num= 3, y_num =3)",
"_____no_output_____"
]
],
[
[
"# 5. Isometric Mapping Correlation with Order",
"_____no_output_____"
]
],
[
[
"with open('../data/03_features/scattering_features_deformation5.pickle', 'rb') as handle:\n scattering_features = pickle.load(handle)\nwith open('../data/03_features/dataset_deformation5.pickle', 'rb') as handle:\n dataset = pickle.load(handle)",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"sc_features = scattering_features.view(scattering_features.shape[0], scattering_features.shape[1] * scattering_features.shape[2] * scattering_features.shape[3])\nX = sc_features.cpu().numpy()\n#standardize\nscaler = StandardScaler()\nX = scaler.fit_transform(X)",
"_____no_output_____"
],
[
"from sklearn.manifold import Isomap\nembedding = Isomap(n_components=2)\nX_transformed = embedding.fit_transform(X[0:500])\ndf = pd.DataFrame(X_transformed)\ndf['order'] = dataset['order']",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"df.corr()",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"from sklearn.manifold import Isomap\ndef visualize_sub_plot_iso(pca_c, x_num= 3, y_num =3):\n fig, axs = plt.subplots(x_num, y_num, figsize=(15,13))\n size =len(sc_features )\n plot_num = x_num * y_num\n frame = int(size/plot_num)\n start = 0\n for i in tqdm(range (x_num)):\n for j in tqdm(range (y_num)): \n final = start + frame\n embedding = Isomap(n_components=2)\n X_transformed = embedding.fit_transform(X[start:final])\n df = pd.DataFrame(X_transformed)\n df['order'] = dataset['order'].iloc[start:final].values\n x= df['order']\n y= df[pca_c]\n start = start + frame\n \n #m, b = np.polyfit(x, y, 1)\n\n axs[i, j].set_ylabel('Iso Map Dimension '+ str(pca_c+1))\n axs[i, j].set_xlabel('Frame Order')\n #axs[i, j].plot(x, m*x + b, color='red')\n axs[i, j].scatter(x,y)\n plt.show()",
"/home/mila/g/gauthies/.conda/envs/ultra/lib/python3.7/site-packages/ipykernel/ipkernel.py:287: DeprecationWarning: `should_run_async` will not call `transform_cell` automatically in the future. Please pass the result to `transformed_cell` argument and any exception that happen during thetransform in `preprocessing_exc_tuple` in IPython 7.17 and above.\n and should_run_async(code)\n"
],
[
"visualize_sub_plot_iso(0, x_num= 3, y_num =3)\n#print('Correlation between order and Pca component 2:', df.corr()['order'][1])",
" 0%| | 0/3 [00:00<?, ?it/s]\n 0%| | 0/3 [00:00<?, ?it/s]\u001b[A\n 33%|███▎ | 1/3 [00:11<00:23, 11.70s/it]\u001b[A\n 67%|██████▋ | 2/3 [00:23<00:11, 11.71s/it]\u001b[A\n100%|██████████| 3/3 [00:35<00:00, 11.71s/it]\u001b[A\n 33%|███▎ | 1/3 [00:35<01:10, 35.12s/it]\n 0%| | 0/3 [00:00<?, ?it/s]\u001b[A\n 33%|███▎ | 1/3 [00:11<00:23, 11.56s/it]\u001b[A\n 67%|██████▋ | 2/3 [00:23<00:11, 11.58s/it]\u001b[A\n100%|██████████| 3/3 [00:35<00:00, 11.82s/it]\u001b[A\n 67%|██████▋ | 2/3 [01:10<00:35, 35.22s/it]\n 0%| | 0/3 [00:00<?, ?it/s]\u001b[A\n 33%|███▎ | 1/3 [00:11<00:23, 11.59s/it]\u001b[A\n 67%|██████▋ | 2/3 [00:23<00:11, 11.62s/it]\u001b[A\n100%|██████████| 3/3 [00:35<00:00, 11.67s/it]\u001b[A\n100%|██████████| 3/3 [01:45<00:00, 35.20s/it]\n"
]
]
] | [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] | [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |