File size: 9,657 Bytes
85339a7 9f80b41 85339a7 e375d80 85339a7 9b5fb3a 85339a7 9984ad0 85339a7 e375d80 85339a7 9984ad0 85339a7 9984ad0 85339a7 12c3d6e e375d80 85339a7 e375d80 85339a7 e375d80 85339a7 e375d80 85339a7 e375d80 85339a7 12c3d6e e375d80 85339a7 12c3d6e 85339a7 12c3d6e e375d80 774ba89 b1772e8 9a69ce4 b1772e8 9a69ce4 12c3d6e 9a69ce4 b1772e8 e02a92e 9a69ce4 e02a92e b1772e8 e375d80 e703bf8 e375d80 85339a7 e703bf8 85339a7 9b64c65 85339a7 c03d5bc e703bf8 85339a7 9b64c65 85339a7 e375d80 8800be8 e703bf8 85339a7 9b64c65 85339a7 e703bf8 85339a7 b574bbe 85339a7 e703bf8 2ee6428 b1772e8 e02a92e b1772e8 2ee6428 e02a92e 85339a7 9f80b41 9984ad0 9f80b41 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 |
---
tags:
- monai
- medical
library_name: monai
license: apache-2.0
---
# Model Overview
A pre-trained model for automated detection of metastases in whole-slide histopathology images.
The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
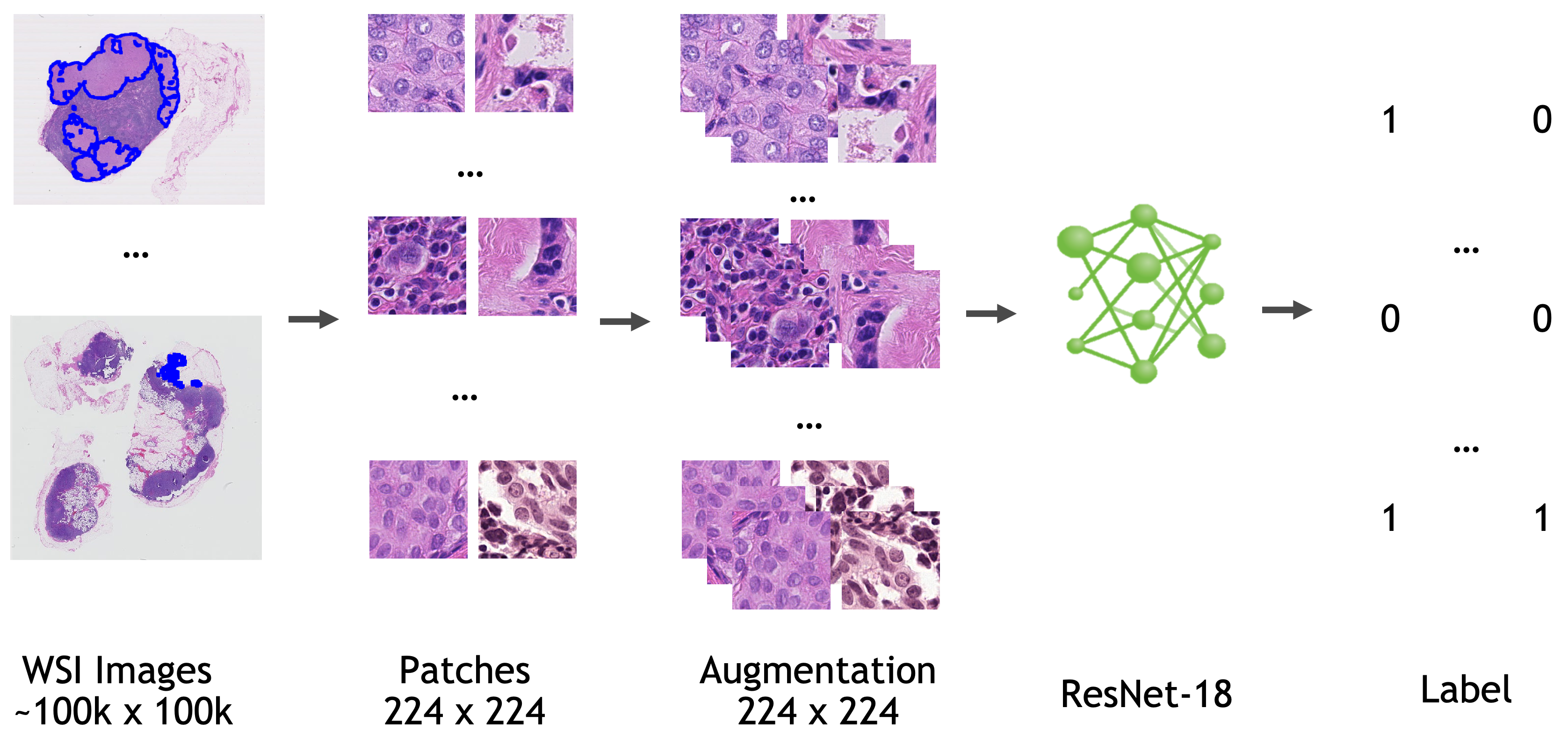
## Data
All the data used to train, validate, and test this model is from [Camelyon-16 Challenge](https://camelyon16.grand-challenge.org/). You can download all the images for "CAMELYON16" data set from various sources listed [here](https://camelyon17.grand-challenge.org/Data/).
Location information for training/validation patches (the location on the whole slide image where patches are extracted) are adopted from [NCRF/coords](https://github.com/baidu-research/NCRF/tree/master/coords).
Annotation information are adopted from [NCRF/jsons](https://github.com/baidu-research/NCRF/tree/master/jsons).
- Target: Tumor
- Task: Detection
- Modality: Histopathology
- Size: 270 WSIs for training/validation, 48 WSIs for testing
### Preprocessing
This bundle expects the training/validation data (whole slide images) reside in a `{dataset_dir}/training/images`. By default `dataset_dir` is pointing to `/workspace/data/medical/pathology/` You can modify `dataset_dir` in the bundle config files to point to a different directory.
To reduce the computation burden during the inference, patches are extracted only where there is tissue and ignoring the background according to a tissue mask. Please also create a directory for prediction output. By default `output_dir` is set to `eval` folder under the bundle root.
Please refer to "Annotation" section of [Camelyon challenge](https://camelyon17.grand-challenge.org/Data/) to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at `/workspace/data/medical/pathology/ground_truths`. But it can be modified in `evaluate_froc.sh`.
## Training configuration
The training was performed with the following:
- Config file: train.config
- GPU: at least 16 GB of GPU memory.
- Actual Model Input: 224 x 224 x 3
- AMP: True
- Optimizer: Novograd
- Learning Rate: 1e-3
- Loss: BCEWithLogitsLoss
- Whole slide image reader: cuCIM (if running on Windows or Mac, please install `OpenSlide` on your system and change `wsi_reader` to "OpenSlide")
### Pretrained Weights
By setting the `"pretrained"` parameter of `TorchVisionFCModel` in the config file to `true`, ImageNet pre-trained weights will be used for training. Please note that these weights are for non-commercial use. Each user is responsible for checking the content of the models/datasets and the applicable licenses and determining if suitable for the intended use. In order to use other pretrained weights, you can use `CheckpointLoader` in train handlers section as the first handler:
```json
{
"_target_": "CheckpointLoader",
"load_path": "$@bundle_root + '/pretrained_resnet18.pth'",
"strict": false,
"load_dict": {
"model_new": "@network"
}
}
```
### Input
The training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
### Output
A probability number of the input patch being tumor or normal.
### Inference on a WSI
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
### Note on determinism
By default this bundle use a deterministic approach to make the results reproducible. However, it comes at a cost of performance loss. Thus if you do not care about reproducibility, you can have a performance gain by replacing `"$monai.utils.set_determinism"` line with `"$setattr(torch.backends.cudnn, 'benchmark', True)"` in initialize section of training configuration (`configs/train.json` and `configs/multi_gpu_train.json` for single GPU and multi-GPU training respectively).
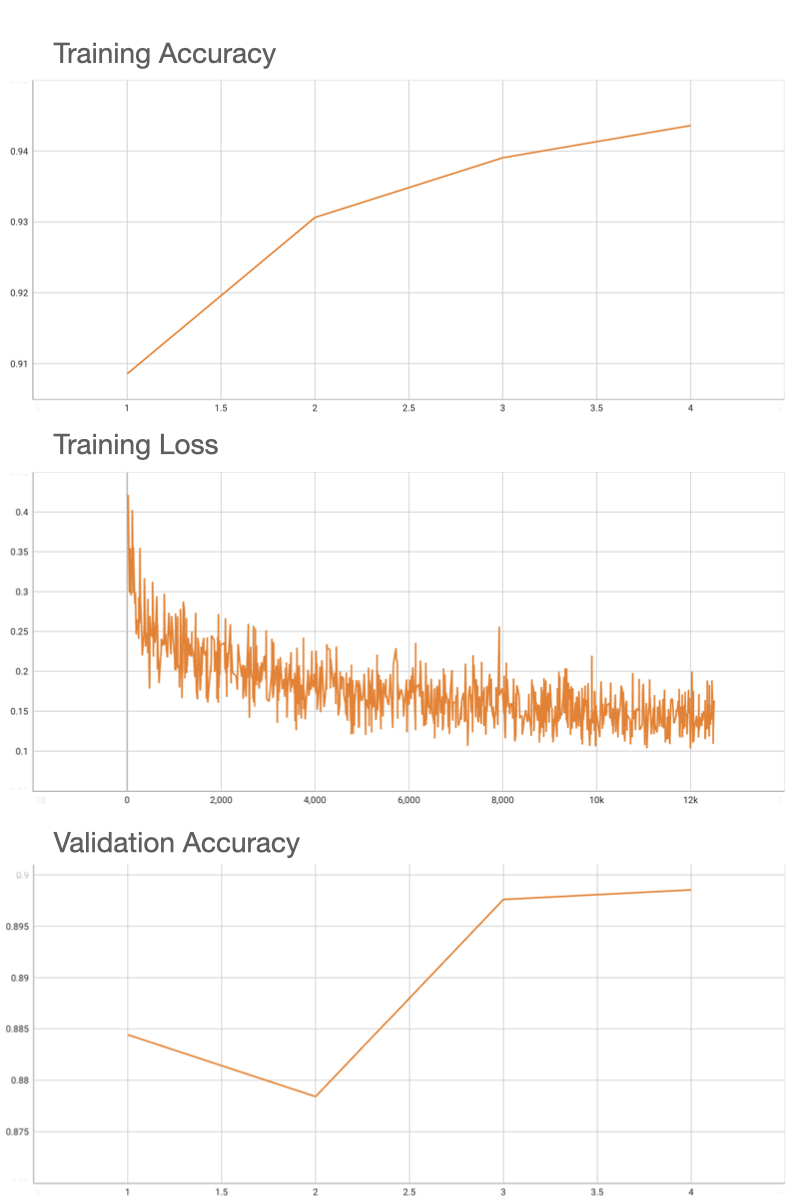
## Performance
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
Using an internal pretrained weights for ResNet18, this model deterministically achieves the 0.90 accuracy on validation patches, and FROC of 0.72 on the 48 Camelyon testing data that have ground truth annotations available.
The `pathology_tumor_detection` bundle supports acceleration with TensorRT. The table below displays the speedup ratios observed on an A100 80G GPU.
Please notice that the benchmark results are tested on one WSI image since the images are too large to benchmark. And the inference time in the end-to-end line stands for one patch of the whole image.
| method | torch_fp32(ms) | torch_amp(ms) | trt_fp32(ms) | trt_fp16(ms) | speedup amp | speedup fp32 | speedup fp16 | amp vs fp16|
| :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: |
| model computation |1.93 | 2.52 | 1.61 | 1.33 | 0.77 | 1.20 | 1.45 | 1.89 |
| end2end |224.97 | 223.50 | 222.65 | 224.03 | 1.01 | 1.01 | 1.00 | 1.00 |
Where:
- `model computation` means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
- `end2end` means run the bundle end-to-end with the TensorRT based model.
- `torch_fp32` and `torch_amp` are for the PyTorch models with or without `amp` mode.
- `trt_fp32` and `trt_fp16` are for the TensorRT based models converted in corresponding precision.
- `speedup amp`, `speedup fp32` and `speedup fp16` are the speedup ratios of corresponding models versus the PyTorch float32 model
- `amp vs fp16` is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
This result is benchmarked under:
- TensorRT: 8.5.3+cuda11.8
- Torch-TensorRT Version: 1.4.0
- CPU Architecture: x86-64
- OS: ubuntu 20.04
- Python version:3.8.10
- CUDA version: 12.0
- GPU models and configuration: A100 80G
## MONAI Bundle Commands
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
#### Execute training
```
python -m monai.bundle run --config_file configs/train.json
```
Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using `--dataset_dir`:
```
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
```
#### Override the `train` config to execute multi-GPU training
```
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
```
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
#### Execute inference
```
CUDA_LAUNCH_BLOCKING=1 python -m monai.bundle run --config_file configs/inference.json
```
#### Evaluate FROC metric
```
cd scripts && source evaluate_froc.sh
```
#### Export checkpoint to TorchScript file
```
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json
```
#### Export checkpoint to TensorRT based models with fp32 or fp16 precision
```
python -m monai.bundle trt_export --net_id network_def --filepath models/model_trt.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.json --precision <fp32/fp16> --dynamic_batchsize "[1, 400, 600]"
```
#### Execute inference with the TensorRT model
```
python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
```
# References
[1] He, Kaiming, et al, "Deep Residual Learning for Image Recognition." In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770-778. 2016. <https://arxiv.org/pdf/1512.03385.pdf>
# License
Copyright (c) MONAI Consortium
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
|