Spaces:
Runtime error
Runtime error
new features added
#7
by
zhihuang
- opened
- app.py +15 -1
- data/img_2d_embedding.csv +3 -0
- data/twitter.asset +2 -2
- data/txt_2d_embedding.csv +3 -0
- home.py +55 -13
- image2image.py +7 -3
- introduction.md +4 -2
- requirements.txt +3 -1
- resources/4x/Fig1.png +3 -0
- resources/4x/Fig1ab.png +3 -0
- resources/4x/Fig1c.png +3 -0
- resources/4x/Fig1d.png +3 -0
- resources/4x/Fig1e.png +3 -0
- resources/4x/Fig1f.png +3 -0
- resources/4x/image_retrieval.png +3 -0
- text2image.py +12 -4
- visualization.py +44 -0
- viz_scripts/calc_img.py +75 -0
app.py
CHANGED
|
@@ -1,18 +1,32 @@
|
|
| 1 |
import home
|
| 2 |
import text2image
|
| 3 |
import image2image
|
|
|
|
| 4 |
import streamlit as st
|
|
|
|
| 5 |
|
| 6 |
|
| 7 |
|
| 8 |
st.set_page_config(layout="wide")
|
| 9 |
|
| 10 |
-
st.sidebar.title("
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 11 |
|
| 12 |
PAGES = {
|
| 13 |
"Introduction": home,
|
| 14 |
"Text to Image": text2image,
|
| 15 |
"Image to Image": image2image,
|
|
|
|
| 16 |
}
|
| 17 |
|
| 18 |
page = st.sidebar.radio("", list(PAGES.keys()))
|
|
|
|
| 1 |
import home
|
| 2 |
import text2image
|
| 3 |
import image2image
|
| 4 |
+
import visualization
|
| 5 |
import streamlit as st
|
| 6 |
+
import streamlit.components.v1 as components
|
| 7 |
|
| 8 |
|
| 9 |
|
| 10 |
st.set_page_config(layout="wide")
|
| 11 |
|
| 12 |
+
st.sidebar.title("WebPLIP")
|
| 13 |
+
|
| 14 |
+
components.html('''
|
| 15 |
+
<!-- Google tag (gtag.js) -->
|
| 16 |
+
<script async src="https://www.googletagmanager.com/gtag/js?id=G-KPF04V95FN"></script>
|
| 17 |
+
<script>
|
| 18 |
+
window.dataLayer = window.dataLayer || [];
|
| 19 |
+
function gtag(){dataLayer.push(arguments);}
|
| 20 |
+
gtag('js', new Date());
|
| 21 |
+
gtag('config', 'G-KPF04V95FN');
|
| 22 |
+
</script>
|
| 23 |
+
''')
|
| 24 |
|
| 25 |
PAGES = {
|
| 26 |
"Introduction": home,
|
| 27 |
"Text to Image": text2image,
|
| 28 |
"Image to Image": image2image,
|
| 29 |
+
"Visualization": visualization,
|
| 30 |
}
|
| 31 |
|
| 32 |
page = st.sidebar.radio("", list(PAGES.keys()))
|
data/img_2d_embedding.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:2c94660a66598254f87494fbb931f01a78feda5452d54c3b3939543a392d2fb7
|
| 3 |
+
size 13600550
|
data/twitter.asset
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
-
size
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:533c5c023e099a2725be0241ca57d6218e37f1f355963ced8d2305270312e428
|
| 3 |
+
size 245669888
|
data/txt_2d_embedding.csv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:d5e5cde958eda7b8e74f327326029df1f7480c3bb2879141bdab8f71dd71fdd4
|
| 3 |
+
size 13659850
|
home.py
CHANGED
|
@@ -1,22 +1,64 @@
|
|
| 1 |
from pathlib import Path
|
| 2 |
import streamlit as st
|
| 3 |
import streamlit.components.v1 as components
|
| 4 |
-
|
|
|
|
|
|
|
|
|
|
| 5 |
|
| 6 |
def read_markdown_file(markdown_file):
|
| 7 |
return Path(markdown_file).read_text()
|
| 8 |
|
| 9 |
|
| 10 |
def app():
|
| 11 |
-
intro_markdown = read_markdown_file("introduction.md")
|
| 12 |
-
st.markdown(intro_markdown, unsafe_allow_html=True)
|
| 13 |
-
|
| 14 |
-
|
| 15 |
-
|
| 16 |
-
|
| 17 |
-
|
| 18 |
-
|
| 19 |
-
|
| 20 |
-
|
| 21 |
-
|
| 22 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
from pathlib import Path
|
| 2 |
import streamlit as st
|
| 3 |
import streamlit.components.v1 as components
|
| 4 |
+
import plotly.figure_factory as ff
|
| 5 |
+
import numpy as np
|
| 6 |
+
import pandas as pd
|
| 7 |
+
from PIL import Image
|
| 8 |
|
| 9 |
def read_markdown_file(markdown_file):
|
| 10 |
return Path(markdown_file).read_text()
|
| 11 |
|
| 12 |
|
| 13 |
def app():
|
| 14 |
+
#intro_markdown = read_markdown_file("introduction.md")
|
| 15 |
+
#st.markdown(intro_markdown, unsafe_allow_html=True)
|
| 16 |
+
st.markdown("# Leveraging medical Twitter to build a visual-language foundation model for pathology")
|
| 17 |
+
|
| 18 |
+
col1, col2 = st.columns([2, 1])
|
| 19 |
+
with col1:
|
| 20 |
+
st.markdown("The lack of annotated publicly available medical images is a major barrier for innovations. At the same time, many de-identified images and much knowledge are shared by clinicians on public forums such as medical Twitter. Here we harness these crowd platforms to curate OpenPath, a large dataset of <b>208,414</b> pathology images paired with natural language descriptions. This is the largest public dataset for pathology images annotated with natural text. We demonstrate the value of this resource by developing PLIP, a multimodal AI with both image and text understanding, which is trained on OpenPath. PLIP achieves state-of-the-art zero-shot and few-short performance for classifying new pathology images across diverse tasks. Moreover, PLIP enables users to retrieve similar cases by either image or natural language search, greatly facilitating knowledge sharing. Our approach demonstrates that publicly shared medical data is a tremendous opportunity that can be harnessed to advance biomedical AI.", unsafe_allow_html=True)
|
| 21 |
+
|
| 22 |
+
fig1ab = Image.open('resources/4x/Fig1ab.png')
|
| 23 |
+
st.image(fig1ab, caption='OpenPath Dataset', output_format='png')
|
| 24 |
+
with col2:
|
| 25 |
+
st.caption('An example of tweet')
|
| 26 |
+
components.html('''
|
| 27 |
+
<blockquote class="twitter-tweet">
|
| 28 |
+
<a href="https://twitter.com/xxx/status/1580753362059788288"></a>
|
| 29 |
+
</blockquote>
|
| 30 |
+
<script async src="https://platform.twitter.com/widgets.js" charset="utf-8">
|
| 31 |
+
</script>
|
| 32 |
+
''',
|
| 33 |
+
height=500)
|
| 34 |
+
|
| 35 |
+
|
| 36 |
+
st.markdown("#### PLIP is trained on the largest public vision–language pathology dataset: OpenPath")
|
| 37 |
+
|
| 38 |
+
col1, col2 = st.columns([1, 1])
|
| 39 |
+
with col1:
|
| 40 |
+
st.markdown("Following the usage policy and guidelines from Twitter and other entities, we established so far the largest public vision–language pathology dataset. To ensure the quality of the data, OpenPath followed rigorous protocols for cohort inclusion and exclusion, including the removal of retweets, sensitive tweets, and non-pathology images, as well as text cleaning.", unsafe_allow_html=True)
|
| 41 |
+
st.markdown("The final OpenPath dataset consists of:", unsafe_allow_html=True)
|
| 42 |
+
st.markdown("- Tweets: 116,504 image–text pairs from Twitter posts (tweets) during Mar. 21, 2006 – Nov. 15, 2022 across 32 pathology subspecialty-specific hashtags;", unsafe_allow_html=True)
|
| 43 |
+
st.markdown("- Replies: 59,869 image–text pairs from the associated replies that received the highest number of likes in the tweet, if applicable;", unsafe_allow_html=True)
|
| 44 |
+
st.markdown("- PathLAION: 32,041 additional image–text pairs from the Internet which are outside from the Twitter community extracted from the LAION dataset.", unsafe_allow_html=True)
|
| 45 |
+
st.markdown("Leveraging the largest publicly available pathology dataset which contains image–text pairs across 32 different pathology subspecialty-specific hashtags, where each image has detailed text descriptions, we fine-tuned a pre-trained CLIP model and proposed a multimodal deep learning model for pathology, PLIP.", unsafe_allow_html=True)
|
| 46 |
+
with col2:
|
| 47 |
+
fig1c = Image.open('resources/4x/Fig1c.png')
|
| 48 |
+
st.image(fig1c, caption='Pathology hashtags in Twitter', output_format='png')
|
| 49 |
+
fig1d = Image.open('resources/4x/Fig1d.png')
|
| 50 |
+
st.image(fig1d, caption='Number of words in sentence', output_format='png')
|
| 51 |
+
|
| 52 |
+
|
| 53 |
+
|
| 54 |
+
st.markdown("#### PLIP is trained with connecting the image and text via contrastive learning")
|
| 55 |
+
|
| 56 |
+
col1, col2 = st.columns([3, 1])
|
| 57 |
+
with col1:
|
| 58 |
+
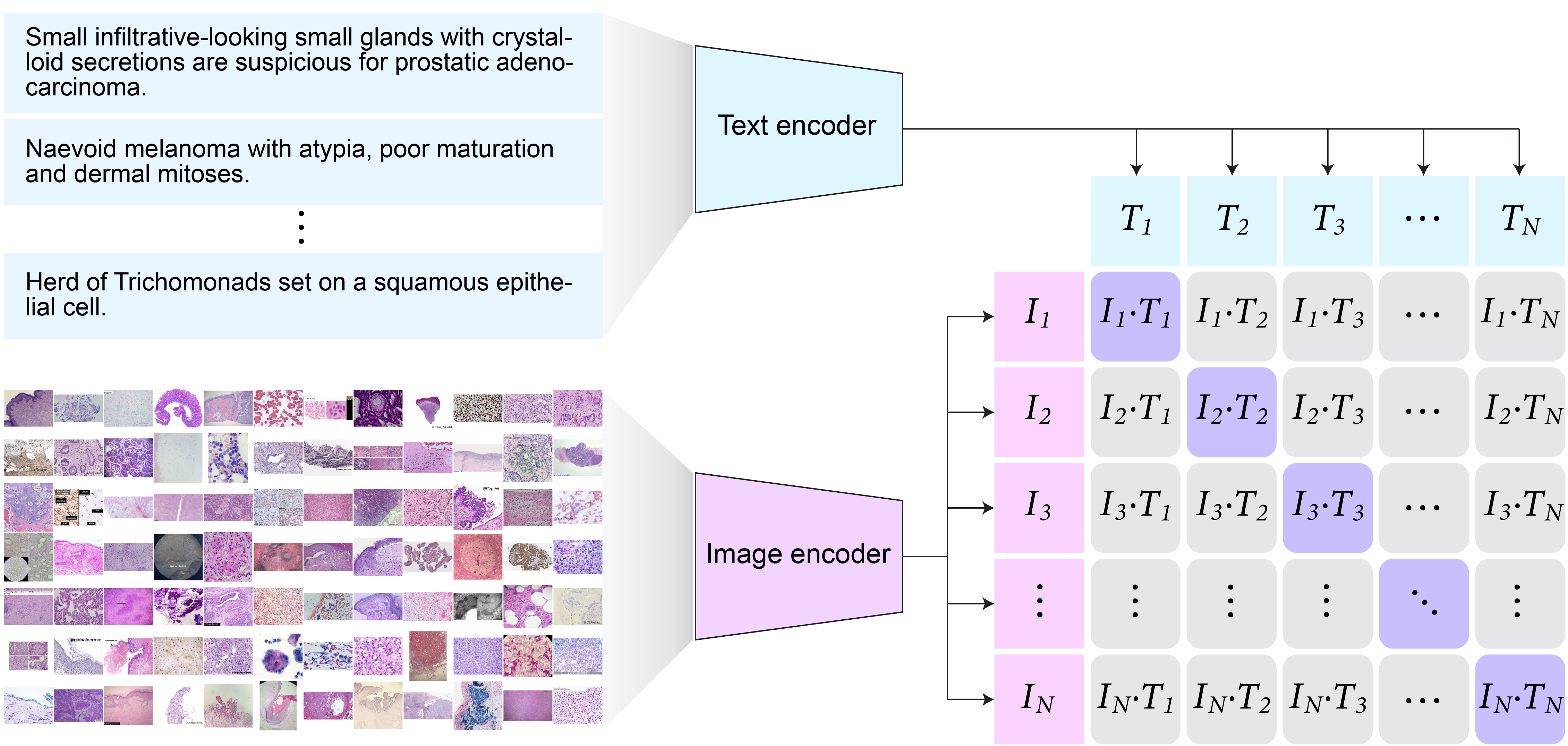
st.markdown("The proposed PLIP model generates two embedding vectors from both the text and image encoders. These vectors were then forced to be similar for each of the paired image and text vectors and dissimilar for non-paired image and text pairs via contrastive learning.", unsafe_allow_html=True)
|
| 59 |
+
fig1e = Image.open('resources/4x/Fig1e.png')
|
| 60 |
+
st.image(fig1e, caption='PLIP training', output_format='png')
|
| 61 |
+
|
| 62 |
+
with col2:
|
| 63 |
+
fig1f = Image.open('resources/4x/Fig1f.png')
|
| 64 |
+
st.image(fig1f, caption='Training illustration', output_format='png')
|
image2image.py
CHANGED
|
@@ -8,6 +8,7 @@ import os
|
|
| 8 |
from io import BytesIO
|
| 9 |
import pickle
|
| 10 |
import base64
|
|
|
|
| 11 |
|
| 12 |
import torch
|
| 13 |
from transformers import (
|
|
@@ -65,15 +66,18 @@ def embed_texts(model, texts, processor):
|
|
| 65 |
def app():
|
| 66 |
st.title('Image to Image Retrieval')
|
| 67 |
st.markdown('#### A pathology image search engine that correlate images with images.')
|
| 68 |
-
|
|
|
|
|
|
|
|
|
|
| 69 |
meta, image_embedding, text_embedding, validation_subset_index = init()
|
| 70 |
model, processor = load_path_clip()
|
| 71 |
|
| 72 |
|
| 73 |
col1, col2 = st.columns(2)
|
| 74 |
with col1:
|
| 75 |
-
data_options = ["All twitter data (
|
| 76 |
-
"Twitter validation data (
|
| 77 |
st.radio(
|
| 78 |
"Choose dataset for image retrieval 👉",
|
| 79 |
key="datapool",
|
|
|
|
| 8 |
from io import BytesIO
|
| 9 |
import pickle
|
| 10 |
import base64
|
| 11 |
+
import datetime
|
| 12 |
|
| 13 |
import torch
|
| 14 |
from transformers import (
|
|
|
|
| 66 |
def app():
|
| 67 |
st.title('Image to Image Retrieval')
|
| 68 |
st.markdown('#### A pathology image search engine that correlate images with images.')
|
| 69 |
+
st.markdown("Image-to-image retrieval can be used to retrieve pathology images that have contents similar to the target image input, with the ability to comprehend the key components from the input image.")
|
| 70 |
+
|
| 71 |
+
st.markdown('#### Demo')
|
| 72 |
+
|
| 73 |
meta, image_embedding, text_embedding, validation_subset_index = init()
|
| 74 |
model, processor = load_path_clip()
|
| 75 |
|
| 76 |
|
| 77 |
col1, col2 = st.columns(2)
|
| 78 |
with col1:
|
| 79 |
+
data_options = ["All twitter data (03/21/2006 — 01/15/2023)",
|
| 80 |
+
"Twitter validation data (11/16/2022 — 01/15/2023)"]
|
| 81 |
st.radio(
|
| 82 |
"Choose dataset for image retrieval 👉",
|
| 83 |
key="datapool",
|
introduction.md
CHANGED
|
@@ -1,4 +1,6 @@
|
|
| 1 |
|
| 2 |
-
#
|
| 3 |
|
| 4 |
-
The
|
|
|
|
|
|
|
|
|
| 1 |
|
| 2 |
+
# Leveraging medical Twitter to build a visual-language foundation model for pathology
|
| 3 |
|
| 4 |
+
The lack of annotated publicly available medical images is a major barrier for innovations. At the same time, many de-identified images and much knowledge are shared by clinicians on public forums such as medical Twitter. Here we harness these crowd platforms to curate OpenPath, a large dataset of 208,414 pathology images paired with natural language descriptions. This is the largest public dataset for pathology images annotated with natural text. We demonstrate the value of this resource by developing PLIP, a multimodal AI with both image and text understanding, which is trained on OpenPath. PLIP achieves state-of-the-art zero-shot and few-short performance for classifying new pathology images across diverse tasks. Moreover, PLIP enables users to retrieve similar cases by either image or natural language search, greatly facilitating knowledge sharing. Our approach demonstrates that publicly shared medical data is a tremendous opportunity that can be harnessed to advance biomedical AI.
|
| 5 |
+
|
| 6 |
+

|
requirements.txt
CHANGED
|
@@ -5,4 +5,6 @@ pandas
|
|
| 5 |
numpy
|
| 6 |
Pillow
|
| 7 |
streamlit==1.19.0
|
| 8 |
-
st_clickable_images
|
|
|
|
|
|
|
|
|
| 5 |
numpy
|
| 6 |
Pillow
|
| 7 |
streamlit==1.19.0
|
| 8 |
+
st_clickable_images
|
| 9 |
+
plotly
|
| 10 |
+
datetime
|
resources/4x/Fig1.png
ADDED

|
Git LFS Details
|
resources/4x/Fig1ab.png
ADDED

|
Git LFS Details
|
resources/4x/Fig1c.png
ADDED

|
Git LFS Details
|
resources/4x/Fig1d.png
ADDED

|
Git LFS Details
|
resources/4x/Fig1e.png
ADDED
|
Git LFS Details
|
resources/4x/Fig1f.png
ADDED

|
Git LFS Details
|
resources/4x/image_retrieval.png
ADDED

|
Git LFS Details
|
text2image.py
CHANGED
|
@@ -64,16 +64,24 @@ def app():
|
|
| 64 |
|
| 65 |
st.title('Text to Image Retrieval')
|
| 66 |
st.markdown('#### A pathology image search engine that correlate texts directly with images.')
|
| 67 |
-
st.caption('Note: The searching query matches images only. The twitter text does not used for searching.')
|
| 68 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 69 |
meta, image_embedding, text_embedding, validation_subset_index = init()
|
| 70 |
model, processor = load_path_clip()
|
| 71 |
|
|
|
|
| 72 |
|
| 73 |
col1, col2 = st.columns(2)
|
| 74 |
with col1:
|
| 75 |
-
data_options = ["All twitter data (
|
| 76 |
-
"Twitter validation data (
|
| 77 |
st.radio(
|
| 78 |
"Choose dataset for image retrieval 👉",
|
| 79 |
key="datapool",
|
|
@@ -81,7 +89,7 @@ def app():
|
|
| 81 |
)
|
| 82 |
with col2:
|
| 83 |
retrieval_options = ["Image only",
|
| 84 |
-
"
|
| 85 |
]
|
| 86 |
st.radio(
|
| 87 |
"Similarity calcuation Mapping input with 👉",
|
|
|
|
| 64 |
|
| 65 |
st.title('Text to Image Retrieval')
|
| 66 |
st.markdown('#### A pathology image search engine that correlate texts directly with images.')
|
|
|
|
| 67 |
|
| 68 |
+
col1, col2 = st.columns([1,1])
|
| 69 |
+
with col1:
|
| 70 |
+
st.markdown("The text-to-image retrieval system can serve as an image search engine, enabling users to match images from multiple queries and retrieve the most relevant image based on a sentence description. This generic system can comprehend semantic and interrelated knowledge, such as “Breast tumor surrounded by fat”.")
|
| 71 |
+
st.markdown("Unlike searching keywords and sentences from Google and indirectly matching the images from the target text, our proposed pathology image retrieval allows direct comparison between input sentences and images.")
|
| 72 |
+
with col2:
|
| 73 |
+
fig1 = Image.open('resources/4x/image_retrieval.png')
|
| 74 |
+
st.image(fig1, caption='Image retrieval from text', width=400, output_format='png')
|
| 75 |
+
|
| 76 |
meta, image_embedding, text_embedding, validation_subset_index = init()
|
| 77 |
model, processor = load_path_clip()
|
| 78 |
|
| 79 |
+
st.markdown('#### Demo')
|
| 80 |
|
| 81 |
col1, col2 = st.columns(2)
|
| 82 |
with col1:
|
| 83 |
+
data_options = ["All twitter data (03/21/2006 — 01/15/2023)",
|
| 84 |
+
"Twitter validation data (11/16/2022 — 01/15/2023)"]
|
| 85 |
st.radio(
|
| 86 |
"Choose dataset for image retrieval 👉",
|
| 87 |
key="datapool",
|
|
|
|
| 89 |
)
|
| 90 |
with col2:
|
| 91 |
retrieval_options = ["Image only",
|
| 92 |
+
"Text and image (beta)",
|
| 93 |
]
|
| 94 |
st.radio(
|
| 95 |
"Similarity calcuation Mapping input with 👉",
|
visualization.py
ADDED
|
@@ -0,0 +1,44 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from pathlib import Path
|
| 2 |
+
import streamlit as st
|
| 3 |
+
import streamlit.components.v1 as components
|
| 4 |
+
import plotly.figure_factory as ff
|
| 5 |
+
import numpy as np
|
| 6 |
+
import pandas as pd
|
| 7 |
+
#from streamlit_plotly_events import plotly_events
|
| 8 |
+
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
def app():
|
| 12 |
+
st.markdown('#### Visualization')
|
| 13 |
+
|
| 14 |
+
img_2d_embed = pd.read_csv('data/img_2d_embedding.csv', index_col=0)
|
| 15 |
+
img_2d_embed = img_2d_embed.sample(frac=0.1, random_state=0)
|
| 16 |
+
|
| 17 |
+
txt_2d_embed = pd.read_csv('data/txt_2d_embedding.csv', index_col=0)
|
| 18 |
+
txt_2d_embed = txt_2d_embed.sample(frac=0.1, random_state=0)
|
| 19 |
+
|
| 20 |
+
col1, col2 = st.columns(2)
|
| 21 |
+
with col1:
|
| 22 |
+
fig1 = ff.create_2d_density(
|
| 23 |
+
x=img_2d_embed['UMAP_1'],
|
| 24 |
+
y=img_2d_embed['UMAP_2'],
|
| 25 |
+
#colors=img_2d_embed['tag'],
|
| 26 |
+
colorscale='Blues', # set the color map
|
| 27 |
+
height=500, # set height of the figure
|
| 28 |
+
width=500, # set width of the figure
|
| 29 |
+
title='Image embedding visualized in 2D UMAP'
|
| 30 |
+
)
|
| 31 |
+
#selected_points = plotly_events(fig1, click_event=True, hover_event=True)
|
| 32 |
+
st.plotly_chart(fig1, use_container_width=True)
|
| 33 |
+
|
| 34 |
+
with col2:
|
| 35 |
+
fig2 = ff.create_2d_density(
|
| 36 |
+
x=txt_2d_embed['UMAP_1'],
|
| 37 |
+
y=txt_2d_embed['UMAP_2'],
|
| 38 |
+
#colors=img_2d_embed['tag'],
|
| 39 |
+
colorscale='Blues', # set the color map
|
| 40 |
+
height=500, # set height of the figure
|
| 41 |
+
width=500, # set width of the figure
|
| 42 |
+
title='Text embedding visualized in 2D UMAP'
|
| 43 |
+
)
|
| 44 |
+
st.plotly_chart(fig2, use_container_width=True)
|
viz_scripts/calc_img.py
ADDED
|
@@ -0,0 +1,75 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
#!/usr/bin/env python3
|
| 2 |
+
# -*- coding: utf-8 -*-
|
| 3 |
+
"""
|
| 4 |
+
Created on Fri Mar 10 21:13:04 2023
|
| 5 |
+
|
| 6 |
+
@author: zhihuang
|
| 7 |
+
"""
|
| 8 |
+
|
| 9 |
+
import pickle
|
| 10 |
+
import os
|
| 11 |
+
import pandas as pd
|
| 12 |
+
import numpy as np
|
| 13 |
+
import umap
|
| 14 |
+
import seaborn as sns
|
| 15 |
+
import matplotlib.pyplot as plt
|
| 16 |
+
opj=os.path.join
|
| 17 |
+
|
| 18 |
+
if __name__ == '__main__':
|
| 19 |
+
dd = '/home/zhihuang/Desktop/webplip/data'
|
| 20 |
+
with open(opj(dd, 'twitter.asset'),'rb') as f:
|
| 21 |
+
data = pickle.load(f)
|
| 22 |
+
|
| 23 |
+
n_neighbors = 15
|
| 24 |
+
random_state = 0
|
| 25 |
+
|
| 26 |
+
reducer = umap.UMAP(n_components=2,
|
| 27 |
+
n_neighbors=n_neighbors,
|
| 28 |
+
min_dist=0.1,
|
| 29 |
+
metric='euclidean',
|
| 30 |
+
random_state=random_state)
|
| 31 |
+
img_2d = reducer.fit(data['image_embedding'])
|
| 32 |
+
img_2d = reducer.transform(data['image_embedding'])
|
| 33 |
+
df_img = pd.DataFrame(np.c_[img_2d, data['meta'].values], columns = ['UMAP_1','UMAP_2'] + list(data['meta'].columns))
|
| 34 |
+
df_img.to_csv(opj(dd, 'img_2d_embedding.csv'))
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
# reducer = umap.UMAP(n_components=2,
|
| 38 |
+
# n_neighbors=n_neighbors,
|
| 39 |
+
# min_dist=0.1,
|
| 40 |
+
# metric='euclidean',
|
| 41 |
+
# random_state=random_state)
|
| 42 |
+
txt_2d = reducer.fit_transform(data['text_embedding'])
|
| 43 |
+
df_txt = pd.DataFrame(np.c_[txt_2d, data['meta'].values], columns = ['UMAP_1','UMAP_2'] + list(data['meta'].columns))
|
| 44 |
+
df_txt.to_csv(opj(dd, 'txt_2d_embedding.csv'))
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
fig, ax = plt.subplots(1,2, figsize=(20,10))
|
| 50 |
+
sns.scatterplot(data=df_img,
|
| 51 |
+
x='UMAP_1',
|
| 52 |
+
y='UMAP_2',
|
| 53 |
+
alpha=0.2,
|
| 54 |
+
ax=ax[0],
|
| 55 |
+
hue='tag'
|
| 56 |
+
)
|
| 57 |
+
|
| 58 |
+
sns.scatterplot(data=df_txt,
|
| 59 |
+
x='UMAP_1',
|
| 60 |
+
y='UMAP_2',
|
| 61 |
+
alpha=0.2,
|
| 62 |
+
ax=ax[1],
|
| 63 |
+
hue='tag'
|
| 64 |
+
)
|
| 65 |
+
|
| 66 |
+
|
| 67 |
+
|
| 68 |
+
|
| 69 |
+
|
| 70 |
+
|
| 71 |
+
|
| 72 |
+
|
| 73 |
+
|
| 74 |
+
|
| 75 |
+
|