url
stringlengths 58
58
| repository_url
stringclasses 1
value | labels_url
stringlengths 72
72
| comments_url
stringlengths 67
67
| events_url
stringlengths 65
65
| html_url
stringlengths 46
48
| id
int64 1.35B
1.52B
| node_id
stringlengths 18
19
| number
int64 571
673
| title
stringlengths 1
172
| user
dict | labels
listlengths 0
2
| state
stringclasses 2
values | locked
bool 1
class | assignee
float64 | assignees
sequencelengths 0
0
| milestone
float64 | comments
sequencelengths 0
30
| created_at
unknown | updated_at
unknown | closed_at
unknown | author_association
stringclasses 1
value | active_lock_reason
float64 | body
stringlengths 28
22.9k
⌀ | reactions
dict | timeline_url
stringlengths 67
67
| performed_via_github_app
float64 | state_reason
stringclasses 2
values | draft
float64 0
0
⌀ | pull_request
dict | is_pull_request
bool 2
classes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
https://api.github.com/repos/deepmind/alphafold/issues/673 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/673/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/673/comments | https://api.github.com/repos/deepmind/alphafold/issues/673/events | https://github.com/deepmind/alphafold/issues/673 | 1,524,110,688 | I_kwDOFoWQLM5a2BVg | 673 | Alphafold-Multimer was run for more than 48h, I still couldn't get the structure of a complex with 4 subunits (total aa length: 2222+689+201+196). | {
"avatar_url": "https://avatars.githubusercontent.com/u/22016970?v=4",
"events_url": "https://api.github.com/users/echoduan/events{/privacy}",
"followers_url": "https://api.github.com/users/echoduan/followers",
"following_url": "https://api.github.com/users/echoduan/following{/other_user}",
"gists_url": "https://api.github.com/users/echoduan/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/echoduan",
"id": 22016970,
"login": "echoduan",
"node_id": "MDQ6VXNlcjIyMDE2OTcw",
"organizations_url": "https://api.github.com/users/echoduan/orgs",
"received_events_url": "https://api.github.com/users/echoduan/received_events",
"repos_url": "https://api.github.com/users/echoduan/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/echoduan/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/echoduan/subscriptions",
"type": "User",
"url": "https://api.github.com/users/echoduan"
} | [] | open | false | null | [] | null | [] | "2023-01-07T20:02:59" | "2023-01-07T20:02:59" | null | NONE | null | Hi, author
python3 docker/run_docker.py \
--fasta_paths=complex1.fasta \
--max_template_date=2022-12-30 \
--model_preset=multimer \
--data_dir=/data --gpu_devices=0,1,2,3 > complex1.log &
1) The program was always calculating the model until now.
I0107 01:49:08.176561 140086208259904 run_docker.py:255] I0107 01:49:08.175831 140502687897408 run_alphafold.py:191] Running model model_1_multimer_v3_pred_0 on complex1
I0107 01:49:08.178550 140086208259904 run_docker.py:255] I0107 01:49:08.178094 140502687897408 model.py:165] Running predict with shape(feat) = {'aatype': (3308,), 'residue_index': (3308,), 'seq_length': (), 'msa': (5799, 3308), 'num_alignments': (), 'template_aatype': (4, 3308), 'template_all_atom_mask': (4, 3308, 37), 'template_all_atom_positions': (4, 3308, 37, 3), 'asym_id': (3308,), 'sym_id': (3308,), 'entity_id': (3308,), 'deletion_matrix': (5799, 3308), 'deletion_mean': (3308,), 'all_atom_mask': (3308, 37), 'all_atom_positions': (3308, 37, 3), 'assembly_num_chains': (), 'entity_mask': (3308,), 'num_templates': (), 'cluster_bias_mask': (5799,), 'bert_mask': (5799, 3308), 'seq_mask': (3308,), 'msa_mask': (5799, 3308)}
2) The instance (p3.8xlarge) has four GPUs (4xNVIDIA V100). All GPUs are available to calculate the model (--gpu_devices=0,1,2,3). But I noticed that 'run_alphafold.py' seems to use four GPUs, but why only the first one GPU memory is fulfilled?

How can I use the four GPUs with maximum memory usage at the same time to speed up the prediction?
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/673/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/673/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/672 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/672/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/672/comments | https://api.github.com/repos/deepmind/alphafold/issues/672/events | https://github.com/deepmind/alphafold/pull/672 | 1,514,648,843 | PR_kwDOFoWQLM5GZVWy | 672 | Fixing zip command issue in Google Colab. | {
"avatar_url": "https://avatars.githubusercontent.com/u/22454783?v=4",
"events_url": "https://api.github.com/users/gmihaila/events{/privacy}",
"followers_url": "https://api.github.com/users/gmihaila/followers",
"following_url": "https://api.github.com/users/gmihaila/following{/other_user}",
"gists_url": "https://api.github.com/users/gmihaila/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/gmihaila",
"id": 22454783,
"login": "gmihaila",
"node_id": "MDQ6VXNlcjIyNDU0Nzgz",
"organizations_url": "https://api.github.com/users/gmihaila/orgs",
"received_events_url": "https://api.github.com/users/gmihaila/received_events",
"repos_url": "https://api.github.com/users/gmihaila/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/gmihaila/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/gmihaila/subscriptions",
"type": "User",
"url": "https://api.github.com/users/gmihaila"
} | [] | open | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/672/checks?check_run_id=10366996221) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request.",
"Added my CLA."
] | "2022-12-30T17:02:08" | "2022-12-30T17:08:29" | null | NONE | null | When running the Google Colab notebook implementation cell `5. Run AlphaFold and download prediction` is throwing the following error:
```
NotImplementedError Traceback (most recent call last)
[<ipython-input-1-bc0091fa34e2>](https://localhost:8080/#) in <module>()
577 sequence = 'AKIGLFYGTQTGVTQTIAESIQQEFGGESIVDLNDIANADASDLNAYDYLIIGCPTWNVGELQSDWEGIYDDLDSVNFQGKKVAYFGAGDQVGYSDNFQDAMGILEEKISSLGSQTVGYWPIEGYDFNESKAVRNNQFVGLAIDEDNQPDLTKNRIKTWVSQLKSEFGL' #@param {type:"string"}
578
--> 579 run_prediction(sequence)
3 frames
[/usr/local/lib/python3.7/dist-packages/google/colab/_system_commands.py](https://localhost:8080/#) in _run_command(cmd, clear_streamed_output)
166 if locale_encoding != _ENCODING:
167 raise NotImplementedError(
--> 168 'A UTF-8 locale is required. Got {}'.format(locale_encoding))
169
170 parent_pty, child_pty = pty.openpty()
NotImplementedError: A UTF-8 locale is required. Got ANSI_X3.4-1968
```
As discussed in the issue: https://github.com/deepmind/alphafold/issues/483
This is more of a Google Colab issue when trying to create the`output_dir ` zip file from the`output_dir ` folder. I solved this by using`shutil ` to create the zip file.
I simply replace the line of code `!zip -q -r {output_dir}.zip {output_dir} ` from cell `5. Run AlphaFold and download prediction` with `shutil.make_archive(output_dir, 'zip', output_dir)`
I also made sure to add `import shutil` at the first cell.
This will create the zip file without using any terminal commands. This fix should not cause any issues in the future since we don't rely on the Google Colab terminal commands anymore.
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/672/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/672/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/672.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/672",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/672.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/672"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/671 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/671/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/671/comments | https://api.github.com/repos/deepmind/alphafold/issues/671/events | https://github.com/deepmind/alphafold/issues/671 | 1,513,313,185 | I_kwDOFoWQLM5aM1Oh | 671 | uniref90 can't download | {
"avatar_url": "https://avatars.githubusercontent.com/u/49931482?v=4",
"events_url": "https://api.github.com/users/hezhiqiang8909/events{/privacy}",
"followers_url": "https://api.github.com/users/hezhiqiang8909/followers",
"following_url": "https://api.github.com/users/hezhiqiang8909/following{/other_user}",
"gists_url": "https://api.github.com/users/hezhiqiang8909/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/hezhiqiang8909",
"id": 49931482,
"login": "hezhiqiang8909",
"node_id": "MDQ6VXNlcjQ5OTMxNDgy",
"organizations_url": "https://api.github.com/users/hezhiqiang8909/orgs",
"received_events_url": "https://api.github.com/users/hezhiqiang8909/received_events",
"repos_url": "https://api.github.com/users/hezhiqiang8909/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/hezhiqiang8909/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/hezhiqiang8909/subscriptions",
"type": "User",
"url": "https://api.github.com/users/hezhiqiang8909"
} | [] | open | false | null | [] | null | [
"Replace ftp://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref90/uniref90.fasta.gz with https://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref90/uniref90. fasta.gz solves the problem of not being able to download\r\n\r\n"
] | "2022-12-29T02:30:48" | "2022-12-29T02:41:03" | null | NONE | null | 
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/671/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/671/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/670 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/670/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/670/comments | https://api.github.com/repos/deepmind/alphafold/issues/670/events | https://github.com/deepmind/alphafold/pull/670 | 1,512,478,836 | PR_kwDOFoWQLM5GSDsg | 670 | Implement case-insensitive mmcif parsing | {
"avatar_url": "https://avatars.githubusercontent.com/u/49076030?v=4",
"events_url": "https://api.github.com/users/SimonKitSangChu/events{/privacy}",
"followers_url": "https://api.github.com/users/SimonKitSangChu/followers",
"following_url": "https://api.github.com/users/SimonKitSangChu/following{/other_user}",
"gists_url": "https://api.github.com/users/SimonKitSangChu/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/SimonKitSangChu",
"id": 49076030,
"login": "SimonKitSangChu",
"node_id": "MDQ6VXNlcjQ5MDc2MDMw",
"organizations_url": "https://api.github.com/users/SimonKitSangChu/orgs",
"received_events_url": "https://api.github.com/users/SimonKitSangChu/received_events",
"repos_url": "https://api.github.com/users/SimonKitSangChu/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/SimonKitSangChu/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/SimonKitSangChu/subscriptions",
"type": "User",
"url": "https://api.github.com/users/SimonKitSangChu"
} | [] | open | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/670/checks?check_run_id=10330182688) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request."
] | "2022-12-28T07:04:19" | "2022-12-28T07:09:03" | null | NONE | null | [Alphafold database](https://github.com/deepmind/alphafold/tree/main/afdb) cif-s are annotated with capitalized PEPTIDE. Parsing these files with the existing case-sensitive function results in empty `mmcif_object`. Suggest switching to case-insensitive parsing. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/670/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/670/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/670.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/670",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/670.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/670"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/669 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/669/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/669/comments | https://api.github.com/repos/deepmind/alphafold/issues/669/events | https://github.com/deepmind/alphafold/issues/669 | 1,511,606,829 | I_kwDOFoWQLM5aGUot | 669 | pip._vendor.urllib3.exceptions.ReadTimeoutError: HTTPSConnectionPool(host='files.pythonhosted.org', port=443): Read timed out. | {
"avatar_url": "https://avatars.githubusercontent.com/u/49931482?v=4",
"events_url": "https://api.github.com/users/hezhiqiang8909/events{/privacy}",
"followers_url": "https://api.github.com/users/hezhiqiang8909/followers",
"following_url": "https://api.github.com/users/hezhiqiang8909/following{/other_user}",
"gists_url": "https://api.github.com/users/hezhiqiang8909/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/hezhiqiang8909",
"id": 49931482,
"login": "hezhiqiang8909",
"node_id": "MDQ6VXNlcjQ5OTMxNDgy",
"organizations_url": "https://api.github.com/users/hezhiqiang8909/orgs",
"received_events_url": "https://api.github.com/users/hezhiqiang8909/received_events",
"repos_url": "https://api.github.com/users/hezhiqiang8909/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/hezhiqiang8909/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/hezhiqiang8909/subscriptions",
"type": "User",
"url": "https://api.github.com/users/hezhiqiang8909"
} | [] | open | false | null | [] | null | [] | "2022-12-27T09:18:35" | "2022-12-27T09:18:35" | null | NONE | null | 
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/669/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/669/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/668 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/668/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/668/comments | https://api.github.com/repos/deepmind/alphafold/issues/668/events | https://github.com/deepmind/alphafold/issues/668 | 1,511,549,702 | I_kwDOFoWQLM5aGGsG | 668 | fatal: unable to access 'https://github.com/soedinglab/hh-suite.git/': gnutls_handshake() failed: The TLS connection was non-properly terminated. | {
"avatar_url": "https://avatars.githubusercontent.com/u/49931482?v=4",
"events_url": "https://api.github.com/users/hezhiqiang8909/events{/privacy}",
"followers_url": "https://api.github.com/users/hezhiqiang8909/followers",
"following_url": "https://api.github.com/users/hezhiqiang8909/following{/other_user}",
"gists_url": "https://api.github.com/users/hezhiqiang8909/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/hezhiqiang8909",
"id": 49931482,
"login": "hezhiqiang8909",
"node_id": "MDQ6VXNlcjQ5OTMxNDgy",
"organizations_url": "https://api.github.com/users/hezhiqiang8909/orgs",
"received_events_url": "https://api.github.com/users/hezhiqiang8909/received_events",
"repos_url": "https://api.github.com/users/hezhiqiang8909/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/hezhiqiang8909/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/hezhiqiang8909/subscriptions",
"type": "User",
"url": "https://api.github.com/users/hezhiqiang8909"
} | [] | open | false | null | [] | null | [] | "2022-12-27T08:09:29" | "2022-12-27T08:09:29" | null | NONE | null | Making a new docker image shows The TLS connection was non-properly terminated.

| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/668/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/668/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/667 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/667/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/667/comments | https://api.github.com/repos/deepmind/alphafold/issues/667/events | https://github.com/deepmind/alphafold/issues/667 | 1,510,736,623 | I_kwDOFoWQLM5aDALv | 667 | CUDA_ERROR_ILLEGAL_ADDRESS error with AlphaFold multimer 2.3.0 | {
"avatar_url": "https://avatars.githubusercontent.com/u/116328913?v=4",
"events_url": "https://api.github.com/users/sz-1002/events{/privacy}",
"followers_url": "https://api.github.com/users/sz-1002/followers",
"following_url": "https://api.github.com/users/sz-1002/following{/other_user}",
"gists_url": "https://api.github.com/users/sz-1002/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/sz-1002",
"id": 116328913,
"login": "sz-1002",
"node_id": "U_kgDOBu8J0Q",
"organizations_url": "https://api.github.com/users/sz-1002/orgs",
"received_events_url": "https://api.github.com/users/sz-1002/received_events",
"repos_url": "https://api.github.com/users/sz-1002/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/sz-1002/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/sz-1002/subscriptions",
"type": "User",
"url": "https://api.github.com/users/sz-1002"
} | [] | open | false | null | [] | null | [
"Hi @sz-1002 \r\nI am having the same issue on the relaxation step.\r\nhttps://github.com/deepmind/alphafold/issues/660\r\n\r\nI tried with newer version of CUDA (11.6) and had the same issue.",
"Hi @leiterenato \r\nThanks for letting me know! Hopefully there will be a solution for this.",
"a different non-relax model issue seems to have been resolved by using cuda 11.8 https://github.com/deepmind/alphafold/issues/646#issuecomment-1364912401\r\n\r\nassuming that doesn't work, can try use_gpu_relax=False, or turn off relax entirely with run_relax=False. we will attempt to address the problem more fully in the new year.",
"We think this is due to jax version change from 0.3.17 to 0.3.25. We don't want to revert jax version though, so are looking for workarounds."
] | "2022-12-26T08:22:37" | "2023-01-05T10:37:26" | null | NONE | null | Hi!
I am trying to run AlphaFold 2.3.0 multimer and encountered this error: `Execution of replica 0 failed: INTERNAL: Failed to load in-memory CUBIN: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered` (see details below). I was wondering if you could help me resolve it? Thank you very much!!
**Machine spec etc.:**
- Ubuntu 20.04 LTS
- GPU: nvidia RTX 3090
- cuda version: tried both 11.1.1 and 11.4.0 (both gave the same error)
- total length of the protein complex: ~2200 aa
When I searched related errors online, it seems there are generally two solutions proposed: (1) change to a newer cuda version [https://github.com/deepmind/dm-haiku/issues/204](url), or (2) disable unified memory [https://github.com/deepmind/alphafold/issues/406](url).
I tried using cuda 11.4.0 instead of 11.1.1 by changing the following lines in Dockerfile, but the same error persists.
```
ARG CUDA=11.1.1 --->>> ARG CUDA=11.4.0
FROM nvidia/cuda:${CUDA}-cudnn8-runtime-ubuntu18.04 --->>> FROM nvidia/cuda:${CUDA}-cudnn8-runtime-ubuntu20.04
conda install -y -c conda-forge cudatoolkit==${CUDA_VERSION} --->>> conda install -y -c "nvidia/label/cuda-11.4.0" cuda-toolkit
```
As for (2) disable unified memory, I am worried that this would give me out of memory error given the size of the protein.
Not sure if this is relevant, but this is a recent problem and prediction for this and other similarly-sized complexes worked fine before (was using v2.2.0 before, and I wonder if this is an issue with e.g. version of jax or jaxlib).
Thank you very much!
**Error message:**
```
I1226 15:42:37.167795 140027647279936 run_docker.py:255] I1226 06:42:37.167222 140635718940480 amber_minimize.py:407] Minimizing protein, attempt 1 of 100.
I1226 15:42:39.806318 140027647279936 run_docker.py:255] I1226 06:42:39.805861 140635718940480 amber_minimize.py:68] Restraining 17790 / 35357 particles.
I1226 15:45:15.867727 140027647279936 run_docker.py:255] I1226 06:45:15.866998 140635718940480 amber_minimize.py:177] alterations info: {'nonstandard_residues': [], 'removed_heterogens': set(), 'missing_residues': {}, 'missing_heavy_atoms': {}, 'missing_terminals': {}, 'Se_in_MET': [], 'removed_chains': {0: []}}
I1226 15:45:42.889597 140027647279936 run_docker.py:255] 2022-12-26 06:45:42.889173: E external/org_tensorflow/tensorflow/compiler/xla/pjrt/pjrt_stream_executor_client.cc:2153] Execution of replica 0 failed: INTERNAL: Failed to load in-memory CUBIN: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered
I1226 15:45:42.899475 140027647279936 run_docker.py:255] Traceback (most recent call last):
I1226 15:45:42.899577 140027647279936 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 432, in <module>
I1226 15:45:42.899646 140027647279936 run_docker.py:255] app.run(main)
I1226 15:45:42.899709 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/absl/app.py", line 312, in run
I1226 15:45:42.899771 140027647279936 run_docker.py:255] _run_main(main, args)
I1226 15:45:42.899834 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/absl/app.py", line 258, in _run_main
I1226 15:45:42.899896 140027647279936 run_docker.py:255] sys.exit(main(argv))
I1226 15:45:42.899957 140027647279936 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 408, in main
I1226 15:45:42.900018 140027647279936 run_docker.py:255] predict_structure(
I1226 15:45:42.900109 140027647279936 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 243, in predict_structure
I1226 15:45:42.900174 140027647279936 run_docker.py:255] relaxed_pdb_str, _, violations = amber_relaxer.process(
I1226 15:45:42.900234 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/relax/relax.py", line 62, in process
I1226 15:45:42.900292 140027647279936 run_docker.py:255] out = amber_minimize.run_pipeline(
I1226 15:45:42.900353 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/relax/amber_minimize.py", line 489, in run_pipeline
I1226 15:45:42.900412 140027647279936 run_docker.py:255] ret.update(get_violation_metrics(prot))
I1226 15:45:42.900472 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/relax/amber_minimize.py", line 357, in get_violation_metrics
I1226 15:45:42.900531 140027647279936 run_docker.py:255] structural_violations, struct_metrics = find_violations(prot)
I1226 15:45:42.900591 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/relax/amber_minimize.py", line 339, in find_violations
I1226 15:45:42.900651 140027647279936 run_docker.py:255] violations = folding.find_structural_violations(
I1226 15:45:42.900712 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/model/folding.py", line 761, in find_structural_violations
I1226 15:45:42.900774 140027647279936 run_docker.py:255] between_residue_clashes = all_atom.between_residue_clash_loss(
I1226 15:45:42.900835 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/model/all_atom.py", line 783, in between_residue_clash_loss
I1226 15:45:42.900898 140027647279936 run_docker.py:255] dists = jnp.sqrt(1e-10 + jnp.sum(
I1226 15:45:42.900959 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/jax/_src/numpy/reductions.py", line 216, in sum
I1226 15:45:42.901019 140027647279936 run_docker.py:255] return _reduce_sum(a, axis=_ensure_optional_axes(axis), dtype=dtype, out=out,
I1226 15:45:42.901078 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/jax/_src/traceback_util.py", line 162, in reraise_with_filtered_traceback
I1226 15:45:42.901138 140027647279936 run_docker.py:255] return fun(*args, **kwargs)
I1226 15:45:42.901199 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/jax/_src/api.py", line 623, in cache_miss
I1226 15:45:42.901261 140027647279936 run_docker.py:255] out_flat = call_bind_continuation(execute(*args_flat))
I1226 15:45:42.901322 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/jax/_src/dispatch.py", line 895, in _execute_compiled
I1226 15:45:42.901383 140027647279936 run_docker.py:255] out_flat = compiled.execute(in_flat)
I1226 15:45:42.901446 140027647279936 run_docker.py:255] jax._src.traceback_util.UnfilteredStackTrace: jaxlib.xla_extension.XlaRuntimeError: INTERNAL: Failed to load in-memory CUBIN: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered
I1226 15:45:42.901507 140027647279936 run_docker.py:255]
I1226 15:45:42.901568 140027647279936 run_docker.py:255] The stack trace below excludes JAX-internal frames.
I1226 15:45:42.901637 140027647279936 run_docker.py:255] The preceding is the original exception that occurred, unmodified.
```
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/667/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/667/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/666 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/666/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/666/comments | https://api.github.com/repos/deepmind/alphafold/issues/666/events | https://github.com/deepmind/alphafold/issues/666 | 1,510,501,796 | I_kwDOFoWQLM5aCG2k | 666 | Colab pro + failing to predict >2000 residues | {
"avatar_url": "https://avatars.githubusercontent.com/u/121411082?v=4",
"events_url": "https://api.github.com/users/HBCH1/events{/privacy}",
"followers_url": "https://api.github.com/users/HBCH1/followers",
"following_url": "https://api.github.com/users/HBCH1/following{/other_user}",
"gists_url": "https://api.github.com/users/HBCH1/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/HBCH1",
"id": 121411082,
"login": "HBCH1",
"node_id": "U_kgDOBzyWCg",
"organizations_url": "https://api.github.com/users/HBCH1/orgs",
"received_events_url": "https://api.github.com/users/HBCH1/received_events",
"repos_url": "https://api.github.com/users/HBCH1/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/HBCH1/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/HBCH1/subscriptions",
"type": "User",
"url": "https://api.github.com/users/HBCH1"
} | [] | open | false | null | [] | null | [] | "2022-12-26T01:55:04" | "2022-12-26T01:55:04" | null | NONE | null | Hi, I am using AlphaFold 2 (Colab pro +) to predict some proteins with long sequences. While it successfully predicted the structures of shorter sequences, it keeps failing with longer ones even tho I upgraded to colab pro + to have higher memory.
Here is what I am getting:
<img width="1234" alt="Alpha fold " src="https://user-images.githubusercontent.com/121411082/209489459-5fcca930-8962-4be4-a175-bea7d17b206c.png">
<img width="904" alt="alpha" src="https://user-images.githubusercontent.com/121411082/209489463-a95d93fa-3a25-48c2-b0b0-bc7f1a743bd9.png">
Is there a way to fix this issue or is AlphaFold 2 not currently able to predict >2000 bp sequences?
Thanks in advance! | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/666/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/666/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/665 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/665/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/665/comments | https://api.github.com/repos/deepmind/alphafold/issues/665/events | https://github.com/deepmind/alphafold/issues/665 | 1,508,351,023 | I_kwDOFoWQLM5Z55wv | 665 | Memory issue when processing large protein. Is my swap not being leveraged | {
"avatar_url": "https://avatars.githubusercontent.com/u/121254015?v=4",
"events_url": "https://api.github.com/users/cheminformaticist/events{/privacy}",
"followers_url": "https://api.github.com/users/cheminformaticist/followers",
"following_url": "https://api.github.com/users/cheminformaticist/following{/other_user}",
"gists_url": "https://api.github.com/users/cheminformaticist/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/cheminformaticist",
"id": 121254015,
"login": "cheminformaticist",
"node_id": "U_kgDOBzowfw",
"organizations_url": "https://api.github.com/users/cheminformaticist/orgs",
"received_events_url": "https://api.github.com/users/cheminformaticist/received_events",
"repos_url": "https://api.github.com/users/cheminformaticist/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/cheminformaticist/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/cheminformaticist/subscriptions",
"type": "User",
"url": "https://api.github.com/users/cheminformaticist"
} | [] | open | false | null | [] | null | [
"Could you try running with `--db_preset=reduced_dbs`?",
"My preference is to use full_dbs, but I will try.",
"I enabled --db_preset=reduced_dbs and ran into the same issue after about 40 minutes. It really seems like swap is not being used... or do I just need to scale up RAM.",
"What stage is it working on when it runs out of RAM?\r\nEdit: This is relevant in part because AlphaFold uses a number of third party resources at several stages, such as in the construction of the MSA or in the relax step."
] | "2022-12-22T18:15:57" | "2023-01-06T21:47:36" | null | NONE | null | I am running predictions for 2500 residue protein on a system with 80GB RAM, 8TB diskspace, and 1TB swap running Alphafold v2.2 via docker. The process fails with the message "RuntimeError: Resource exhausted: Out of memory while trying to allocate 51935261664 bytes." The ram was upgraded to 128GB, but the job dies with the same error.
docker stats: shows the MEM LIMIT is not including available swap. I am likely mistaken, but I thought it would... Is my swap not being leveraged by the container? How can I modify the docker image, such that when launched the container leverages swap (which I previously thought was available by default)? If I am off target, any advice would be appreciated...
Note: I have another system with 512GB RAM that no problem completing the calculations for the same protein. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/665/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/665/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/664 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/664/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/664/comments | https://api.github.com/repos/deepmind/alphafold/issues/664/events | https://github.com/deepmind/alphafold/issues/664 | 1,507,971,670 | I_kwDOFoWQLM5Z4dJW | 664 | Generating multiple (>10) models from one sequence | {
"avatar_url": "https://avatars.githubusercontent.com/u/121240930?v=4",
"events_url": "https://api.github.com/users/cgovaert/events{/privacy}",
"followers_url": "https://api.github.com/users/cgovaert/followers",
"following_url": "https://api.github.com/users/cgovaert/following{/other_user}",
"gists_url": "https://api.github.com/users/cgovaert/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/cgovaert",
"id": 121240930,
"login": "cgovaert",
"node_id": "U_kgDOBzn9Yg",
"organizations_url": "https://api.github.com/users/cgovaert/orgs",
"received_events_url": "https://api.github.com/users/cgovaert/received_events",
"repos_url": "https://api.github.com/users/cgovaert/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/cgovaert/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/cgovaert/subscriptions",
"type": "User",
"url": "https://api.github.com/users/cgovaert"
} | [] | open | false | null | [] | null | [
"While you certainly can do this with AlphaFold, you will probably only explore \"near-native\" conformations with it, because of what it is trained to produce (crystal structures). If you want to explore conformational space more thoroughly, I would recommend using molecular dynamics.\r\nIf you would like to use AlphaFold to investigate where the flexible region likely is, some studies have indicated that segments with lower pLDDT scores tend to be more flexible, and at least with 25 structures (typical output for multimers), this has matched where the variation occurs for me.\r\nFor doing this with AlphaFold, the easiest way would be to run in multimer mode (which, according to the latest update notes, is best to do even for monomers if you know the stoichiometry) and change the num_multimer_predictions_per_model value. As there are 5 models, pass the number of structures you want / 5 as the value to this flag in the command line. In other words, for 100 output structures, add --num_multimer_predictions_per_model=20 to your command line arguments. \r\n",
"Great idea. will do. To be more precise, I do want to run MD, I want to see if there is/are other conformation of a subdomain that we believe changes upon ligand binding. Thanks"
] | "2022-12-22T14:17:35" | "2022-12-22T16:15:23" | null | NONE | null | Hi all
I'm new to the field. I would like to generate between 100 and 50 models for my target sequence to explore conformal flexibility of a specific region. This region is often (comparing prediction on my target and various orthologs) predicted with lower confidence, and sometimes with different conformation. We have experimental evidence that this domain may be mobile.
Thus I want to generate many models from the same target sequence to identify possible conformational diversity.
We can run Alphafold locally but I'm not sure how to paramtyrize such a run.
Can anybody help here ?
Thank a bunch
Cedric
—
Prof. Cedric Govaerts, Ph.D.
Universite Libre de Bruxelles
Campus Plaine. Phone :+32 2 650 53 77
Building BC, Room 1C4 203
Boulevard du Triomphe, Acces 2
1050 Brussels
Belgium
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/664/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/664/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/663 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/663/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/663/comments | https://api.github.com/repos/deepmind/alphafold/issues/663/events | https://github.com/deepmind/alphafold/issues/663 | 1,506,058,944 | I_kwDOFoWQLM5ZxKLA | 663 | Problems with the new AlphaFold Colab Version | {
"avatar_url": "https://avatars.githubusercontent.com/u/61228596?v=4",
"events_url": "https://api.github.com/users/RosaLuzia/events{/privacy}",
"followers_url": "https://api.github.com/users/RosaLuzia/followers",
"following_url": "https://api.github.com/users/RosaLuzia/following{/other_user}",
"gists_url": "https://api.github.com/users/RosaLuzia/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/RosaLuzia",
"id": 61228596,
"login": "RosaLuzia",
"node_id": "MDQ6VXNlcjYxMjI4NTk2",
"organizations_url": "https://api.github.com/users/RosaLuzia/orgs",
"received_events_url": "https://api.github.com/users/RosaLuzia/received_events",
"repos_url": "https://api.github.com/users/RosaLuzia/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/RosaLuzia/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/RosaLuzia/subscriptions",
"type": "User",
"url": "https://api.github.com/users/RosaLuzia"
} | [
{
"color": "E99695",
"default": false,
"description": "Something isn't working",
"id": 3094826889,
"name": "error report",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODg5",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/error%20report"
},
{
"color": "FEF2C0",
"default": false,
"description": "AlphaFold colab issue",
"id": 3313914804,
"name": "colab",
"node_id": "MDU6TGFiZWwzMzEzOTE0ODA0",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/colab"
}
] | open | false | null | [] | null | [
"Is this affecting only one or two of your structures, or is there a widespread degradation in quality?\r\n\r\nMonomer models should be unchanged by v.2.3.0, but the msa databases used to generate the inputs to the monomer model have been updated. it is possible that for some targets this can reduce the quality of the msa, but for most targets it should be the same or better. Have the sizes of the msa results reduced for the structures where you are having problems?\r\n\r\nFor this particular target, we found that running with the full local installation, rather than in the colab, gave better results. The full installation uses templates and a larger bfd database, both of which can improve quality."
] | "2022-12-21T10:18:24" | "2022-12-21T17:28:15" | null | NONE | null | I run AlphaFold Colab with a lot of similar peptides which are characterized by alpha helices and beta sheets. I always get nice structures with the right assumed conformation, but since the new version of AlphaFold Colab is released structure prediction went wrong. I attach two predictions of the same sequence, one is predicted with an older version and the other with the recent version. Any suggestion how to fix the problem?
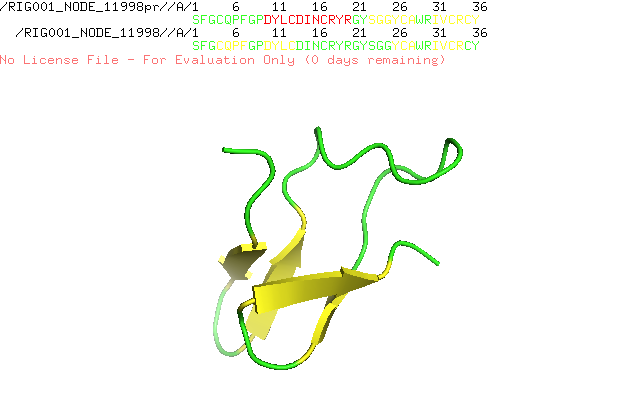
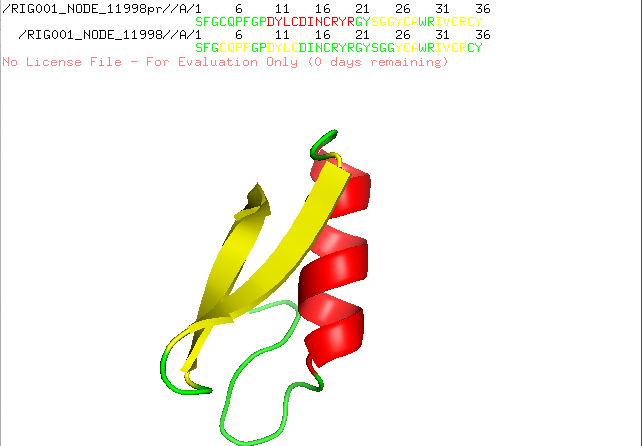
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/663/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/663/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/662 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/662/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/662/comments | https://api.github.com/repos/deepmind/alphafold/issues/662/events | https://github.com/deepmind/alphafold/issues/662 | 1,505,387,450 | I_kwDOFoWQLM5ZumO6 | 662 | Calculate mean and per residue pLDDT scores of AlphaFold's model PDBs (post-prediction) | {
"avatar_url": "https://avatars.githubusercontent.com/u/98129594?v=4",
"events_url": "https://api.github.com/users/meh47336/events{/privacy}",
"followers_url": "https://api.github.com/users/meh47336/followers",
"following_url": "https://api.github.com/users/meh47336/following{/other_user}",
"gists_url": "https://api.github.com/users/meh47336/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/meh47336",
"id": 98129594,
"login": "meh47336",
"node_id": "U_kgDOBdlWug",
"organizations_url": "https://api.github.com/users/meh47336/orgs",
"received_events_url": "https://api.github.com/users/meh47336/received_events",
"repos_url": "https://api.github.com/users/meh47336/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/meh47336/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/meh47336/subscriptions",
"type": "User",
"url": "https://api.github.com/users/meh47336"
} | [] | closed | false | null | [] | null | [
"If the pdb files come from a non-Alphafold model, any confidence metrics will be output by that other model.\r\n\r\nOne has to run the alphafold model to get alphafold confidence metrics, since those metrics are the models own measure of its certainty.",
"If you have the .npz files from the AlphaFold variant, you might be able to build something analogous to pLDDT scores based on that. The pdb simply does not have any of the information needed to estimate error in a quantified way.",
"The PDB are created by AlphaFold however it's just a Singularity container composed of the ML parts of AF (no MSA, etc). More specifically it's on Summit at OLCF (I asked the question here because things are a bit more active over here.\r\n\r\nThe output at Summit gives a pkl file and a PDB. The b-factor column of the PDBs contains all zero values rather than pLDDT scores. Any idea why this would not have been calculated or am I just missing something?",
"The per-residue pLDDT values are stored in a numpy array within the .pkl output!"
] | "2022-12-20T22:21:02" | "2022-12-23T17:45:01" | "2022-12-23T17:45:00" | NONE | null | Hello,
I've found the lddt script here but am unsure how to implement it outside of the AlphaFold. Recently I've obtained many predictions using a variant of AlphaFold, however it did not include pLDDT scores in the PDB outputs. Is there a way to calculate the scores retroactively using the PDB outputs?
Thank you for any help or suggestions, | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/662/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/662/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/661 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/661/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/661/comments | https://api.github.com/repos/deepmind/alphafold/issues/661/events | https://github.com/deepmind/alphafold/issues/661 | 1,505,333,292 | I_kwDOFoWQLM5ZuZAs | 661 | HHblits error "sequences in merged A3M file do not all have the same number of columns," | {
"avatar_url": "https://avatars.githubusercontent.com/u/5776199?v=4",
"events_url": "https://api.github.com/users/ajbordner/events{/privacy}",
"followers_url": "https://api.github.com/users/ajbordner/followers",
"following_url": "https://api.github.com/users/ajbordner/following{/other_user}",
"gists_url": "https://api.github.com/users/ajbordner/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ajbordner",
"id": 5776199,
"login": "ajbordner",
"node_id": "MDQ6VXNlcjU3NzYxOTk=",
"organizations_url": "https://api.github.com/users/ajbordner/orgs",
"received_events_url": "https://api.github.com/users/ajbordner/received_events",
"repos_url": "https://api.github.com/users/ajbordner/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ajbordner/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ajbordner/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ajbordner"
} | [
{
"color": "BFDADC",
"default": false,
"description": "Third-party tool issue",
"id": 3313961085,
"name": "third-party tool",
"node_id": "MDU6TGFiZWwzMzEzOTYxMDg1",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/third-party%20tool"
}
] | open | false | null | [] | null | [
"The uniref30 db has been updated in v2.3.0 which can cause different behaviour for this hhblits search. HHBlits is sensitive and the new db can cause errors for some targets which did not error before.\r\n\r\nA suggested workaround is to try with the db_preset flag set to 'reduced_dbs'.",
"There seems to be another issue causing the hhblits error. The hhblits command that AlphaFold runs completes without an error when run outside of the docker container. The same version of hhblits as in the Dockerfile (3.3.0), same database files, and input sequence file were used for the test.\r\n\r\nhhblits -i <FASTA input sequence file> -cpu 4 -oa3m /tmp/output.a3m -o /dev/null -n 3 -e 0.001 -maxseq 1000000 -realign_max 100000 -maxfilt 100000 -min_prefilter_hits 1000 -d <alphafold data directory>/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt -d <alphafold data directory>/uniref30/UniRef30_2021_03\r\n",
"we tried running hhblits directly too, but got the same error that you (and we) see when running the target in docker.\r\n\r\ncan you double check that your hhblits run was with the exact same sequence? and did it also give warnings about the titin results (ours did)?\r\n\r\nassuming it was the same sequence, something funny might be happening with ram usage, but im afraid using reduced_dbs is the only workaround for now.",
"I now also get the same error messages. I must have run hhblits for only 1 db since it only generates errors when run with both dbs. Thank you for your help."
] | "2022-12-20T21:35:55" | "2023-01-04T22:13:29" | null | NONE | null | AlphaFold is crashing due to an HHblits error in v2.3.0 but not in v2.2.4. This happens, for example, with the input sequence:
>chain_A
LEVQVPEDPVVALVGTDATLCCSFSPEPGFSLAQLNLIWQLTDTKQLVHSFAEGQDQGSAYANRTALFPDLLAQGNASLRLQRVRVADEGSFTCFVSIRDFGSAAVSLQVAAPYSKPSMTLEPNKDLRPGDTVTITCSSYQGYPEAEVFWQDGQGVPLTGNVTTSQMANEQGLFDVHSILRVVLGANGTYSCLVRNPVLQQDAHSSVTITPQRSPTGAVEVQVPEDPVVALVGTDATLRCSFSPEPGFSLAQLNLIWQLTDTKQLVHSFTEGRDQGSAYANRTALFPDLLAQGNASLRLQRVRVADEGSFTCFVSIRDFGSAAVSLQVAAPYSKPSMTLEPNKDLRPGDTVTITCSSYRGYPEAEVFWQDGQGVPLTGNVTTSQMANEQGLFDVHSVLRVVLGANGTYSCLVRNPVLQQDAHGSVTITGQPMTFPPEA
There are many long titin sequences in HHblits hits, which may be causing the issue. I have included an excerpt of the output log.
[test.log](https://github.com/deepmind/alphafold/files/10272808/test.log)
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/661/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/661/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/660 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/660/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/660/comments | https://api.github.com/repos/deepmind/alphafold/issues/660/events | https://github.com/deepmind/alphafold/issues/660 | 1,505,155,550 | I_kwDOFoWQLM5Zttne | 660 | [BUG] Relaxation not working for Multimers on v2.3.0 | {
"avatar_url": "https://avatars.githubusercontent.com/u/277946?v=4",
"events_url": "https://api.github.com/users/leiterenato/events{/privacy}",
"followers_url": "https://api.github.com/users/leiterenato/followers",
"following_url": "https://api.github.com/users/leiterenato/following{/other_user}",
"gists_url": "https://api.github.com/users/leiterenato/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/leiterenato",
"id": 277946,
"login": "leiterenato",
"node_id": "MDQ6VXNlcjI3Nzk0Ng==",
"organizations_url": "https://api.github.com/users/leiterenato/orgs",
"received_events_url": "https://api.github.com/users/leiterenato/received_events",
"repos_url": "https://api.github.com/users/leiterenato/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/leiterenato/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/leiterenato/subscriptions",
"type": "User",
"url": "https://api.github.com/users/leiterenato"
} | [] | open | false | null | [] | null | [
"we are looking into this, but might not come back with something until after the holiday period.\r\n\r\ncan i check: if you run inference+relax all in v2.2.4, rather than inference and then separately relax, it does work?",
"Hi @joshabramson \r\n\r\nI tried to run v2.3.0 using the `run_docker.py` from `https://github.com/deepmind/alphafold/tree/main/docker/run_docker.py`.\r\n\r\nI tried with both full_bfd and reduced_bfd without any success.\r\n\r\nFirst try:\r\n```\r\nnohup python3 docker/run_docker.py \\\r\n --fasta_paths=/home/renatoleite/workspace/vertex-ai-alphafold-inference-pipeline/sequences/1S78.fasta \\\r\n --output_dir=/home/renatoleite/workspace/outputs \\\r\n --max_template_date=2030-01-01 \\\r\n --model_preset=multimer \\\r\n --data_dir=/mnt/nfs/alphafold \\\r\n --num_multimer_predictions_per_model=1 &\r\n```\r\nSecond try:\r\n```\r\nnohup python3 docker/run_docker.py \\\r\n --fasta_paths=/home/renatoleite/workspace/vertex-ai-alphafold-inference-pipeline/sequences/1S78.fasta \\\r\n --output_dir=/home/renatoleite/workspace/outputs \\\r\n --max_template_date=2030-01-01 \\\r\n --model_preset=multimer \\\r\n --data_dir=/mnt/nfs/alphafold \\\r\n --num_multimer_predictions_per_model=1 \\\r\n --db_preset=reduced_dbs & \r\n```\r\n\r\nI also tried with my pipeline (separated prediction and relaxation), but got the same error.\r\nhttps://github.com/GoogleCloudPlatform/vertex-ai-alphafold-inference-pipeline\r\n\r\nFor v2.2.4 it worked with both solutions (separated and inference+relax).\r\n\r\n"
] | "2022-12-20T19:46:41" | "2022-12-23T19:58:21" | null | NONE | null | Relaxation step is failing for multimers using AlphaFold v2.3.0.
Monomers are working fine.
Error:
```
jaxlib.xla_extension.XlaRuntimeError: INTERNAL: Failed to load in-memory CUBIN: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered
E external/org_tensorflow/tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:1043] could not synchronize on CUDA context: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered
```
I tested the relaxation with up to 8 x A100 GPUs, but it failed (apparently there is a memory leak somewhere).
If I run the inference with v2.3.0 and relaxation with v2.2.4, it works fine.
I attached a sequence that returned this error (changed the suffix to txt to work on github UI).
[1S78.txt](https://github.com/deepmind/alphafold/files/10271938/1S78.txt) | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/660/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/660/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/659 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/659/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/659/comments | https://api.github.com/repos/deepmind/alphafold/issues/659/events | https://github.com/deepmind/alphafold/issues/659 | 1,501,878,693 | I_kwDOFoWQLM5ZhNml | 659 | error when running alphafold multimer | {
"avatar_url": "https://avatars.githubusercontent.com/u/86952016?v=4",
"events_url": "https://api.github.com/users/lamyankin/events{/privacy}",
"followers_url": "https://api.github.com/users/lamyankin/followers",
"following_url": "https://api.github.com/users/lamyankin/following{/other_user}",
"gists_url": "https://api.github.com/users/lamyankin/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/lamyankin",
"id": 86952016,
"login": "lamyankin",
"node_id": "MDQ6VXNlcjg2OTUyMDE2",
"organizations_url": "https://api.github.com/users/lamyankin/orgs",
"received_events_url": "https://api.github.com/users/lamyankin/received_events",
"repos_url": "https://api.github.com/users/lamyankin/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/lamyankin/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/lamyankin/subscriptions",
"type": "User",
"url": "https://api.github.com/users/lamyankin"
} | [] | open | false | null | [] | null | [
"Hi @lamyankin \r\nI had the same issue with a different file from `pdb_mmcif`. \r\nI solved this problem by downloading a newer version of the database.",
"Hi, glad to hear your reply!\r\nDid you download the newer pdb_mmcif database or all databases ?",
"I downloaded the full database again (but I don't think it is necessary)."
] | "2022-12-18T12:02:11" | "2022-12-20T13:16:10" | null | NONE | null | I am running the alphafold multimer to predict a complex composed of A and B and the problem occurs , here are the error message:
1218 18:46:51.980194 139874231673472 run_docker.py:255] Traceback (most recent call last):
I1218 18:46:51.980485 139874231673472 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 422, in <module>
I1218 18:46:51.980680 139874231673472 run_docker.py:255] app.run(main)
I1218 18:46:51.980863 139874231673472 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 312, in run
I1218 18:46:51.981044 139874231673472 run_docker.py:255] _run_main(main, args)
I1218 18:46:51.981219 139874231673472 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 258, in _run_main
I1218 18:46:51.981395 139874231673472 run_docker.py:255] sys.exit(main(argv))
I1218 18:46:51.981570 139874231673472 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 406, in main
I1218 18:46:51.981745 139874231673472 run_docker.py:255] random_seed=random_seed)
I1218 18:46:51.981920 139874231673472 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 174, in predict_structure
I1218 18:46:51.982094 139874231673472 run_docker.py:255] msa_output_dir=msa_output_dir)
I1218 18:46:51.982327 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/pipeline_multimer.py", line 269, in process
I1218 18:46:51.982502 139874231673472 run_docker.py:255] is_homomer_or_monomer=is_homomer_or_monomer)
I1218 18:46:51.982620 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/pipeline_multimer.py", line 214, in _process_single_chain
I1218 18:46:51.982703 139874231673472 run_docker.py:255] msa_output_dir=chain_msa_output_dir)
I1218 18:46:51.982784 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/pipeline.py", line 225, in process
I1218 18:46:51.982866 139874231673472 run_docker.py:255] hits=pdb_template_hits)
I1218 18:46:51.982964 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/templates.py", line 968, in get_templates
I1218 18:46:51.983068 139874231673472 run_docker.py:255] kalign_binary_path=self._kalign_binary_path)
I1218 18:46:51.983152 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/templates.py", line 737, in _process_single_hit
I1218 18:46:51.983233 139874231673472 run_docker.py:255] cif_string = _read_file(cif_path)
I1218 18:46:51.983314 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/templates.py", line 681, in _read_file
I1218 18:46:51.983396 139874231673472 run_docker.py:255] with open(path, 'r') as f:
I1218 18:46:51.983479 139874231673472 run_docker.py:255] FileNotFoundError: [Errno 2] No such file or directory: '/mnt/template_mmcif_dir/7u0h.cif'
Then I have try searching the 7u0h.cif in the /path/alphafold/pdb_mmcif/mmcif_files, but i didn't found it.
What should i do to solve this problem? looking forward to your reply. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/659/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/659/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/658 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/658/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/658/comments | https://api.github.com/repos/deepmind/alphafold/issues/658/events | https://github.com/deepmind/alphafold/issues/658 | 1,501,730,455 | I_kwDOFoWQLM5ZgpaX | 658 | Different alphafold version get different results | {
"avatar_url": "https://avatars.githubusercontent.com/u/86952016?v=4",
"events_url": "https://api.github.com/users/lamyankin/events{/privacy}",
"followers_url": "https://api.github.com/users/lamyankin/followers",
"following_url": "https://api.github.com/users/lamyankin/following{/other_user}",
"gists_url": "https://api.github.com/users/lamyankin/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/lamyankin",
"id": 86952016,
"login": "lamyankin",
"node_id": "MDQ6VXNlcjg2OTUyMDE2",
"organizations_url": "https://api.github.com/users/lamyankin/orgs",
"received_events_url": "https://api.github.com/users/lamyankin/received_events",
"repos_url": "https://api.github.com/users/lamyankin/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/lamyankin/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/lamyankin/subscriptions",
"type": "User",
"url": "https://api.github.com/users/lamyankin"
} | [] | open | false | null | [] | null | [
"@lamyankin \r\nWhen you run the multimer system you use 5 different models and, possibly, 5 different runs for each model (each one with a different seed).\r\nTo make sure you are comparing the same results you need to check the model name and seed used to run the inference.\r\n\r\nThere are v3 (from v2.3.0) and v2 (from v2.x.x) models.\r\nI did some tests and the v3 performs much better.\r\n\r\n"
] | "2022-12-18T04:02:45" | "2022-12-19T20:13:33" | null | NONE | null |
I run the alphafold in different version(one is v2.2.4, another is earlier )and get different results using the model_preset of multimer. As shown in the picture:

This is a complex composed of Cas9 and SNRPD2 (red) proteins. The predicted struture of cas9 is nearly the same in both results, as well as SNRPD2. However, the binding site of the cas9 and SNRPD2 is different in two results.
I want to know if it's because of the different versions of alphafold or because of my command?
here is my command:
nohup python3 docker/run_docker.py \
--fasta_paths=/home/zwf/alphafold/hpl1.fasta,/home/zwf/alphafold/SNRPD2.fasta \
--model_preset=multimer \
--docker_user=0 \
--db_preset=full_dbs \
--output_dir=/data/alphafold_outdir \
--max_template_date=2021-12-10 \
--data_dir=/data/alphalfold &> /home/zwf/alphafold/SNRPD2.out& | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/658/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/658/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/657 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/657/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/657/comments | https://api.github.com/repos/deepmind/alphafold/issues/657/events | https://github.com/deepmind/alphafold/issues/657 | 1,501,713,262 | I_kwDOFoWQLM5ZglNu | 657 | Backwards compatibility of AlphaFold 2.3.0 database with prevous versions of the software | {
"avatar_url": "https://avatars.githubusercontent.com/u/1719485?v=4",
"events_url": "https://api.github.com/users/panchyni/events{/privacy}",
"followers_url": "https://api.github.com/users/panchyni/followers",
"following_url": "https://api.github.com/users/panchyni/following{/other_user}",
"gists_url": "https://api.github.com/users/panchyni/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/panchyni",
"id": 1719485,
"login": "panchyni",
"node_id": "MDQ6VXNlcjE3MTk0ODU=",
"organizations_url": "https://api.github.com/users/panchyni/orgs",
"received_events_url": "https://api.github.com/users/panchyni/received_events",
"repos_url": "https://api.github.com/users/panchyni/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/panchyni/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/panchyni/subscriptions",
"type": "User",
"url": "https://api.github.com/users/panchyni"
} | [] | open | false | null | [] | null | [] | "2022-12-18T02:48:07" | "2022-12-18T02:48:07" | null | NONE | null | I am currently working with researchers at my university to update the version and database of AlphaFold on our university's computing cluster. From discussions we had prior to the release of 2.3.0, I had been told that updating the UniProt and PDB databases shouldn't affect the reproducibility of results or functionality of prior versions, given the common file structure and the ability to control the age of models used with input arguments. However, in the 2.3.0 update I noticed that the uniref30 database uses a slightly different folder structure and no longer contains *_db links and *_db.index files.
As such, I was hoping you could tell me if previous versions of AlphaFold depend on these files or are the *ffdata and *ffindex files sufficient? Ideally, I would like to avoid maintaining multiple databases and/or update scripts running on our system, so it would be ideal if I could simply make a directory in the format of the previous uniprot30 directory and soft link the necessary contents of uniref30 there so that we can maintain a single directory which support all versions of AlphaFold on our cluster. If the *db/db.index file are necessary for prior versions or there is some other conflict I am missing, I would appreciate any additional information you could give me.
I understand this is more an issue with the community need of our computing cluster than your software, but any information you could provide about the significance or lack their of regarding this change would save me a lot time and possibly errors trying to test it myself. Thank you for you time and consideration. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/657/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/657/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/656 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/656/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/656/comments | https://api.github.com/repos/deepmind/alphafold/issues/656/events | https://github.com/deepmind/alphafold/issues/656 | 1,498,941,741 | I_kwDOFoWQLM5ZWAkt | 656 | uniref30_database_path issue | {
"avatar_url": "https://avatars.githubusercontent.com/u/106186885?v=4",
"events_url": "https://api.github.com/users/limh25/events{/privacy}",
"followers_url": "https://api.github.com/users/limh25/followers",
"following_url": "https://api.github.com/users/limh25/following{/other_user}",
"gists_url": "https://api.github.com/users/limh25/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/limh25",
"id": 106186885,
"login": "limh25",
"node_id": "U_kgDOBlRIhQ",
"organizations_url": "https://api.github.com/users/limh25/orgs",
"received_events_url": "https://api.github.com/users/limh25/received_events",
"repos_url": "https://api.github.com/users/limh25/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/limh25/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/limh25/subscriptions",
"type": "User",
"url": "https://api.github.com/users/limh25"
} | [] | closed | false | null | [] | null | [
"You need to rebuild after pulling the newest version \r\n\r\n```docker build -f docker/Dockerfile -t alphafold .```",
"Hi @AChatzigoulas\r\nThank you for your help.\r\nI still have a problem in building the docker image.\r\n\r\n`Step 11/18 : RUN wget -q -P /app/alphafold/alphafold/common/ https://git.scicore.unibas.ch/schwede/openstructure/-/raw/7102c63615b64735c4941278d92b554ec94415f8/modules/mol/alg/src/stereo_chemical_props.txt\r\n ---> Running in 9b77342609c3\r\nThe command '/bin/bash -o pipefail -c wget -q -P /app/alphafold/alphafold/common/ https://git.scicore.unibas.ch/schwede/openstructure/-/raw/7102c63615b64735c4941278d92b554ec94415f8/modules/mol/alg/src/stereo_chemical_props.txt' returned a non-zero code: 5`\r\n\r\nDo you know the cause or the solution?\r\n\r\nThanks.",
"This is an SSL verification failure. Sadly, I do not know the cause or the solution for this one.",
"I was able to solve by modifying the Dockerfile\r\n\r\n```\r\nRUN wget --no-check-certificate -q -P /app/alphafold/alphafold/common/ \\\r\n https://git.scicore.unibas.ch/schwede/openstructure/-/raw/7102c63615b64735c4941278d92b554ec94415f8/modules/mol/alg/src/stereo_chemical_props.txt\r\n```\r\n\r\nThe \"--no-check-certificate\" option solved the issue.\r\n\r\nThanks."
] | "2022-12-15T19:05:05" | "2022-12-20T18:46:02" | "2022-12-20T18:46:01" | NONE | null | Hi,
I am trying to run the new ver. 2.3.0 for multimer predictions.
I had no issue with previous versions, including 2.2.4.
I tried to use direct download from the releases, and I also tried to use from git clone repository.
In both cases, I get an error message and the job terminates.
`I1215 18:59:41.374994 140456684652352 run_docker.py:255] FATAL Flags parsing error: Unknown command line flag 'uniref30_database_path'. Did you mean: uniref90_database_path ?`
It seems the directories are correctly mounted.
`I1215 18:59:37.418998 140456684652352 run_docker.py:113] Mounting /home/ec2-user/input_fasta/part1 -> /mnt/fasta_path_0
I1215 18:59:37.419240 140456684652352 run_docker.py:113] Mounting /disk1/databases/uniref90 -> /mnt/uniref90_database_path
I1215 18:59:37.419375 140456684652352 run_docker.py:113] Mounting /disk1/databases/mgnify -> /mnt/mgnify_database_path
I1215 18:59:37.419479 140456684652352 run_docker.py:113] Mounting /disk1/databases -> /mnt/data_dir
I1215 18:59:37.419581 140456684652352 run_docker.py:113] Mounting /disk1/databases/pdb_mmcif/mmcif_files -> /mnt/template_mmcif_dir
I1215 18:59:37.419708 140456684652352 run_docker.py:113] Mounting /disk1/databases/pdb_mmcif -> /mnt/obsolete_pdbs_path
I1215 18:59:37.419818 140456684652352 run_docker.py:113] Mounting /disk1/databases/uniprot -> /mnt/uniprot_database_path
I1215 18:59:37.419928 140456684652352 run_docker.py:113] Mounting /disk1/databases/pdb_seqres -> /mnt/pdb_seqres_database_path
I1215 18:59:37.420024 140456684652352 run_docker.py:113] Mounting /disk1/databases/uniref30/UniRef30_2021_03 -> /mnt/uniref30_database_path
I1215 18:59:37.420170 140456684652352 run_docker.py:113] Mounting /disk1/databases/bfd -> /mnt/bfd_database_path
I1215 18:59:41.374994 140456684652352 run_docker.py:255] FATAL Flags parsing error: Unknown command line flag 'uniref30_database_path'. Did you mean: uniref90_database_path ?
I1215 18:59:41.375221 140456684652352 run_docker.py:255] Pass --helpshort or --helpfull to see help on flags.`
I updated the databases:
- mgnify
- params
- pdb_mmcif
- pdb_seqres
- uniref30
- uniref90
Please help.
| {
"+1": 1,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 1,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/656/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/656/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/655 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/655/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/655/comments | https://api.github.com/repos/deepmind/alphafold/issues/655/events | https://github.com/deepmind/alphafold/issues/655 | 1,493,200,209 | I_kwDOFoWQLM5ZAG1R | 655 | A new problem occurred while I was running the Alphafold | {
"avatar_url": "https://avatars.githubusercontent.com/u/119162846?v=4",
"events_url": "https://api.github.com/users/pujialin/events{/privacy}",
"followers_url": "https://api.github.com/users/pujialin/followers",
"following_url": "https://api.github.com/users/pujialin/following{/other_user}",
"gists_url": "https://api.github.com/users/pujialin/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/pujialin",
"id": 119162846,
"login": "pujialin",
"node_id": "U_kgDOBxpH3g",
"organizations_url": "https://api.github.com/users/pujialin/orgs",
"received_events_url": "https://api.github.com/users/pujialin/received_events",
"repos_url": "https://api.github.com/users/pujialin/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/pujialin/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/pujialin/subscriptions",
"type": "User",
"url": "https://api.github.com/users/pujialin"
} | [
{
"color": "FEF2C0",
"default": false,
"description": "AlphaFold colab issue",
"id": 3313914804,
"name": "colab",
"node_id": "MDU6TGFiZWwzMzEzOTE0ODA0",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/colab"
}
] | open | false | null | [] | null | [
"I think this was caused by the changes in v2.3.0 which happened recently - your Colab notebook likely got out of sync with the code. Could you try running in a fresh notebook?\r\n https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb"
] | "2022-12-13T02:21:05" | "2022-12-13T10:32:36" | null | NONE | null | When I entered the amino acid sequence, the program reported an error:
AttributeError Traceback (most recent call last)
[<ipython-input-4-6962bd6ebd6c>](https://localhost:8080/#) in <module>
23
24 # Validate the input.
---> 25 sequences, model_type_to_use = notebook_utils.validate_input(
26 input_sequences=input_sequences,
27 min_length=MIN_SINGLE_SEQUENCE_LENGTH,
AttributeError: module 'alphafold.notebooks.notebook_utils' has no attribute 'validate_input'
I don't know what's going on, I hope everyone can help me. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/655/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/655/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/654 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/654/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/654/comments | https://api.github.com/repos/deepmind/alphafold/issues/654/events | https://github.com/deepmind/alphafold/issues/654 | 1,491,848,370 | I_kwDOFoWQLM5Y68yy | 654 | Add explicit `git pull` step in README? | {
"avatar_url": "https://avatars.githubusercontent.com/u/46537483?v=4",
"events_url": "https://api.github.com/users/ulupo/events{/privacy}",
"followers_url": "https://api.github.com/users/ulupo/followers",
"following_url": "https://api.github.com/users/ulupo/following{/other_user}",
"gists_url": "https://api.github.com/users/ulupo/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ulupo",
"id": 46537483,
"login": "ulupo",
"node_id": "MDQ6VXNlcjQ2NTM3NDgz",
"organizations_url": "https://api.github.com/users/ulupo/orgs",
"received_events_url": "https://api.github.com/users/ulupo/received_events",
"repos_url": "https://api.github.com/users/ulupo/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ulupo/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ulupo/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ulupo"
} | [] | open | false | null | [] | null | [] | "2022-12-12T13:08:33" | "2022-12-12T13:08:33" | null | NONE | null | From the README > "Updating existing installation":
https://github.com/deepmind/alphafold/blob/569eb4fea3733b979cb0442750b875759dd5ecc0/README.md?plain=1#L190-L191
I may have misunderstood, but wouldn't it be helpful to explicitly add the instruction to `git pull` if fetching returns no errors/displays merge conflicts? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/654/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/654/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/653 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/653/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/653/comments | https://api.github.com/repos/deepmind/alphafold/issues/653/events | https://github.com/deepmind/alphafold/issues/653 | 1,488,537,284 | I_kwDOFoWQLM5YuUbE | 653 | run_docker.py failure caused by module not found error | {
"avatar_url": "https://avatars.githubusercontent.com/u/120262538?v=4",
"events_url": "https://api.github.com/users/ailiqiangCSU/events{/privacy}",
"followers_url": "https://api.github.com/users/ailiqiangCSU/followers",
"following_url": "https://api.github.com/users/ailiqiangCSU/following{/other_user}",
"gists_url": "https://api.github.com/users/ailiqiangCSU/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ailiqiangCSU",
"id": 120262538,
"login": "ailiqiangCSU",
"node_id": "U_kgDOBysPig",
"organizations_url": "https://api.github.com/users/ailiqiangCSU/orgs",
"received_events_url": "https://api.github.com/users/ailiqiangCSU/received_events",
"repos_url": "https://api.github.com/users/ailiqiangCSU/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ailiqiangCSU/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ailiqiangCSU/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ailiqiangCSU"
} | [
{
"color": "ffffff",
"default": false,
"description": "An issue with Docker",
"id": 3094826898,
"name": "docker",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODk4",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/docker"
},
{
"color": "F9D0C4",
"default": false,
"description": "Issue setting up AlphaFold",
"id": 3317500161,
"name": "setup",
"node_id": "MDU6TGFiZWwzMzE3NTAwMTYx",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/setup"
}
] | open | false | null | [] | null | [] | "2022-12-10T14:33:09" | "2022-12-11T14:25:32" | null | NONE | null | When I run the run_docker.py with the following command,
python3 docker/run_docker.py \
--fasta_paths=/data/xiaoai/alphafold2_software/alphafold-main/T1082.fasta \
--max_template_date=2020-05-14 \
--model_preset=monomer \
--db_preset=full_dbs \
--data_dir=/data/xiaoai/alphafold2_software/download_file
I got this error message:
I1210 22:19:52.657300 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/alphafold-main -> /mnt/fasta_path_0
I1210 22:19:52.657422 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/uniref90 -> /mnt/uniref90_database_path
I1210 22:19:52.657488 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/mgnify -> /mnt/mgnify_database_path
I1210 22:19:52.657539 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file -> /mnt/data_dir
I1210 22:19:52.657588 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/pdb_mmcif/mmcif_files -> /mnt/template_mmcif_dir
I1210 22:19:52.657644 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/pdb_mmcif -> /mnt/obsolete_pdbs_path
I1210 22:19:52.657705 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/pdb70 -> /mnt/pdb70_database_path
I1210 22:19:52.657769 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/uniclust30/uniclust30_2018_08 -> /mnt/uniclust30_database_path
I1210 22:19:52.657833 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/bfd -> /mnt/bfd_database_path
I1210 22:19:53.050984 139724585183040 run_docker.py:255] Traceback (most recent call last):
I1210 22:19:53.051107 139724585183040 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 26, in <module>
I1210 22:19:53.051178 139724585183040 run_docker.py:255] from absl import app
I1210 22:19:53.051237 139724585183040 run_docker.py:255] ModuleNotFoundError: No module named 'absl'
but I find that absl-py already exist in conda list and python
I import absl in python without any error
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/653/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/653/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/652 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/652/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/652/comments | https://api.github.com/repos/deepmind/alphafold/issues/652/events | https://github.com/deepmind/alphafold/pull/652 | 1,485,120,206 | PR_kwDOFoWQLM5E0Pxk | 652 | Fix exception causes in data/templates.py | {
"avatar_url": "https://avatars.githubusercontent.com/u/56778?v=4",
"events_url": "https://api.github.com/users/cool-RR/events{/privacy}",
"followers_url": "https://api.github.com/users/cool-RR/followers",
"following_url": "https://api.github.com/users/cool-RR/following{/other_user}",
"gists_url": "https://api.github.com/users/cool-RR/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/cool-RR",
"id": 56778,
"login": "cool-RR",
"node_id": "MDQ6VXNlcjU2Nzc4",
"organizations_url": "https://api.github.com/users/cool-RR/orgs",
"received_events_url": "https://api.github.com/users/cool-RR/received_events",
"repos_url": "https://api.github.com/users/cool-RR/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/cool-RR/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/cool-RR/subscriptions",
"type": "User",
"url": "https://api.github.com/users/cool-RR"
} | [] | open | false | null | [] | null | [] | "2022-12-08T17:49:31" | "2022-12-08T17:49:31" | null | NONE | null | More context about this change here: https://blog.ram.rachum.com/post/621791438475296768/improving-python-exception-chaining-with | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/652/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/652/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/652.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/652",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/652.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/652"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/651 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/651/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/651/comments | https://api.github.com/repos/deepmind/alphafold/issues/651/events | https://github.com/deepmind/alphafold/issues/651 | 1,482,138,994 | I_kwDOFoWQLM5YV6Vy | 651 | GPU ran out of memory! | {
"avatar_url": "https://avatars.githubusercontent.com/u/5677674?v=4",
"events_url": "https://api.github.com/users/shahryary/events{/privacy}",
"followers_url": "https://api.github.com/users/shahryary/followers",
"following_url": "https://api.github.com/users/shahryary/following{/other_user}",
"gists_url": "https://api.github.com/users/shahryary/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/shahryary",
"id": 5677674,
"login": "shahryary",
"node_id": "MDQ6VXNlcjU2Nzc2NzQ=",
"organizations_url": "https://api.github.com/users/shahryary/orgs",
"received_events_url": "https://api.github.com/users/shahryary/received_events",
"repos_url": "https://api.github.com/users/shahryary/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/shahryary/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/shahryary/subscriptions",
"type": "User",
"url": "https://api.github.com/users/shahryary"
} | [
{
"color": "A65D8A",
"default": false,
"description": "Out of memory error message",
"id": 3938926450,
"name": "out of memory",
"node_id": "LA_kwDOFoWQLM7qxz9y",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/out%20of%20memory"
}
] | closed | false | null | [] | null | [
"We improved the GPU usage for AlphaFold-Multimer in [v2.3.0](https://github.com/deepmind/alphafold/releases/tag/v2.3.0) - hopefully this addresses your issue. Closing this issue now, feel free to reopen if you still encounter this in v2.3.0."
] | "2022-12-07T15:06:44" | "2022-12-13T11:55:11" | "2022-12-13T11:55:11" | NONE | null | `Allocator (GPU_0_bfc) ran out of memory trying to allocate 163.17GiB (rounded to 175203296000) requested by op`
I've been using the Alphafold for more than a year, and the GPU problem is annoying many users to run complex/ long sequences.
Alphafold is not supporting multi GPUs, and it's only using one GPU core - although, in the documentation, they mentioned it would attempt to use all available GPU cores!
I tried to run Alphafold(conda-based, docker/ singularity) in different instances from Amazon, G5.48x /G4 series. Still, I couldn't get results because only one GPU core taking into account instead of all of them.
The most significant target users of Alphafold are scientists who are more interested in running 'real' long samples rather than just a few lines of sequences. Without supporting multi-GPU mode, the tools are hopeless. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/651/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/651/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/650 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/650/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/650/comments | https://api.github.com/repos/deepmind/alphafold/issues/650/events | https://github.com/deepmind/alphafold/issues/650 | 1,476,046,364 | I_kwDOFoWQLM5X-q4c | 650 | jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead. | {
"avatar_url": "https://avatars.githubusercontent.com/u/108690998?v=4",
"events_url": "https://api.github.com/users/MasterNivek/events{/privacy}",
"followers_url": "https://api.github.com/users/MasterNivek/followers",
"following_url": "https://api.github.com/users/MasterNivek/following{/other_user}",
"gists_url": "https://api.github.com/users/MasterNivek/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/MasterNivek",
"id": 108690998,
"login": "MasterNivek",
"node_id": "U_kgDOBnp-Ng",
"organizations_url": "https://api.github.com/users/MasterNivek/orgs",
"received_events_url": "https://api.github.com/users/MasterNivek/received_events",
"repos_url": "https://api.github.com/users/MasterNivek/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/MasterNivek/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/MasterNivek/subscriptions",
"type": "User",
"url": "https://api.github.com/users/MasterNivek"
} | [] | closed | false | null | [] | null | [
"solved in pr #630 \r\n\r\njust replace all tree_utils\r\n",
"Thanks, fixed in v2.3.0."
] | "2022-12-05T08:55:33" | "2022-12-13T11:55:30" | "2022-12-13T11:55:30" | NONE | null | any one knows of a solution please

| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/650/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/650/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/649 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/649/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/649/comments | https://api.github.com/repos/deepmind/alphafold/issues/649/events | https://github.com/deepmind/alphafold/issues/649 | 1,472,724,200 | I_kwDOFoWQLM5Xx_zo | 649 | AlphaFold multimer has import error in Step 5 | {
"avatar_url": "https://avatars.githubusercontent.com/u/65905482?v=4",
"events_url": "https://api.github.com/users/Isabel-ql/events{/privacy}",
"followers_url": "https://api.github.com/users/Isabel-ql/followers",
"following_url": "https://api.github.com/users/Isabel-ql/following{/other_user}",
"gists_url": "https://api.github.com/users/Isabel-ql/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Isabel-ql",
"id": 65905482,
"login": "Isabel-ql",
"node_id": "MDQ6VXNlcjY1OTA1NDgy",
"organizations_url": "https://api.github.com/users/Isabel-ql/orgs",
"received_events_url": "https://api.github.com/users/Isabel-ql/received_events",
"repos_url": "https://api.github.com/users/Isabel-ql/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Isabel-ql/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Isabel-ql/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Isabel-ql"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"Hi thanks for raising this. Its a duplicate of https://github.com/deepmind/alphafold/issues/648 so closing"
] | "2022-12-02T11:32:40" | "2022-12-02T14:56:42" | "2022-12-02T14:56:42" | NONE | null | Hi,
I tried to predict structures of complexes with AlphaFold Multimer using the link: https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharinga
It worked well before, but a problem appeared yesterday. In step 5, "Search against genetic database", there's an import error (See below). I tried with different browsers, the problem still exists.
Is anybody know how to fix the problem? Thanks very much!
Cheers,
Qun
<img width="1381" alt="AlphaFold Multimer_step5" src="https://user-images.githubusercontent.com/65905482/205283476-bb018f25-61b6-4391-8a66-022dc90acce7.png">
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/649/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/649/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/648 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/648/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/648/comments | https://api.github.com/repos/deepmind/alphafold/issues/648/events | https://github.com/deepmind/alphafold/issues/648 | 1,470,885,359 | I_kwDOFoWQLM5Xq-3v | 648 | ImportError: Python version mismatch: module was compiled for Python 3.7, but the interpreter version is incompatible: 3.8.15 (default, Oct 12 2022, 19:14:39) [GCC 7.5.0]. | {
"avatar_url": "https://avatars.githubusercontent.com/u/119576148?v=4",
"events_url": "https://api.github.com/users/itheod04/events{/privacy}",
"followers_url": "https://api.github.com/users/itheod04/followers",
"following_url": "https://api.github.com/users/itheod04/following{/other_user}",
"gists_url": "https://api.github.com/users/itheod04/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/itheod04",
"id": 119576148,
"login": "itheod04",
"node_id": "U_kgDOByCWVA",
"organizations_url": "https://api.github.com/users/itheod04/orgs",
"received_events_url": "https://api.github.com/users/itheod04/received_events",
"repos_url": "https://api.github.com/users/itheod04/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/itheod04/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/itheod04/subscriptions",
"type": "User",
"url": "https://api.github.com/users/itheod04"
} | [] | closed | false | null | [] | null | [
"I have the same issue :/",
"Same issue ",
"+1",
"Hi thanks for raising this. We've updated to 3.8 in the latest [commit](https://github.com/deepmind/alphafold/commit/4494af848e41f3acd51a190d4c78d38cb1a7f85d) to match colab"
] | "2022-12-01T08:23:26" | "2022-12-02T14:30:31" | "2022-12-02T14:30:30" | NONE | null | a) ImportError: Python version mismatch: module was compiled for Python 3.7, but the interpreter version is incompatible: 3.8.15 (default, Oct 12 2022, 19:14:39)
[GCC 7.5.0].

b) ImportError: Traceback (most recent call last):
File "/opt/conda/lib/python3.7/site-packages/tensorflow/python/pywrap_tensorflow.py", line 62, in <module>
from tensorflow.python._pywrap_tensorflow_internal import *
ImportError: Python version mismatch: module was compiled for Python 3.7, but the interpreter version is incompatible: 3.8.15 (default, Oct 12 2022, 19:14:39)

[GCC 7.5.0].
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/648/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/648/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/647 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/647/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/647/comments | https://api.github.com/repos/deepmind/alphafold/issues/647/events | https://github.com/deepmind/alphafold/pull/647 | 1,470,257,031 | PR_kwDOFoWQLM5EAN2K | 647 | Code of Conduct | {
"avatar_url": "https://avatars.githubusercontent.com/u/91554112?v=4",
"events_url": "https://api.github.com/users/achawlaa/events{/privacy}",
"followers_url": "https://api.github.com/users/achawlaa/followers",
"following_url": "https://api.github.com/users/achawlaa/following{/other_user}",
"gists_url": "https://api.github.com/users/achawlaa/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/achawlaa",
"id": 91554112,
"login": "achawlaa",
"node_id": "U_kgDOBXUBQA",
"organizations_url": "https://api.github.com/users/achawlaa/orgs",
"received_events_url": "https://api.github.com/users/achawlaa/received_events",
"repos_url": "https://api.github.com/users/achawlaa/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/achawlaa/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/achawlaa/subscriptions",
"type": "User",
"url": "https://api.github.com/users/achawlaa"
} | [] | open | false | null | [] | null | [] | "2022-11-30T21:47:13" | "2022-11-30T22:37:00" | null | NONE | null | Code of Conduct is a very important document that clearly demonstrates the expectations for behavior for the participants of this project. The adoption and enforcement of a code of conduct helps structure a positive social atmosphere for a community to grow. Alphafold currently does not have a code of conduct page so I wanted to create one for it. Please see COD.md for the code of conduct page I constructed for Alphafold. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/647/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/647/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/647.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/647",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/647.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/647"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/646 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/646/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/646/comments | https://api.github.com/repos/deepmind/alphafold/issues/646/events | https://github.com/deepmind/alphafold/issues/646 | 1,468,059,827 | I_kwDOFoWQLM5XgNCz | 646 | A problem occurred while I was running the Alphafold | {
"avatar_url": "https://avatars.githubusercontent.com/u/119414515?v=4",
"events_url": "https://api.github.com/users/jhyeonv/events{/privacy}",
"followers_url": "https://api.github.com/users/jhyeonv/followers",
"following_url": "https://api.github.com/users/jhyeonv/following{/other_user}",
"gists_url": "https://api.github.com/users/jhyeonv/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/jhyeonv",
"id": 119414515,
"login": "jhyeonv",
"node_id": "U_kgDOBx4e8w",
"organizations_url": "https://api.github.com/users/jhyeonv/orgs",
"received_events_url": "https://api.github.com/users/jhyeonv/received_events",
"repos_url": "https://api.github.com/users/jhyeonv/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/jhyeonv/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/jhyeonv/subscriptions",
"type": "User",
"url": "https://api.github.com/users/jhyeonv"
} | [
{
"color": "000000",
"default": false,
"description": "",
"id": 3356911328,
"name": "cuda",
"node_id": "MDU6TGFiZWwzMzU2OTExMzI4",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/cuda"
}
] | open | false | null | [] | null | [
"I got this same error since CUDA moved to v.12. I think there will need to be an update on the side of the developer team here or a downgrade in system drivers. ",
"does this error persist when using AlphaFold v2.3.0?",
"@joshabramson I think so. I did a fresh build with the newest dockerfile. Here's the output where the error picks up:\r\n```\r\nI1223 20:04:51.789108 139688861775680 run_docker.py:255] I1224 01:04:51.788743 139711970813760 run_alphafold.py:191] Running model model_1_pred_0 on ECU03_1140\r\nI1223 20:04:54.797782 139688861775680 run_docker.py:255] I1224 01:04:54.797097 139711970813760 model.py:165] Running predict with shape(feat) = {'aatype': (4, 117), 'residue_index': (4, 117), 'seq_length': (4,), 'template_aatype': (4, 4, 117), 'template_all_atom_masks': (4, 4, 117, 37), 'template_all_atom_positions': (4, 4, 117, 37, 3), 'template_sum_probs': (4, 4, 1), 'is_distillation': (4,), 'seq_mask': (4, 117), 'msa_mask': (4, 508, 117), 'msa_row_mask': (4, 508), 'random_crop_to_size_seed': (4, 2), 'template_mask': (4, 4), 'template_pseudo_beta': (4, 4, 117, 3), 'template_pseudo_beta_mask': (4, 4, 117), 'atom14_atom_exists': (4, 117, 14), 'residx_atom14_to_atom37': (4, 117, 14), 'residx_atom37_to_atom14': (4, 117, 37), 'atom37_atom_exists': (4, 117, 37), 'extra_msa': (4, 5120, 117), 'extra_msa_mask': (4, 5120, 117), 'extra_msa_row_mask': (4, 5120), 'bert_mask': (4, 508, 117), 'true_msa': (4, 508, 117), 'extra_has_deletion': (4, 5120, 117), 'extra_deletion_value': (4, 5120, 117), 'msa_feat': (4, 508, 117, 49), 'target_feat': (4, 117, 22)}\r\nI1223 20:04:56.327672 139688861775680 run_docker.py:255] 2022-12-24 01:04:56.327116: W external/org_tensorflow/tensorflow/compiler/xla/stream_executor/gpu/asm_compiler.cc:231] Falling back to the CUDA driver for PTX compilation; ptxas does not support CC 8.9\r\nI1223 20:04:56.327813 139688861775680 run_docker.py:255] 2022-12-24 01:04:56.327156: W external/org_tensorflow/tensorflow/compiler/xla/stream_executor/gpu/asm_compiler.cc:234] Used ptxas at ptxas\r\nI1223 20:04:56.355589 139688861775680 run_docker.py:255] 2022-12-24 01:04:56.355236: E external/org_tensorflow/tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:628] failed to get PTX kernel \"shift_right_logical\" from module: CUDA_ERROR_NOT_FOUND: named symbol not found\r\nI1223 20:04:56.355756 139688861775680 run_docker.py:255] 2022-12-24 01:04:56.355273: E external/org_tensorflow/tensorflow/compiler/xla/pjrt/pjrt_stream_executor_client.cc:2153] Execution of replica 0 failed: INTERNAL: Could not find the corresponding function\r\nI1223 20:04:56.480186 139688861775680 run_docker.py:255] Traceback (most recent call last):\r\nI1223 20:04:56.480370 139688861775680 run_docker.py:255] File \"/app/alphafold/run_alphafold.py\", line 432, in <module>\r\nI1223 20:04:56.480440 139688861775680 run_docker.py:255] app.run(main)\r\nI1223 20:04:56.480502 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/absl/app.py\", line 312, in run\r\nI1223 20:04:56.480558 139688861775680 run_docker.py:255] _run_main(main, args)\r\nI1223 20:04:56.480613 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/absl/app.py\", line 258, in _run_main\r\nI1223 20:04:56.480718 139688861775680 run_docker.py:255] sys.exit(main(argv))\r\nI1223 20:04:56.480788 139688861775680 run_docker.py:255] File \"/app/alphafold/run_alphafold.py\", line 408, in main\r\nI1223 20:04:56.480844 139688861775680 run_docker.py:255] predict_structure(\r\nI1223 20:04:56.480898 139688861775680 run_docker.py:255] File \"/app/alphafold/run_alphafold.py\", line 199, in predict_structure\r\nI1223 20:04:56.480951 139688861775680 run_docker.py:255] prediction_result = model_runner.predict(processed_feature_dict,\r\nI1223 20:04:56.481002 139688861775680 run_docker.py:255] File \"/app/alphafold/alphafold/model/model.py\", line 167, in predict\r\nI1223 20:04:56.481052 139688861775680 run_docker.py:255] result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)\r\nI1223 20:04:56.481102 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/random.py\", line 132, in PRNGKey\r\nI1223 20:04:56.481152 139688861775680 run_docker.py:255] key = prng.seed_with_impl(impl, seed)\r\nI1223 20:04:56.481202 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 267, in seed_with_impl\r\nI1223 20:04:56.481253 139688861775680 run_docker.py:255] return random_seed(seed, impl=impl)\r\nI1223 20:04:56.481304 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 580, in random_seed\r\nI1223 20:04:56.481354 139688861775680 run_docker.py:255] return random_seed_p.bind(seeds_arr, impl=impl)\r\nI1223 20:04:56.481404 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 329, in bind\r\nI1223 20:04:56.481456 139688861775680 run_docker.py:255] return self.bind_with_trace(find_top_trace(args), args, params)\r\nI1223 20:04:56.481508 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 332, in bind_with_trace\r\nI1223 20:04:56.481559 139688861775680 run_docker.py:255] out = trace.process_primitive(self, map(trace.full_raise, args), params)\r\nI1223 20:04:56.481609 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 712, in process_primitive\r\nI1223 20:04:56.481659 139688861775680 run_docker.py:255] return primitive.impl(*tracers, **params)\r\nI1223 20:04:56.481709 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 592, in random_seed_impl\r\nI1223 20:04:56.481759 139688861775680 run_docker.py:255] base_arr = random_seed_impl_base(seeds, impl=impl)\r\nI1223 20:04:56.481808 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 597, in random_seed_impl_base\r\nI1223 20:04:56.481858 139688861775680 run_docker.py:255] return seed(seeds)\r\nI1223 20:04:56.481911 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 832, in threefry_seed\r\nI1223 20:04:56.481950 139688861775680 run_docker.py:255] lax.shift_right_logical(seed, lax_internal._const(seed, 32)))\r\nI1223 20:04:56.481987 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/lax/lax.py\", line 515, in shift_right_logical\r\nI1223 20:04:56.482024 139688861775680 run_docker.py:255] return shift_right_logical_p.bind(x, y)\r\nI1223 20:04:56.482062 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 329, in bind\r\nI1223 20:04:56.482100 139688861775680 run_docker.py:255] return self.bind_with_trace(find_top_trace(args), args, params)\r\nI1223 20:04:56.482138 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 332, in bind_with_trace\r\nI1223 20:04:56.482177 139688861775680 run_docker.py:255] out = trace.process_primitive(self, map(trace.full_raise, args), params)\r\nI1223 20:04:56.482217 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 712, in process_primitive\r\nI1223 20:04:56.482254 139688861775680 run_docker.py:255] return primitive.impl(*tracers, **params)\r\nI1223 20:04:56.482291 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/dispatch.py\", line 115, in apply_primitive\r\nI1223 20:04:56.482333 139688861775680 run_docker.py:255] return compiled_fun(*args)\r\nI1223 20:04:56.482372 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/dispatch.py\", line 200, in <lambda>\r\nI1223 20:04:56.482409 139688861775680 run_docker.py:255] return lambda *args, **kw: compiled(*args, **kw)[0]\r\nI1223 20:04:56.482447 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/dispatch.py\", line 895, in _execute_compiled\r\nI1223 20:04:56.482484 139688861775680 run_docker.py:255] out_flat = compiled.execute(in_flat)\r\nI1223 20:04:56.482520 139688861775680 run_docker.py:255] jaxlib.xla_extension.XlaRuntimeError: INTERNAL: Could not find the corresponding function\r\n```",
"@joshabramson I was able to get alphafold to run by substituting nvidia/cuda:11.8.0-cudnn8-devel-ubuntu20.04 into the dockerfile. "
] | "2022-11-29T13:22:15" | "2022-12-26T05:56:10" | null | NONE | null | Hi.
A problem occurred while I was running the Alphafold.
Could I ask for help on how to solve it?
Please check the command below and if you need any information about that plase let me know.
```
jhyeon@jhyeon-Ubuntu:~/Desktop/data/SW/alphafold$ sudo python3 docker/run_docker.py --fasta_paths=T.fa --max_template_date=2021-12-31 --data_dir=/mnt/8THDD/data/db/AFDB/ --model_preset=monomer --output_dir=/home/jhyeon/Desktop/data/alphatest
[sudo] password for jhyeon:
I1129 20:06:44.587526 140678175391744 run_docker.py:113] Mounting /home/jhyeon/Desktop/data/SW/alphafold -> /mnt/fasta_path_0
I1129 20:06:44.998342 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/uniref90 -> /mnt/uniref90_database_path
I1129 20:06:45.015172 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/mgnify -> /mnt/mgnify_database_path
I1129 20:06:45.015434 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB -> /mnt/data_dir
I1129 20:06:45.039951 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/pdb_mmcif/mmcif_files -> /mnt/template_mmcif_dir
I1129 20:06:45.040283 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/pdb_mmcif -> /mnt/obsolete_pdbs_path
I1129 20:06:45.058208 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/pdb70 -> /mnt/pdb70_database_path
I1129 20:06:45.099748 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/uniclust30/uniclust30_2018_08 -> /mnt/uniclust30_database_path
I1129 20:06:45.100375 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/bfd -> /mnt/bfd_database_path
I1129 20:06:47.216925 140678175391744 run_docker.py:255] /opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:37: FutureWarning: jax.tree_structure is deprecated, and will be removed in a future release. Use jax.tree_util.tree_structure instead.
I1129 20:06:47.217015 140678175391744 run_docker.py:255] PyTreeDef = type(jax.tree_structure(None))
I1129 20:06:47.668879 140678175391744 run_docker.py:255] I1129 11:06:47.668288 140554574919488 templates.py:857] Using precomputed obsolete pdbs /mnt/obsolete_pdbs_path/obsolete.dat.
I1129 20:06:50.314509 140678175391744 run_docker.py:255] I1129 11:06:50.314045 140554574919488 xla_bridge.py:353] Unable to initialize backend 'tpu_driver': NOT_FOUND: Unable to find driver in registry given worker:
I1129 20:06:50.395502 140678175391744 run_docker.py:255] I1129 11:06:50.394992 140554574919488 xla_bridge.py:353] Unable to initialize backend 'rocm': NOT_FOUND: Could not find registered platform with name: "rocm". Available platform names are: Host CUDA Interpreter
I1129 20:06:50.395585 140678175391744 run_docker.py:255] I1129 11:06:50.395253 140554574919488 xla_bridge.py:353] Unable to initialize backend 'tpu': module 'jaxlib.xla_extension' has no attribute 'get_tpu_client'
I1129 20:06:50.395616 140678175391744 run_docker.py:255] I1129 11:06:50.395327 140554574919488 xla_bridge.py:353] Unable to initialize backend 'plugin': xla_extension has no attributes named get_plugin_device_client. Compile TensorFlow with //tensorflow/compiler/xla/python:enable_plugin_device set to true (defaults to false) to enable this.
I1129 20:07:01.872005 140678175391744 run_docker.py:255] I1129 11:07:01.871499 140554574919488 run_alphafold.py:377] Have 5 models: ['model_1_pred_0', 'model_2_pred_0', 'model_3_pred_0', 'model_4_pred_0', 'model_5_pred_0']
I1129 20:07:01.872115 140678175391744 run_docker.py:255] I1129 11:07:01.871589 140554574919488 run_alphafold.py:393] Using random seed 776916752095554131 for the data pipeline
I1129 20:07:01.872142 140678175391744 run_docker.py:255] I1129 11:07:01.871697 140554574919488 run_alphafold.py:161] Predicting T
I1129 20:07:01.872171 140678175391744 run_docker.py:255] I1129 11:07:01.871899 140554574919488 jackhmmer.py:133] Launching subprocess "/usr/bin/jackhmmer -o /dev/null -A /tmp/tmpyb0_qtih/output.sto --noali --F1 0.0005 --F2 5e-05 --F3 5e-07 --incE 0.0001 -E 0.0001 --cpu 8 -N 1 /mnt/fasta_path_0/T.fa /mnt/uniref90_database_path/uniref90.fasta"
I1129 20:07:01.908833 140678175391744 run_docker.py:255] I1129 11:07:01.908337 140554574919488 utils.py:36] Started Jackhmmer (uniref90.fasta) query
I1129 20:17:08.123620 140678175391744 run_docker.py:255] I1129 11:17:08.122851 140554574919488 utils.py:40] Finished Jackhmmer (uniref90.fasta) query in 606.214 seconds
I1129 20:17:08.124002 140678175391744 run_docker.py:255] I1129 11:17:08.123401 140554574919488 jackhmmer.py:133] Launching subprocess "/usr/bin/jackhmmer -o /dev/null -A /tmp/tmp3h2swg2s/output.sto --noali --F1 0.0005 --F2 5e-05 --F3 5e-07 --incE 0.0001 -E 0.0001 --cpu 8 -N 1 /mnt/fasta_path_0/T.fa /mnt/mgnify_database_path/mgy_clusters_2018_12.fa"
I1129 20:17:08.153515 140678175391744 run_docker.py:255] I1129 11:17:08.152948 140554574919488 utils.py:36] Started Jackhmmer (mgy_clusters_2018_12.fa) query
I1129 20:25:33.106717 140678175391744 run_docker.py:255] I1129 11:25:33.105955 140554574919488 utils.py:40] Finished Jackhmmer (mgy_clusters_2018_12.fa) query in 504.953 seconds
I1129 20:25:33.107178 140678175391744 run_docker.py:255] I1129 11:25:33.106705 140554574919488 hhsearch.py:85] Launching subprocess "/usr/bin/hhsearch -i /tmp/tmpi641el8l/query.a3m -o /tmp/tmpi641el8l/output.hhr -maxseq 1000000 -d /mnt/pdb70_database_path/pdb70"
I1129 20:25:33.138576 140678175391744 run_docker.py:255] I1129 11:25:33.138024 140554574919488 utils.py:36] Started HHsearch query
I1129 20:26:54.573565 140678175391744 run_docker.py:255] I1129 11:26:54.573027 140554574919488 utils.py:40] Finished HHsearch query in 81.435 seconds
I1129 20:26:54.582471 140678175391744 run_docker.py:255] I1129 11:26:54.581876 140554574919488 hhblits.py:128] Launching subprocess "/usr/bin/hhblits -i /mnt/fasta_path_0/T.fa -cpu 4 -oa3m /tmp/tmp0adsap7y/output.a3m -o /dev/null -n 3 -e 0.001 -maxseq 1000000 -realign_max 100000 -maxfilt 100000 -min_prefilter_hits 1000 -d /mnt/bfd_database_path/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt -d /mnt/uniclust30_database_path/uniclust30_2018_08"
I1129 20:26:54.614019 140678175391744 run_docker.py:255] I1129 11:26:54.613526 140554574919488 utils.py:36] Started HHblits query
I1129 20:39:47.502100 140678175391744 run_docker.py:255] I1129 11:39:47.501456 140554574919488 utils.py:40] Finished HHblits query in 772.888 seconds
I1129 20:39:47.511255 140678175391744 run_docker.py:255] I1129 11:39:47.510757 140554574919488 templates.py:878] Searching for template for: MAAAAAAAAAAAAAAAAAAAAAAAAA
I1129 20:39:49.477572 140678175391744 run_docker.py:255] I1129 11:39:49.476987 140554574919488 templates.py:268] Found an exact template match 4d10_F.
I1129 20:39:49.481890 140678175391744 run_docker.py:255] I1129 11:39:49.481489 140554574919488 templates.py:913] Skipped invalid hit 4D10_F COP9 SIGNALOSOME COMPLEX SUBUNIT 1; SIGNALING PROTEIN; 3.8A {HOMO SAPIENS}, error: None, warning: 4d10_F (sum_probs: 0.0, rank: 1): feature extracting errors: Template all atom mask was all zeros: 4d10_F. Residue range: 4-13, mmCIF parsing errors: {}
I1129 20:39:54.642663 140678175391744 run_docker.py:255] I1129 11:39:54.642101 140554574919488 templates.py:268] Found an exact template match 4v7h_BQ.
I1129 20:39:56.860277 140678175391744 run_docker.py:255] I1129 11:39:56.859714 140554574919488 templates.py:268] Found an exact template match 6j3y_W.
I1129 20:39:56.861339 140678175391744 run_docker.py:255] I1129 11:39:56.860958 140554574919488 templates.py:913] Skipped invalid hit 6J3Y_W Photosystem II reaction center protein; Photosystem, ELECTRON TRANSPORT; HET: LMG, HEM, DGD, SQD, LMU, BCR, CLA, OEX, PHO, PL9, LHG, A86; 3.3A {Chaetoceros gracilis}, error: None, warning: 6j3y_W (sum_probs: 0.0, rank: 3): feature extracting errors: Template all atom mask was all zeros: 6j3y_W. Residue range: 0-18, mmCIF parsing errors: {}
I1129 20:39:59.248605 140678175391744 run_docker.py:255] I1129 11:39:59.247956 140554574919488 templates.py:268] Found an exact template match 6j3z_w.
I1129 20:39:59.249619 140678175391744 run_docker.py:255] I1129 11:39:59.249154 140554574919488 templates.py:913] Skipped invalid hit 6J3Z_w Photosystem II reaction center protein; Photosystem, ELECTRON TRANSPORT; HET: LMG, HEM, DGD, SQD, LMU, BCR, CLA, OEX, PHO, PL9, LHG, A86; 3.6A {Chaetoceros gracilis}, error: None, warning: 6j3z_w (sum_probs: 0.0, rank: 4): feature extracting errors: Template all atom mask was all zeros: 6j3z_w. Residue range: 0-18, mmCIF parsing errors: {}
I1129 20:39:59.369462 140678175391744 run_docker.py:255] I1129 11:39:59.369046 140554574919488 templates.py:268] Found an exact template match 1m0u_B.
I1129 20:39:59.372590 140678175391744 run_docker.py:255] I1129 11:39:59.372143 140554574919488 templates.py:913] Skipped invalid hit 1M0U_B GST2 gene product (E.C.2.5.1.18); GST, Flight Muscle Protein, Sigma; HET: SO4, GSH; 1.75A {Drosophila melanogaster} SCOP: a.45.1.1, c.47.1.5, error: None, warning: 1m0u_B (sum_probs: 0.0, rank: 5): feature extracting errors: Template all atom mask was all zeros: 1m0u_B. Residue range: 0-23, mmCIF parsing errors: {}
I1129 20:39:59.973728 140678175391744 run_docker.py:255] I1129 11:39:59.973092 140554574919488 templates.py:268] Found an exact template match 1kn7_A.
I1129 20:40:02.077867 140678175391744 run_docker.py:255] I1129 11:40:02.077314 140554574919488 templates.py:268] Found an exact template match 6rfq_8.
I1129 20:40:04.344768 140678175391744 run_docker.py:255] I1129 11:40:04.336282 140554574919488 templates.py:268] Found an exact template match 6rfr_8.
I1129 20:40:08.787640 140678175391744 run_docker.py:255] I1129 11:40:08.786539 140554574919488 templates.py:268] Found an exact template match 6t59_s3.
I1129 20:40:08.790601 140678175391744 run_docker.py:255] I1129 11:40:08.790089 140554574919488 templates.py:913] Skipped invalid hit 6T59_s3 Ribosomal protein L8, uL3, uL4; TUBULIN, nascent chain-associated complex, ribosome-nascent; HET: MG; 3.11A {Oryctolagus cuniculus}, error: None, warning: 6t59_s3 (sum_probs: 0.0, rank: 9): feature extracting errors: Template all atom mask was all zeros: 6t59_s3. Residue range: 273-297, mmCIF parsing errors: {}
I1129 20:40:09.027597 140678175391744 run_docker.py:255] I1129 11:40:09.027103 140554574919488 templates.py:268] Found an exact template match 3lpj_B.
I1129 20:40:09.033292 140678175391744 run_docker.py:255] I1129 11:40:09.032789 140554574919488 templates.py:913] Skipped invalid hit 3LPJ_B Structure of BACE Bound to; Alzheimer's, Aspartyl protease, Hydrolase; HET: TLA, Z75; 1.79A {Homo sapiens}, error: None, warning: 3lpj_B (sum_probs: 0.0, rank: 10): feature extracting errors: Template all atom mask was all zeros: 3lpj_B. Residue range: 0-24, mmCIF parsing errors: {}
I1129 20:40:09.033350 140678175391744 run_docker.py:255] I1129 11:40:09.033004 140554574919488 pipeline.py:234] Uniref90 MSA size: 1 sequences.
I1129 20:40:09.033376 140678175391744 run_docker.py:255] I1129 11:40:09.033061 140554574919488 pipeline.py:235] BFD MSA size: 1 sequences.
I1129 20:40:09.033398 140678175391744 run_docker.py:255] I1129 11:40:09.033080 140554574919488 pipeline.py:236] MGnify MSA size: 1 sequences.
I1129 20:40:09.033423 140678175391744 run_docker.py:255] I1129 11:40:09.033095 140554574919488 pipeline.py:238] Final (deduplicated) MSA size: 1 sequences.
I1129 20:40:09.033465 140678175391744 run_docker.py:255] I1129 11:40:09.033219 140554574919488 pipeline.py:241] Total number of templates (NB: this can include bad templates and is later filtered to top 4): 4.
I1129 20:40:09.041668 140678175391744 run_docker.py:255] I1129 11:40:09.041206 140554574919488 run_alphafold.py:190] Running model model_1_pred_0 on T
I1129 20:40:10.478550 140678175391744 run_docker.py:255] I1129 11:40:10.478168 140554574919488 model.py:166] Running predict with shape(feat) = {'aatype': (4, 26), 'residue_index': (4, 26), 'seq_length': (4,), 'template_aatype': (4, 4, 26), 'template_all_atom_masks': (4, 4, 26, 37), 'template_all_atom_positions': (4, 4, 26, 37, 3), 'template_sum_probs': (4, 4, 1), 'is_distillation': (4,), 'seq_mask': (4, 26), 'msa_mask': (4, 508, 26), 'msa_row_mask': (4, 508), 'random_crop_to_size_seed': (4, 2), 'template_mask': (4, 4), 'template_pseudo_beta': (4, 4, 26, 3), 'template_pseudo_beta_mask': (4, 4, 26), 'atom14_atom_exists': (4, 26, 14), 'residx_atom14_to_atom37': (4, 26, 14), 'residx_atom37_to_atom14': (4, 26, 37), 'atom37_atom_exists': (4, 26, 37), 'extra_msa': (4, 5120, 26), 'extra_msa_mask': (4, 5120, 26), 'extra_msa_row_mask': (4, 5120), 'bert_mask': (4, 508, 26), 'true_msa': (4, 508, 26), 'extra_has_deletion': (4, 5120, 26), 'extra_deletion_value': (4, 5120, 26), 'msa_feat': (4, 508, 26, 49), 'target_feat': (4, 26, 22)}
I1129 20:40:10.573517 140678175391744 run_docker.py:255] 2022-11-29 11:40:10.572368: W external/org_tensorflow/tensorflow/compiler/xla/stream_executor/gpu/asm_compiler.cc:231] Falling back to the CUDA driver for PTX compilation; ptxas does not support CC 8.9
I1129 20:40:10.573782 140678175391744 run_docker.py:255] 2022-11-29 11:40:10.572425: W external/org_tensorflow/tensorflow/compiler/xla/stream_executor/gpu/asm_compiler.cc:234] Used ptxas at ptxas
I1129 20:40:10.586611 140678175391744 run_docker.py:255] 2022-11-29 11:40:10.585689: E external/org_tensorflow/tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:628] failed to get PTX kernel "shift_right_logical" from module: CUDA_ERROR_NOT_FOUND: named symbol not found
I1129 20:40:10.586843 140678175391744 run_docker.py:255] 2022-11-29 11:40:10.585779: E external/org_tensorflow/tensorflow/compiler/xla/pjrt/pjrt_stream_executor_client.cc:2153] Execution of replica 0 failed: INTERNAL: Could not find the corresponding function
I1129 20:40:10.594201 140678175391744 run_docker.py:255] Traceback (most recent call last):
I1129 20:40:10.594406 140678175391744 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 422, in <module>
I1129 20:40:10.594515 140678175391744 run_docker.py:255] app.run(main)
I1129 20:40:10.594610 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 312, in run
I1129 20:40:10.594703 140678175391744 run_docker.py:255] _run_main(main, args)
I1129 20:40:10.594786 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 258, in _run_main
I1129 20:40:10.594868 140678175391744 run_docker.py:255] sys.exit(main(argv))
I1129 20:40:10.594947 140678175391744 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 406, in main
I1129 20:40:10.595050 140678175391744 run_docker.py:255] random_seed=random_seed)
I1129 20:40:10.595142 140678175391744 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 199, in predict_structure
I1129 20:40:10.595224 140678175391744 run_docker.py:255] random_seed=model_random_seed)
I1129 20:40:10.595304 140678175391744 run_docker.py:255] File "/app/alphafold/alphafold/model/model.py", line 167, in predict
I1129 20:40:10.595379 140678175391744 run_docker.py:255] result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)
I1129 20:40:10.595466 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/random.py", line 132, in PRNGKey
I1129 20:40:10.595538 140678175391744 run_docker.py:255] key = prng.seed_with_impl(impl, seed)
I1129 20:40:10.595611 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 267, in seed_with_impl
I1129 20:40:10.595727 140678175391744 run_docker.py:255] return random_seed(seed, impl=impl)
I1129 20:40:10.595801 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 580, in random_seed
I1129 20:40:10.595872 140678175391744 run_docker.py:255] return random_seed_p.bind(seeds_arr, impl=impl)
I1129 20:40:10.595945 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 329, in bind
I1129 20:40:10.596016 140678175391744 run_docker.py:255] return self.bind_with_trace(find_top_trace(args), args, params)
I1129 20:40:10.596088 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 332, in bind_with_trace
I1129 20:40:10.596163 140678175391744 run_docker.py:255] out = trace.process_primitive(self, map(trace.full_raise, args), params)
I1129 20:40:10.596238 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 712, in process_primitive
I1129 20:40:10.596311 140678175391744 run_docker.py:255] return primitive.impl(*tracers, **params)
I1129 20:40:10.596384 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 592, in random_seed_impl
I1129 20:40:10.596455 140678175391744 run_docker.py:255] base_arr = random_seed_impl_base(seeds, impl=impl)
I1129 20:40:10.596517 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 597, in random_seed_impl_base
I1129 20:40:10.596581 140678175391744 run_docker.py:255] return seed(seeds)
I1129 20:40:10.596646 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 832, in threefry_seed
I1129 20:40:10.596710 140678175391744 run_docker.py:255] lax.shift_right_logical(seed, lax_internal._const(seed, 32)))
I1129 20:40:10.596774 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/lax/lax.py", line 515, in shift_right_logical
I1129 20:40:10.596839 140678175391744 run_docker.py:255] return shift_right_logical_p.bind(x, y)
I1129 20:40:10.596904 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 329, in bind
I1129 20:40:10.596969 140678175391744 run_docker.py:255] return self.bind_with_trace(find_top_trace(args), args, params)
I1129 20:40:10.597032 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 332, in bind_with_trace
I1129 20:40:10.597097 140678175391744 run_docker.py:255] out = trace.process_primitive(self, map(trace.full_raise, args), params)
I1129 20:40:10.597160 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 712, in process_primitive
I1129 20:40:10.597218 140678175391744 run_docker.py:255] return primitive.impl(*tracers, **params)
I1129 20:40:10.597279 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/dispatch.py", line 115, in apply_primitive
I1129 20:40:10.597339 140678175391744 run_docker.py:255] return compiled_fun(*args)
I1129 20:40:10.597402 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/dispatch.py", line 200, in <lambda>
I1129 20:40:10.597465 140678175391744 run_docker.py:255] return lambda *args, **kw: compiled(*args, **kw)[0]
I1129 20:40:10.597525 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/dispatch.py", line 895, in _execute_compiled
I1129 20:40:10.597588 140678175391744 run_docker.py:255] out_flat = compiled.execute(in_flat)
I1129 20:40:10.597648 140678175391744 run_docker.py:255] jaxlib.xla_extension.XlaRuntimeError: INTERNAL: Could not find the corresponding function
``` | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/646/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/646/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/645 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/645/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/645/comments | https://api.github.com/repos/deepmind/alphafold/issues/645/events | https://github.com/deepmind/alphafold/issues/645 | 1,467,828,543 | I_kwDOFoWQLM5XfUk_ | 645 | Running Alphafold using unpublished Pdb as a custom template | {
"avatar_url": "https://avatars.githubusercontent.com/u/119045747?v=4",
"events_url": "https://api.github.com/users/Shashankhare/events{/privacy}",
"followers_url": "https://api.github.com/users/Shashankhare/followers",
"following_url": "https://api.github.com/users/Shashankhare/following{/other_user}",
"gists_url": "https://api.github.com/users/Shashankhare/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Shashankhare",
"id": 119045747,
"login": "Shashankhare",
"node_id": "U_kgDOBxh-cw",
"organizations_url": "https://api.github.com/users/Shashankhare/orgs",
"received_events_url": "https://api.github.com/users/Shashankhare/received_events",
"repos_url": "https://api.github.com/users/Shashankhare/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Shashankhare/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Shashankhare/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Shashankhare"
} | [] | open | false | null | [] | null | [] | "2022-11-29T10:34:07" | "2022-11-29T10:34:07" | null | NONE | null | Hi,
I am trying to run Alphafold using a custom template. The template I am using as input is not published yet but I have both realspace-refined Pdb and cif files. When I am trying to run Alphafold with the unpublished Pdb, I get an error saying "no templates found”. I am wondering if is it possible to run Alphafold using unpublished Pdb as a custom template.
Regards,
Shashank. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/645/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/645/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/644 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/644/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/644/comments | https://api.github.com/repos/deepmind/alphafold/issues/644/events | https://github.com/deepmind/alphafold/pull/644 | 1,465,995,763 | PR_kwDOFoWQLM5Dx7Y_ | 644 | Update Readme.md File: Added license badge, Forks count badge, Watchers Count Badge | {
"avatar_url": "https://avatars.githubusercontent.com/u/101518890?v=4",
"events_url": "https://api.github.com/users/dhakalnirajan/events{/privacy}",
"followers_url": "https://api.github.com/users/dhakalnirajan/followers",
"following_url": "https://api.github.com/users/dhakalnirajan/following{/other_user}",
"gists_url": "https://api.github.com/users/dhakalnirajan/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dhakalnirajan",
"id": 101518890,
"login": "dhakalnirajan",
"node_id": "U_kgDOBg0OKg",
"organizations_url": "https://api.github.com/users/dhakalnirajan/orgs",
"received_events_url": "https://api.github.com/users/dhakalnirajan/received_events",
"repos_url": "https://api.github.com/users/dhakalnirajan/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dhakalnirajan/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dhakalnirajan/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dhakalnirajan"
} | [] | open | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/644/checks?check_run_id=9739001347) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request."
] | "2022-11-28T08:15:56" | "2022-12-24T14:09:36" | null | NONE | null | I have added license badge that redirects to the license page when clicked.
I have added GitHub repository Fork count badge that tells the visitors how many times the repository has been forked.
I have added GitHub Watchers Count Badge that displays the number of watchers on the repository. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/644/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/644/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/644.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/644",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/644.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/644"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/643 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/643/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/643/comments | https://api.github.com/repos/deepmind/alphafold/issues/643/events | https://github.com/deepmind/alphafold/pull/643 | 1,465,964,347 | PR_kwDOFoWQLM5Dx0mJ | 643 | Readme.md file update: Added License badge, Forks count badge and Watchers Count badge, | {
"avatar_url": "https://avatars.githubusercontent.com/u/101518890?v=4",
"events_url": "https://api.github.com/users/dhakalnirajan/events{/privacy}",
"followers_url": "https://api.github.com/users/dhakalnirajan/followers",
"following_url": "https://api.github.com/users/dhakalnirajan/following{/other_user}",
"gists_url": "https://api.github.com/users/dhakalnirajan/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dhakalnirajan",
"id": 101518890,
"login": "dhakalnirajan",
"node_id": "U_kgDOBg0OKg",
"organizations_url": "https://api.github.com/users/dhakalnirajan/orgs",
"received_events_url": "https://api.github.com/users/dhakalnirajan/received_events",
"repos_url": "https://api.github.com/users/dhakalnirajan/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dhakalnirajan/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dhakalnirajan/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dhakalnirajan"
} | [] | closed | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/643/checks?check_run_id=9738583833) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request."
] | "2022-11-28T07:51:06" | "2022-11-28T08:00:38" | "2022-11-28T08:00:38" | NONE | null | I have added license badge that redirects to the license page when clicked.
I have added GitHub repository Fork count badge that tells the visitors how many times the repository has been forked.
I have added GitHub Watchers Count Badge that displays the number of watchers on the repository. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/643/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/643/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/643.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/643",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/643.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/643"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/642 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/642/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/642/comments | https://api.github.com/repos/deepmind/alphafold/issues/642/events | https://github.com/deepmind/alphafold/issues/642 | 1,465,935,691 | I_kwDOFoWQLM5XYGdL | 642 | Can I customize the database? | {
"avatar_url": "https://avatars.githubusercontent.com/u/47802187?v=4",
"events_url": "https://api.github.com/users/dahaigui/events{/privacy}",
"followers_url": "https://api.github.com/users/dahaigui/followers",
"following_url": "https://api.github.com/users/dahaigui/following{/other_user}",
"gists_url": "https://api.github.com/users/dahaigui/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dahaigui",
"id": 47802187,
"login": "dahaigui",
"node_id": "MDQ6VXNlcjQ3ODAyMTg3",
"organizations_url": "https://api.github.com/users/dahaigui/orgs",
"received_events_url": "https://api.github.com/users/dahaigui/received_events",
"repos_url": "https://api.github.com/users/dahaigui/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dahaigui/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dahaigui/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dahaigui"
} | [] | open | false | null | [] | null | [] | "2022-11-28T07:18:50" | "2022-11-28T07:18:50" | null | NONE | null | My protein sequence is from a mutant library, is it possible to speed up my protein structure prediction by changing the dataset? For example, without using the BFD dataset, if it is possible, how should I implement it? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/642/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/642/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/641 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/641/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/641/comments | https://api.github.com/repos/deepmind/alphafold/issues/641/events | https://github.com/deepmind/alphafold/pull/641 | 1,465,540,000 | PR_kwDOFoWQLM5Dwbxe | 641 | Bugfix: Updated the AlphaFold notebook to make it compatible with newer versions of Google Colab (fixes the jax_experimental_name_stack error) | {
"avatar_url": "https://avatars.githubusercontent.com/u/40127722?v=4",
"events_url": "https://api.github.com/users/anupam-tiwari/events{/privacy}",
"followers_url": "https://api.github.com/users/anupam-tiwari/followers",
"following_url": "https://api.github.com/users/anupam-tiwari/following{/other_user}",
"gists_url": "https://api.github.com/users/anupam-tiwari/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/anupam-tiwari",
"id": 40127722,
"login": "anupam-tiwari",
"node_id": "MDQ6VXNlcjQwMTI3NzIy",
"organizations_url": "https://api.github.com/users/anupam-tiwari/orgs",
"received_events_url": "https://api.github.com/users/anupam-tiwari/received_events",
"repos_url": "https://api.github.com/users/anupam-tiwari/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/anupam-tiwari/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/anupam-tiwari/subscriptions",
"type": "User",
"url": "https://api.github.com/users/anupam-tiwari"
} | [] | closed | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/641/checks?check_run_id=9731462268) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request.",
"Hi thanks for providing this PR. Its no longer needed as we have updated Jax and Haiku, so I will close this."
] | "2022-11-27T18:43:37" | "2022-11-30T18:38:29" | "2022-11-30T18:38:29" | NONE | null | Since the newest Release of Google Colab(reference: https://colab.research.google.com/notebooks/relnotes.ipynb) the notebook has moved from Jax 0.3.17 to 0.3.25 which has Deleted the jax_experimental_name_stack config option(reference: https://jax.readthedocs.io/en/latest/changelog.html) This is causing the notebook to crash at the "5. Run AlphaFold and download prediction" step by throwing a config error as shown below:

This pull request solves the issue by downgrading the Jax and Jaxlib to 0.3.17 and 0.3.15[cuda] in the first stage of running the notebook and thus making the notebook functional | {
"+1": 1,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 1,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/641/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/641/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/641.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/641",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/641.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/641"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/640 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/640/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/640/comments | https://api.github.com/repos/deepmind/alphafold/issues/640/events | https://github.com/deepmind/alphafold/issues/640 | 1,465,309,598 | I_kwDOFoWQLM5XVtme | 640 | Alphafold Multimer Results | {
"avatar_url": "https://avatars.githubusercontent.com/u/119135711?v=4",
"events_url": "https://api.github.com/users/dozicauf/events{/privacy}",
"followers_url": "https://api.github.com/users/dozicauf/followers",
"following_url": "https://api.github.com/users/dozicauf/following{/other_user}",
"gists_url": "https://api.github.com/users/dozicauf/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dozicauf",
"id": 119135711,
"login": "dozicauf",
"node_id": "U_kgDOBxnd3w",
"organizations_url": "https://api.github.com/users/dozicauf/orgs",
"received_events_url": "https://api.github.com/users/dozicauf/received_events",
"repos_url": "https://api.github.com/users/dozicauf/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dozicauf/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dozicauf/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dozicauf"
} | [] | open | false | null | [] | null | [] | "2022-11-27T02:24:02" | "2022-11-27T02:24:02" | null | NONE | null | Hi,
I am wondering if anyone knows a website with Alphafold multimer predicted structures. The reason why I am asking this is that I am interested in exploring the PAE confusion matrix of different oligomeric structures. Also, is anyone aware of any code/rule by which we could say that a particular protein forms a dimer let's say? I know that we can determine that qualitatively by looking at the PAE and myb by looking at the contacts between chains that are less than 8A, but is there a rule for example of how many of such contact points we need to have or any other quantifiable parameter that could be used to determine whether a protein forms a particular oligomer based on the PAE. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/640/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/640/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/639 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/639/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/639/comments | https://api.github.com/repos/deepmind/alphafold/issues/639/events | https://github.com/deepmind/alphafold/issues/639 | 1,465,168,586 | I_kwDOFoWQLM5XVLLK | 639 | AlphaFold multimer has name error in the last step | {
"avatar_url": "https://avatars.githubusercontent.com/u/65905482?v=4",
"events_url": "https://api.github.com/users/Isabel-ql/events{/privacy}",
"followers_url": "https://api.github.com/users/Isabel-ql/followers",
"following_url": "https://api.github.com/users/Isabel-ql/following{/other_user}",
"gists_url": "https://api.github.com/users/Isabel-ql/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Isabel-ql",
"id": 65905482,
"login": "Isabel-ql",
"node_id": "MDQ6VXNlcjY1OTA1NDgy",
"organizations_url": "https://api.github.com/users/Isabel-ql/orgs",
"received_events_url": "https://api.github.com/users/Isabel-ql/received_events",
"repos_url": "https://api.github.com/users/Isabel-ql/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Isabel-ql/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Isabel-ql/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Isabel-ql"
} | [] | closed | false | null | [] | null | [
"Did you run all the cells properly, especially the second one \"2. Download AlphaFold\"?",
"> Did you run all the cells properly, especially the second one \"2. Download AlphaFold\"?\r\n\r\nYes, the other steps are all successful. Only the last step failed. \r\n<img width=\"719\" alt=\"AlphaFold Multimer_Step2\" src=\"https://user-images.githubusercontent.com/65905482/204095350-b0e68050-8404-4846-bb36-1c44df44d31d.png\">\r\n",
"Did you get it running? if now try this version: https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharing",
"> Did you get it running? if now try this version: https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharing\r\n\r\nThe old one is still problematic, but the new link you sent me works! I just finished the prediction. Thanks so much! ",
"Hi thanks for this. This may have been due to the colab kernel restarting, so that whilst the previous cells had run correctly they have since had their output cleared. When this happens, the colab needs to be rerun from the beginning. ",
"> Did you get it running? if now try this version: https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharing\r\n\r\nHi, a new problem appeared with the new link. In step 5, \"Search against genetic database\", there's an import error. See below. The problem started from yesterday, and still exists now. How to fix it? Thanks!\r\n\r\nCheers,\r\nQun\r\n\r\n<img width=\"1381\" alt=\"AlphaFold Multimer_step5\" src=\"https://user-images.githubusercontent.com/65905482/205282261-502402d8-f5b4-4742-a10d-51d8628cca67.png\">\r\n"
] | "2022-11-26T14:24:51" | "2022-12-02T11:25:16" | "2022-11-30T18:35:18" | NONE | null | Hi,
I have tried to use AlphaFold-Multimer to predict the structures of complexes, I did the prediction from the following link:
https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb
However, problem appeared in the last step, "Run AlphaFold and download prediction". The notice said "NameError: name 'model_type_to_use' is not defined". How to solve this problem?
Thanks very much!
Cheers,
Qun
<img width="1515" alt="AlphaFold Multimer_NameError" src="https://user-images.githubusercontent.com/65905482/204093603-ca29246b-1a42-466f-9a52-2b860e54a732.png">
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/639/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/639/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/638 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/638/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/638/comments | https://api.github.com/repos/deepmind/alphafold/issues/638/events | https://github.com/deepmind/alphafold/issues/638 | 1,463,803,012 | I_kwDOFoWQLM5XP9yE | 638 | Alphafold broken on Colab | {
"avatar_url": "https://avatars.githubusercontent.com/u/18224663?v=4",
"events_url": "https://api.github.com/users/danjaco/events{/privacy}",
"followers_url": "https://api.github.com/users/danjaco/followers",
"following_url": "https://api.github.com/users/danjaco/following{/other_user}",
"gists_url": "https://api.github.com/users/danjaco/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/danjaco",
"id": 18224663,
"login": "danjaco",
"node_id": "MDQ6VXNlcjE4MjI0NjYz",
"organizations_url": "https://api.github.com/users/danjaco/orgs",
"received_events_url": "https://api.github.com/users/danjaco/received_events",
"repos_url": "https://api.github.com/users/danjaco/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/danjaco/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/danjaco/subscriptions",
"type": "User",
"url": "https://api.github.com/users/danjaco"
} | [] | closed | false | null | [] | null | [
"Colab is also broken here\r\n\r\n\r\n",
"Hi Dan, thanks for raising this. The python dependencies in colab change over time, which means that the environment that the code runs in may diverge from alphafold, even if the alphafold code itself is stable. We will update the requirements when this happens. The most recent commit updated jax and haiku to match the colab versions, so this issue should be fixed",
"sorry, there is just a new error\r\n\r\n\r\n\r\n",
"I have an error:\r\n\r\n",
"Hi this error is from a Phenix notebook that isn't maintained by DeepMind. Please raise this issue in the Phenix repo. Thanks!"
] | "2022-11-24T20:54:55" | "2022-12-05T23:05:20" | "2022-11-30T18:32:15" | NONE | null | This issue is reported by others earlier, but I rase this separately as a more permanent solution is needed. I've come to rely on Colab version of Alphafold but a couple of times recently it's been down for over a week without us having any idea when it would be fixed
Would it be possible to make a second version available, that is known to be stable, so that if a new rollout fails there is something to fall back to? Or some other solution that could make it more reliable?
Thanks,
Dan | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/638/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/638/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/637 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/637/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/637/comments | https://api.github.com/repos/deepmind/alphafold/issues/637/events | https://github.com/deepmind/alphafold/issues/637 | 1,462,050,712 | I_kwDOFoWQLM5XJR-Y | 637 | Alpha Fold crash | {
"avatar_url": "https://avatars.githubusercontent.com/u/118919803?v=4",
"events_url": "https://api.github.com/users/HPN2/events{/privacy}",
"followers_url": "https://api.github.com/users/HPN2/followers",
"following_url": "https://api.github.com/users/HPN2/following{/other_user}",
"gists_url": "https://api.github.com/users/HPN2/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/HPN2",
"id": 118919803,
"login": "HPN2",
"node_id": "U_kgDOBxaSew",
"organizations_url": "https://api.github.com/users/HPN2/orgs",
"received_events_url": "https://api.github.com/users/HPN2/received_events",
"repos_url": "https://api.github.com/users/HPN2/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/HPN2/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/HPN2/subscriptions",
"type": "User",
"url": "https://api.github.com/users/HPN2"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"Same error here but unsure how to solve it",
"This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime",
"> This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime\r\n\r\nHey, I have been struggeling to apply your solution. Could you elaborate further on when and where to insert this command?",
"Thanks for raising this. Closing as this is a duplicate of https://github.com/deepmind/alphafold/issues/635."
] | "2022-11-23T16:22:49" | "2022-11-28T20:09:13" | "2022-11-28T20:09:12" | NONE | null | After running AF Colab multimer for weeks, it started to get unstable this week. Recently it has crashed multiple times. Each when starting task 5 (Run AlphaFold and download prediction) the system crashes within no time although I am Prolab+ user.
The following or similar error message occurs, which is similar to what other users feedback:
```
/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:144: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.
leaves, treedef = jax.tree_flatten(tree)
/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:145: FutureWarning: jax.tree_unflatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_unflatten instead.
return jax.tree_unflatten(treedef, leaves)
---------------------------------------------------------------------------
UnfilteredStackTrace Traceback (most recent call last)
[<ipython-input-5-ca4ee2dc266d>](https://localhost:8080/) in <module>
44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)
---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))
46
________________________________________
23 frames
________________________________________
UnfilteredStackTrace: AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'
The stack trace below excludes JAX-internal frames.
The preceding is the original exception that occurred, unmodified.
--------------------
The above exception was the direct cause of the following exception:
AttributeError Traceback (most recent call last)
[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/) in wrapped(self, *args, **kwargs)
406 f = functools.partial(unbound_method, self)
407 f = functools.partial(run_interceptors, f, method_name, self)
--> 408 if jax.config.jax_experimental_name_stack and module_name:
409 local_module_name = module_name.split("/")[-1]
410 f = jax.named_call(f, name=local_module_name)
AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'
``` | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/637/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/637/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/636 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/636/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/636/comments | https://api.github.com/repos/deepmind/alphafold/issues/636/events | https://github.com/deepmind/alphafold/issues/636 | 1,462,018,485 | I_kwDOFoWQLM5XJKG1 | 636 | Running without MSA features | {
"avatar_url": "https://avatars.githubusercontent.com/u/10798127?v=4",
"events_url": "https://api.github.com/users/Gabriel-Ducrocq/events{/privacy}",
"followers_url": "https://api.github.com/users/Gabriel-Ducrocq/followers",
"following_url": "https://api.github.com/users/Gabriel-Ducrocq/following{/other_user}",
"gists_url": "https://api.github.com/users/Gabriel-Ducrocq/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Gabriel-Ducrocq",
"id": 10798127,
"login": "Gabriel-Ducrocq",
"node_id": "MDQ6VXNlcjEwNzk4MTI3",
"organizations_url": "https://api.github.com/users/Gabriel-Ducrocq/orgs",
"received_events_url": "https://api.github.com/users/Gabriel-Ducrocq/received_events",
"repos_url": "https://api.github.com/users/Gabriel-Ducrocq/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Gabriel-Ducrocq/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Gabriel-Ducrocq/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Gabriel-Ducrocq"
} | [] | open | false | null | [] | null | [] | "2022-11-23T16:04:01" | "2022-11-23T16:04:01" | null | NONE | null | Hello,
I modified alphafold so that it takes my own custom template features. I would like to modify it again so that it runs without MSAs features. Is there any convenient way to do this ?
Does setting all the MSAs features to 0 is equivalent to no MSAs features ?
Thank you,
Gabriel. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/636/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/636/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/635 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/635/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/635/comments | https://api.github.com/repos/deepmind/alphafold/issues/635/events | https://github.com/deepmind/alphafold/issues/635 | 1,460,435,030 | I_kwDOFoWQLM5XDHhW | 635 | AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack' | {
"avatar_url": "https://avatars.githubusercontent.com/u/118849987?v=4",
"events_url": "https://api.github.com/users/zhangkaka1123/events{/privacy}",
"followers_url": "https://api.github.com/users/zhangkaka1123/followers",
"following_url": "https://api.github.com/users/zhangkaka1123/following{/other_user}",
"gists_url": "https://api.github.com/users/zhangkaka1123/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/zhangkaka1123",
"id": 118849987,
"login": "zhangkaka1123",
"node_id": "U_kgDOBxWBww",
"organizations_url": "https://api.github.com/users/zhangkaka1123/orgs",
"received_events_url": "https://api.github.com/users/zhangkaka1123/received_events",
"repos_url": "https://api.github.com/users/zhangkaka1123/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/zhangkaka1123/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/zhangkaka1123/subscriptions",
"type": "User",
"url": "https://api.github.com/users/zhangkaka1123"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"Hi! I'm getting the same error. It seems like Colab version of AlphaFold uses JAX v0.3.25. `jax.config.jax_experimental_name_stack` was removed in this version. If you manage to use older version of JAX with this Colab notebook, I guess it would work.",
"Hi! I'm also receiving this error. The AlphaFold `requirements.txt` file which is installed via pip3 in cell 2 specifies JAX 0.3.17, so I'm unsure why the version changes (running `print(jax.__version__)` at the start of cell 5 prints `0.3.25`)",
"+1 using a monomer.\r\n\r\nFull error message \r\n\r\n",
"Similar problem and it only started this week. ",
"Hello. Same issue since november 18.",
"I believe I have the same issue, first noticed 2-3 days ago with DeepMind's AlphaFold Colab (now running AF2.2.4), when I try to run it in multimer form, without exceeding sequence size limits. Things proceed well through construction of MSAs, but as it starts to run Model_1_multimer, it crashes with the following error message (below). Thx in advance for your kind help!\r\n\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:144: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.\r\n leaves, treedef = jax.tree_flatten(tree)\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:145: FutureWarning: jax.tree_unflatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_unflatten instead.\r\n return jax.tree_unflatten(treedef, leaves)\r\n---------------------------------------------------------------------------\r\nUnfilteredStackTrace Traceback (most recent call last)\r\n<ipython-input-5-b59661ebfbb4> in <module>\r\n 44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)\r\n---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))\r\n 46 \r\n\r\n23 frames\r\nUnfilteredStackTrace: AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'\r\n\r\nThe stack trace below excludes JAX-internal frames.\r\nThe preceding is the original exception that occurred, unmodified.\r\n--------------------\r\nThe above exception was the direct cause of the following exception:\r\n\r\nAttributeError Traceback (most recent call last)\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py in wrapped(self, *args, **kwargs)\r\n 406 f = functools.partial(unbound_method, self)\r\n 407 f = functools.partial(run_interceptors, f, method_name, self)\r\n--> 408 if jax.config.jax_experimental_name_stack and module_name:\r\n 409 local_module_name = module_name.split(\"/\")[-1]\r\n 410 f = jax.named_call(f, name=local_module_name)\r\n\r\nAttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'",
"This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime",
"Many thanks for the information. I thought s.th. must have happened.\nBest wishes,\n Heinz\n\n\n\nDr Heinz Peter Nasheuer\n\nPersonal Professor\n\nSchool of Biological and Chemical Sciences\n\nBiochemistry, Room 110\n\nArts & Science Building, Main Concourse\n\nUniversity of Galway\n\nDistillery Road\n\nGalway, H91 TK33, Ireland\n\n\nPhone: ++353-91-49 2430\n\n________________________________\nFrom: anupam-tiwari ***@***.***>\nSent: 26 November 2022 14:58\nTo: deepmind/alphafold ***@***.***>\nCc: Nasheuer, Heinz-Peter ***@***.***>; Comment ***@***.***>\nSubject: Re: [deepmind/alphafold] AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack' (Issue #635)\n\nEXTERNAL EMAIL: This email originated outside the University of Galway. Do not open attachments or click on links unless you believe the content is safe.\nRÍOMHPHOST SEACHTRACH: Níor tháinig an ríomhphost seo ó Ollscoil na Gaillimhe. Ná hoscail ceangaltáin agus ná cliceáil ar naisc mura gcreideann tú go bhfuil an t-ábhar sábháilte.\n\n\nThis is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime\n\n—\nReply to this email directly, view it on GitHub<https://github.com/deepmind/alphafold/issues/635#issuecomment-1328060690>, or unsubscribe<https://github.com/notifications/unsubscribe-auth/A4LJE65LQUP4OSICZQPNDUDWKIQQJANCNFSM6AAAAAASIFO6AQ>.\nYou are receiving this because you commented.Message ID: ***@***.***>\n",
"> This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime\r\n\r\nHey, I have been struggeling to apply your solution. Could you elaborate further on when and where to insert this command?",
"I can't get it to work either. jax version appears to already 0.3.17 while jaxlib is 0.3.25. But I can't seem to install 0.3.17.\r\n\r\n\r\n\r\n",
"The earliest version I was able to downgrade jaxlib to is 0.3.20, however this still leads to the original error.",
"> > This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime\r\n> \r\n> Hey, I have been struggeling to apply your solution. Could you elaborate further on when and where to insert this command?\r\n\r\nSame I have been having issue where to run the command that was provided. If someone could help with this, that would be great!",
"https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharing \r\nhere you go guys, I made a modified notebook, the first step involves jax and jaxlib step, let me know how it goes! ",
"Yes, it seems to work. Thank you!\r\n\r\nI don't quite understand why you downgrade *before* downloading the packages, but it works. ",
"I tried to include this in the downloading package step, but to use the downgraded version we have to restart the runtime which in turn messes up the step-1 and step-2 code, hence I made it a separate step",
"Thanks for doing this!\r\n\r\nI tried but now I got a different error message. \r\n\r\n\r\nNameError Traceback (most recent call last)\r\n<ipython-input-2-15b941f78883><https://localhost:8080/> in <module>\r\n 18\r\n 19 # --- Run the model ---\r\n---> 20 if model_type_to_use == notebook_utils.ModelType.MONOMER:\r\n 21 model_names = config.MODEL_PRESETS['monomer'] + ('model_2_ptm',)\r\n 22 elif model_type_to_use == notebook_utils.ModelType.MULTIMER:\r\n\r\nNameError: name 'model_type_to_use' is not defined\r\n\r\n\r\nJust to confirm I used the multimer model with 2 sequences and they were run fine through the MSA step.",
"> Hello. Same issue since november 18.\r\n\r\nAgree Nov 18th in the morning everything was working fine and in the new week it started.",
"Thanks for this! Closing as this is a duplicate of https://github.com/deepmind/alphafold/issues/634",
"I appreciate all the comments! The issue has been solved in the new version! I just run two chain prediction successfully. Many thanks!"
] | "2022-11-22T19:24:16" | "2022-11-28T20:58:33" | "2022-11-28T20:13:53" | NONE | null | Hello,
I am predicting protein multimers using AlphaFold2 however it always failed and showed something like the following:
UnfilteredStackTrace Traceback (most recent call last)
[<ipython-input-5-ca4ee2dc266d>](https://localhost:8080/#) in <module>
44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)
---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))
46
23 frames
UnfilteredStackTrace: AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'
The stack trace below excludes JAX-internal frames.
The preceding is the original exception that occurred, unmodified.
--------------------
The above exception was the direct cause of the following exception:
AttributeError Traceback (most recent call last)
[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in wrapped(self, *args, **kwargs)
406 f = functools.partial(unbound_method, self)
407 f = functools.partial(run_interceptors, f, method_name, self)
--> 408 if jax.config.jax_experimental_name_stack and module_name:
409 local_module_name = module_name.split("/")[-1]
410 f = jax.named_call(f, name=local_module_name)
AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'
It shows "Attribute error" but I have no idea about coding to solve this issue.
I would really appreciate if someone knows what is going on.
Tons of thanks!
Kaka | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/635/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/635/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/634 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/634/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/634/comments | https://api.github.com/repos/deepmind/alphafold/issues/634/events | https://github.com/deepmind/alphafold/issues/634 | 1,460,318,554 | I_kwDOFoWQLM5XCrFa | 634 | AlphaFold Colab crashing at cell 5 | {
"avatar_url": "https://avatars.githubusercontent.com/u/25184542?v=4",
"events_url": "https://api.github.com/users/mprokhorov/events{/privacy}",
"followers_url": "https://api.github.com/users/mprokhorov/followers",
"following_url": "https://api.github.com/users/mprokhorov/following{/other_user}",
"gists_url": "https://api.github.com/users/mprokhorov/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/mprokhorov",
"id": 25184542,
"login": "mprokhorov",
"node_id": "MDQ6VXNlcjI1MTg0NTQy",
"organizations_url": "https://api.github.com/users/mprokhorov/orgs",
"received_events_url": "https://api.github.com/users/mprokhorov/received_events",
"repos_url": "https://api.github.com/users/mprokhorov/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/mprokhorov/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/mprokhorov/subscriptions",
"type": "User",
"url": "https://api.github.com/users/mprokhorov"
} | [] | closed | false | null | [] | null | [
"This is the full Stack Trace:\r\n```\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:144: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.\r\n leaves, treedef = jax.tree_flatten(tree)\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:145: FutureWarning: jax.tree_unflatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_unflatten instead.\r\n return jax.tree_unflatten(treedef, leaves)\r\n---------------------------------------------------------------------------\r\nUnfilteredStackTrace Traceback (most recent call last)\r\n[<ipython-input-5-ca4ee2dc266d>](https://localhost:8080/#) in <module>\r\n 44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)\r\n---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))\r\n 46 \r\n\r\n23 frames\r\n[/opt/conda/lib/python3.7/site-packages/alphafold/model/model.py](https://localhost:8080/#) in predict(self, feat, random_seed)\r\n 166 tree.map_structure(lambda x: x.shape, feat))\r\n--> 167 result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)\r\n 168 \r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/traceback_util.py](https://localhost:8080/#) in reraise_with_filtered_traceback(*args, **kwargs)\r\n 161 try:\r\n--> 162 return fun(*args, **kwargs)\r\n 163 except Exception as e:\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/api.py](https://localhost:8080/#) in cache_miss(*args, **kwargs)\r\n 621 jax.config.jax_debug_nans or jax.config.jax_debug_infs):\r\n--> 622 execute = dispatch._xla_call_impl_lazy(fun_, *tracers, **params)\r\n 623 out_flat = call_bind_continuation(execute(*args_flat))\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/dispatch.py](https://localhost:8080/#) in _xla_call_impl_lazy(***failed resolving arguments***)\r\n 236 return xla_callable(fun, device, backend, name, donated_invars, keep_unused,\r\n--> 237 *arg_specs)\r\n 238 \r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/linear_util.py](https://localhost:8080/#) in memoized_fun(fun, *args)\r\n 302 else:\r\n--> 303 ans = call(fun, *args)\r\n 304 cache[key] = (ans, fun.stores)\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/dispatch.py](https://localhost:8080/#) in _xla_callable_uncached(fun, device, backend, name, donated_invars, keep_unused, *arg_specs)\r\n 359 return lower_xla_callable(fun, device, backend, name, donated_invars, False,\r\n--> 360 keep_unused, *arg_specs).compile().unsafe_call\r\n 361 \r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/profiler.py](https://localhost:8080/#) in wrapper(*args, **kwargs)\r\n 313 with TraceAnnotation(name, **decorator_kwargs):\r\n--> 314 return func(*args, **kwargs)\r\n 315 return wrapper\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/dispatch.py](https://localhost:8080/#) in lower_xla_callable(fun, device, backend, name, donated_invars, always_lower, keep_unused, *arg_specs)\r\n 445 jaxpr, out_type, consts = pe.trace_to_jaxpr_final2(\r\n--> 446 fun, pe.debug_info_final(fun, \"jit\"))\r\n 447 out_avals, kept_outputs = util.unzip2(out_type)\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/profiler.py](https://localhost:8080/#) in wrapper(*args, **kwargs)\r\n 313 with TraceAnnotation(name, **decorator_kwargs):\r\n--> 314 return func(*args, **kwargs)\r\n 315 return wrapper\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/interpreters/partial_eval.py](https://localhost:8080/#) in trace_to_jaxpr_final2(fun, debug_info)\r\n 2076 with core.new_sublevel():\r\n-> 2077 jaxpr, out_type, consts = trace_to_subjaxpr_dynamic2(fun, main, debug_info)\r\n 2078 del fun, main\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/interpreters/partial_eval.py](https://localhost:8080/#) in trace_to_subjaxpr_dynamic2(fun, main, debug_info)\r\n 2026 in_tracers_ = [t for t, keep in zip(in_tracers, keep_inputs) if keep]\r\n-> 2027 ans = fun.call_wrapped(*in_tracers_)\r\n 2028 out_tracers = map(trace.full_raise, ans)\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/linear_util.py](https://localhost:8080/#) in call_wrapped(self, *args, **kwargs)\r\n 166 try:\r\n--> 167 ans = self.f(*args, **dict(self.params, **kwargs))\r\n 168 except:\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/transform.py](https://localhost:8080/#) in apply_fn(params, *args, **kwargs)\r\n 126 \r\n--> 127 out, state = f.apply(params, {}, *args, **kwargs)\r\n 128 if state:\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/transform.py](https://localhost:8080/#) in apply_fn(params, state, rng, *args, **kwargs)\r\n 353 try:\r\n--> 354 out = f(*args, **kwargs)\r\n 355 except jax.errors.UnexpectedTracerError as e:\r\n\r\n[/opt/conda/lib/python3.7/site-packages/alphafold/model/model.py](https://localhost:8080/#) in _forward_fn(batch)\r\n 81 def _forward_fn(batch):\r\n---> 82 model = modules.AlphaFold(self.config.model)\r\n 83 return model(\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in __call__(cls, *args, **kwargs)\r\n 120 init = wrap_method(\"__init__\", cls.__init__)\r\n--> 121 init(module, *args, **kwargs)\r\n 122 \r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in wrapped(self, *args, **kwargs)\r\n 407 f = functools.partial(run_interceptors, f, method_name, self)\r\n--> 408 if jax.config.jax_experimental_name_stack and module_name:\r\n 409 local_module_name = module_name.split(\"/\")[-1]\r\n\r\nUnfilteredStackTrace: AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'\r\n\r\nThe stack trace below excludes JAX-internal frames.\r\nThe preceding is the original exception that occurred, unmodified.\r\n\r\n--------------------\r\n\r\nThe above exception was the direct cause of the following exception:\r\n\r\nAttributeError Traceback (most recent call last)\r\n[<ipython-input-5-ca4ee2dc266d>](https://localhost:8080/#) in <module>\r\n 43 model_runner = model.RunModel(cfg, params)\r\n 44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)\r\n---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))\r\n 46 \r\n 47 mean_plddt = prediction['plddt'].mean()\r\n\r\n[/opt/conda/lib/python3.7/site-packages/alphafold/model/model.py](https://localhost:8080/#) in predict(self, feat, random_seed)\r\n 165 logging.info('Running predict with shape(feat) = %s',\r\n 166 tree.map_structure(lambda x: x.shape, feat))\r\n--> 167 result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)\r\n 168 \r\n 169 # This block is to ensure benchmark timings are accurate. Some blocking is\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/transform.py](https://localhost:8080/#) in apply_fn(params, *args, **kwargs)\r\n 125 \"name (e.g. `f.apply(.., state=my_state)`)\")\r\n 126 \r\n--> 127 out, state = f.apply(params, {}, *args, **kwargs)\r\n 128 if state:\r\n 129 raise ValueError(\"If your transformed function uses `hk.{get,set}_state` \"\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/transform.py](https://localhost:8080/#) in apply_fn(params, state, rng, *args, **kwargs)\r\n 352 with base.new_context(params=params, state=state, rng=rng) as ctx:\r\n 353 try:\r\n--> 354 out = f(*args, **kwargs)\r\n 355 except jax.errors.UnexpectedTracerError as e:\r\n 356 raise jax.errors.UnexpectedTracerError(unexpected_tracer_hint) from e\r\n\r\n[/opt/conda/lib/python3.7/site-packages/alphafold/model/model.py](https://localhost:8080/#) in _forward_fn(batch)\r\n 80 else:\r\n 81 def _forward_fn(batch):\r\n---> 82 model = modules.AlphaFold(self.config.model)\r\n 83 return model(\r\n 84 batch,\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in __call__(cls, *args, **kwargs)\r\n 119 # Now attempt to initialize the object.\r\n 120 init = wrap_method(\"__init__\", cls.__init__)\r\n--> 121 init(module, *args, **kwargs)\r\n 122 \r\n 123 if (config.get_config().module_auto_repr and\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in wrapped(self, *args, **kwargs)\r\n 406 f = functools.partial(unbound_method, self)\r\n 407 f = functools.partial(run_interceptors, f, method_name, self)\r\n--> 408 if jax.config.jax_experimental_name_stack and module_name:\r\n 409 local_module_name = module_name.split(\"/\")[-1]\r\n 410 f = jax.named_call(f, name=local_module_name)\r\n\r\nAttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'\r\n```",
"This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime",
"Thanks for raising this. This has been fixed in the latest commit."
] | "2022-11-22T17:58:00" | "2022-11-28T21:02:53" | "2022-11-28T21:02:53" | NONE | null | Today I've tried to run Colab version of AlphaFold v2.2.4, but it crashed every time while executing cell number 5. I'm getting these errors:
**AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'**
**'Config' object has no attribute 'jax_experimental_name_stack'** | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/634/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/634/timeline | null | completed | null | null | false |
End of preview. Expand
in Dataset Viewer.
- Downloads last month
- 2