url
stringlengths 58
58
| repository_url
stringclasses 1
value | labels_url
stringlengths 72
72
| comments_url
stringlengths 67
67
| events_url
stringlengths 65
65
| html_url
stringlengths 46
48
| id
int64 1.35B
1.52B
| node_id
stringlengths 18
19
| number
int64 571
673
| title
stringlengths 1
172
| user
dict | labels
listlengths 0
2
| state
stringclasses 2
values | locked
bool 1
class | assignee
float64 | assignees
sequencelengths 0
0
| milestone
float64 | comments
sequencelengths 0
30
| created_at
unknown | updated_at
unknown | closed_at
unknown | author_association
stringclasses 1
value | active_lock_reason
float64 | body
stringlengths 28
22.9k
⌀ | reactions
dict | timeline_url
stringlengths 67
67
| performed_via_github_app
float64 | state_reason
stringclasses 2
values | draft
float64 0
0
⌀ | pull_request
dict | is_pull_request
bool 2
classes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
https://api.github.com/repos/deepmind/alphafold/issues/673 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/673/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/673/comments | https://api.github.com/repos/deepmind/alphafold/issues/673/events | https://github.com/deepmind/alphafold/issues/673 | 1,524,110,688 | I_kwDOFoWQLM5a2BVg | 673 | Alphafold-Multimer was run for more than 48h, I still couldn't get the structure of a complex with 4 subunits (total aa length: 2222+689+201+196). | {
"avatar_url": "https://avatars.githubusercontent.com/u/22016970?v=4",
"events_url": "https://api.github.com/users/echoduan/events{/privacy}",
"followers_url": "https://api.github.com/users/echoduan/followers",
"following_url": "https://api.github.com/users/echoduan/following{/other_user}",
"gists_url": "https://api.github.com/users/echoduan/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/echoduan",
"id": 22016970,
"login": "echoduan",
"node_id": "MDQ6VXNlcjIyMDE2OTcw",
"organizations_url": "https://api.github.com/users/echoduan/orgs",
"received_events_url": "https://api.github.com/users/echoduan/received_events",
"repos_url": "https://api.github.com/users/echoduan/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/echoduan/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/echoduan/subscriptions",
"type": "User",
"url": "https://api.github.com/users/echoduan"
} | [] | open | false | null | [] | null | [] | "2023-01-07T20:02:59Z" | "2023-01-07T20:02:59Z" | null | NONE | null | Hi, author
python3 docker/run_docker.py \
--fasta_paths=complex1.fasta \
--max_template_date=2022-12-30 \
--model_preset=multimer \
--data_dir=/data --gpu_devices=0,1,2,3 > complex1.log &
1) The program was always calculating the model until now.
I0107 01:49:08.176561 140086208259904 run_docker.py:255] I0107 01:49:08.175831 140502687897408 run_alphafold.py:191] Running model model_1_multimer_v3_pred_0 on complex1
I0107 01:49:08.178550 140086208259904 run_docker.py:255] I0107 01:49:08.178094 140502687897408 model.py:165] Running predict with shape(feat) = {'aatype': (3308,), 'residue_index': (3308,), 'seq_length': (), 'msa': (5799, 3308), 'num_alignments': (), 'template_aatype': (4, 3308), 'template_all_atom_mask': (4, 3308, 37), 'template_all_atom_positions': (4, 3308, 37, 3), 'asym_id': (3308,), 'sym_id': (3308,), 'entity_id': (3308,), 'deletion_matrix': (5799, 3308), 'deletion_mean': (3308,), 'all_atom_mask': (3308, 37), 'all_atom_positions': (3308, 37, 3), 'assembly_num_chains': (), 'entity_mask': (3308,), 'num_templates': (), 'cluster_bias_mask': (5799,), 'bert_mask': (5799, 3308), 'seq_mask': (3308,), 'msa_mask': (5799, 3308)}
2) The instance (p3.8xlarge) has four GPUs (4xNVIDIA V100). All GPUs are available to calculate the model (--gpu_devices=0,1,2,3). But I noticed that 'run_alphafold.py' seems to use four GPUs, but why only the first one GPU memory is fulfilled?

How can I use the four GPUs with maximum memory usage at the same time to speed up the prediction?
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/673/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/673/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/672 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/672/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/672/comments | https://api.github.com/repos/deepmind/alphafold/issues/672/events | https://github.com/deepmind/alphafold/pull/672 | 1,514,648,843 | PR_kwDOFoWQLM5GZVWy | 672 | Fixing zip command issue in Google Colab. | {
"avatar_url": "https://avatars.githubusercontent.com/u/22454783?v=4",
"events_url": "https://api.github.com/users/gmihaila/events{/privacy}",
"followers_url": "https://api.github.com/users/gmihaila/followers",
"following_url": "https://api.github.com/users/gmihaila/following{/other_user}",
"gists_url": "https://api.github.com/users/gmihaila/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/gmihaila",
"id": 22454783,
"login": "gmihaila",
"node_id": "MDQ6VXNlcjIyNDU0Nzgz",
"organizations_url": "https://api.github.com/users/gmihaila/orgs",
"received_events_url": "https://api.github.com/users/gmihaila/received_events",
"repos_url": "https://api.github.com/users/gmihaila/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/gmihaila/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/gmihaila/subscriptions",
"type": "User",
"url": "https://api.github.com/users/gmihaila"
} | [] | open | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/672/checks?check_run_id=10366996221) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request.",
"Added my CLA."
] | "2022-12-30T17:02:08Z" | "2022-12-30T17:08:29Z" | null | NONE | null | When running the Google Colab notebook implementation cell `5. Run AlphaFold and download prediction` is throwing the following error:
```
NotImplementedError Traceback (most recent call last)
[<ipython-input-1-bc0091fa34e2>](https://localhost:8080/#) in <module>()
577 sequence = 'AKIGLFYGTQTGVTQTIAESIQQEFGGESIVDLNDIANADASDLNAYDYLIIGCPTWNVGELQSDWEGIYDDLDSVNFQGKKVAYFGAGDQVGYSDNFQDAMGILEEKISSLGSQTVGYWPIEGYDFNESKAVRNNQFVGLAIDEDNQPDLTKNRIKTWVSQLKSEFGL' #@param {type:"string"}
578
--> 579 run_prediction(sequence)
3 frames
[/usr/local/lib/python3.7/dist-packages/google/colab/_system_commands.py](https://localhost:8080/#) in _run_command(cmd, clear_streamed_output)
166 if locale_encoding != _ENCODING:
167 raise NotImplementedError(
--> 168 'A UTF-8 locale is required. Got {}'.format(locale_encoding))
169
170 parent_pty, child_pty = pty.openpty()
NotImplementedError: A UTF-8 locale is required. Got ANSI_X3.4-1968
```
As discussed in the issue: https://github.com/deepmind/alphafold/issues/483
This is more of a Google Colab issue when trying to create the`output_dir ` zip file from the`output_dir ` folder. I solved this by using`shutil ` to create the zip file.
I simply replace the line of code `!zip -q -r {output_dir}.zip {output_dir} ` from cell `5. Run AlphaFold and download prediction` with `shutil.make_archive(output_dir, 'zip', output_dir)`
I also made sure to add `import shutil` at the first cell.
This will create the zip file without using any terminal commands. This fix should not cause any issues in the future since we don't rely on the Google Colab terminal commands anymore.
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/672/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/672/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/672.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/672",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/672.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/672"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/671 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/671/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/671/comments | https://api.github.com/repos/deepmind/alphafold/issues/671/events | https://github.com/deepmind/alphafold/issues/671 | 1,513,313,185 | I_kwDOFoWQLM5aM1Oh | 671 | uniref90 can't download | {
"avatar_url": "https://avatars.githubusercontent.com/u/49931482?v=4",
"events_url": "https://api.github.com/users/hezhiqiang8909/events{/privacy}",
"followers_url": "https://api.github.com/users/hezhiqiang8909/followers",
"following_url": "https://api.github.com/users/hezhiqiang8909/following{/other_user}",
"gists_url": "https://api.github.com/users/hezhiqiang8909/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/hezhiqiang8909",
"id": 49931482,
"login": "hezhiqiang8909",
"node_id": "MDQ6VXNlcjQ5OTMxNDgy",
"organizations_url": "https://api.github.com/users/hezhiqiang8909/orgs",
"received_events_url": "https://api.github.com/users/hezhiqiang8909/received_events",
"repos_url": "https://api.github.com/users/hezhiqiang8909/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/hezhiqiang8909/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/hezhiqiang8909/subscriptions",
"type": "User",
"url": "https://api.github.com/users/hezhiqiang8909"
} | [] | open | false | null | [] | null | [
"Replace ftp://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref90/uniref90.fasta.gz with https://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref90/uniref90. fasta.gz solves the problem of not being able to download\r\n\r\n"
] | "2022-12-29T02:30:48Z" | "2022-12-29T02:41:03Z" | null | NONE | null | 
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/671/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/671/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/670 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/670/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/670/comments | https://api.github.com/repos/deepmind/alphafold/issues/670/events | https://github.com/deepmind/alphafold/pull/670 | 1,512,478,836 | PR_kwDOFoWQLM5GSDsg | 670 | Implement case-insensitive mmcif parsing | {
"avatar_url": "https://avatars.githubusercontent.com/u/49076030?v=4",
"events_url": "https://api.github.com/users/SimonKitSangChu/events{/privacy}",
"followers_url": "https://api.github.com/users/SimonKitSangChu/followers",
"following_url": "https://api.github.com/users/SimonKitSangChu/following{/other_user}",
"gists_url": "https://api.github.com/users/SimonKitSangChu/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/SimonKitSangChu",
"id": 49076030,
"login": "SimonKitSangChu",
"node_id": "MDQ6VXNlcjQ5MDc2MDMw",
"organizations_url": "https://api.github.com/users/SimonKitSangChu/orgs",
"received_events_url": "https://api.github.com/users/SimonKitSangChu/received_events",
"repos_url": "https://api.github.com/users/SimonKitSangChu/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/SimonKitSangChu/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/SimonKitSangChu/subscriptions",
"type": "User",
"url": "https://api.github.com/users/SimonKitSangChu"
} | [] | open | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/670/checks?check_run_id=10330182688) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request."
] | "2022-12-28T07:04:19Z" | "2022-12-28T07:09:03Z" | null | NONE | null | [Alphafold database](https://github.com/deepmind/alphafold/tree/main/afdb) cif-s are annotated with capitalized PEPTIDE. Parsing these files with the existing case-sensitive function results in empty `mmcif_object`. Suggest switching to case-insensitive parsing. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/670/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/670/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/670.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/670",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/670.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/670"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/669 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/669/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/669/comments | https://api.github.com/repos/deepmind/alphafold/issues/669/events | https://github.com/deepmind/alphafold/issues/669 | 1,511,606,829 | I_kwDOFoWQLM5aGUot | 669 | pip._vendor.urllib3.exceptions.ReadTimeoutError: HTTPSConnectionPool(host='files.pythonhosted.org', port=443): Read timed out. | {
"avatar_url": "https://avatars.githubusercontent.com/u/49931482?v=4",
"events_url": "https://api.github.com/users/hezhiqiang8909/events{/privacy}",
"followers_url": "https://api.github.com/users/hezhiqiang8909/followers",
"following_url": "https://api.github.com/users/hezhiqiang8909/following{/other_user}",
"gists_url": "https://api.github.com/users/hezhiqiang8909/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/hezhiqiang8909",
"id": 49931482,
"login": "hezhiqiang8909",
"node_id": "MDQ6VXNlcjQ5OTMxNDgy",
"organizations_url": "https://api.github.com/users/hezhiqiang8909/orgs",
"received_events_url": "https://api.github.com/users/hezhiqiang8909/received_events",
"repos_url": "https://api.github.com/users/hezhiqiang8909/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/hezhiqiang8909/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/hezhiqiang8909/subscriptions",
"type": "User",
"url": "https://api.github.com/users/hezhiqiang8909"
} | [] | open | false | null | [] | null | [] | "2022-12-27T09:18:35Z" | "2022-12-27T09:18:35Z" | null | NONE | null | 
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/669/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/669/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/668 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/668/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/668/comments | https://api.github.com/repos/deepmind/alphafold/issues/668/events | https://github.com/deepmind/alphafold/issues/668 | 1,511,549,702 | I_kwDOFoWQLM5aGGsG | 668 | fatal: unable to access 'https://github.com/soedinglab/hh-suite.git/': gnutls_handshake() failed: The TLS connection was non-properly terminated. | {
"avatar_url": "https://avatars.githubusercontent.com/u/49931482?v=4",
"events_url": "https://api.github.com/users/hezhiqiang8909/events{/privacy}",
"followers_url": "https://api.github.com/users/hezhiqiang8909/followers",
"following_url": "https://api.github.com/users/hezhiqiang8909/following{/other_user}",
"gists_url": "https://api.github.com/users/hezhiqiang8909/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/hezhiqiang8909",
"id": 49931482,
"login": "hezhiqiang8909",
"node_id": "MDQ6VXNlcjQ5OTMxNDgy",
"organizations_url": "https://api.github.com/users/hezhiqiang8909/orgs",
"received_events_url": "https://api.github.com/users/hezhiqiang8909/received_events",
"repos_url": "https://api.github.com/users/hezhiqiang8909/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/hezhiqiang8909/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/hezhiqiang8909/subscriptions",
"type": "User",
"url": "https://api.github.com/users/hezhiqiang8909"
} | [] | open | false | null | [] | null | [] | "2022-12-27T08:09:29Z" | "2022-12-27T08:09:29Z" | null | NONE | null | Making a new docker image shows The TLS connection was non-properly terminated.

| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/668/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/668/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/667 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/667/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/667/comments | https://api.github.com/repos/deepmind/alphafold/issues/667/events | https://github.com/deepmind/alphafold/issues/667 | 1,510,736,623 | I_kwDOFoWQLM5aDALv | 667 | CUDA_ERROR_ILLEGAL_ADDRESS error with AlphaFold multimer 2.3.0 | {
"avatar_url": "https://avatars.githubusercontent.com/u/116328913?v=4",
"events_url": "https://api.github.com/users/sz-1002/events{/privacy}",
"followers_url": "https://api.github.com/users/sz-1002/followers",
"following_url": "https://api.github.com/users/sz-1002/following{/other_user}",
"gists_url": "https://api.github.com/users/sz-1002/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/sz-1002",
"id": 116328913,
"login": "sz-1002",
"node_id": "U_kgDOBu8J0Q",
"organizations_url": "https://api.github.com/users/sz-1002/orgs",
"received_events_url": "https://api.github.com/users/sz-1002/received_events",
"repos_url": "https://api.github.com/users/sz-1002/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/sz-1002/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/sz-1002/subscriptions",
"type": "User",
"url": "https://api.github.com/users/sz-1002"
} | [] | open | false | null | [] | null | [
"Hi @sz-1002 \r\nI am having the same issue on the relaxation step.\r\nhttps://github.com/deepmind/alphafold/issues/660\r\n\r\nI tried with newer version of CUDA (11.6) and had the same issue.",
"Hi @leiterenato \r\nThanks for letting me know! Hopefully there will be a solution for this.",
"a different non-relax model issue seems to have been resolved by using cuda 11.8 https://github.com/deepmind/alphafold/issues/646#issuecomment-1364912401\r\n\r\nassuming that doesn't work, can try use_gpu_relax=False, or turn off relax entirely with run_relax=False. we will attempt to address the problem more fully in the new year.",
"We think this is due to jax version change from 0.3.17 to 0.3.25. We don't want to revert jax version though, so are looking for workarounds."
] | "2022-12-26T08:22:37Z" | "2023-01-05T10:37:26Z" | null | NONE | null | Hi!
I am trying to run AlphaFold 2.3.0 multimer and encountered this error: `Execution of replica 0 failed: INTERNAL: Failed to load in-memory CUBIN: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered` (see details below). I was wondering if you could help me resolve it? Thank you very much!!
**Machine spec etc.:**
- Ubuntu 20.04 LTS
- GPU: nvidia RTX 3090
- cuda version: tried both 11.1.1 and 11.4.0 (both gave the same error)
- total length of the protein complex: ~2200 aa
When I searched related errors online, it seems there are generally two solutions proposed: (1) change to a newer cuda version [https://github.com/deepmind/dm-haiku/issues/204](url), or (2) disable unified memory [https://github.com/deepmind/alphafold/issues/406](url).
I tried using cuda 11.4.0 instead of 11.1.1 by changing the following lines in Dockerfile, but the same error persists.
```
ARG CUDA=11.1.1 --->>> ARG CUDA=11.4.0
FROM nvidia/cuda:${CUDA}-cudnn8-runtime-ubuntu18.04 --->>> FROM nvidia/cuda:${CUDA}-cudnn8-runtime-ubuntu20.04
conda install -y -c conda-forge cudatoolkit==${CUDA_VERSION} --->>> conda install -y -c "nvidia/label/cuda-11.4.0" cuda-toolkit
```
As for (2) disable unified memory, I am worried that this would give me out of memory error given the size of the protein.
Not sure if this is relevant, but this is a recent problem and prediction for this and other similarly-sized complexes worked fine before (was using v2.2.0 before, and I wonder if this is an issue with e.g. version of jax or jaxlib).
Thank you very much!
**Error message:**
```
I1226 15:42:37.167795 140027647279936 run_docker.py:255] I1226 06:42:37.167222 140635718940480 amber_minimize.py:407] Minimizing protein, attempt 1 of 100.
I1226 15:42:39.806318 140027647279936 run_docker.py:255] I1226 06:42:39.805861 140635718940480 amber_minimize.py:68] Restraining 17790 / 35357 particles.
I1226 15:45:15.867727 140027647279936 run_docker.py:255] I1226 06:45:15.866998 140635718940480 amber_minimize.py:177] alterations info: {'nonstandard_residues': [], 'removed_heterogens': set(), 'missing_residues': {}, 'missing_heavy_atoms': {}, 'missing_terminals': {}, 'Se_in_MET': [], 'removed_chains': {0: []}}
I1226 15:45:42.889597 140027647279936 run_docker.py:255] 2022-12-26 06:45:42.889173: E external/org_tensorflow/tensorflow/compiler/xla/pjrt/pjrt_stream_executor_client.cc:2153] Execution of replica 0 failed: INTERNAL: Failed to load in-memory CUBIN: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered
I1226 15:45:42.899475 140027647279936 run_docker.py:255] Traceback (most recent call last):
I1226 15:45:42.899577 140027647279936 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 432, in <module>
I1226 15:45:42.899646 140027647279936 run_docker.py:255] app.run(main)
I1226 15:45:42.899709 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/absl/app.py", line 312, in run
I1226 15:45:42.899771 140027647279936 run_docker.py:255] _run_main(main, args)
I1226 15:45:42.899834 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/absl/app.py", line 258, in _run_main
I1226 15:45:42.899896 140027647279936 run_docker.py:255] sys.exit(main(argv))
I1226 15:45:42.899957 140027647279936 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 408, in main
I1226 15:45:42.900018 140027647279936 run_docker.py:255] predict_structure(
I1226 15:45:42.900109 140027647279936 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 243, in predict_structure
I1226 15:45:42.900174 140027647279936 run_docker.py:255] relaxed_pdb_str, _, violations = amber_relaxer.process(
I1226 15:45:42.900234 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/relax/relax.py", line 62, in process
I1226 15:45:42.900292 140027647279936 run_docker.py:255] out = amber_minimize.run_pipeline(
I1226 15:45:42.900353 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/relax/amber_minimize.py", line 489, in run_pipeline
I1226 15:45:42.900412 140027647279936 run_docker.py:255] ret.update(get_violation_metrics(prot))
I1226 15:45:42.900472 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/relax/amber_minimize.py", line 357, in get_violation_metrics
I1226 15:45:42.900531 140027647279936 run_docker.py:255] structural_violations, struct_metrics = find_violations(prot)
I1226 15:45:42.900591 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/relax/amber_minimize.py", line 339, in find_violations
I1226 15:45:42.900651 140027647279936 run_docker.py:255] violations = folding.find_structural_violations(
I1226 15:45:42.900712 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/model/folding.py", line 761, in find_structural_violations
I1226 15:45:42.900774 140027647279936 run_docker.py:255] between_residue_clashes = all_atom.between_residue_clash_loss(
I1226 15:45:42.900835 140027647279936 run_docker.py:255] File "/app/alphafold/alphafold/model/all_atom.py", line 783, in between_residue_clash_loss
I1226 15:45:42.900898 140027647279936 run_docker.py:255] dists = jnp.sqrt(1e-10 + jnp.sum(
I1226 15:45:42.900959 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/jax/_src/numpy/reductions.py", line 216, in sum
I1226 15:45:42.901019 140027647279936 run_docker.py:255] return _reduce_sum(a, axis=_ensure_optional_axes(axis), dtype=dtype, out=out,
I1226 15:45:42.901078 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/jax/_src/traceback_util.py", line 162, in reraise_with_filtered_traceback
I1226 15:45:42.901138 140027647279936 run_docker.py:255] return fun(*args, **kwargs)
I1226 15:45:42.901199 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/jax/_src/api.py", line 623, in cache_miss
I1226 15:45:42.901261 140027647279936 run_docker.py:255] out_flat = call_bind_continuation(execute(*args_flat))
I1226 15:45:42.901322 140027647279936 run_docker.py:255] File "/opt/conda/lib/python3.8/site-packages/jax/_src/dispatch.py", line 895, in _execute_compiled
I1226 15:45:42.901383 140027647279936 run_docker.py:255] out_flat = compiled.execute(in_flat)
I1226 15:45:42.901446 140027647279936 run_docker.py:255] jax._src.traceback_util.UnfilteredStackTrace: jaxlib.xla_extension.XlaRuntimeError: INTERNAL: Failed to load in-memory CUBIN: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered
I1226 15:45:42.901507 140027647279936 run_docker.py:255]
I1226 15:45:42.901568 140027647279936 run_docker.py:255] The stack trace below excludes JAX-internal frames.
I1226 15:45:42.901637 140027647279936 run_docker.py:255] The preceding is the original exception that occurred, unmodified.
```
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/667/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/667/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/666 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/666/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/666/comments | https://api.github.com/repos/deepmind/alphafold/issues/666/events | https://github.com/deepmind/alphafold/issues/666 | 1,510,501,796 | I_kwDOFoWQLM5aCG2k | 666 | Colab pro + failing to predict >2000 residues | {
"avatar_url": "https://avatars.githubusercontent.com/u/121411082?v=4",
"events_url": "https://api.github.com/users/HBCH1/events{/privacy}",
"followers_url": "https://api.github.com/users/HBCH1/followers",
"following_url": "https://api.github.com/users/HBCH1/following{/other_user}",
"gists_url": "https://api.github.com/users/HBCH1/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/HBCH1",
"id": 121411082,
"login": "HBCH1",
"node_id": "U_kgDOBzyWCg",
"organizations_url": "https://api.github.com/users/HBCH1/orgs",
"received_events_url": "https://api.github.com/users/HBCH1/received_events",
"repos_url": "https://api.github.com/users/HBCH1/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/HBCH1/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/HBCH1/subscriptions",
"type": "User",
"url": "https://api.github.com/users/HBCH1"
} | [] | open | false | null | [] | null | [] | "2022-12-26T01:55:04Z" | "2022-12-26T01:55:04Z" | null | NONE | null | Hi, I am using AlphaFold 2 (Colab pro +) to predict some proteins with long sequences. While it successfully predicted the structures of shorter sequences, it keeps failing with longer ones even tho I upgraded to colab pro + to have higher memory.
Here is what I am getting:
<img width="1234" alt="Alpha fold " src="https://user-images.githubusercontent.com/121411082/209489459-5fcca930-8962-4be4-a175-bea7d17b206c.png">
<img width="904" alt="alpha" src="https://user-images.githubusercontent.com/121411082/209489463-a95d93fa-3a25-48c2-b0b0-bc7f1a743bd9.png">
Is there a way to fix this issue or is AlphaFold 2 not currently able to predict >2000 bp sequences?
Thanks in advance! | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/666/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/666/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/665 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/665/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/665/comments | https://api.github.com/repos/deepmind/alphafold/issues/665/events | https://github.com/deepmind/alphafold/issues/665 | 1,508,351,023 | I_kwDOFoWQLM5Z55wv | 665 | Memory issue when processing large protein. Is my swap not being leveraged | {
"avatar_url": "https://avatars.githubusercontent.com/u/121254015?v=4",
"events_url": "https://api.github.com/users/cheminformaticist/events{/privacy}",
"followers_url": "https://api.github.com/users/cheminformaticist/followers",
"following_url": "https://api.github.com/users/cheminformaticist/following{/other_user}",
"gists_url": "https://api.github.com/users/cheminformaticist/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/cheminformaticist",
"id": 121254015,
"login": "cheminformaticist",
"node_id": "U_kgDOBzowfw",
"organizations_url": "https://api.github.com/users/cheminformaticist/orgs",
"received_events_url": "https://api.github.com/users/cheminformaticist/received_events",
"repos_url": "https://api.github.com/users/cheminformaticist/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/cheminformaticist/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/cheminformaticist/subscriptions",
"type": "User",
"url": "https://api.github.com/users/cheminformaticist"
} | [] | open | false | null | [] | null | [
"Could you try running with `--db_preset=reduced_dbs`?",
"My preference is to use full_dbs, but I will try.",
"I enabled --db_preset=reduced_dbs and ran into the same issue after about 40 minutes. It really seems like swap is not being used... or do I just need to scale up RAM.",
"What stage is it working on when it runs out of RAM?\r\nEdit: This is relevant in part because AlphaFold uses a number of third party resources at several stages, such as in the construction of the MSA or in the relax step."
] | "2022-12-22T18:15:57Z" | "2023-01-06T21:47:36Z" | null | NONE | null | I am running predictions for 2500 residue protein on a system with 80GB RAM, 8TB diskspace, and 1TB swap running Alphafold v2.2 via docker. The process fails with the message "RuntimeError: Resource exhausted: Out of memory while trying to allocate 51935261664 bytes." The ram was upgraded to 128GB, but the job dies with the same error.
docker stats: shows the MEM LIMIT is not including available swap. I am likely mistaken, but I thought it would... Is my swap not being leveraged by the container? How can I modify the docker image, such that when launched the container leverages swap (which I previously thought was available by default)? If I am off target, any advice would be appreciated...
Note: I have another system with 512GB RAM that no problem completing the calculations for the same protein. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/665/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/665/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/664 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/664/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/664/comments | https://api.github.com/repos/deepmind/alphafold/issues/664/events | https://github.com/deepmind/alphafold/issues/664 | 1,507,971,670 | I_kwDOFoWQLM5Z4dJW | 664 | Generating multiple (>10) models from one sequence | {
"avatar_url": "https://avatars.githubusercontent.com/u/121240930?v=4",
"events_url": "https://api.github.com/users/cgovaert/events{/privacy}",
"followers_url": "https://api.github.com/users/cgovaert/followers",
"following_url": "https://api.github.com/users/cgovaert/following{/other_user}",
"gists_url": "https://api.github.com/users/cgovaert/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/cgovaert",
"id": 121240930,
"login": "cgovaert",
"node_id": "U_kgDOBzn9Yg",
"organizations_url": "https://api.github.com/users/cgovaert/orgs",
"received_events_url": "https://api.github.com/users/cgovaert/received_events",
"repos_url": "https://api.github.com/users/cgovaert/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/cgovaert/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/cgovaert/subscriptions",
"type": "User",
"url": "https://api.github.com/users/cgovaert"
} | [] | open | false | null | [] | null | [
"While you certainly can do this with AlphaFold, you will probably only explore \"near-native\" conformations with it, because of what it is trained to produce (crystal structures). If you want to explore conformational space more thoroughly, I would recommend using molecular dynamics.\r\nIf you would like to use AlphaFold to investigate where the flexible region likely is, some studies have indicated that segments with lower pLDDT scores tend to be more flexible, and at least with 25 structures (typical output for multimers), this has matched where the variation occurs for me.\r\nFor doing this with AlphaFold, the easiest way would be to run in multimer mode (which, according to the latest update notes, is best to do even for monomers if you know the stoichiometry) and change the num_multimer_predictions_per_model value. As there are 5 models, pass the number of structures you want / 5 as the value to this flag in the command line. In other words, for 100 output structures, add --num_multimer_predictions_per_model=20 to your command line arguments. \r\n",
"Great idea. will do. To be more precise, I do want to run MD, I want to see if there is/are other conformation of a subdomain that we believe changes upon ligand binding. Thanks"
] | "2022-12-22T14:17:35Z" | "2022-12-22T16:15:23Z" | null | NONE | null | Hi all
I'm new to the field. I would like to generate between 100 and 50 models for my target sequence to explore conformal flexibility of a specific region. This region is often (comparing prediction on my target and various orthologs) predicted with lower confidence, and sometimes with different conformation. We have experimental evidence that this domain may be mobile.
Thus I want to generate many models from the same target sequence to identify possible conformational diversity.
We can run Alphafold locally but I'm not sure how to paramtyrize such a run.
Can anybody help here ?
Thank a bunch
Cedric
—
Prof. Cedric Govaerts, Ph.D.
Universite Libre de Bruxelles
Campus Plaine. Phone :+32 2 650 53 77
Building BC, Room 1C4 203
Boulevard du Triomphe, Acces 2
1050 Brussels
Belgium
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/664/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/664/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/663 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/663/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/663/comments | https://api.github.com/repos/deepmind/alphafold/issues/663/events | https://github.com/deepmind/alphafold/issues/663 | 1,506,058,944 | I_kwDOFoWQLM5ZxKLA | 663 | Problems with the new AlphaFold Colab Version | {
"avatar_url": "https://avatars.githubusercontent.com/u/61228596?v=4",
"events_url": "https://api.github.com/users/RosaLuzia/events{/privacy}",
"followers_url": "https://api.github.com/users/RosaLuzia/followers",
"following_url": "https://api.github.com/users/RosaLuzia/following{/other_user}",
"gists_url": "https://api.github.com/users/RosaLuzia/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/RosaLuzia",
"id": 61228596,
"login": "RosaLuzia",
"node_id": "MDQ6VXNlcjYxMjI4NTk2",
"organizations_url": "https://api.github.com/users/RosaLuzia/orgs",
"received_events_url": "https://api.github.com/users/RosaLuzia/received_events",
"repos_url": "https://api.github.com/users/RosaLuzia/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/RosaLuzia/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/RosaLuzia/subscriptions",
"type": "User",
"url": "https://api.github.com/users/RosaLuzia"
} | [
{
"color": "E99695",
"default": false,
"description": "Something isn't working",
"id": 3094826889,
"name": "error report",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODg5",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/error%20report"
},
{
"color": "FEF2C0",
"default": false,
"description": "AlphaFold colab issue",
"id": 3313914804,
"name": "colab",
"node_id": "MDU6TGFiZWwzMzEzOTE0ODA0",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/colab"
}
] | open | false | null | [] | null | [
"Is this affecting only one or two of your structures, or is there a widespread degradation in quality?\r\n\r\nMonomer models should be unchanged by v.2.3.0, but the msa databases used to generate the inputs to the monomer model have been updated. it is possible that for some targets this can reduce the quality of the msa, but for most targets it should be the same or better. Have the sizes of the msa results reduced for the structures where you are having problems?\r\n\r\nFor this particular target, we found that running with the full local installation, rather than in the colab, gave better results. The full installation uses templates and a larger bfd database, both of which can improve quality."
] | "2022-12-21T10:18:24Z" | "2022-12-21T17:28:15Z" | null | NONE | null | I run AlphaFold Colab with a lot of similar peptides which are characterized by alpha helices and beta sheets. I always get nice structures with the right assumed conformation, but since the new version of AlphaFold Colab is released structure prediction went wrong. I attach two predictions of the same sequence, one is predicted with an older version and the other with the recent version. Any suggestion how to fix the problem?
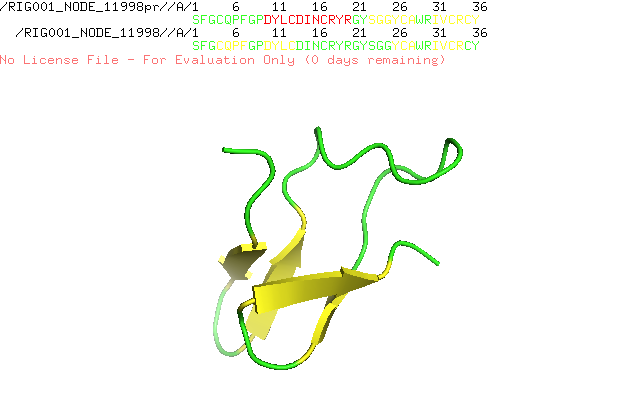
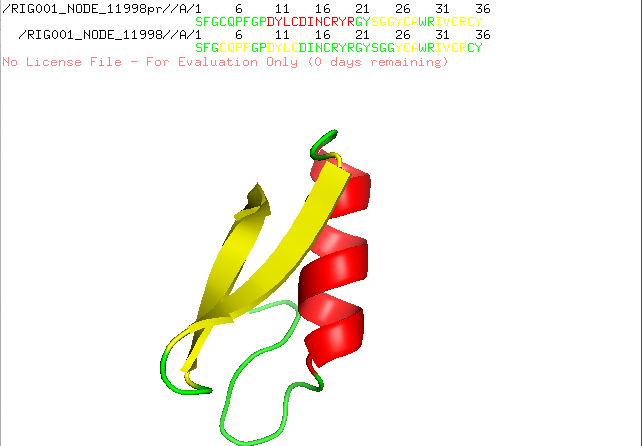
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/663/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/663/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/662 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/662/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/662/comments | https://api.github.com/repos/deepmind/alphafold/issues/662/events | https://github.com/deepmind/alphafold/issues/662 | 1,505,387,450 | I_kwDOFoWQLM5ZumO6 | 662 | Calculate mean and per residue pLDDT scores of AlphaFold's model PDBs (post-prediction) | {
"avatar_url": "https://avatars.githubusercontent.com/u/98129594?v=4",
"events_url": "https://api.github.com/users/meh47336/events{/privacy}",
"followers_url": "https://api.github.com/users/meh47336/followers",
"following_url": "https://api.github.com/users/meh47336/following{/other_user}",
"gists_url": "https://api.github.com/users/meh47336/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/meh47336",
"id": 98129594,
"login": "meh47336",
"node_id": "U_kgDOBdlWug",
"organizations_url": "https://api.github.com/users/meh47336/orgs",
"received_events_url": "https://api.github.com/users/meh47336/received_events",
"repos_url": "https://api.github.com/users/meh47336/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/meh47336/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/meh47336/subscriptions",
"type": "User",
"url": "https://api.github.com/users/meh47336"
} | [] | closed | false | null | [] | null | [
"If the pdb files come from a non-Alphafold model, any confidence metrics will be output by that other model.\r\n\r\nOne has to run the alphafold model to get alphafold confidence metrics, since those metrics are the models own measure of its certainty.",
"If you have the .npz files from the AlphaFold variant, you might be able to build something analogous to pLDDT scores based on that. The pdb simply does not have any of the information needed to estimate error in a quantified way.",
"The PDB are created by AlphaFold however it's just a Singularity container composed of the ML parts of AF (no MSA, etc). More specifically it's on Summit at OLCF (I asked the question here because things are a bit more active over here.\r\n\r\nThe output at Summit gives a pkl file and a PDB. The b-factor column of the PDBs contains all zero values rather than pLDDT scores. Any idea why this would not have been calculated or am I just missing something?",
"The per-residue pLDDT values are stored in a numpy array within the .pkl output!"
] | "2022-12-20T22:21:02Z" | "2022-12-23T17:45:01Z" | "2022-12-23T17:45:00Z" | NONE | null | Hello,
I've found the lddt script here but am unsure how to implement it outside of the AlphaFold. Recently I've obtained many predictions using a variant of AlphaFold, however it did not include pLDDT scores in the PDB outputs. Is there a way to calculate the scores retroactively using the PDB outputs?
Thank you for any help or suggestions, | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/662/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/662/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/661 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/661/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/661/comments | https://api.github.com/repos/deepmind/alphafold/issues/661/events | https://github.com/deepmind/alphafold/issues/661 | 1,505,333,292 | I_kwDOFoWQLM5ZuZAs | 661 | HHblits error "sequences in merged A3M file do not all have the same number of columns," | {
"avatar_url": "https://avatars.githubusercontent.com/u/5776199?v=4",
"events_url": "https://api.github.com/users/ajbordner/events{/privacy}",
"followers_url": "https://api.github.com/users/ajbordner/followers",
"following_url": "https://api.github.com/users/ajbordner/following{/other_user}",
"gists_url": "https://api.github.com/users/ajbordner/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ajbordner",
"id": 5776199,
"login": "ajbordner",
"node_id": "MDQ6VXNlcjU3NzYxOTk=",
"organizations_url": "https://api.github.com/users/ajbordner/orgs",
"received_events_url": "https://api.github.com/users/ajbordner/received_events",
"repos_url": "https://api.github.com/users/ajbordner/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ajbordner/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ajbordner/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ajbordner"
} | [
{
"color": "BFDADC",
"default": false,
"description": "Third-party tool issue",
"id": 3313961085,
"name": "third-party tool",
"node_id": "MDU6TGFiZWwzMzEzOTYxMDg1",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/third-party%20tool"
}
] | open | false | null | [] | null | [
"The uniref30 db has been updated in v2.3.0 which can cause different behaviour for this hhblits search. HHBlits is sensitive and the new db can cause errors for some targets which did not error before.\r\n\r\nA suggested workaround is to try with the db_preset flag set to 'reduced_dbs'.",
"There seems to be another issue causing the hhblits error. The hhblits command that AlphaFold runs completes without an error when run outside of the docker container. The same version of hhblits as in the Dockerfile (3.3.0), same database files, and input sequence file were used for the test.\r\n\r\nhhblits -i <FASTA input sequence file> -cpu 4 -oa3m /tmp/output.a3m -o /dev/null -n 3 -e 0.001 -maxseq 1000000 -realign_max 100000 -maxfilt 100000 -min_prefilter_hits 1000 -d <alphafold data directory>/bfd/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt -d <alphafold data directory>/uniref30/UniRef30_2021_03\r\n",
"we tried running hhblits directly too, but got the same error that you (and we) see when running the target in docker.\r\n\r\ncan you double check that your hhblits run was with the exact same sequence? and did it also give warnings about the titin results (ours did)?\r\n\r\nassuming it was the same sequence, something funny might be happening with ram usage, but im afraid using reduced_dbs is the only workaround for now.",
"I now also get the same error messages. I must have run hhblits for only 1 db since it only generates errors when run with both dbs. Thank you for your help."
] | "2022-12-20T21:35:55Z" | "2023-01-04T22:13:29Z" | null | NONE | null | AlphaFold is crashing due to an HHblits error in v2.3.0 but not in v2.2.4. This happens, for example, with the input sequence:
>chain_A
LEVQVPEDPVVALVGTDATLCCSFSPEPGFSLAQLNLIWQLTDTKQLVHSFAEGQDQGSAYANRTALFPDLLAQGNASLRLQRVRVADEGSFTCFVSIRDFGSAAVSLQVAAPYSKPSMTLEPNKDLRPGDTVTITCSSYQGYPEAEVFWQDGQGVPLTGNVTTSQMANEQGLFDVHSILRVVLGANGTYSCLVRNPVLQQDAHSSVTITPQRSPTGAVEVQVPEDPVVALVGTDATLRCSFSPEPGFSLAQLNLIWQLTDTKQLVHSFTEGRDQGSAYANRTALFPDLLAQGNASLRLQRVRVADEGSFTCFVSIRDFGSAAVSLQVAAPYSKPSMTLEPNKDLRPGDTVTITCSSYRGYPEAEVFWQDGQGVPLTGNVTTSQMANEQGLFDVHSVLRVVLGANGTYSCLVRNPVLQQDAHGSVTITGQPMTFPPEA
There are many long titin sequences in HHblits hits, which may be causing the issue. I have included an excerpt of the output log.
[test.log](https://github.com/deepmind/alphafold/files/10272808/test.log)
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/661/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/661/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/660 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/660/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/660/comments | https://api.github.com/repos/deepmind/alphafold/issues/660/events | https://github.com/deepmind/alphafold/issues/660 | 1,505,155,550 | I_kwDOFoWQLM5Zttne | 660 | [BUG] Relaxation not working for Multimers on v2.3.0 | {
"avatar_url": "https://avatars.githubusercontent.com/u/277946?v=4",
"events_url": "https://api.github.com/users/leiterenato/events{/privacy}",
"followers_url": "https://api.github.com/users/leiterenato/followers",
"following_url": "https://api.github.com/users/leiterenato/following{/other_user}",
"gists_url": "https://api.github.com/users/leiterenato/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/leiterenato",
"id": 277946,
"login": "leiterenato",
"node_id": "MDQ6VXNlcjI3Nzk0Ng==",
"organizations_url": "https://api.github.com/users/leiterenato/orgs",
"received_events_url": "https://api.github.com/users/leiterenato/received_events",
"repos_url": "https://api.github.com/users/leiterenato/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/leiterenato/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/leiterenato/subscriptions",
"type": "User",
"url": "https://api.github.com/users/leiterenato"
} | [] | open | false | null | [] | null | [
"we are looking into this, but might not come back with something until after the holiday period.\r\n\r\ncan i check: if you run inference+relax all in v2.2.4, rather than inference and then separately relax, it does work?",
"Hi @joshabramson \r\n\r\nI tried to run v2.3.0 using the `run_docker.py` from `https://github.com/deepmind/alphafold/tree/main/docker/run_docker.py`.\r\n\r\nI tried with both full_bfd and reduced_bfd without any success.\r\n\r\nFirst try:\r\n```\r\nnohup python3 docker/run_docker.py \\\r\n --fasta_paths=/home/renatoleite/workspace/vertex-ai-alphafold-inference-pipeline/sequences/1S78.fasta \\\r\n --output_dir=/home/renatoleite/workspace/outputs \\\r\n --max_template_date=2030-01-01 \\\r\n --model_preset=multimer \\\r\n --data_dir=/mnt/nfs/alphafold \\\r\n --num_multimer_predictions_per_model=1 &\r\n```\r\nSecond try:\r\n```\r\nnohup python3 docker/run_docker.py \\\r\n --fasta_paths=/home/renatoleite/workspace/vertex-ai-alphafold-inference-pipeline/sequences/1S78.fasta \\\r\n --output_dir=/home/renatoleite/workspace/outputs \\\r\n --max_template_date=2030-01-01 \\\r\n --model_preset=multimer \\\r\n --data_dir=/mnt/nfs/alphafold \\\r\n --num_multimer_predictions_per_model=1 \\\r\n --db_preset=reduced_dbs & \r\n```\r\n\r\nI also tried with my pipeline (separated prediction and relaxation), but got the same error.\r\nhttps://github.com/GoogleCloudPlatform/vertex-ai-alphafold-inference-pipeline\r\n\r\nFor v2.2.4 it worked with both solutions (separated and inference+relax).\r\n\r\n"
] | "2022-12-20T19:46:41Z" | "2022-12-23T19:58:21Z" | null | NONE | null | Relaxation step is failing for multimers using AlphaFold v2.3.0.
Monomers are working fine.
Error:
```
jaxlib.xla_extension.XlaRuntimeError: INTERNAL: Failed to load in-memory CUBIN: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered
E external/org_tensorflow/tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:1043] could not synchronize on CUDA context: CUDA_ERROR_ILLEGAL_ADDRESS: an illegal memory access was encountered
```
I tested the relaxation with up to 8 x A100 GPUs, but it failed (apparently there is a memory leak somewhere).
If I run the inference with v2.3.0 and relaxation with v2.2.4, it works fine.
I attached a sequence that returned this error (changed the suffix to txt to work on github UI).
[1S78.txt](https://github.com/deepmind/alphafold/files/10271938/1S78.txt) | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/660/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/660/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/659 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/659/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/659/comments | https://api.github.com/repos/deepmind/alphafold/issues/659/events | https://github.com/deepmind/alphafold/issues/659 | 1,501,878,693 | I_kwDOFoWQLM5ZhNml | 659 | error when running alphafold multimer | {
"avatar_url": "https://avatars.githubusercontent.com/u/86952016?v=4",
"events_url": "https://api.github.com/users/lamyankin/events{/privacy}",
"followers_url": "https://api.github.com/users/lamyankin/followers",
"following_url": "https://api.github.com/users/lamyankin/following{/other_user}",
"gists_url": "https://api.github.com/users/lamyankin/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/lamyankin",
"id": 86952016,
"login": "lamyankin",
"node_id": "MDQ6VXNlcjg2OTUyMDE2",
"organizations_url": "https://api.github.com/users/lamyankin/orgs",
"received_events_url": "https://api.github.com/users/lamyankin/received_events",
"repos_url": "https://api.github.com/users/lamyankin/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/lamyankin/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/lamyankin/subscriptions",
"type": "User",
"url": "https://api.github.com/users/lamyankin"
} | [] | open | false | null | [] | null | [
"Hi @lamyankin \r\nI had the same issue with a different file from `pdb_mmcif`. \r\nI solved this problem by downloading a newer version of the database.",
"Hi, glad to hear your reply!\r\nDid you download the newer pdb_mmcif database or all databases ?",
"I downloaded the full database again (but I don't think it is necessary)."
] | "2022-12-18T12:02:11Z" | "2022-12-20T13:16:10Z" | null | NONE | null | I am running the alphafold multimer to predict a complex composed of A and B and the problem occurs , here are the error message:
1218 18:46:51.980194 139874231673472 run_docker.py:255] Traceback (most recent call last):
I1218 18:46:51.980485 139874231673472 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 422, in <module>
I1218 18:46:51.980680 139874231673472 run_docker.py:255] app.run(main)
I1218 18:46:51.980863 139874231673472 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 312, in run
I1218 18:46:51.981044 139874231673472 run_docker.py:255] _run_main(main, args)
I1218 18:46:51.981219 139874231673472 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 258, in _run_main
I1218 18:46:51.981395 139874231673472 run_docker.py:255] sys.exit(main(argv))
I1218 18:46:51.981570 139874231673472 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 406, in main
I1218 18:46:51.981745 139874231673472 run_docker.py:255] random_seed=random_seed)
I1218 18:46:51.981920 139874231673472 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 174, in predict_structure
I1218 18:46:51.982094 139874231673472 run_docker.py:255] msa_output_dir=msa_output_dir)
I1218 18:46:51.982327 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/pipeline_multimer.py", line 269, in process
I1218 18:46:51.982502 139874231673472 run_docker.py:255] is_homomer_or_monomer=is_homomer_or_monomer)
I1218 18:46:51.982620 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/pipeline_multimer.py", line 214, in _process_single_chain
I1218 18:46:51.982703 139874231673472 run_docker.py:255] msa_output_dir=chain_msa_output_dir)
I1218 18:46:51.982784 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/pipeline.py", line 225, in process
I1218 18:46:51.982866 139874231673472 run_docker.py:255] hits=pdb_template_hits)
I1218 18:46:51.982964 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/templates.py", line 968, in get_templates
I1218 18:46:51.983068 139874231673472 run_docker.py:255] kalign_binary_path=self._kalign_binary_path)
I1218 18:46:51.983152 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/templates.py", line 737, in _process_single_hit
I1218 18:46:51.983233 139874231673472 run_docker.py:255] cif_string = _read_file(cif_path)
I1218 18:46:51.983314 139874231673472 run_docker.py:255] File "/app/alphafold/alphafold/data/templates.py", line 681, in _read_file
I1218 18:46:51.983396 139874231673472 run_docker.py:255] with open(path, 'r') as f:
I1218 18:46:51.983479 139874231673472 run_docker.py:255] FileNotFoundError: [Errno 2] No such file or directory: '/mnt/template_mmcif_dir/7u0h.cif'
Then I have try searching the 7u0h.cif in the /path/alphafold/pdb_mmcif/mmcif_files, but i didn't found it.
What should i do to solve this problem? looking forward to your reply. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/659/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/659/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/658 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/658/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/658/comments | https://api.github.com/repos/deepmind/alphafold/issues/658/events | https://github.com/deepmind/alphafold/issues/658 | 1,501,730,455 | I_kwDOFoWQLM5ZgpaX | 658 | Different alphafold version get different results | {
"avatar_url": "https://avatars.githubusercontent.com/u/86952016?v=4",
"events_url": "https://api.github.com/users/lamyankin/events{/privacy}",
"followers_url": "https://api.github.com/users/lamyankin/followers",
"following_url": "https://api.github.com/users/lamyankin/following{/other_user}",
"gists_url": "https://api.github.com/users/lamyankin/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/lamyankin",
"id": 86952016,
"login": "lamyankin",
"node_id": "MDQ6VXNlcjg2OTUyMDE2",
"organizations_url": "https://api.github.com/users/lamyankin/orgs",
"received_events_url": "https://api.github.com/users/lamyankin/received_events",
"repos_url": "https://api.github.com/users/lamyankin/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/lamyankin/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/lamyankin/subscriptions",
"type": "User",
"url": "https://api.github.com/users/lamyankin"
} | [] | open | false | null | [] | null | [
"@lamyankin \r\nWhen you run the multimer system you use 5 different models and, possibly, 5 different runs for each model (each one with a different seed).\r\nTo make sure you are comparing the same results you need to check the model name and seed used to run the inference.\r\n\r\nThere are v3 (from v2.3.0) and v2 (from v2.x.x) models.\r\nI did some tests and the v3 performs much better.\r\n\r\n"
] | "2022-12-18T04:02:45Z" | "2022-12-19T20:13:33Z" | null | NONE | null |
I run the alphafold in different version(one is v2.2.4, another is earlier )and get different results using the model_preset of multimer. As shown in the picture:

This is a complex composed of Cas9 and SNRPD2 (red) proteins. The predicted struture of cas9 is nearly the same in both results, as well as SNRPD2. However, the binding site of the cas9 and SNRPD2 is different in two results.
I want to know if it's because of the different versions of alphafold or because of my command?
here is my command:
nohup python3 docker/run_docker.py \
--fasta_paths=/home/zwf/alphafold/hpl1.fasta,/home/zwf/alphafold/SNRPD2.fasta \
--model_preset=multimer \
--docker_user=0 \
--db_preset=full_dbs \
--output_dir=/data/alphafold_outdir \
--max_template_date=2021-12-10 \
--data_dir=/data/alphalfold &> /home/zwf/alphafold/SNRPD2.out& | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/658/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/658/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/657 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/657/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/657/comments | https://api.github.com/repos/deepmind/alphafold/issues/657/events | https://github.com/deepmind/alphafold/issues/657 | 1,501,713,262 | I_kwDOFoWQLM5ZglNu | 657 | Backwards compatibility of AlphaFold 2.3.0 database with prevous versions of the software | {
"avatar_url": "https://avatars.githubusercontent.com/u/1719485?v=4",
"events_url": "https://api.github.com/users/panchyni/events{/privacy}",
"followers_url": "https://api.github.com/users/panchyni/followers",
"following_url": "https://api.github.com/users/panchyni/following{/other_user}",
"gists_url": "https://api.github.com/users/panchyni/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/panchyni",
"id": 1719485,
"login": "panchyni",
"node_id": "MDQ6VXNlcjE3MTk0ODU=",
"organizations_url": "https://api.github.com/users/panchyni/orgs",
"received_events_url": "https://api.github.com/users/panchyni/received_events",
"repos_url": "https://api.github.com/users/panchyni/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/panchyni/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/panchyni/subscriptions",
"type": "User",
"url": "https://api.github.com/users/panchyni"
} | [] | open | false | null | [] | null | [] | "2022-12-18T02:48:07Z" | "2022-12-18T02:48:07Z" | null | NONE | null | I am currently working with researchers at my university to update the version and database of AlphaFold on our university's computing cluster. From discussions we had prior to the release of 2.3.0, I had been told that updating the UniProt and PDB databases shouldn't affect the reproducibility of results or functionality of prior versions, given the common file structure and the ability to control the age of models used with input arguments. However, in the 2.3.0 update I noticed that the uniref30 database uses a slightly different folder structure and no longer contains *_db links and *_db.index files.
As such, I was hoping you could tell me if previous versions of AlphaFold depend on these files or are the *ffdata and *ffindex files sufficient? Ideally, I would like to avoid maintaining multiple databases and/or update scripts running on our system, so it would be ideal if I could simply make a directory in the format of the previous uniprot30 directory and soft link the necessary contents of uniref30 there so that we can maintain a single directory which support all versions of AlphaFold on our cluster. If the *db/db.index file are necessary for prior versions or there is some other conflict I am missing, I would appreciate any additional information you could give me.
I understand this is more an issue with the community need of our computing cluster than your software, but any information you could provide about the significance or lack their of regarding this change would save me a lot time and possibly errors trying to test it myself. Thank you for you time and consideration. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/657/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/657/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/656 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/656/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/656/comments | https://api.github.com/repos/deepmind/alphafold/issues/656/events | https://github.com/deepmind/alphafold/issues/656 | 1,498,941,741 | I_kwDOFoWQLM5ZWAkt | 656 | uniref30_database_path issue | {
"avatar_url": "https://avatars.githubusercontent.com/u/106186885?v=4",
"events_url": "https://api.github.com/users/limh25/events{/privacy}",
"followers_url": "https://api.github.com/users/limh25/followers",
"following_url": "https://api.github.com/users/limh25/following{/other_user}",
"gists_url": "https://api.github.com/users/limh25/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/limh25",
"id": 106186885,
"login": "limh25",
"node_id": "U_kgDOBlRIhQ",
"organizations_url": "https://api.github.com/users/limh25/orgs",
"received_events_url": "https://api.github.com/users/limh25/received_events",
"repos_url": "https://api.github.com/users/limh25/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/limh25/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/limh25/subscriptions",
"type": "User",
"url": "https://api.github.com/users/limh25"
} | [] | closed | false | null | [] | null | [
"You need to rebuild after pulling the newest version \r\n\r\n```docker build -f docker/Dockerfile -t alphafold .```",
"Hi @AChatzigoulas\r\nThank you for your help.\r\nI still have a problem in building the docker image.\r\n\r\n`Step 11/18 : RUN wget -q -P /app/alphafold/alphafold/common/ https://git.scicore.unibas.ch/schwede/openstructure/-/raw/7102c63615b64735c4941278d92b554ec94415f8/modules/mol/alg/src/stereo_chemical_props.txt\r\n ---> Running in 9b77342609c3\r\nThe command '/bin/bash -o pipefail -c wget -q -P /app/alphafold/alphafold/common/ https://git.scicore.unibas.ch/schwede/openstructure/-/raw/7102c63615b64735c4941278d92b554ec94415f8/modules/mol/alg/src/stereo_chemical_props.txt' returned a non-zero code: 5`\r\n\r\nDo you know the cause or the solution?\r\n\r\nThanks.",
"This is an SSL verification failure. Sadly, I do not know the cause or the solution for this one.",
"I was able to solve by modifying the Dockerfile\r\n\r\n```\r\nRUN wget --no-check-certificate -q -P /app/alphafold/alphafold/common/ \\\r\n https://git.scicore.unibas.ch/schwede/openstructure/-/raw/7102c63615b64735c4941278d92b554ec94415f8/modules/mol/alg/src/stereo_chemical_props.txt\r\n```\r\n\r\nThe \"--no-check-certificate\" option solved the issue.\r\n\r\nThanks."
] | "2022-12-15T19:05:05Z" | "2022-12-20T18:46:02Z" | "2022-12-20T18:46:01Z" | NONE | null | Hi,
I am trying to run the new ver. 2.3.0 for multimer predictions.
I had no issue with previous versions, including 2.2.4.
I tried to use direct download from the releases, and I also tried to use from git clone repository.
In both cases, I get an error message and the job terminates.
`I1215 18:59:41.374994 140456684652352 run_docker.py:255] FATAL Flags parsing error: Unknown command line flag 'uniref30_database_path'. Did you mean: uniref90_database_path ?`
It seems the directories are correctly mounted.
`I1215 18:59:37.418998 140456684652352 run_docker.py:113] Mounting /home/ec2-user/input_fasta/part1 -> /mnt/fasta_path_0
I1215 18:59:37.419240 140456684652352 run_docker.py:113] Mounting /disk1/databases/uniref90 -> /mnt/uniref90_database_path
I1215 18:59:37.419375 140456684652352 run_docker.py:113] Mounting /disk1/databases/mgnify -> /mnt/mgnify_database_path
I1215 18:59:37.419479 140456684652352 run_docker.py:113] Mounting /disk1/databases -> /mnt/data_dir
I1215 18:59:37.419581 140456684652352 run_docker.py:113] Mounting /disk1/databases/pdb_mmcif/mmcif_files -> /mnt/template_mmcif_dir
I1215 18:59:37.419708 140456684652352 run_docker.py:113] Mounting /disk1/databases/pdb_mmcif -> /mnt/obsolete_pdbs_path
I1215 18:59:37.419818 140456684652352 run_docker.py:113] Mounting /disk1/databases/uniprot -> /mnt/uniprot_database_path
I1215 18:59:37.419928 140456684652352 run_docker.py:113] Mounting /disk1/databases/pdb_seqres -> /mnt/pdb_seqres_database_path
I1215 18:59:37.420024 140456684652352 run_docker.py:113] Mounting /disk1/databases/uniref30/UniRef30_2021_03 -> /mnt/uniref30_database_path
I1215 18:59:37.420170 140456684652352 run_docker.py:113] Mounting /disk1/databases/bfd -> /mnt/bfd_database_path
I1215 18:59:41.374994 140456684652352 run_docker.py:255] FATAL Flags parsing error: Unknown command line flag 'uniref30_database_path'. Did you mean: uniref90_database_path ?
I1215 18:59:41.375221 140456684652352 run_docker.py:255] Pass --helpshort or --helpfull to see help on flags.`
I updated the databases:
- mgnify
- params
- pdb_mmcif
- pdb_seqres
- uniref30
- uniref90
Please help.
| {
"+1": 1,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 1,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/656/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/656/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/655 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/655/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/655/comments | https://api.github.com/repos/deepmind/alphafold/issues/655/events | https://github.com/deepmind/alphafold/issues/655 | 1,493,200,209 | I_kwDOFoWQLM5ZAG1R | 655 | A new problem occurred while I was running the Alphafold | {
"avatar_url": "https://avatars.githubusercontent.com/u/119162846?v=4",
"events_url": "https://api.github.com/users/pujialin/events{/privacy}",
"followers_url": "https://api.github.com/users/pujialin/followers",
"following_url": "https://api.github.com/users/pujialin/following{/other_user}",
"gists_url": "https://api.github.com/users/pujialin/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/pujialin",
"id": 119162846,
"login": "pujialin",
"node_id": "U_kgDOBxpH3g",
"organizations_url": "https://api.github.com/users/pujialin/orgs",
"received_events_url": "https://api.github.com/users/pujialin/received_events",
"repos_url": "https://api.github.com/users/pujialin/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/pujialin/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/pujialin/subscriptions",
"type": "User",
"url": "https://api.github.com/users/pujialin"
} | [
{
"color": "FEF2C0",
"default": false,
"description": "AlphaFold colab issue",
"id": 3313914804,
"name": "colab",
"node_id": "MDU6TGFiZWwzMzEzOTE0ODA0",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/colab"
}
] | open | false | null | [] | null | [
"I think this was caused by the changes in v2.3.0 which happened recently - your Colab notebook likely got out of sync with the code. Could you try running in a fresh notebook?\r\n https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb"
] | "2022-12-13T02:21:05Z" | "2022-12-13T10:32:36Z" | null | NONE | null | When I entered the amino acid sequence, the program reported an error:
AttributeError Traceback (most recent call last)
[<ipython-input-4-6962bd6ebd6c>](https://localhost:8080/#) in <module>
23
24 # Validate the input.
---> 25 sequences, model_type_to_use = notebook_utils.validate_input(
26 input_sequences=input_sequences,
27 min_length=MIN_SINGLE_SEQUENCE_LENGTH,
AttributeError: module 'alphafold.notebooks.notebook_utils' has no attribute 'validate_input'
I don't know what's going on, I hope everyone can help me. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/655/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/655/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/654 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/654/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/654/comments | https://api.github.com/repos/deepmind/alphafold/issues/654/events | https://github.com/deepmind/alphafold/issues/654 | 1,491,848,370 | I_kwDOFoWQLM5Y68yy | 654 | Add explicit `git pull` step in README? | {
"avatar_url": "https://avatars.githubusercontent.com/u/46537483?v=4",
"events_url": "https://api.github.com/users/ulupo/events{/privacy}",
"followers_url": "https://api.github.com/users/ulupo/followers",
"following_url": "https://api.github.com/users/ulupo/following{/other_user}",
"gists_url": "https://api.github.com/users/ulupo/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ulupo",
"id": 46537483,
"login": "ulupo",
"node_id": "MDQ6VXNlcjQ2NTM3NDgz",
"organizations_url": "https://api.github.com/users/ulupo/orgs",
"received_events_url": "https://api.github.com/users/ulupo/received_events",
"repos_url": "https://api.github.com/users/ulupo/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ulupo/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ulupo/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ulupo"
} | [] | open | false | null | [] | null | [] | "2022-12-12T13:08:33Z" | "2022-12-12T13:08:33Z" | null | NONE | null | From the README > "Updating existing installation":
https://github.com/deepmind/alphafold/blob/569eb4fea3733b979cb0442750b875759dd5ecc0/README.md?plain=1#L190-L191
I may have misunderstood, but wouldn't it be helpful to explicitly add the instruction to `git pull` if fetching returns no errors/displays merge conflicts? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/654/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/654/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/653 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/653/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/653/comments | https://api.github.com/repos/deepmind/alphafold/issues/653/events | https://github.com/deepmind/alphafold/issues/653 | 1,488,537,284 | I_kwDOFoWQLM5YuUbE | 653 | run_docker.py failure caused by module not found error | {
"avatar_url": "https://avatars.githubusercontent.com/u/120262538?v=4",
"events_url": "https://api.github.com/users/ailiqiangCSU/events{/privacy}",
"followers_url": "https://api.github.com/users/ailiqiangCSU/followers",
"following_url": "https://api.github.com/users/ailiqiangCSU/following{/other_user}",
"gists_url": "https://api.github.com/users/ailiqiangCSU/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ailiqiangCSU",
"id": 120262538,
"login": "ailiqiangCSU",
"node_id": "U_kgDOBysPig",
"organizations_url": "https://api.github.com/users/ailiqiangCSU/orgs",
"received_events_url": "https://api.github.com/users/ailiqiangCSU/received_events",
"repos_url": "https://api.github.com/users/ailiqiangCSU/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ailiqiangCSU/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ailiqiangCSU/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ailiqiangCSU"
} | [
{
"color": "ffffff",
"default": false,
"description": "An issue with Docker",
"id": 3094826898,
"name": "docker",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODk4",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/docker"
},
{
"color": "F9D0C4",
"default": false,
"description": "Issue setting up AlphaFold",
"id": 3317500161,
"name": "setup",
"node_id": "MDU6TGFiZWwzMzE3NTAwMTYx",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/setup"
}
] | open | false | null | [] | null | [] | "2022-12-10T14:33:09Z" | "2022-12-11T14:25:32Z" | null | NONE | null | When I run the run_docker.py with the following command,
python3 docker/run_docker.py \
--fasta_paths=/data/xiaoai/alphafold2_software/alphafold-main/T1082.fasta \
--max_template_date=2020-05-14 \
--model_preset=monomer \
--db_preset=full_dbs \
--data_dir=/data/xiaoai/alphafold2_software/download_file
I got this error message:
I1210 22:19:52.657300 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/alphafold-main -> /mnt/fasta_path_0
I1210 22:19:52.657422 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/uniref90 -> /mnt/uniref90_database_path
I1210 22:19:52.657488 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/mgnify -> /mnt/mgnify_database_path
I1210 22:19:52.657539 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file -> /mnt/data_dir
I1210 22:19:52.657588 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/pdb_mmcif/mmcif_files -> /mnt/template_mmcif_dir
I1210 22:19:52.657644 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/pdb_mmcif -> /mnt/obsolete_pdbs_path
I1210 22:19:52.657705 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/pdb70 -> /mnt/pdb70_database_path
I1210 22:19:52.657769 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/uniclust30/uniclust30_2018_08 -> /mnt/uniclust30_database_path
I1210 22:19:52.657833 139724585183040 run_docker.py:113] Mounting /data/xiaoai/alphafold2_software/download_file/bfd -> /mnt/bfd_database_path
I1210 22:19:53.050984 139724585183040 run_docker.py:255] Traceback (most recent call last):
I1210 22:19:53.051107 139724585183040 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 26, in <module>
I1210 22:19:53.051178 139724585183040 run_docker.py:255] from absl import app
I1210 22:19:53.051237 139724585183040 run_docker.py:255] ModuleNotFoundError: No module named 'absl'
but I find that absl-py already exist in conda list and python
I import absl in python without any error
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/653/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/653/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/652 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/652/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/652/comments | https://api.github.com/repos/deepmind/alphafold/issues/652/events | https://github.com/deepmind/alphafold/pull/652 | 1,485,120,206 | PR_kwDOFoWQLM5E0Pxk | 652 | Fix exception causes in data/templates.py | {
"avatar_url": "https://avatars.githubusercontent.com/u/56778?v=4",
"events_url": "https://api.github.com/users/cool-RR/events{/privacy}",
"followers_url": "https://api.github.com/users/cool-RR/followers",
"following_url": "https://api.github.com/users/cool-RR/following{/other_user}",
"gists_url": "https://api.github.com/users/cool-RR/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/cool-RR",
"id": 56778,
"login": "cool-RR",
"node_id": "MDQ6VXNlcjU2Nzc4",
"organizations_url": "https://api.github.com/users/cool-RR/orgs",
"received_events_url": "https://api.github.com/users/cool-RR/received_events",
"repos_url": "https://api.github.com/users/cool-RR/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/cool-RR/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/cool-RR/subscriptions",
"type": "User",
"url": "https://api.github.com/users/cool-RR"
} | [] | open | false | null | [] | null | [] | "2022-12-08T17:49:31Z" | "2022-12-08T17:49:31Z" | null | NONE | null | More context about this change here: https://blog.ram.rachum.com/post/621791438475296768/improving-python-exception-chaining-with | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/652/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/652/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/652.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/652",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/652.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/652"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/651 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/651/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/651/comments | https://api.github.com/repos/deepmind/alphafold/issues/651/events | https://github.com/deepmind/alphafold/issues/651 | 1,482,138,994 | I_kwDOFoWQLM5YV6Vy | 651 | GPU ran out of memory! | {
"avatar_url": "https://avatars.githubusercontent.com/u/5677674?v=4",
"events_url": "https://api.github.com/users/shahryary/events{/privacy}",
"followers_url": "https://api.github.com/users/shahryary/followers",
"following_url": "https://api.github.com/users/shahryary/following{/other_user}",
"gists_url": "https://api.github.com/users/shahryary/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/shahryary",
"id": 5677674,
"login": "shahryary",
"node_id": "MDQ6VXNlcjU2Nzc2NzQ=",
"organizations_url": "https://api.github.com/users/shahryary/orgs",
"received_events_url": "https://api.github.com/users/shahryary/received_events",
"repos_url": "https://api.github.com/users/shahryary/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/shahryary/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/shahryary/subscriptions",
"type": "User",
"url": "https://api.github.com/users/shahryary"
} | [
{
"color": "A65D8A",
"default": false,
"description": "Out of memory error message",
"id": 3938926450,
"name": "out of memory",
"node_id": "LA_kwDOFoWQLM7qxz9y",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/out%20of%20memory"
}
] | closed | false | null | [] | null | [
"We improved the GPU usage for AlphaFold-Multimer in [v2.3.0](https://github.com/deepmind/alphafold/releases/tag/v2.3.0) - hopefully this addresses your issue. Closing this issue now, feel free to reopen if you still encounter this in v2.3.0."
] | "2022-12-07T15:06:44Z" | "2022-12-13T11:55:11Z" | "2022-12-13T11:55:11Z" | NONE | null | `Allocator (GPU_0_bfc) ran out of memory trying to allocate 163.17GiB (rounded to 175203296000) requested by op`
I've been using the Alphafold for more than a year, and the GPU problem is annoying many users to run complex/ long sequences.
Alphafold is not supporting multi GPUs, and it's only using one GPU core - although, in the documentation, they mentioned it would attempt to use all available GPU cores!
I tried to run Alphafold(conda-based, docker/ singularity) in different instances from Amazon, G5.48x /G4 series. Still, I couldn't get results because only one GPU core taking into account instead of all of them.
The most significant target users of Alphafold are scientists who are more interested in running 'real' long samples rather than just a few lines of sequences. Without supporting multi-GPU mode, the tools are hopeless. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/651/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/651/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/650 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/650/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/650/comments | https://api.github.com/repos/deepmind/alphafold/issues/650/events | https://github.com/deepmind/alphafold/issues/650 | 1,476,046,364 | I_kwDOFoWQLM5X-q4c | 650 | jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead. | {
"avatar_url": "https://avatars.githubusercontent.com/u/108690998?v=4",
"events_url": "https://api.github.com/users/MasterNivek/events{/privacy}",
"followers_url": "https://api.github.com/users/MasterNivek/followers",
"following_url": "https://api.github.com/users/MasterNivek/following{/other_user}",
"gists_url": "https://api.github.com/users/MasterNivek/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/MasterNivek",
"id": 108690998,
"login": "MasterNivek",
"node_id": "U_kgDOBnp-Ng",
"organizations_url": "https://api.github.com/users/MasterNivek/orgs",
"received_events_url": "https://api.github.com/users/MasterNivek/received_events",
"repos_url": "https://api.github.com/users/MasterNivek/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/MasterNivek/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/MasterNivek/subscriptions",
"type": "User",
"url": "https://api.github.com/users/MasterNivek"
} | [] | closed | false | null | [] | null | [
"solved in pr #630 \r\n\r\njust replace all tree_utils\r\n",
"Thanks, fixed in v2.3.0."
] | "2022-12-05T08:55:33Z" | "2022-12-13T11:55:30Z" | "2022-12-13T11:55:30Z" | NONE | null | any one knows of a solution please

| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/650/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/650/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/649 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/649/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/649/comments | https://api.github.com/repos/deepmind/alphafold/issues/649/events | https://github.com/deepmind/alphafold/issues/649 | 1,472,724,200 | I_kwDOFoWQLM5Xx_zo | 649 | AlphaFold multimer has import error in Step 5 | {
"avatar_url": "https://avatars.githubusercontent.com/u/65905482?v=4",
"events_url": "https://api.github.com/users/Isabel-ql/events{/privacy}",
"followers_url": "https://api.github.com/users/Isabel-ql/followers",
"following_url": "https://api.github.com/users/Isabel-ql/following{/other_user}",
"gists_url": "https://api.github.com/users/Isabel-ql/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Isabel-ql",
"id": 65905482,
"login": "Isabel-ql",
"node_id": "MDQ6VXNlcjY1OTA1NDgy",
"organizations_url": "https://api.github.com/users/Isabel-ql/orgs",
"received_events_url": "https://api.github.com/users/Isabel-ql/received_events",
"repos_url": "https://api.github.com/users/Isabel-ql/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Isabel-ql/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Isabel-ql/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Isabel-ql"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"Hi thanks for raising this. Its a duplicate of https://github.com/deepmind/alphafold/issues/648 so closing"
] | "2022-12-02T11:32:40Z" | "2022-12-02T14:56:42Z" | "2022-12-02T14:56:42Z" | NONE | null | Hi,
I tried to predict structures of complexes with AlphaFold Multimer using the link: https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharinga
It worked well before, but a problem appeared yesterday. In step 5, "Search against genetic database", there's an import error (See below). I tried with different browsers, the problem still exists.
Is anybody know how to fix the problem? Thanks very much!
Cheers,
Qun
<img width="1381" alt="AlphaFold Multimer_step5" src="https://user-images.githubusercontent.com/65905482/205283476-bb018f25-61b6-4391-8a66-022dc90acce7.png">
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/649/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/649/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/648 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/648/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/648/comments | https://api.github.com/repos/deepmind/alphafold/issues/648/events | https://github.com/deepmind/alphafold/issues/648 | 1,470,885,359 | I_kwDOFoWQLM5Xq-3v | 648 | ImportError: Python version mismatch: module was compiled for Python 3.7, but the interpreter version is incompatible: 3.8.15 (default, Oct 12 2022, 19:14:39) [GCC 7.5.0]. | {
"avatar_url": "https://avatars.githubusercontent.com/u/119576148?v=4",
"events_url": "https://api.github.com/users/itheod04/events{/privacy}",
"followers_url": "https://api.github.com/users/itheod04/followers",
"following_url": "https://api.github.com/users/itheod04/following{/other_user}",
"gists_url": "https://api.github.com/users/itheod04/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/itheod04",
"id": 119576148,
"login": "itheod04",
"node_id": "U_kgDOByCWVA",
"organizations_url": "https://api.github.com/users/itheod04/orgs",
"received_events_url": "https://api.github.com/users/itheod04/received_events",
"repos_url": "https://api.github.com/users/itheod04/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/itheod04/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/itheod04/subscriptions",
"type": "User",
"url": "https://api.github.com/users/itheod04"
} | [] | closed | false | null | [] | null | [
"I have the same issue :/",
"Same issue ",
"+1",
"Hi thanks for raising this. We've updated to 3.8 in the latest [commit](https://github.com/deepmind/alphafold/commit/4494af848e41f3acd51a190d4c78d38cb1a7f85d) to match colab"
] | "2022-12-01T08:23:26Z" | "2022-12-02T14:30:31Z" | "2022-12-02T14:30:30Z" | NONE | null | a) ImportError: Python version mismatch: module was compiled for Python 3.7, but the interpreter version is incompatible: 3.8.15 (default, Oct 12 2022, 19:14:39)
[GCC 7.5.0].

b) ImportError: Traceback (most recent call last):
File "/opt/conda/lib/python3.7/site-packages/tensorflow/python/pywrap_tensorflow.py", line 62, in <module>
from tensorflow.python._pywrap_tensorflow_internal import *
ImportError: Python version mismatch: module was compiled for Python 3.7, but the interpreter version is incompatible: 3.8.15 (default, Oct 12 2022, 19:14:39)

[GCC 7.5.0].
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/648/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/648/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/647 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/647/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/647/comments | https://api.github.com/repos/deepmind/alphafold/issues/647/events | https://github.com/deepmind/alphafold/pull/647 | 1,470,257,031 | PR_kwDOFoWQLM5EAN2K | 647 | Code of Conduct | {
"avatar_url": "https://avatars.githubusercontent.com/u/91554112?v=4",
"events_url": "https://api.github.com/users/achawlaa/events{/privacy}",
"followers_url": "https://api.github.com/users/achawlaa/followers",
"following_url": "https://api.github.com/users/achawlaa/following{/other_user}",
"gists_url": "https://api.github.com/users/achawlaa/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/achawlaa",
"id": 91554112,
"login": "achawlaa",
"node_id": "U_kgDOBXUBQA",
"organizations_url": "https://api.github.com/users/achawlaa/orgs",
"received_events_url": "https://api.github.com/users/achawlaa/received_events",
"repos_url": "https://api.github.com/users/achawlaa/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/achawlaa/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/achawlaa/subscriptions",
"type": "User",
"url": "https://api.github.com/users/achawlaa"
} | [] | open | false | null | [] | null | [] | "2022-11-30T21:47:13Z" | "2022-11-30T22:37:00Z" | null | NONE | null | Code of Conduct is a very important document that clearly demonstrates the expectations for behavior for the participants of this project. The adoption and enforcement of a code of conduct helps structure a positive social atmosphere for a community to grow. Alphafold currently does not have a code of conduct page so I wanted to create one for it. Please see COD.md for the code of conduct page I constructed for Alphafold. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/647/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/647/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/647.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/647",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/647.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/647"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/646 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/646/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/646/comments | https://api.github.com/repos/deepmind/alphafold/issues/646/events | https://github.com/deepmind/alphafold/issues/646 | 1,468,059,827 | I_kwDOFoWQLM5XgNCz | 646 | A problem occurred while I was running the Alphafold | {
"avatar_url": "https://avatars.githubusercontent.com/u/119414515?v=4",
"events_url": "https://api.github.com/users/jhyeonv/events{/privacy}",
"followers_url": "https://api.github.com/users/jhyeonv/followers",
"following_url": "https://api.github.com/users/jhyeonv/following{/other_user}",
"gists_url": "https://api.github.com/users/jhyeonv/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/jhyeonv",
"id": 119414515,
"login": "jhyeonv",
"node_id": "U_kgDOBx4e8w",
"organizations_url": "https://api.github.com/users/jhyeonv/orgs",
"received_events_url": "https://api.github.com/users/jhyeonv/received_events",
"repos_url": "https://api.github.com/users/jhyeonv/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/jhyeonv/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/jhyeonv/subscriptions",
"type": "User",
"url": "https://api.github.com/users/jhyeonv"
} | [
{
"color": "000000",
"default": false,
"description": "",
"id": 3356911328,
"name": "cuda",
"node_id": "MDU6TGFiZWwzMzU2OTExMzI4",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/cuda"
}
] | open | false | null | [] | null | [
"I got this same error since CUDA moved to v.12. I think there will need to be an update on the side of the developer team here or a downgrade in system drivers. ",
"does this error persist when using AlphaFold v2.3.0?",
"@joshabramson I think so. I did a fresh build with the newest dockerfile. Here's the output where the error picks up:\r\n```\r\nI1223 20:04:51.789108 139688861775680 run_docker.py:255] I1224 01:04:51.788743 139711970813760 run_alphafold.py:191] Running model model_1_pred_0 on ECU03_1140\r\nI1223 20:04:54.797782 139688861775680 run_docker.py:255] I1224 01:04:54.797097 139711970813760 model.py:165] Running predict with shape(feat) = {'aatype': (4, 117), 'residue_index': (4, 117), 'seq_length': (4,), 'template_aatype': (4, 4, 117), 'template_all_atom_masks': (4, 4, 117, 37), 'template_all_atom_positions': (4, 4, 117, 37, 3), 'template_sum_probs': (4, 4, 1), 'is_distillation': (4,), 'seq_mask': (4, 117), 'msa_mask': (4, 508, 117), 'msa_row_mask': (4, 508), 'random_crop_to_size_seed': (4, 2), 'template_mask': (4, 4), 'template_pseudo_beta': (4, 4, 117, 3), 'template_pseudo_beta_mask': (4, 4, 117), 'atom14_atom_exists': (4, 117, 14), 'residx_atom14_to_atom37': (4, 117, 14), 'residx_atom37_to_atom14': (4, 117, 37), 'atom37_atom_exists': (4, 117, 37), 'extra_msa': (4, 5120, 117), 'extra_msa_mask': (4, 5120, 117), 'extra_msa_row_mask': (4, 5120), 'bert_mask': (4, 508, 117), 'true_msa': (4, 508, 117), 'extra_has_deletion': (4, 5120, 117), 'extra_deletion_value': (4, 5120, 117), 'msa_feat': (4, 508, 117, 49), 'target_feat': (4, 117, 22)}\r\nI1223 20:04:56.327672 139688861775680 run_docker.py:255] 2022-12-24 01:04:56.327116: W external/org_tensorflow/tensorflow/compiler/xla/stream_executor/gpu/asm_compiler.cc:231] Falling back to the CUDA driver for PTX compilation; ptxas does not support CC 8.9\r\nI1223 20:04:56.327813 139688861775680 run_docker.py:255] 2022-12-24 01:04:56.327156: W external/org_tensorflow/tensorflow/compiler/xla/stream_executor/gpu/asm_compiler.cc:234] Used ptxas at ptxas\r\nI1223 20:04:56.355589 139688861775680 run_docker.py:255] 2022-12-24 01:04:56.355236: E external/org_tensorflow/tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:628] failed to get PTX kernel \"shift_right_logical\" from module: CUDA_ERROR_NOT_FOUND: named symbol not found\r\nI1223 20:04:56.355756 139688861775680 run_docker.py:255] 2022-12-24 01:04:56.355273: E external/org_tensorflow/tensorflow/compiler/xla/pjrt/pjrt_stream_executor_client.cc:2153] Execution of replica 0 failed: INTERNAL: Could not find the corresponding function\r\nI1223 20:04:56.480186 139688861775680 run_docker.py:255] Traceback (most recent call last):\r\nI1223 20:04:56.480370 139688861775680 run_docker.py:255] File \"/app/alphafold/run_alphafold.py\", line 432, in <module>\r\nI1223 20:04:56.480440 139688861775680 run_docker.py:255] app.run(main)\r\nI1223 20:04:56.480502 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/absl/app.py\", line 312, in run\r\nI1223 20:04:56.480558 139688861775680 run_docker.py:255] _run_main(main, args)\r\nI1223 20:04:56.480613 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/absl/app.py\", line 258, in _run_main\r\nI1223 20:04:56.480718 139688861775680 run_docker.py:255] sys.exit(main(argv))\r\nI1223 20:04:56.480788 139688861775680 run_docker.py:255] File \"/app/alphafold/run_alphafold.py\", line 408, in main\r\nI1223 20:04:56.480844 139688861775680 run_docker.py:255] predict_structure(\r\nI1223 20:04:56.480898 139688861775680 run_docker.py:255] File \"/app/alphafold/run_alphafold.py\", line 199, in predict_structure\r\nI1223 20:04:56.480951 139688861775680 run_docker.py:255] prediction_result = model_runner.predict(processed_feature_dict,\r\nI1223 20:04:56.481002 139688861775680 run_docker.py:255] File \"/app/alphafold/alphafold/model/model.py\", line 167, in predict\r\nI1223 20:04:56.481052 139688861775680 run_docker.py:255] result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)\r\nI1223 20:04:56.481102 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/random.py\", line 132, in PRNGKey\r\nI1223 20:04:56.481152 139688861775680 run_docker.py:255] key = prng.seed_with_impl(impl, seed)\r\nI1223 20:04:56.481202 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 267, in seed_with_impl\r\nI1223 20:04:56.481253 139688861775680 run_docker.py:255] return random_seed(seed, impl=impl)\r\nI1223 20:04:56.481304 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 580, in random_seed\r\nI1223 20:04:56.481354 139688861775680 run_docker.py:255] return random_seed_p.bind(seeds_arr, impl=impl)\r\nI1223 20:04:56.481404 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 329, in bind\r\nI1223 20:04:56.481456 139688861775680 run_docker.py:255] return self.bind_with_trace(find_top_trace(args), args, params)\r\nI1223 20:04:56.481508 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 332, in bind_with_trace\r\nI1223 20:04:56.481559 139688861775680 run_docker.py:255] out = trace.process_primitive(self, map(trace.full_raise, args), params)\r\nI1223 20:04:56.481609 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 712, in process_primitive\r\nI1223 20:04:56.481659 139688861775680 run_docker.py:255] return primitive.impl(*tracers, **params)\r\nI1223 20:04:56.481709 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 592, in random_seed_impl\r\nI1223 20:04:56.481759 139688861775680 run_docker.py:255] base_arr = random_seed_impl_base(seeds, impl=impl)\r\nI1223 20:04:56.481808 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 597, in random_seed_impl_base\r\nI1223 20:04:56.481858 139688861775680 run_docker.py:255] return seed(seeds)\r\nI1223 20:04:56.481911 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/prng.py\", line 832, in threefry_seed\r\nI1223 20:04:56.481950 139688861775680 run_docker.py:255] lax.shift_right_logical(seed, lax_internal._const(seed, 32)))\r\nI1223 20:04:56.481987 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/lax/lax.py\", line 515, in shift_right_logical\r\nI1223 20:04:56.482024 139688861775680 run_docker.py:255] return shift_right_logical_p.bind(x, y)\r\nI1223 20:04:56.482062 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 329, in bind\r\nI1223 20:04:56.482100 139688861775680 run_docker.py:255] return self.bind_with_trace(find_top_trace(args), args, params)\r\nI1223 20:04:56.482138 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 332, in bind_with_trace\r\nI1223 20:04:56.482177 139688861775680 run_docker.py:255] out = trace.process_primitive(self, map(trace.full_raise, args), params)\r\nI1223 20:04:56.482217 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/core.py\", line 712, in process_primitive\r\nI1223 20:04:56.482254 139688861775680 run_docker.py:255] return primitive.impl(*tracers, **params)\r\nI1223 20:04:56.482291 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/dispatch.py\", line 115, in apply_primitive\r\nI1223 20:04:56.482333 139688861775680 run_docker.py:255] return compiled_fun(*args)\r\nI1223 20:04:56.482372 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/dispatch.py\", line 200, in <lambda>\r\nI1223 20:04:56.482409 139688861775680 run_docker.py:255] return lambda *args, **kw: compiled(*args, **kw)[0]\r\nI1223 20:04:56.482447 139688861775680 run_docker.py:255] File \"/opt/conda/lib/python3.8/site-packages/jax/_src/dispatch.py\", line 895, in _execute_compiled\r\nI1223 20:04:56.482484 139688861775680 run_docker.py:255] out_flat = compiled.execute(in_flat)\r\nI1223 20:04:56.482520 139688861775680 run_docker.py:255] jaxlib.xla_extension.XlaRuntimeError: INTERNAL: Could not find the corresponding function\r\n```",
"@joshabramson I was able to get alphafold to run by substituting nvidia/cuda:11.8.0-cudnn8-devel-ubuntu20.04 into the dockerfile. "
] | "2022-11-29T13:22:15Z" | "2022-12-26T05:56:10Z" | null | NONE | null | Hi.
A problem occurred while I was running the Alphafold.
Could I ask for help on how to solve it?
Please check the command below and if you need any information about that plase let me know.
```
jhyeon@jhyeon-Ubuntu:~/Desktop/data/SW/alphafold$ sudo python3 docker/run_docker.py --fasta_paths=T.fa --max_template_date=2021-12-31 --data_dir=/mnt/8THDD/data/db/AFDB/ --model_preset=monomer --output_dir=/home/jhyeon/Desktop/data/alphatest
[sudo] password for jhyeon:
I1129 20:06:44.587526 140678175391744 run_docker.py:113] Mounting /home/jhyeon/Desktop/data/SW/alphafold -> /mnt/fasta_path_0
I1129 20:06:44.998342 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/uniref90 -> /mnt/uniref90_database_path
I1129 20:06:45.015172 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/mgnify -> /mnt/mgnify_database_path
I1129 20:06:45.015434 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB -> /mnt/data_dir
I1129 20:06:45.039951 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/pdb_mmcif/mmcif_files -> /mnt/template_mmcif_dir
I1129 20:06:45.040283 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/pdb_mmcif -> /mnt/obsolete_pdbs_path
I1129 20:06:45.058208 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/pdb70 -> /mnt/pdb70_database_path
I1129 20:06:45.099748 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/uniclust30/uniclust30_2018_08 -> /mnt/uniclust30_database_path
I1129 20:06:45.100375 140678175391744 run_docker.py:113] Mounting /mnt/8THDD/data/db/AFDB/bfd -> /mnt/bfd_database_path
I1129 20:06:47.216925 140678175391744 run_docker.py:255] /opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:37: FutureWarning: jax.tree_structure is deprecated, and will be removed in a future release. Use jax.tree_util.tree_structure instead.
I1129 20:06:47.217015 140678175391744 run_docker.py:255] PyTreeDef = type(jax.tree_structure(None))
I1129 20:06:47.668879 140678175391744 run_docker.py:255] I1129 11:06:47.668288 140554574919488 templates.py:857] Using precomputed obsolete pdbs /mnt/obsolete_pdbs_path/obsolete.dat.
I1129 20:06:50.314509 140678175391744 run_docker.py:255] I1129 11:06:50.314045 140554574919488 xla_bridge.py:353] Unable to initialize backend 'tpu_driver': NOT_FOUND: Unable to find driver in registry given worker:
I1129 20:06:50.395502 140678175391744 run_docker.py:255] I1129 11:06:50.394992 140554574919488 xla_bridge.py:353] Unable to initialize backend 'rocm': NOT_FOUND: Could not find registered platform with name: "rocm". Available platform names are: Host CUDA Interpreter
I1129 20:06:50.395585 140678175391744 run_docker.py:255] I1129 11:06:50.395253 140554574919488 xla_bridge.py:353] Unable to initialize backend 'tpu': module 'jaxlib.xla_extension' has no attribute 'get_tpu_client'
I1129 20:06:50.395616 140678175391744 run_docker.py:255] I1129 11:06:50.395327 140554574919488 xla_bridge.py:353] Unable to initialize backend 'plugin': xla_extension has no attributes named get_plugin_device_client. Compile TensorFlow with //tensorflow/compiler/xla/python:enable_plugin_device set to true (defaults to false) to enable this.
I1129 20:07:01.872005 140678175391744 run_docker.py:255] I1129 11:07:01.871499 140554574919488 run_alphafold.py:377] Have 5 models: ['model_1_pred_0', 'model_2_pred_0', 'model_3_pred_0', 'model_4_pred_0', 'model_5_pred_0']
I1129 20:07:01.872115 140678175391744 run_docker.py:255] I1129 11:07:01.871589 140554574919488 run_alphafold.py:393] Using random seed 776916752095554131 for the data pipeline
I1129 20:07:01.872142 140678175391744 run_docker.py:255] I1129 11:07:01.871697 140554574919488 run_alphafold.py:161] Predicting T
I1129 20:07:01.872171 140678175391744 run_docker.py:255] I1129 11:07:01.871899 140554574919488 jackhmmer.py:133] Launching subprocess "/usr/bin/jackhmmer -o /dev/null -A /tmp/tmpyb0_qtih/output.sto --noali --F1 0.0005 --F2 5e-05 --F3 5e-07 --incE 0.0001 -E 0.0001 --cpu 8 -N 1 /mnt/fasta_path_0/T.fa /mnt/uniref90_database_path/uniref90.fasta"
I1129 20:07:01.908833 140678175391744 run_docker.py:255] I1129 11:07:01.908337 140554574919488 utils.py:36] Started Jackhmmer (uniref90.fasta) query
I1129 20:17:08.123620 140678175391744 run_docker.py:255] I1129 11:17:08.122851 140554574919488 utils.py:40] Finished Jackhmmer (uniref90.fasta) query in 606.214 seconds
I1129 20:17:08.124002 140678175391744 run_docker.py:255] I1129 11:17:08.123401 140554574919488 jackhmmer.py:133] Launching subprocess "/usr/bin/jackhmmer -o /dev/null -A /tmp/tmp3h2swg2s/output.sto --noali --F1 0.0005 --F2 5e-05 --F3 5e-07 --incE 0.0001 -E 0.0001 --cpu 8 -N 1 /mnt/fasta_path_0/T.fa /mnt/mgnify_database_path/mgy_clusters_2018_12.fa"
I1129 20:17:08.153515 140678175391744 run_docker.py:255] I1129 11:17:08.152948 140554574919488 utils.py:36] Started Jackhmmer (mgy_clusters_2018_12.fa) query
I1129 20:25:33.106717 140678175391744 run_docker.py:255] I1129 11:25:33.105955 140554574919488 utils.py:40] Finished Jackhmmer (mgy_clusters_2018_12.fa) query in 504.953 seconds
I1129 20:25:33.107178 140678175391744 run_docker.py:255] I1129 11:25:33.106705 140554574919488 hhsearch.py:85] Launching subprocess "/usr/bin/hhsearch -i /tmp/tmpi641el8l/query.a3m -o /tmp/tmpi641el8l/output.hhr -maxseq 1000000 -d /mnt/pdb70_database_path/pdb70"
I1129 20:25:33.138576 140678175391744 run_docker.py:255] I1129 11:25:33.138024 140554574919488 utils.py:36] Started HHsearch query
I1129 20:26:54.573565 140678175391744 run_docker.py:255] I1129 11:26:54.573027 140554574919488 utils.py:40] Finished HHsearch query in 81.435 seconds
I1129 20:26:54.582471 140678175391744 run_docker.py:255] I1129 11:26:54.581876 140554574919488 hhblits.py:128] Launching subprocess "/usr/bin/hhblits -i /mnt/fasta_path_0/T.fa -cpu 4 -oa3m /tmp/tmp0adsap7y/output.a3m -o /dev/null -n 3 -e 0.001 -maxseq 1000000 -realign_max 100000 -maxfilt 100000 -min_prefilter_hits 1000 -d /mnt/bfd_database_path/bfd_metaclust_clu_complete_id30_c90_final_seq.sorted_opt -d /mnt/uniclust30_database_path/uniclust30_2018_08"
I1129 20:26:54.614019 140678175391744 run_docker.py:255] I1129 11:26:54.613526 140554574919488 utils.py:36] Started HHblits query
I1129 20:39:47.502100 140678175391744 run_docker.py:255] I1129 11:39:47.501456 140554574919488 utils.py:40] Finished HHblits query in 772.888 seconds
I1129 20:39:47.511255 140678175391744 run_docker.py:255] I1129 11:39:47.510757 140554574919488 templates.py:878] Searching for template for: MAAAAAAAAAAAAAAAAAAAAAAAAA
I1129 20:39:49.477572 140678175391744 run_docker.py:255] I1129 11:39:49.476987 140554574919488 templates.py:268] Found an exact template match 4d10_F.
I1129 20:39:49.481890 140678175391744 run_docker.py:255] I1129 11:39:49.481489 140554574919488 templates.py:913] Skipped invalid hit 4D10_F COP9 SIGNALOSOME COMPLEX SUBUNIT 1; SIGNALING PROTEIN; 3.8A {HOMO SAPIENS}, error: None, warning: 4d10_F (sum_probs: 0.0, rank: 1): feature extracting errors: Template all atom mask was all zeros: 4d10_F. Residue range: 4-13, mmCIF parsing errors: {}
I1129 20:39:54.642663 140678175391744 run_docker.py:255] I1129 11:39:54.642101 140554574919488 templates.py:268] Found an exact template match 4v7h_BQ.
I1129 20:39:56.860277 140678175391744 run_docker.py:255] I1129 11:39:56.859714 140554574919488 templates.py:268] Found an exact template match 6j3y_W.
I1129 20:39:56.861339 140678175391744 run_docker.py:255] I1129 11:39:56.860958 140554574919488 templates.py:913] Skipped invalid hit 6J3Y_W Photosystem II reaction center protein; Photosystem, ELECTRON TRANSPORT; HET: LMG, HEM, DGD, SQD, LMU, BCR, CLA, OEX, PHO, PL9, LHG, A86; 3.3A {Chaetoceros gracilis}, error: None, warning: 6j3y_W (sum_probs: 0.0, rank: 3): feature extracting errors: Template all atom mask was all zeros: 6j3y_W. Residue range: 0-18, mmCIF parsing errors: {}
I1129 20:39:59.248605 140678175391744 run_docker.py:255] I1129 11:39:59.247956 140554574919488 templates.py:268] Found an exact template match 6j3z_w.
I1129 20:39:59.249619 140678175391744 run_docker.py:255] I1129 11:39:59.249154 140554574919488 templates.py:913] Skipped invalid hit 6J3Z_w Photosystem II reaction center protein; Photosystem, ELECTRON TRANSPORT; HET: LMG, HEM, DGD, SQD, LMU, BCR, CLA, OEX, PHO, PL9, LHG, A86; 3.6A {Chaetoceros gracilis}, error: None, warning: 6j3z_w (sum_probs: 0.0, rank: 4): feature extracting errors: Template all atom mask was all zeros: 6j3z_w. Residue range: 0-18, mmCIF parsing errors: {}
I1129 20:39:59.369462 140678175391744 run_docker.py:255] I1129 11:39:59.369046 140554574919488 templates.py:268] Found an exact template match 1m0u_B.
I1129 20:39:59.372590 140678175391744 run_docker.py:255] I1129 11:39:59.372143 140554574919488 templates.py:913] Skipped invalid hit 1M0U_B GST2 gene product (E.C.2.5.1.18); GST, Flight Muscle Protein, Sigma; HET: SO4, GSH; 1.75A {Drosophila melanogaster} SCOP: a.45.1.1, c.47.1.5, error: None, warning: 1m0u_B (sum_probs: 0.0, rank: 5): feature extracting errors: Template all atom mask was all zeros: 1m0u_B. Residue range: 0-23, mmCIF parsing errors: {}
I1129 20:39:59.973728 140678175391744 run_docker.py:255] I1129 11:39:59.973092 140554574919488 templates.py:268] Found an exact template match 1kn7_A.
I1129 20:40:02.077867 140678175391744 run_docker.py:255] I1129 11:40:02.077314 140554574919488 templates.py:268] Found an exact template match 6rfq_8.
I1129 20:40:04.344768 140678175391744 run_docker.py:255] I1129 11:40:04.336282 140554574919488 templates.py:268] Found an exact template match 6rfr_8.
I1129 20:40:08.787640 140678175391744 run_docker.py:255] I1129 11:40:08.786539 140554574919488 templates.py:268] Found an exact template match 6t59_s3.
I1129 20:40:08.790601 140678175391744 run_docker.py:255] I1129 11:40:08.790089 140554574919488 templates.py:913] Skipped invalid hit 6T59_s3 Ribosomal protein L8, uL3, uL4; TUBULIN, nascent chain-associated complex, ribosome-nascent; HET: MG; 3.11A {Oryctolagus cuniculus}, error: None, warning: 6t59_s3 (sum_probs: 0.0, rank: 9): feature extracting errors: Template all atom mask was all zeros: 6t59_s3. Residue range: 273-297, mmCIF parsing errors: {}
I1129 20:40:09.027597 140678175391744 run_docker.py:255] I1129 11:40:09.027103 140554574919488 templates.py:268] Found an exact template match 3lpj_B.
I1129 20:40:09.033292 140678175391744 run_docker.py:255] I1129 11:40:09.032789 140554574919488 templates.py:913] Skipped invalid hit 3LPJ_B Structure of BACE Bound to; Alzheimer's, Aspartyl protease, Hydrolase; HET: TLA, Z75; 1.79A {Homo sapiens}, error: None, warning: 3lpj_B (sum_probs: 0.0, rank: 10): feature extracting errors: Template all atom mask was all zeros: 3lpj_B. Residue range: 0-24, mmCIF parsing errors: {}
I1129 20:40:09.033350 140678175391744 run_docker.py:255] I1129 11:40:09.033004 140554574919488 pipeline.py:234] Uniref90 MSA size: 1 sequences.
I1129 20:40:09.033376 140678175391744 run_docker.py:255] I1129 11:40:09.033061 140554574919488 pipeline.py:235] BFD MSA size: 1 sequences.
I1129 20:40:09.033398 140678175391744 run_docker.py:255] I1129 11:40:09.033080 140554574919488 pipeline.py:236] MGnify MSA size: 1 sequences.
I1129 20:40:09.033423 140678175391744 run_docker.py:255] I1129 11:40:09.033095 140554574919488 pipeline.py:238] Final (deduplicated) MSA size: 1 sequences.
I1129 20:40:09.033465 140678175391744 run_docker.py:255] I1129 11:40:09.033219 140554574919488 pipeline.py:241] Total number of templates (NB: this can include bad templates and is later filtered to top 4): 4.
I1129 20:40:09.041668 140678175391744 run_docker.py:255] I1129 11:40:09.041206 140554574919488 run_alphafold.py:190] Running model model_1_pred_0 on T
I1129 20:40:10.478550 140678175391744 run_docker.py:255] I1129 11:40:10.478168 140554574919488 model.py:166] Running predict with shape(feat) = {'aatype': (4, 26), 'residue_index': (4, 26), 'seq_length': (4,), 'template_aatype': (4, 4, 26), 'template_all_atom_masks': (4, 4, 26, 37), 'template_all_atom_positions': (4, 4, 26, 37, 3), 'template_sum_probs': (4, 4, 1), 'is_distillation': (4,), 'seq_mask': (4, 26), 'msa_mask': (4, 508, 26), 'msa_row_mask': (4, 508), 'random_crop_to_size_seed': (4, 2), 'template_mask': (4, 4), 'template_pseudo_beta': (4, 4, 26, 3), 'template_pseudo_beta_mask': (4, 4, 26), 'atom14_atom_exists': (4, 26, 14), 'residx_atom14_to_atom37': (4, 26, 14), 'residx_atom37_to_atom14': (4, 26, 37), 'atom37_atom_exists': (4, 26, 37), 'extra_msa': (4, 5120, 26), 'extra_msa_mask': (4, 5120, 26), 'extra_msa_row_mask': (4, 5120), 'bert_mask': (4, 508, 26), 'true_msa': (4, 508, 26), 'extra_has_deletion': (4, 5120, 26), 'extra_deletion_value': (4, 5120, 26), 'msa_feat': (4, 508, 26, 49), 'target_feat': (4, 26, 22)}
I1129 20:40:10.573517 140678175391744 run_docker.py:255] 2022-11-29 11:40:10.572368: W external/org_tensorflow/tensorflow/compiler/xla/stream_executor/gpu/asm_compiler.cc:231] Falling back to the CUDA driver for PTX compilation; ptxas does not support CC 8.9
I1129 20:40:10.573782 140678175391744 run_docker.py:255] 2022-11-29 11:40:10.572425: W external/org_tensorflow/tensorflow/compiler/xla/stream_executor/gpu/asm_compiler.cc:234] Used ptxas at ptxas
I1129 20:40:10.586611 140678175391744 run_docker.py:255] 2022-11-29 11:40:10.585689: E external/org_tensorflow/tensorflow/compiler/xla/stream_executor/cuda/cuda_driver.cc:628] failed to get PTX kernel "shift_right_logical" from module: CUDA_ERROR_NOT_FOUND: named symbol not found
I1129 20:40:10.586843 140678175391744 run_docker.py:255] 2022-11-29 11:40:10.585779: E external/org_tensorflow/tensorflow/compiler/xla/pjrt/pjrt_stream_executor_client.cc:2153] Execution of replica 0 failed: INTERNAL: Could not find the corresponding function
I1129 20:40:10.594201 140678175391744 run_docker.py:255] Traceback (most recent call last):
I1129 20:40:10.594406 140678175391744 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 422, in <module>
I1129 20:40:10.594515 140678175391744 run_docker.py:255] app.run(main)
I1129 20:40:10.594610 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 312, in run
I1129 20:40:10.594703 140678175391744 run_docker.py:255] _run_main(main, args)
I1129 20:40:10.594786 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 258, in _run_main
I1129 20:40:10.594868 140678175391744 run_docker.py:255] sys.exit(main(argv))
I1129 20:40:10.594947 140678175391744 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 406, in main
I1129 20:40:10.595050 140678175391744 run_docker.py:255] random_seed=random_seed)
I1129 20:40:10.595142 140678175391744 run_docker.py:255] File "/app/alphafold/run_alphafold.py", line 199, in predict_structure
I1129 20:40:10.595224 140678175391744 run_docker.py:255] random_seed=model_random_seed)
I1129 20:40:10.595304 140678175391744 run_docker.py:255] File "/app/alphafold/alphafold/model/model.py", line 167, in predict
I1129 20:40:10.595379 140678175391744 run_docker.py:255] result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)
I1129 20:40:10.595466 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/random.py", line 132, in PRNGKey
I1129 20:40:10.595538 140678175391744 run_docker.py:255] key = prng.seed_with_impl(impl, seed)
I1129 20:40:10.595611 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 267, in seed_with_impl
I1129 20:40:10.595727 140678175391744 run_docker.py:255] return random_seed(seed, impl=impl)
I1129 20:40:10.595801 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 580, in random_seed
I1129 20:40:10.595872 140678175391744 run_docker.py:255] return random_seed_p.bind(seeds_arr, impl=impl)
I1129 20:40:10.595945 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 329, in bind
I1129 20:40:10.596016 140678175391744 run_docker.py:255] return self.bind_with_trace(find_top_trace(args), args, params)
I1129 20:40:10.596088 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 332, in bind_with_trace
I1129 20:40:10.596163 140678175391744 run_docker.py:255] out = trace.process_primitive(self, map(trace.full_raise, args), params)
I1129 20:40:10.596238 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 712, in process_primitive
I1129 20:40:10.596311 140678175391744 run_docker.py:255] return primitive.impl(*tracers, **params)
I1129 20:40:10.596384 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 592, in random_seed_impl
I1129 20:40:10.596455 140678175391744 run_docker.py:255] base_arr = random_seed_impl_base(seeds, impl=impl)
I1129 20:40:10.596517 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 597, in random_seed_impl_base
I1129 20:40:10.596581 140678175391744 run_docker.py:255] return seed(seeds)
I1129 20:40:10.596646 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/prng.py", line 832, in threefry_seed
I1129 20:40:10.596710 140678175391744 run_docker.py:255] lax.shift_right_logical(seed, lax_internal._const(seed, 32)))
I1129 20:40:10.596774 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/lax/lax.py", line 515, in shift_right_logical
I1129 20:40:10.596839 140678175391744 run_docker.py:255] return shift_right_logical_p.bind(x, y)
I1129 20:40:10.596904 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 329, in bind
I1129 20:40:10.596969 140678175391744 run_docker.py:255] return self.bind_with_trace(find_top_trace(args), args, params)
I1129 20:40:10.597032 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 332, in bind_with_trace
I1129 20:40:10.597097 140678175391744 run_docker.py:255] out = trace.process_primitive(self, map(trace.full_raise, args), params)
I1129 20:40:10.597160 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/core.py", line 712, in process_primitive
I1129 20:40:10.597218 140678175391744 run_docker.py:255] return primitive.impl(*tracers, **params)
I1129 20:40:10.597279 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/dispatch.py", line 115, in apply_primitive
I1129 20:40:10.597339 140678175391744 run_docker.py:255] return compiled_fun(*args)
I1129 20:40:10.597402 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/dispatch.py", line 200, in <lambda>
I1129 20:40:10.597465 140678175391744 run_docker.py:255] return lambda *args, **kw: compiled(*args, **kw)[0]
I1129 20:40:10.597525 140678175391744 run_docker.py:255] File "/opt/conda/lib/python3.7/site-packages/jax/_src/dispatch.py", line 895, in _execute_compiled
I1129 20:40:10.597588 140678175391744 run_docker.py:255] out_flat = compiled.execute(in_flat)
I1129 20:40:10.597648 140678175391744 run_docker.py:255] jaxlib.xla_extension.XlaRuntimeError: INTERNAL: Could not find the corresponding function
``` | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/646/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/646/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/645 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/645/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/645/comments | https://api.github.com/repos/deepmind/alphafold/issues/645/events | https://github.com/deepmind/alphafold/issues/645 | 1,467,828,543 | I_kwDOFoWQLM5XfUk_ | 645 | Running Alphafold using unpublished Pdb as a custom template | {
"avatar_url": "https://avatars.githubusercontent.com/u/119045747?v=4",
"events_url": "https://api.github.com/users/Shashankhare/events{/privacy}",
"followers_url": "https://api.github.com/users/Shashankhare/followers",
"following_url": "https://api.github.com/users/Shashankhare/following{/other_user}",
"gists_url": "https://api.github.com/users/Shashankhare/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Shashankhare",
"id": 119045747,
"login": "Shashankhare",
"node_id": "U_kgDOBxh-cw",
"organizations_url": "https://api.github.com/users/Shashankhare/orgs",
"received_events_url": "https://api.github.com/users/Shashankhare/received_events",
"repos_url": "https://api.github.com/users/Shashankhare/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Shashankhare/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Shashankhare/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Shashankhare"
} | [] | open | false | null | [] | null | [] | "2022-11-29T10:34:07Z" | "2022-11-29T10:34:07Z" | null | NONE | null | Hi,
I am trying to run Alphafold using a custom template. The template I am using as input is not published yet but I have both realspace-refined Pdb and cif files. When I am trying to run Alphafold with the unpublished Pdb, I get an error saying "no templates found”. I am wondering if is it possible to run Alphafold using unpublished Pdb as a custom template.
Regards,
Shashank. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/645/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/645/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/644 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/644/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/644/comments | https://api.github.com/repos/deepmind/alphafold/issues/644/events | https://github.com/deepmind/alphafold/pull/644 | 1,465,995,763 | PR_kwDOFoWQLM5Dx7Y_ | 644 | Update Readme.md File: Added license badge, Forks count badge, Watchers Count Badge | {
"avatar_url": "https://avatars.githubusercontent.com/u/101518890?v=4",
"events_url": "https://api.github.com/users/dhakalnirajan/events{/privacy}",
"followers_url": "https://api.github.com/users/dhakalnirajan/followers",
"following_url": "https://api.github.com/users/dhakalnirajan/following{/other_user}",
"gists_url": "https://api.github.com/users/dhakalnirajan/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dhakalnirajan",
"id": 101518890,
"login": "dhakalnirajan",
"node_id": "U_kgDOBg0OKg",
"organizations_url": "https://api.github.com/users/dhakalnirajan/orgs",
"received_events_url": "https://api.github.com/users/dhakalnirajan/received_events",
"repos_url": "https://api.github.com/users/dhakalnirajan/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dhakalnirajan/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dhakalnirajan/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dhakalnirajan"
} | [] | open | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/644/checks?check_run_id=9739001347) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request."
] | "2022-11-28T08:15:56Z" | "2022-12-24T14:09:36Z" | null | NONE | null | I have added license badge that redirects to the license page when clicked.
I have added GitHub repository Fork count badge that tells the visitors how many times the repository has been forked.
I have added GitHub Watchers Count Badge that displays the number of watchers on the repository. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/644/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/644/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/644.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/644",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/644.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/644"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/643 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/643/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/643/comments | https://api.github.com/repos/deepmind/alphafold/issues/643/events | https://github.com/deepmind/alphafold/pull/643 | 1,465,964,347 | PR_kwDOFoWQLM5Dx0mJ | 643 | Readme.md file update: Added License badge, Forks count badge and Watchers Count badge, | {
"avatar_url": "https://avatars.githubusercontent.com/u/101518890?v=4",
"events_url": "https://api.github.com/users/dhakalnirajan/events{/privacy}",
"followers_url": "https://api.github.com/users/dhakalnirajan/followers",
"following_url": "https://api.github.com/users/dhakalnirajan/following{/other_user}",
"gists_url": "https://api.github.com/users/dhakalnirajan/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dhakalnirajan",
"id": 101518890,
"login": "dhakalnirajan",
"node_id": "U_kgDOBg0OKg",
"organizations_url": "https://api.github.com/users/dhakalnirajan/orgs",
"received_events_url": "https://api.github.com/users/dhakalnirajan/received_events",
"repos_url": "https://api.github.com/users/dhakalnirajan/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dhakalnirajan/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dhakalnirajan/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dhakalnirajan"
} | [] | closed | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/643/checks?check_run_id=9738583833) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request."
] | "2022-11-28T07:51:06Z" | "2022-11-28T08:00:38Z" | "2022-11-28T08:00:38Z" | NONE | null | I have added license badge that redirects to the license page when clicked.
I have added GitHub repository Fork count badge that tells the visitors how many times the repository has been forked.
I have added GitHub Watchers Count Badge that displays the number of watchers on the repository. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/643/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/643/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/643.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/643",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/643.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/643"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/642 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/642/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/642/comments | https://api.github.com/repos/deepmind/alphafold/issues/642/events | https://github.com/deepmind/alphafold/issues/642 | 1,465,935,691 | I_kwDOFoWQLM5XYGdL | 642 | Can I customize the database? | {
"avatar_url": "https://avatars.githubusercontent.com/u/47802187?v=4",
"events_url": "https://api.github.com/users/dahaigui/events{/privacy}",
"followers_url": "https://api.github.com/users/dahaigui/followers",
"following_url": "https://api.github.com/users/dahaigui/following{/other_user}",
"gists_url": "https://api.github.com/users/dahaigui/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dahaigui",
"id": 47802187,
"login": "dahaigui",
"node_id": "MDQ6VXNlcjQ3ODAyMTg3",
"organizations_url": "https://api.github.com/users/dahaigui/orgs",
"received_events_url": "https://api.github.com/users/dahaigui/received_events",
"repos_url": "https://api.github.com/users/dahaigui/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dahaigui/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dahaigui/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dahaigui"
} | [] | open | false | null | [] | null | [] | "2022-11-28T07:18:50Z" | "2022-11-28T07:18:50Z" | null | NONE | null | My protein sequence is from a mutant library, is it possible to speed up my protein structure prediction by changing the dataset? For example, without using the BFD dataset, if it is possible, how should I implement it? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/642/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/642/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/641 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/641/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/641/comments | https://api.github.com/repos/deepmind/alphafold/issues/641/events | https://github.com/deepmind/alphafold/pull/641 | 1,465,540,000 | PR_kwDOFoWQLM5Dwbxe | 641 | Bugfix: Updated the AlphaFold notebook to make it compatible with newer versions of Google Colab (fixes the jax_experimental_name_stack error) | {
"avatar_url": "https://avatars.githubusercontent.com/u/40127722?v=4",
"events_url": "https://api.github.com/users/anupam-tiwari/events{/privacy}",
"followers_url": "https://api.github.com/users/anupam-tiwari/followers",
"following_url": "https://api.github.com/users/anupam-tiwari/following{/other_user}",
"gists_url": "https://api.github.com/users/anupam-tiwari/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/anupam-tiwari",
"id": 40127722,
"login": "anupam-tiwari",
"node_id": "MDQ6VXNlcjQwMTI3NzIy",
"organizations_url": "https://api.github.com/users/anupam-tiwari/orgs",
"received_events_url": "https://api.github.com/users/anupam-tiwari/received_events",
"repos_url": "https://api.github.com/users/anupam-tiwari/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/anupam-tiwari/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/anupam-tiwari/subscriptions",
"type": "User",
"url": "https://api.github.com/users/anupam-tiwari"
} | [] | closed | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/641/checks?check_run_id=9731462268) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request.",
"Hi thanks for providing this PR. Its no longer needed as we have updated Jax and Haiku, so I will close this."
] | "2022-11-27T18:43:37Z" | "2022-11-30T18:38:29Z" | "2022-11-30T18:38:29Z" | NONE | null | Since the newest Release of Google Colab(reference: https://colab.research.google.com/notebooks/relnotes.ipynb) the notebook has moved from Jax 0.3.17 to 0.3.25 which has Deleted the jax_experimental_name_stack config option(reference: https://jax.readthedocs.io/en/latest/changelog.html) This is causing the notebook to crash at the "5. Run AlphaFold and download prediction" step by throwing a config error as shown below:

This pull request solves the issue by downgrading the Jax and Jaxlib to 0.3.17 and 0.3.15[cuda] in the first stage of running the notebook and thus making the notebook functional | {
"+1": 1,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 1,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/641/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/641/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/641.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/641",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/641.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/641"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/640 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/640/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/640/comments | https://api.github.com/repos/deepmind/alphafold/issues/640/events | https://github.com/deepmind/alphafold/issues/640 | 1,465,309,598 | I_kwDOFoWQLM5XVtme | 640 | Alphafold Multimer Results | {
"avatar_url": "https://avatars.githubusercontent.com/u/119135711?v=4",
"events_url": "https://api.github.com/users/dozicauf/events{/privacy}",
"followers_url": "https://api.github.com/users/dozicauf/followers",
"following_url": "https://api.github.com/users/dozicauf/following{/other_user}",
"gists_url": "https://api.github.com/users/dozicauf/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dozicauf",
"id": 119135711,
"login": "dozicauf",
"node_id": "U_kgDOBxnd3w",
"organizations_url": "https://api.github.com/users/dozicauf/orgs",
"received_events_url": "https://api.github.com/users/dozicauf/received_events",
"repos_url": "https://api.github.com/users/dozicauf/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dozicauf/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dozicauf/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dozicauf"
} | [] | open | false | null | [] | null | [] | "2022-11-27T02:24:02Z" | "2022-11-27T02:24:02Z" | null | NONE | null | Hi,
I am wondering if anyone knows a website with Alphafold multimer predicted structures. The reason why I am asking this is that I am interested in exploring the PAE confusion matrix of different oligomeric structures. Also, is anyone aware of any code/rule by which we could say that a particular protein forms a dimer let's say? I know that we can determine that qualitatively by looking at the PAE and myb by looking at the contacts between chains that are less than 8A, but is there a rule for example of how many of such contact points we need to have or any other quantifiable parameter that could be used to determine whether a protein forms a particular oligomer based on the PAE. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/640/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/640/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/639 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/639/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/639/comments | https://api.github.com/repos/deepmind/alphafold/issues/639/events | https://github.com/deepmind/alphafold/issues/639 | 1,465,168,586 | I_kwDOFoWQLM5XVLLK | 639 | AlphaFold multimer has name error in the last step | {
"avatar_url": "https://avatars.githubusercontent.com/u/65905482?v=4",
"events_url": "https://api.github.com/users/Isabel-ql/events{/privacy}",
"followers_url": "https://api.github.com/users/Isabel-ql/followers",
"following_url": "https://api.github.com/users/Isabel-ql/following{/other_user}",
"gists_url": "https://api.github.com/users/Isabel-ql/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Isabel-ql",
"id": 65905482,
"login": "Isabel-ql",
"node_id": "MDQ6VXNlcjY1OTA1NDgy",
"organizations_url": "https://api.github.com/users/Isabel-ql/orgs",
"received_events_url": "https://api.github.com/users/Isabel-ql/received_events",
"repos_url": "https://api.github.com/users/Isabel-ql/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Isabel-ql/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Isabel-ql/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Isabel-ql"
} | [] | closed | false | null | [] | null | [
"Did you run all the cells properly, especially the second one \"2. Download AlphaFold\"?",
"> Did you run all the cells properly, especially the second one \"2. Download AlphaFold\"?\r\n\r\nYes, the other steps are all successful. Only the last step failed. \r\n<img width=\"719\" alt=\"AlphaFold Multimer_Step2\" src=\"https://user-images.githubusercontent.com/65905482/204095350-b0e68050-8404-4846-bb36-1c44df44d31d.png\">\r\n",
"Did you get it running? if now try this version: https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharing",
"> Did you get it running? if now try this version: https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharing\r\n\r\nThe old one is still problematic, but the new link you sent me works! I just finished the prediction. Thanks so much! ",
"Hi thanks for this. This may have been due to the colab kernel restarting, so that whilst the previous cells had run correctly they have since had their output cleared. When this happens, the colab needs to be rerun from the beginning. ",
"> Did you get it running? if now try this version: https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharing\r\n\r\nHi, a new problem appeared with the new link. In step 5, \"Search against genetic database\", there's an import error. See below. The problem started from yesterday, and still exists now. How to fix it? Thanks!\r\n\r\nCheers,\r\nQun\r\n\r\n<img width=\"1381\" alt=\"AlphaFold Multimer_step5\" src=\"https://user-images.githubusercontent.com/65905482/205282261-502402d8-f5b4-4742-a10d-51d8628cca67.png\">\r\n"
] | "2022-11-26T14:24:51Z" | "2022-12-02T11:25:16Z" | "2022-11-30T18:35:18Z" | NONE | null | Hi,
I have tried to use AlphaFold-Multimer to predict the structures of complexes, I did the prediction from the following link:
https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb
However, problem appeared in the last step, "Run AlphaFold and download prediction". The notice said "NameError: name 'model_type_to_use' is not defined". How to solve this problem?
Thanks very much!
Cheers,
Qun
<img width="1515" alt="AlphaFold Multimer_NameError" src="https://user-images.githubusercontent.com/65905482/204093603-ca29246b-1a42-466f-9a52-2b860e54a732.png">
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/639/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/639/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/638 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/638/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/638/comments | https://api.github.com/repos/deepmind/alphafold/issues/638/events | https://github.com/deepmind/alphafold/issues/638 | 1,463,803,012 | I_kwDOFoWQLM5XP9yE | 638 | Alphafold broken on Colab | {
"avatar_url": "https://avatars.githubusercontent.com/u/18224663?v=4",
"events_url": "https://api.github.com/users/danjaco/events{/privacy}",
"followers_url": "https://api.github.com/users/danjaco/followers",
"following_url": "https://api.github.com/users/danjaco/following{/other_user}",
"gists_url": "https://api.github.com/users/danjaco/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/danjaco",
"id": 18224663,
"login": "danjaco",
"node_id": "MDQ6VXNlcjE4MjI0NjYz",
"organizations_url": "https://api.github.com/users/danjaco/orgs",
"received_events_url": "https://api.github.com/users/danjaco/received_events",
"repos_url": "https://api.github.com/users/danjaco/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/danjaco/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/danjaco/subscriptions",
"type": "User",
"url": "https://api.github.com/users/danjaco"
} | [] | closed | false | null | [] | null | [
"Colab is also broken here\r\n\r\n\r\n",
"Hi Dan, thanks for raising this. The python dependencies in colab change over time, which means that the environment that the code runs in may diverge from alphafold, even if the alphafold code itself is stable. We will update the requirements when this happens. The most recent commit updated jax and haiku to match the colab versions, so this issue should be fixed",
"sorry, there is just a new error\r\n\r\n\r\n\r\n",
"I have an error:\r\n\r\n",
"Hi this error is from a Phenix notebook that isn't maintained by DeepMind. Please raise this issue in the Phenix repo. Thanks!"
] | "2022-11-24T20:54:55Z" | "2022-12-05T23:05:20Z" | "2022-11-30T18:32:15Z" | NONE | null | This issue is reported by others earlier, but I rase this separately as a more permanent solution is needed. I've come to rely on Colab version of Alphafold but a couple of times recently it's been down for over a week without us having any idea when it would be fixed
Would it be possible to make a second version available, that is known to be stable, so that if a new rollout fails there is something to fall back to? Or some other solution that could make it more reliable?
Thanks,
Dan | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/638/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/638/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/637 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/637/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/637/comments | https://api.github.com/repos/deepmind/alphafold/issues/637/events | https://github.com/deepmind/alphafold/issues/637 | 1,462,050,712 | I_kwDOFoWQLM5XJR-Y | 637 | Alpha Fold crash | {
"avatar_url": "https://avatars.githubusercontent.com/u/118919803?v=4",
"events_url": "https://api.github.com/users/HPN2/events{/privacy}",
"followers_url": "https://api.github.com/users/HPN2/followers",
"following_url": "https://api.github.com/users/HPN2/following{/other_user}",
"gists_url": "https://api.github.com/users/HPN2/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/HPN2",
"id": 118919803,
"login": "HPN2",
"node_id": "U_kgDOBxaSew",
"organizations_url": "https://api.github.com/users/HPN2/orgs",
"received_events_url": "https://api.github.com/users/HPN2/received_events",
"repos_url": "https://api.github.com/users/HPN2/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/HPN2/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/HPN2/subscriptions",
"type": "User",
"url": "https://api.github.com/users/HPN2"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"Same error here but unsure how to solve it",
"This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime",
"> This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime\r\n\r\nHey, I have been struggeling to apply your solution. Could you elaborate further on when and where to insert this command?",
"Thanks for raising this. Closing as this is a duplicate of https://github.com/deepmind/alphafold/issues/635."
] | "2022-11-23T16:22:49Z" | "2022-11-28T20:09:13Z" | "2022-11-28T20:09:12Z" | NONE | null | After running AF Colab multimer for weeks, it started to get unstable this week. Recently it has crashed multiple times. Each when starting task 5 (Run AlphaFold and download prediction) the system crashes within no time although I am Prolab+ user.
The following or similar error message occurs, which is similar to what other users feedback:
```
/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:144: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.
leaves, treedef = jax.tree_flatten(tree)
/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:145: FutureWarning: jax.tree_unflatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_unflatten instead.
return jax.tree_unflatten(treedef, leaves)
---------------------------------------------------------------------------
UnfilteredStackTrace Traceback (most recent call last)
[<ipython-input-5-ca4ee2dc266d>](https://localhost:8080/) in <module>
44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)
---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))
46
________________________________________
23 frames
________________________________________
UnfilteredStackTrace: AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'
The stack trace below excludes JAX-internal frames.
The preceding is the original exception that occurred, unmodified.
--------------------
The above exception was the direct cause of the following exception:
AttributeError Traceback (most recent call last)
[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/) in wrapped(self, *args, **kwargs)
406 f = functools.partial(unbound_method, self)
407 f = functools.partial(run_interceptors, f, method_name, self)
--> 408 if jax.config.jax_experimental_name_stack and module_name:
409 local_module_name = module_name.split("/")[-1]
410 f = jax.named_call(f, name=local_module_name)
AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'
``` | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/637/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/637/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/636 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/636/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/636/comments | https://api.github.com/repos/deepmind/alphafold/issues/636/events | https://github.com/deepmind/alphafold/issues/636 | 1,462,018,485 | I_kwDOFoWQLM5XJKG1 | 636 | Running without MSA features | {
"avatar_url": "https://avatars.githubusercontent.com/u/10798127?v=4",
"events_url": "https://api.github.com/users/Gabriel-Ducrocq/events{/privacy}",
"followers_url": "https://api.github.com/users/Gabriel-Ducrocq/followers",
"following_url": "https://api.github.com/users/Gabriel-Ducrocq/following{/other_user}",
"gists_url": "https://api.github.com/users/Gabriel-Ducrocq/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Gabriel-Ducrocq",
"id": 10798127,
"login": "Gabriel-Ducrocq",
"node_id": "MDQ6VXNlcjEwNzk4MTI3",
"organizations_url": "https://api.github.com/users/Gabriel-Ducrocq/orgs",
"received_events_url": "https://api.github.com/users/Gabriel-Ducrocq/received_events",
"repos_url": "https://api.github.com/users/Gabriel-Ducrocq/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Gabriel-Ducrocq/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Gabriel-Ducrocq/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Gabriel-Ducrocq"
} | [] | open | false | null | [] | null | [] | "2022-11-23T16:04:01Z" | "2022-11-23T16:04:01Z" | null | NONE | null | Hello,
I modified alphafold so that it takes my own custom template features. I would like to modify it again so that it runs without MSAs features. Is there any convenient way to do this ?
Does setting all the MSAs features to 0 is equivalent to no MSAs features ?
Thank you,
Gabriel. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/636/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/636/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/635 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/635/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/635/comments | https://api.github.com/repos/deepmind/alphafold/issues/635/events | https://github.com/deepmind/alphafold/issues/635 | 1,460,435,030 | I_kwDOFoWQLM5XDHhW | 635 | AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack' | {
"avatar_url": "https://avatars.githubusercontent.com/u/118849987?v=4",
"events_url": "https://api.github.com/users/zhangkaka1123/events{/privacy}",
"followers_url": "https://api.github.com/users/zhangkaka1123/followers",
"following_url": "https://api.github.com/users/zhangkaka1123/following{/other_user}",
"gists_url": "https://api.github.com/users/zhangkaka1123/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/zhangkaka1123",
"id": 118849987,
"login": "zhangkaka1123",
"node_id": "U_kgDOBxWBww",
"organizations_url": "https://api.github.com/users/zhangkaka1123/orgs",
"received_events_url": "https://api.github.com/users/zhangkaka1123/received_events",
"repos_url": "https://api.github.com/users/zhangkaka1123/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/zhangkaka1123/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/zhangkaka1123/subscriptions",
"type": "User",
"url": "https://api.github.com/users/zhangkaka1123"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"Hi! I'm getting the same error. It seems like Colab version of AlphaFold uses JAX v0.3.25. `jax.config.jax_experimental_name_stack` was removed in this version. If you manage to use older version of JAX with this Colab notebook, I guess it would work.",
"Hi! I'm also receiving this error. The AlphaFold `requirements.txt` file which is installed via pip3 in cell 2 specifies JAX 0.3.17, so I'm unsure why the version changes (running `print(jax.__version__)` at the start of cell 5 prints `0.3.25`)",
"+1 using a monomer.\r\n\r\nFull error message \r\n\r\n",
"Similar problem and it only started this week. ",
"Hello. Same issue since november 18.",
"I believe I have the same issue, first noticed 2-3 days ago with DeepMind's AlphaFold Colab (now running AF2.2.4), when I try to run it in multimer form, without exceeding sequence size limits. Things proceed well through construction of MSAs, but as it starts to run Model_1_multimer, it crashes with the following error message (below). Thx in advance for your kind help!\r\n\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:144: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.\r\n leaves, treedef = jax.tree_flatten(tree)\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:145: FutureWarning: jax.tree_unflatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_unflatten instead.\r\n return jax.tree_unflatten(treedef, leaves)\r\n---------------------------------------------------------------------------\r\nUnfilteredStackTrace Traceback (most recent call last)\r\n<ipython-input-5-b59661ebfbb4> in <module>\r\n 44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)\r\n---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))\r\n 46 \r\n\r\n23 frames\r\nUnfilteredStackTrace: AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'\r\n\r\nThe stack trace below excludes JAX-internal frames.\r\nThe preceding is the original exception that occurred, unmodified.\r\n--------------------\r\nThe above exception was the direct cause of the following exception:\r\n\r\nAttributeError Traceback (most recent call last)\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py in wrapped(self, *args, **kwargs)\r\n 406 f = functools.partial(unbound_method, self)\r\n 407 f = functools.partial(run_interceptors, f, method_name, self)\r\n--> 408 if jax.config.jax_experimental_name_stack and module_name:\r\n 409 local_module_name = module_name.split(\"/\")[-1]\r\n 410 f = jax.named_call(f, name=local_module_name)\r\n\r\nAttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'",
"This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime",
"Many thanks for the information. I thought s.th. must have happened.\nBest wishes,\n Heinz\n\n\n\nDr Heinz Peter Nasheuer\n\nPersonal Professor\n\nSchool of Biological and Chemical Sciences\n\nBiochemistry, Room 110\n\nArts & Science Building, Main Concourse\n\nUniversity of Galway\n\nDistillery Road\n\nGalway, H91 TK33, Ireland\n\n\nPhone: ++353-91-49 2430\n\n________________________________\nFrom: anupam-tiwari ***@***.***>\nSent: 26 November 2022 14:58\nTo: deepmind/alphafold ***@***.***>\nCc: Nasheuer, Heinz-Peter ***@***.***>; Comment ***@***.***>\nSubject: Re: [deepmind/alphafold] AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack' (Issue #635)\n\nEXTERNAL EMAIL: This email originated outside the University of Galway. Do not open attachments or click on links unless you believe the content is safe.\nRÍOMHPHOST SEACHTRACH: Níor tháinig an ríomhphost seo ó Ollscoil na Gaillimhe. Ná hoscail ceangaltáin agus ná cliceáil ar naisc mura gcreideann tú go bhfuil an t-ábhar sábháilte.\n\n\nThis is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime\n\n—\nReply to this email directly, view it on GitHub<https://github.com/deepmind/alphafold/issues/635#issuecomment-1328060690>, or unsubscribe<https://github.com/notifications/unsubscribe-auth/A4LJE65LQUP4OSICZQPNDUDWKIQQJANCNFSM6AAAAAASIFO6AQ>.\nYou are receiving this because you commented.Message ID: ***@***.***>\n",
"> This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime\r\n\r\nHey, I have been struggeling to apply your solution. Could you elaborate further on when and where to insert this command?",
"I can't get it to work either. jax version appears to already 0.3.17 while jaxlib is 0.3.25. But I can't seem to install 0.3.17.\r\n\r\n\r\n\r\n",
"The earliest version I was able to downgrade jaxlib to is 0.3.20, however this still leads to the original error.",
"> > This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime\r\n> \r\n> Hey, I have been struggeling to apply your solution. Could you elaborate further on when and where to insert this command?\r\n\r\nSame I have been having issue where to run the command that was provided. If someone could help with this, that would be great!",
"https://colab.research.google.com/drive/1J1G8OC6LZwT6Tqdz4uLRmnVzj7WWgUg1?usp=sharing \r\nhere you go guys, I made a modified notebook, the first step involves jax and jaxlib step, let me know how it goes! ",
"Yes, it seems to work. Thank you!\r\n\r\nI don't quite understand why you downgrade *before* downloading the packages, but it works. ",
"I tried to include this in the downloading package step, but to use the downgraded version we have to restart the runtime which in turn messes up the step-1 and step-2 code, hence I made it a separate step",
"Thanks for doing this!\r\n\r\nI tried but now I got a different error message. \r\n\r\n\r\nNameError Traceback (most recent call last)\r\n<ipython-input-2-15b941f78883><https://localhost:8080/> in <module>\r\n 18\r\n 19 # --- Run the model ---\r\n---> 20 if model_type_to_use == notebook_utils.ModelType.MONOMER:\r\n 21 model_names = config.MODEL_PRESETS['monomer'] + ('model_2_ptm',)\r\n 22 elif model_type_to_use == notebook_utils.ModelType.MULTIMER:\r\n\r\nNameError: name 'model_type_to_use' is not defined\r\n\r\n\r\nJust to confirm I used the multimer model with 2 sequences and they were run fine through the MSA step.",
"> Hello. Same issue since november 18.\r\n\r\nAgree Nov 18th in the morning everything was working fine and in the new week it started.",
"Thanks for this! Closing as this is a duplicate of https://github.com/deepmind/alphafold/issues/634",
"I appreciate all the comments! The issue has been solved in the new version! I just run two chain prediction successfully. Many thanks!"
] | "2022-11-22T19:24:16Z" | "2022-11-28T20:58:33Z" | "2022-11-28T20:13:53Z" | NONE | null | Hello,
I am predicting protein multimers using AlphaFold2 however it always failed and showed something like the following:
UnfilteredStackTrace Traceback (most recent call last)
[<ipython-input-5-ca4ee2dc266d>](https://localhost:8080/#) in <module>
44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)
---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))
46
23 frames
UnfilteredStackTrace: AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'
The stack trace below excludes JAX-internal frames.
The preceding is the original exception that occurred, unmodified.
--------------------
The above exception was the direct cause of the following exception:
AttributeError Traceback (most recent call last)
[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in wrapped(self, *args, **kwargs)
406 f = functools.partial(unbound_method, self)
407 f = functools.partial(run_interceptors, f, method_name, self)
--> 408 if jax.config.jax_experimental_name_stack and module_name:
409 local_module_name = module_name.split("/")[-1]
410 f = jax.named_call(f, name=local_module_name)
AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'
It shows "Attribute error" but I have no idea about coding to solve this issue.
I would really appreciate if someone knows what is going on.
Tons of thanks!
Kaka | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/635/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/635/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/634 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/634/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/634/comments | https://api.github.com/repos/deepmind/alphafold/issues/634/events | https://github.com/deepmind/alphafold/issues/634 | 1,460,318,554 | I_kwDOFoWQLM5XCrFa | 634 | AlphaFold Colab crashing at cell 5 | {
"avatar_url": "https://avatars.githubusercontent.com/u/25184542?v=4",
"events_url": "https://api.github.com/users/mprokhorov/events{/privacy}",
"followers_url": "https://api.github.com/users/mprokhorov/followers",
"following_url": "https://api.github.com/users/mprokhorov/following{/other_user}",
"gists_url": "https://api.github.com/users/mprokhorov/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/mprokhorov",
"id": 25184542,
"login": "mprokhorov",
"node_id": "MDQ6VXNlcjI1MTg0NTQy",
"organizations_url": "https://api.github.com/users/mprokhorov/orgs",
"received_events_url": "https://api.github.com/users/mprokhorov/received_events",
"repos_url": "https://api.github.com/users/mprokhorov/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/mprokhorov/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/mprokhorov/subscriptions",
"type": "User",
"url": "https://api.github.com/users/mprokhorov"
} | [] | closed | false | null | [] | null | [
"This is the full Stack Trace:\r\n```\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:144: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.\r\n leaves, treedef = jax.tree_flatten(tree)\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:145: FutureWarning: jax.tree_unflatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_unflatten instead.\r\n return jax.tree_unflatten(treedef, leaves)\r\n---------------------------------------------------------------------------\r\nUnfilteredStackTrace Traceback (most recent call last)\r\n[<ipython-input-5-ca4ee2dc266d>](https://localhost:8080/#) in <module>\r\n 44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)\r\n---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))\r\n 46 \r\n\r\n23 frames\r\n[/opt/conda/lib/python3.7/site-packages/alphafold/model/model.py](https://localhost:8080/#) in predict(self, feat, random_seed)\r\n 166 tree.map_structure(lambda x: x.shape, feat))\r\n--> 167 result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)\r\n 168 \r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/traceback_util.py](https://localhost:8080/#) in reraise_with_filtered_traceback(*args, **kwargs)\r\n 161 try:\r\n--> 162 return fun(*args, **kwargs)\r\n 163 except Exception as e:\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/api.py](https://localhost:8080/#) in cache_miss(*args, **kwargs)\r\n 621 jax.config.jax_debug_nans or jax.config.jax_debug_infs):\r\n--> 622 execute = dispatch._xla_call_impl_lazy(fun_, *tracers, **params)\r\n 623 out_flat = call_bind_continuation(execute(*args_flat))\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/dispatch.py](https://localhost:8080/#) in _xla_call_impl_lazy(***failed resolving arguments***)\r\n 236 return xla_callable(fun, device, backend, name, donated_invars, keep_unused,\r\n--> 237 *arg_specs)\r\n 238 \r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/linear_util.py](https://localhost:8080/#) in memoized_fun(fun, *args)\r\n 302 else:\r\n--> 303 ans = call(fun, *args)\r\n 304 cache[key] = (ans, fun.stores)\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/dispatch.py](https://localhost:8080/#) in _xla_callable_uncached(fun, device, backend, name, donated_invars, keep_unused, *arg_specs)\r\n 359 return lower_xla_callable(fun, device, backend, name, donated_invars, False,\r\n--> 360 keep_unused, *arg_specs).compile().unsafe_call\r\n 361 \r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/profiler.py](https://localhost:8080/#) in wrapper(*args, **kwargs)\r\n 313 with TraceAnnotation(name, **decorator_kwargs):\r\n--> 314 return func(*args, **kwargs)\r\n 315 return wrapper\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/dispatch.py](https://localhost:8080/#) in lower_xla_callable(fun, device, backend, name, donated_invars, always_lower, keep_unused, *arg_specs)\r\n 445 jaxpr, out_type, consts = pe.trace_to_jaxpr_final2(\r\n--> 446 fun, pe.debug_info_final(fun, \"jit\"))\r\n 447 out_avals, kept_outputs = util.unzip2(out_type)\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/_src/profiler.py](https://localhost:8080/#) in wrapper(*args, **kwargs)\r\n 313 with TraceAnnotation(name, **decorator_kwargs):\r\n--> 314 return func(*args, **kwargs)\r\n 315 return wrapper\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/interpreters/partial_eval.py](https://localhost:8080/#) in trace_to_jaxpr_final2(fun, debug_info)\r\n 2076 with core.new_sublevel():\r\n-> 2077 jaxpr, out_type, consts = trace_to_subjaxpr_dynamic2(fun, main, debug_info)\r\n 2078 del fun, main\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/interpreters/partial_eval.py](https://localhost:8080/#) in trace_to_subjaxpr_dynamic2(fun, main, debug_info)\r\n 2026 in_tracers_ = [t for t, keep in zip(in_tracers, keep_inputs) if keep]\r\n-> 2027 ans = fun.call_wrapped(*in_tracers_)\r\n 2028 out_tracers = map(trace.full_raise, ans)\r\n\r\n[/usr/local/lib/python3.7/dist-packages/jax/linear_util.py](https://localhost:8080/#) in call_wrapped(self, *args, **kwargs)\r\n 166 try:\r\n--> 167 ans = self.f(*args, **dict(self.params, **kwargs))\r\n 168 except:\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/transform.py](https://localhost:8080/#) in apply_fn(params, *args, **kwargs)\r\n 126 \r\n--> 127 out, state = f.apply(params, {}, *args, **kwargs)\r\n 128 if state:\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/transform.py](https://localhost:8080/#) in apply_fn(params, state, rng, *args, **kwargs)\r\n 353 try:\r\n--> 354 out = f(*args, **kwargs)\r\n 355 except jax.errors.UnexpectedTracerError as e:\r\n\r\n[/opt/conda/lib/python3.7/site-packages/alphafold/model/model.py](https://localhost:8080/#) in _forward_fn(batch)\r\n 81 def _forward_fn(batch):\r\n---> 82 model = modules.AlphaFold(self.config.model)\r\n 83 return model(\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in __call__(cls, *args, **kwargs)\r\n 120 init = wrap_method(\"__init__\", cls.__init__)\r\n--> 121 init(module, *args, **kwargs)\r\n 122 \r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in wrapped(self, *args, **kwargs)\r\n 407 f = functools.partial(run_interceptors, f, method_name, self)\r\n--> 408 if jax.config.jax_experimental_name_stack and module_name:\r\n 409 local_module_name = module_name.split(\"/\")[-1]\r\n\r\nUnfilteredStackTrace: AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'\r\n\r\nThe stack trace below excludes JAX-internal frames.\r\nThe preceding is the original exception that occurred, unmodified.\r\n\r\n--------------------\r\n\r\nThe above exception was the direct cause of the following exception:\r\n\r\nAttributeError Traceback (most recent call last)\r\n[<ipython-input-5-ca4ee2dc266d>](https://localhost:8080/#) in <module>\r\n 43 model_runner = model.RunModel(cfg, params)\r\n 44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)\r\n---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))\r\n 46 \r\n 47 mean_plddt = prediction['plddt'].mean()\r\n\r\n[/opt/conda/lib/python3.7/site-packages/alphafold/model/model.py](https://localhost:8080/#) in predict(self, feat, random_seed)\r\n 165 logging.info('Running predict with shape(feat) = %s',\r\n 166 tree.map_structure(lambda x: x.shape, feat))\r\n--> 167 result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)\r\n 168 \r\n 169 # This block is to ensure benchmark timings are accurate. Some blocking is\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/transform.py](https://localhost:8080/#) in apply_fn(params, *args, **kwargs)\r\n 125 \"name (e.g. `f.apply(.., state=my_state)`)\")\r\n 126 \r\n--> 127 out, state = f.apply(params, {}, *args, **kwargs)\r\n 128 if state:\r\n 129 raise ValueError(\"If your transformed function uses `hk.{get,set}_state` \"\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/transform.py](https://localhost:8080/#) in apply_fn(params, state, rng, *args, **kwargs)\r\n 352 with base.new_context(params=params, state=state, rng=rng) as ctx:\r\n 353 try:\r\n--> 354 out = f(*args, **kwargs)\r\n 355 except jax.errors.UnexpectedTracerError as e:\r\n 356 raise jax.errors.UnexpectedTracerError(unexpected_tracer_hint) from e\r\n\r\n[/opt/conda/lib/python3.7/site-packages/alphafold/model/model.py](https://localhost:8080/#) in _forward_fn(batch)\r\n 80 else:\r\n 81 def _forward_fn(batch):\r\n---> 82 model = modules.AlphaFold(self.config.model)\r\n 83 return model(\r\n 84 batch,\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in __call__(cls, *args, **kwargs)\r\n 119 # Now attempt to initialize the object.\r\n 120 init = wrap_method(\"__init__\", cls.__init__)\r\n--> 121 init(module, *args, **kwargs)\r\n 122 \r\n 123 if (config.get_config().module_auto_repr and\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/module.py](https://localhost:8080/#) in wrapped(self, *args, **kwargs)\r\n 406 f = functools.partial(unbound_method, self)\r\n 407 f = functools.partial(run_interceptors, f, method_name, self)\r\n--> 408 if jax.config.jax_experimental_name_stack and module_name:\r\n 409 local_module_name = module_name.split(\"/\")[-1]\r\n 410 f = jax.named_call(f, name=local_module_name)\r\n\r\nAttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'\r\n```",
"This is an upgrade from google, the newest version of colab by default takes up jax 0.3.25 (https://colab.research.google.com/notebooks/relnotes.ipynb#scrollTo=iOWrh41ocOca) which has no support for 'jax_experimental_name_stack', I have solved this issue by downgrading colab jax to 0.3.17, you can do it by adding this command: !pip install --upgrade \"jax[cuda]==0.3.17\" -f https://storage.googleapis.com/jax-releases/jax_cuda_releases.html and then restarting the runtime",
"Thanks for raising this. This has been fixed in the latest commit."
] | "2022-11-22T17:58:00Z" | "2022-11-28T21:02:53Z" | "2022-11-28T21:02:53Z" | NONE | null | Today I've tried to run Colab version of AlphaFold v2.2.4, but it crashed every time while executing cell number 5. I'm getting these errors:
**AttributeError: 'Config' object has no attribute 'jax_experimental_name_stack'**
**'Config' object has no attribute 'jax_experimental_name_stack'** | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/634/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/634/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/633 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/633/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/633/comments | https://api.github.com/repos/deepmind/alphafold/issues/633/events | https://github.com/deepmind/alphafold/issues/633 | 1,456,975,182 | I_kwDOFoWQLM5W161O | 633 | A question about running time on MSA | {
"avatar_url": "https://avatars.githubusercontent.com/u/25558295?v=4",
"events_url": "https://api.github.com/users/YiningWang2/events{/privacy}",
"followers_url": "https://api.github.com/users/YiningWang2/followers",
"following_url": "https://api.github.com/users/YiningWang2/following{/other_user}",
"gists_url": "https://api.github.com/users/YiningWang2/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/YiningWang2",
"id": 25558295,
"login": "YiningWang2",
"node_id": "MDQ6VXNlcjI1NTU4Mjk1",
"organizations_url": "https://api.github.com/users/YiningWang2/orgs",
"received_events_url": "https://api.github.com/users/YiningWang2/received_events",
"repos_url": "https://api.github.com/users/YiningWang2/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/YiningWang2/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/YiningWang2/subscriptions",
"type": "User",
"url": "https://api.github.com/users/YiningWang2"
} | [] | open | false | null | [] | null | [] | "2022-11-20T17:07:47Z" | "2022-11-20T17:07:47Z" | null | NONE | null | Hello, recently i am curious about the search time when i generate MSAs. The same database I used, i fount different sequence has quite different search time. And the difference is not strongly correlative with the sequence length. I want to know what cause different speed, can you help me? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/633/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/633/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/632 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/632/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/632/comments | https://api.github.com/repos/deepmind/alphafold/issues/632/events | https://github.com/deepmind/alphafold/pull/632 | 1,453,752,049 | PR_kwDOFoWQLM5DIuJ6 | 632 | fix comment | {
"avatar_url": "https://avatars.githubusercontent.com/u/22633385?v=4",
"events_url": "https://api.github.com/users/eltociear/events{/privacy}",
"followers_url": "https://api.github.com/users/eltociear/followers",
"following_url": "https://api.github.com/users/eltociear/following{/other_user}",
"gists_url": "https://api.github.com/users/eltociear/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/eltociear",
"id": 22633385,
"login": "eltociear",
"node_id": "MDQ6VXNlcjIyNjMzMzg1",
"organizations_url": "https://api.github.com/users/eltociear/orgs",
"received_events_url": "https://api.github.com/users/eltociear/received_events",
"repos_url": "https://api.github.com/users/eltociear/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/eltociear/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/eltociear/subscriptions",
"type": "User",
"url": "https://api.github.com/users/eltociear"
} | [] | open | false | null | [] | null | [] | "2022-11-17T17:56:38Z" | "2022-11-17T17:56:38Z" | null | NONE | null | dihedral-angle-definiting -> dihedral-angle-defining | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/632/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/632/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/632.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/632",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/632.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/632"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/631 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/631/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/631/comments | https://api.github.com/repos/deepmind/alphafold/issues/631/events | https://github.com/deepmind/alphafold/issues/631 | 1,451,358,060 | I_kwDOFoWQLM5Wgfds | 631 | How to accelerate protein prediction | {
"avatar_url": "https://avatars.githubusercontent.com/u/47802187?v=4",
"events_url": "https://api.github.com/users/dahaigui/events{/privacy}",
"followers_url": "https://api.github.com/users/dahaigui/followers",
"following_url": "https://api.github.com/users/dahaigui/following{/other_user}",
"gists_url": "https://api.github.com/users/dahaigui/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/dahaigui",
"id": 47802187,
"login": "dahaigui",
"node_id": "MDQ6VXNlcjQ3ODAyMTg3",
"organizations_url": "https://api.github.com/users/dahaigui/orgs",
"received_events_url": "https://api.github.com/users/dahaigui/received_events",
"repos_url": "https://api.github.com/users/dahaigui/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/dahaigui/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/dahaigui/subscriptions",
"type": "User",
"url": "https://api.github.com/users/dahaigui"
} | [] | open | false | null | [] | null | [
"This is addressed in the \"Inferencing many proteins\" section of the README. The short answer is not exactly, but you can reduce the runtime for similar proteins by keeping the network a fixed size, and a bulk inference script could be built on the RunModel.predict method.",
"This is good to know. Could you please elaborate with an example please? Or point me to a resource where I can find more about this?",
"@smturzo \r\n> This is good to know. Could you please elaborate with an example please? Or point me to a resource where I can find more about this?\r\n\r\nThe reference protein seq is \r\n\\>CsaV3_7G025510\r\nMIGRLRMNHCVPDFEMADDFSLPTFSSLTRPRKSSLPDDDVMELLWQNGQVVTHSQNQRSFRKSPPSKFDVSIPQEQAATREIRPSTQLEEHHELFMQEDEMASWLNYPLVEDHNFCSDLLFPAITAPLCANPQPDIRPSATATLTLTPRPPIPPCRRPEVQTSVQFSRNKATVESEPSNSKVMVRESTVVDSCDTPSVGPESRASEMARRKLVEVVNGGGVRYEIARGSDGVRGASVGGDGIGEKEMMTCEMTVTSSPGGSSASAEPACPKLAVDDRKRKGRALDDTECQSEDVEYESADPKKQLRGSTSTKRSRAAEVHNLSERRRRDRINEKMKALQELIPRCNKTDKASMLDEAIEYLKTLQLQVQMMSMGCGMMPMMFPGVQQYLPPPMGMGMGMGMEMGMNRPMMQFHNLLAGSNL**P**MQAGATAAAHLGPRFPLPPFAMPPVPGNDPSRAQAMNNQPDPMANSVGTQNTTPPSVLGFPDSYQQFLSSTQMQFHMTQALQNQHPVQLNTSRPCTSRGPENRDNHQSG\r\n\r\nOne of the proteins in our mutant library differs from the reference genome by only one amino acid, with the following sequence\r\n\\>Csa_Mutant_125_7G025510\r\nMIGRLRMNHCVPDFEMADDFSLPTFSSLTRPRKSSLPDDDVMELLWQNGQVVTHSQNQRSFRKSPPSKFDVSIPQEQAATREIRPSTQLEEHHELFMQEDEMASWLNYPLVEDHNFCSDLLFPAITAPLCANPQPDIRPSATATLTLTPRPPIPPCRRPEVQTSVQFSRNKATVESEPSNSKVMVRESTVVDSCDTPSVGPESRASEMARRKLVEVVNGGGVRYEIARGSDGVRGASVGGDGIGEKEMMTCEMTVTSSPGGSSASAEPACPKLAVDDRKRKGRALDDTECQSEDVEYESADPKKQLRGSTSTKRSRAAEVHNLSERRRRDRINEKMKALQELIPRCNKTDKASMLDEAIEYLKTLQLQVQMMSMGCGMMPMMFPGVQQYLPPPMGMGMGMGMEMGMNRPMMQFHNLLAGSNL**K**MQAGATAAAHLGPRFPLPPFAMPPVPGNDPSRAQAMNNQPDPMANSVGTQNTTPPSVLGFPDSYQQFLSSTQMQFHMTQALQNQHPVQLNTSRPCTSRGPENRDNHQSG\r\n\r\n"
] | "2022-11-16T10:52:49Z" | "2022-11-17T08:37:53Z" | null | NONE | null | We have built a mutant library for cucumber and have completed resequencing some of the individual plants, finding a number of proteins with non-synonymous mutations. We have translated these proteins and hope to predict the protein structure using AlphaFold2. These proteins have minimal differences from the original protein sequences, is it possible to reduce the MSA time to speed up the protein structure prediction? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/631/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/631/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/630 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/630/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/630/comments | https://api.github.com/repos/deepmind/alphafold/issues/630/events | https://github.com/deepmind/alphafold/pull/630 | 1,451,031,795 | PR_kwDOFoWQLM5C_n8g | 630 | fix: jax moved tree_util. fix deprecated warnning | {
"avatar_url": "https://avatars.githubusercontent.com/u/29097382?v=4",
"events_url": "https://api.github.com/users/jinmingyi1998/events{/privacy}",
"followers_url": "https://api.github.com/users/jinmingyi1998/followers",
"following_url": "https://api.github.com/users/jinmingyi1998/following{/other_user}",
"gists_url": "https://api.github.com/users/jinmingyi1998/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/jinmingyi1998",
"id": 29097382,
"login": "jinmingyi1998",
"node_id": "MDQ6VXNlcjI5MDk3Mzgy",
"organizations_url": "https://api.github.com/users/jinmingyi1998/orgs",
"received_events_url": "https://api.github.com/users/jinmingyi1998/received_events",
"repos_url": "https://api.github.com/users/jinmingyi1998/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/jinmingyi1998/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/jinmingyi1998/subscriptions",
"type": "User",
"url": "https://api.github.com/users/jinmingyi1998"
} | [] | closed | false | null | [] | null | [
"start from jax - 0.3.15",
"Thanks, fixed in v2.3.0.",
"\r\n`\r\n File \"<stdin>\", line 1, in <module>\r\nAttributeError: module 'jax.tree_util' has no attribute 'tree_multimap'\r\n`\r\n\r\nI dont think it is fixed?\r\n\r\nUsing \r\n`jax==0.3.25\r\njaxlib==0.3.25+cuda11.cudnn82`",
"> `File \"<stdin>\", line 1, in <module> AttributeError: module 'jax.tree_util' has no attribute 'tree_multimap'`\r\n\r\nthis pr is not for your error.\r\n\r\nI think you need to pull the latest code. there is not \"tree_multimap\" in the code"
] | "2022-11-16T07:28:04Z" | "2022-12-20T02:11:39Z" | "2022-12-13T11:55:55Z" | NONE | null | https://github.com/google/jax/blob/c1192f383750d5d64a69c19914b7481239b9a327/jax/__init__.py#L120
jax remove `jax.tree_xxx` . use `jax.tree_util.tree_xxx` instead | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/630/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/630/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/630.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/630",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/630.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/630"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/629 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/629/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/629/comments | https://api.github.com/repos/deepmind/alphafold/issues/629/events | https://github.com/deepmind/alphafold/issues/629 | 1,450,676,329 | I_kwDOFoWQLM5Wd5Bp | 629 | Pickle output result_model_1_multimer_v2_pred_0.pkl has a JAX dependency | {
"avatar_url": "https://avatars.githubusercontent.com/u/8163725?v=4",
"events_url": "https://api.github.com/users/tomgoddard/events{/privacy}",
"followers_url": "https://api.github.com/users/tomgoddard/followers",
"following_url": "https://api.github.com/users/tomgoddard/following{/other_user}",
"gists_url": "https://api.github.com/users/tomgoddard/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/tomgoddard",
"id": 8163725,
"login": "tomgoddard",
"node_id": "MDQ6VXNlcjgxNjM3MjU=",
"organizations_url": "https://api.github.com/users/tomgoddard/orgs",
"received_events_url": "https://api.github.com/users/tomgoddard/received_events",
"repos_url": "https://api.github.com/users/tomgoddard/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/tomgoddard/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/tomgoddard/subscriptions",
"type": "User",
"url": "https://api.github.com/users/tomgoddard"
} | [] | open | false | null | [] | null | [
"[result_model_1_multimer_v2_pred_0.zip](https://github.com/deepmind/alphafold/files/10017395/result_model_1_multimer_v2_pred_0.zip)\r\n",
"you can fix this by installing jax[cpu] into your environment. At least that work with my visualizer",
"Yes installing jax and jaxlib can work. But this effects thousands of users of ChimeraX and we don't want to ship jax and jaxlib with our application in order to open a file. I'm not sure what jax _Device_Array got pickled but it is not the PAE data that ChimeraX is extracting from the file. I suspect pickling the _Device_Array was an accidental change in AlphaFold. I have hacked around the problem in our upcoming ChimeraX 1.5 release my making a fake jax module and trying again if loading the pickle file fails.\r\n\r\nMy purpose in reporting this is so that the pickle files can be more easily used by other developers, as they were in AlphaFold 2.2.0.",
"Hi, I think what's get pickled is the distogram. I am also not sure, if it \"depickles\" properly. \r\n\r\n`print(metadata['distogram']) ->\r\n{'bin_edges': DeviceArray([ 2.3125 , 2.625 , 2.9375 , 3.25 , 3.5625 ,\r\n 3.875 , 4.1875 , 4.5 , 4.8125 , 5.125 ,\r\n 5.4375 , 5.75 , 6.0625 , 6.375 , 6.6875 ,\r\n 7. , 7.3125 , 7.625 , 7.9375 , 8.25 ,\r\n 8.5625 , 8.875 , 9.1875 , 9.5 , 9.812499,\r\n 10.125 , 10.4375 , 10.75 , 11.0625 , 11.375 ,\r\n 11.6875 , 12. , 12.3125 , 12.625 , 12.9375 ,\r\n 13.25 , 13.5625 , 13.875 , 14.1875 , 14.499999,\r\n 14.8125 , 15.125 , 15.4375 , 15.75 , 16.0625 ,\r\n 16.375 , 16.6875 , 16.999998, 17.312498, 17.625 ,\r\n 17.9375 , 18.25 , 18.5625 , 18.875 , 19.1875 ,\r\n 19.5 , 19.8125 , 20.125 , 20.437498, 20.75 ,\r\n 21.0625 , 21.375 , 21.6875 ], dtype=float32), 'logits': DeviceArray([[[ 7.99458694e+01, -1.86893845e+01, -2.46103077e+01, ...,\r\n -1.09830141e+00, 1.10635986e+01, 2.55225778e+00],\r\n [ 5.37824154e+00, 1.02621212e+01, -1.24734116e+00, ...,\r\n -8.33254433e+00, -6.89190483e+00, 7.95750475e+00],\r\n [ 1.54681420e+00, 3.07494116e+00, 3.35776448e-01, ...,\r\n -9.31102562e+00, -6.93648672e+00, 5.46216071e-01],\r\n ...,\r\n [-1.10818462e+01, -9.99354744e+00, -1.05438919e+01, ...,\r\n -1.65992379e+00, -2.05721903e+00, 4.57334995e+00],\r\n [-1.16069813e+01, -1.08642635e+01, -1.08378592e+01, ...,`\r\n\r\nPAE is unaffected "
] | "2022-11-16T01:45:24Z" | "2022-11-29T15:47:31Z" | null | NONE | null | In AlphaFold 2.2.4 the pickled per-structure output files from a multimer prediction such as
result_model_1_multimer_v2_pred_0.pkl
now contains a JAX dependency (apparently a jax DeviceArray structure was pickled). This prevents the .pkl data from being read by Python interpreters that do not have jax installed. This prevents accessing PAE data in the .pkl file when the file is moved to another machine for analysis where the Python does not have jax and jaxlib installed. This significantly reduces the portability of the files and appears like it was not intended. AlphaFold 2.2.0 multimer .pkl output does not have the JAX dependency.
Attempting to load the .pkl data without jax gives the following jax ModuleNotFoundError.
```
$ python3
Python 3.7.5 (default, Nov 26 2019, 14:12:06)
[Clang 11.0.0 (clang-1100.0.33.12)] on darwin
Type "help", "copyright", "credits" or "license" for more information.
>>> import pickle
>>> with open('result_model_1_multimer_v2_pred_0.pkl', 'rb') as f:
... d = pickle.load(f)
...
Traceback (most recent call last):
File "<stdin>", line 2, in <module>
ModuleNotFoundError: No module named 'jax'
```
I suspect that adding a jax DeviceArray to the .pkl output was probably an accident added in AlphaFold 2.2.4 (or possibly in 2.2.3, 2.2.2 or 2.2.1). It would help analysis of the AlphaFold predictions to remove this jax data from the .pkl file.
I develop the ChimeraX molecular visualization which supports AlphaFold PAE error analysis which was broken by the added jax dependency for reading the .pkl files.
I'll attach an example "result_model_1_multimer_v2_pred_0.pkl" from AlphaFold 2.2.4 that exhibits the problem.
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/629/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/629/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/628 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/628/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/628/comments | https://api.github.com/repos/deepmind/alphafold/issues/628/events | https://github.com/deepmind/alphafold/pull/628 | 1,447,548,434 | PR_kwDOFoWQLM5Cz7_T | 628 | fix(sec): upgrade tensorflow-cpu to 2.9.1 | {
"avatar_url": "https://avatars.githubusercontent.com/u/101684156?v=4",
"events_url": "https://api.github.com/users/chncaption/events{/privacy}",
"followers_url": "https://api.github.com/users/chncaption/followers",
"following_url": "https://api.github.com/users/chncaption/following{/other_user}",
"gists_url": "https://api.github.com/users/chncaption/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/chncaption",
"id": 101684156,
"login": "chncaption",
"node_id": "U_kgDOBg-TvA",
"organizations_url": "https://api.github.com/users/chncaption/orgs",
"received_events_url": "https://api.github.com/users/chncaption/received_events",
"repos_url": "https://api.github.com/users/chncaption/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/chncaption/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/chncaption/subscriptions",
"type": "User",
"url": "https://api.github.com/users/chncaption"
} | [] | open | false | null | [] | null | [] | "2022-11-14T07:41:28Z" | "2022-11-14T07:41:28Z" | null | NONE | null | ### What happened?
There are 1 security vulnerabilities found in tensorflow-cpu 2.9.0
- [CVE-2022-36000](https://www.oscs1024.com/hd/CVE-2022-36000)
### What did I do?
Upgrade tensorflow-cpu from 2.9.0 to 2.9.1 for vulnerability fix
### What did you expect to happen?
Ideally, no insecure libs should be used.
### The specification of the pull request
[PR Specification](https://www.oscs1024.com/docs/pr-specification/) from OSCS | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/628/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/628/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/628.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/628",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/628.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/628"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/627 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/627/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/627/comments | https://api.github.com/repos/deepmind/alphafold/issues/627/events | https://github.com/deepmind/alphafold/issues/627 | 1,445,004,258 | I_kwDOFoWQLM5WIQPi | 627 | An error occurred while downloading the database | {
"avatar_url": "https://avatars.githubusercontent.com/u/92859882?v=4",
"events_url": "https://api.github.com/users/Caiguoyu/events{/privacy}",
"followers_url": "https://api.github.com/users/Caiguoyu/followers",
"following_url": "https://api.github.com/users/Caiguoyu/following{/other_user}",
"gists_url": "https://api.github.com/users/Caiguoyu/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Caiguoyu",
"id": 92859882,
"login": "Caiguoyu",
"node_id": "U_kgDOBYjt6g",
"organizations_url": "https://api.github.com/users/Caiguoyu/orgs",
"received_events_url": "https://api.github.com/users/Caiguoyu/received_events",
"repos_url": "https://api.github.com/users/Caiguoyu/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Caiguoyu/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Caiguoyu/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Caiguoyu"
} | [] | open | false | null | [] | null | [] | "2022-11-11T06:36:08Z" | "2022-11-11T06:36:08Z" | null | NONE | null | When I download the database, I get the following error: download_db.sh:行 82: 2358338 段错误 (核心已转储) wget -P "$pdb70" "http://wwwuser.gwdg.de/~compbiol/data/hhsuite/databases/hhsuite_dbs/old-releases/${pdb70_filename}" , How do I fix this error? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/627/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/627/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/626 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/626/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/626/comments | https://api.github.com/repos/deepmind/alphafold/issues/626/events | https://github.com/deepmind/alphafold/issues/626 | 1,443,987,462 | I_kwDOFoWQLM5WEYAG | 626 | Erros while predicting the structures | {
"avatar_url": "https://avatars.githubusercontent.com/u/112063013?v=4",
"events_url": "https://api.github.com/users/jeid94/events{/privacy}",
"followers_url": "https://api.github.com/users/jeid94/followers",
"following_url": "https://api.github.com/users/jeid94/following{/other_user}",
"gists_url": "https://api.github.com/users/jeid94/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/jeid94",
"id": 112063013,
"login": "jeid94",
"node_id": "U_kgDOBq3yJQ",
"organizations_url": "https://api.github.com/users/jeid94/orgs",
"received_events_url": "https://api.github.com/users/jeid94/received_events",
"repos_url": "https://api.github.com/users/jeid94/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/jeid94/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/jeid94/subscriptions",
"type": "User",
"url": "https://api.github.com/users/jeid94"
} | [] | closed | false | null | [] | null | [
"Hi! This looks like its in ColabFold so I would suggest raising a question there. It does look like you need to rerun the earlier cells to define cf_af. Thanks!",
"Thank you so much for your help\r\n"
] | "2022-11-10T14:18:29Z" | "2022-11-10T14:54:44Z" | "2022-11-10T14:26:02Z" | NONE | null | Dear alphafold developers,
I faced some errors while trying to predict protein structures. Finally, I obtained a predicted structure so alphafold ran normally. I am just wondering if these errors will have a major effect on the prediction or not?
Thank you so much for your help,
Best regrards,
Here are the errors :
NameError Traceback (most recent call last)
<ipython-input-3-d068b61051d4> in <module>
40 # --- Search against genetic databases ---
41
---> 42 I = cf_af.prep_msa(I, msa_method, add_custom_msa, msa_format,
43 pair_mode, pair_cov, pair_qid, TMP_DIR=TMP_DIR)
44 mod_I = I
NameError: name 'cf_af' is not defined
NameError Traceback (most recent call last)
[<ipython-input-4-bdca699a292d>](https://localhost:8080/#) in <module>
11 #@markdown - `qid` minimum sequence identity with query (%)
12
---> 13 mod_I = cf_af.prep_filter(I, trim, trim_inverse, cov, qid)
14
15 if I["msas"] != mod_I["msas"]:
NameError: name 'cf_af' is not defined
NameError Traceback (most recent call last)
[<ipython-input-2-f9c7eceac359>](https://localhost:8080/#) in <module>
16 #@markdown - For example, **sequence:**`ABC:DEF`, **homooligomer:** `2:1`, the first protein `ABC` will be modeled as a homodimer (2 copies) and second `DEF` a monomer (1 copy).
17
---> 18 I = cf_af.prep_inputs(sequence, jobname, homooligomer, clean=IN_COLAB)
NameError: name 'cf_af' is not defined
NameError Traceback (most recent call last)
[<ipython-input-5-74c943a59072>](https://localhost:8080/#) in <module>
38
39 # prep input features
---> 40 feature_dict = cf_af.prep_feats(mod_I, clean=IN_COLAB)
41 Ls_plot = feature_dict["Ls"]
42
NameError: name 'cf_af' is not defined | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/626/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/626/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/625 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/625/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/625/comments | https://api.github.com/repos/deepmind/alphafold/issues/625/events | https://github.com/deepmind/alphafold/issues/625 | 1,443,096,059 | I_kwDOFoWQLM5WA-X7 | 625 | `template_domain_names` missing in features.pkl on multimer mode. | {
"avatar_url": "https://avatars.githubusercontent.com/u/21062531?v=4",
"events_url": "https://api.github.com/users/eunos-1128/events{/privacy}",
"followers_url": "https://api.github.com/users/eunos-1128/followers",
"following_url": "https://api.github.com/users/eunos-1128/following{/other_user}",
"gists_url": "https://api.github.com/users/eunos-1128/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/eunos-1128",
"id": 21062531,
"login": "eunos-1128",
"node_id": "MDQ6VXNlcjIxMDYyNTMx",
"organizations_url": "https://api.github.com/users/eunos-1128/orgs",
"received_events_url": "https://api.github.com/users/eunos-1128/received_events",
"repos_url": "https://api.github.com/users/eunos-1128/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/eunos-1128/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/eunos-1128/subscriptions",
"type": "User",
"url": "https://api.github.com/users/eunos-1128"
} | [] | open | false | null | [] | null | [] | "2022-11-10T01:52:18Z" | "2022-12-06T12:06:12Z" | null | NONE | null | I tried to check out `features.pkl` generated by running AlphaFold to find which templates are used.
When `--model_preset=monomer` is used I could find `template_domain_names` as a key name of the dictionary unpickled, but not upon `--model_preset=multimer`.
```python
import pickle
with open("../features.pkl", "rb") as f:
features = pickle.load(f)
print('template_domain_name' in features) # => False
```
```python
features.keys()
# => `template_domain_names` not found
# dict_keys(['aatype', 'residue_index', 'seq_length', 'msa', 'num_alignments', 'template_aatype', 'template_all_atom_mask', 'template_all_atom_positions', 'asym_id', 'sym_id', 'entity_id', 'deletion_matrix', 'deletion_mean', 'all_atom_mask', 'all_atom_positions', 'assembly_num_chains', 'entity_mask', 'num_templates', 'cluster_bias_mask', 'bert_mask', 'seq_mask', 'msa_mask'])
```
When running with multimer mode, templates are not used for prediction models? (I could see things like template names in docker log file, though.)
Otherwise, running on multimer mode generates no data related to templates used for prediction while it makes use of templates' information?
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/625/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/625/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/624 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/624/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/624/comments | https://api.github.com/repos/deepmind/alphafold/issues/624/events | https://github.com/deepmind/alphafold/issues/624 | 1,442,158,157 | I_kwDOFoWQLM5V9ZZN | 624 | AlphaFold multimer mode crashes with lower case protein sequences but not upper case protein sequences | {
"avatar_url": "https://avatars.githubusercontent.com/u/683829?v=4",
"events_url": "https://api.github.com/users/doricke/events{/privacy}",
"followers_url": "https://api.github.com/users/doricke/followers",
"following_url": "https://api.github.com/users/doricke/following{/other_user}",
"gists_url": "https://api.github.com/users/doricke/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/doricke",
"id": 683829,
"login": "doricke",
"node_id": "MDQ6VXNlcjY4MzgyOQ==",
"organizations_url": "https://api.github.com/users/doricke/orgs",
"received_events_url": "https://api.github.com/users/doricke/received_events",
"repos_url": "https://api.github.com/users/doricke/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/doricke/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/doricke/subscriptions",
"type": "User",
"url": "https://api.github.com/users/doricke"
} | [] | closed | false | null | [] | null | [
"This is by design, the sequences should be in all-uppercase letters, although I agree that AlphaFold should fail earlier and with a nicer error message."
] | "2022-11-09T14:00:18Z" | "2022-12-13T11:58:10Z" | "2022-12-13T11:58:10Z" | NONE | null | In multimer mode, the following test sequence runs without error:
```
>HBA_HUMAN_2a Hemoglobin subunit alpha OS=Homo sapiens OX=9606 GN=HBA2 PE=1 SV=2
MALSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHFDLS
HGSAQVKGHGKKVADALTNAVAHVDDMPNALSALSDLHAHKLRVDPVNFK
LLSHCLLVTLAAHLPAEFTPAVHASLDKFLASVSTVLTSKYR
>HBB_HUMAN_2a Hemoglobin subunit beta OS=Homo sapiens OX=9606 GN=HBB PE=1 SV=2
MAHLTPEEKSAVTALWGKVNVDEVGGEALGRLLVVYPWTQRFFESFGDLS
TPDAVMGNPKVKAHGKKVLGAFSDGLAHLDNLKGTFATLSELHCDKLHVD
PENFRLLGNVLVCVLAHHFGKEFTPPVQAAYQKVVAGVANALAHKYH
>HBA_HUMAN_2b Hemoglobin subunit alpha OS=Homo sapiens OX=9606 GN=HBA2 PE=1 SV=2
MALSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHFDLS
HGSAQVKGHGKKVADALTNAVAHVDDMPNALSALSDLHAHKLRVDPVNFK
LLSHCLLVTLAAHLPAEFTPAVHASLDKFLASVSTVLTSKYR
>HBB_HUMAN_2b Hemoglobin subunit beta OS=Homo sapiens OX=9606 GN=HBB PE=1 SV=2
MAHLTPEEKSAVTALWGKVNVDEVGGEALGRLLVVYPWTQRFFESFGDLS
TPDAVMGNPKVKAHGKKVLGAFSDGLAHLDNLKGTFATLSELHCDKLHVD
PENFRLLGNVLVCVLAHHFGKEFTPPVQAAYQKVVAGVANALAHKYH
```
The same sequence, but just with lower case amino acids, causes AlphaFold to crash:
```
>HBA_HUMAN_2a Hemoglobin subunit alpha OS=Homo sapiens OX=9606 GN=HBA2 PE=1 SV=2
malspadktnvkaawgkvgahageygaealermflsfpttktyfphfdls
hgsaqvkghgkkvadaltnavahvddmpnalsalsdlhahklrvdpvnfk
llshcllvtlaahlpaeftpavhasldkflasvstvltskyr
>HBB_HUMAN_2a Hemoglobin subunit beta OS=Homo sapiens OX=9606 GN=HBB PE=1 SV=2
mahltpeeksavtalwgkvnvdevggealgrllvvypwtqrffesfgdls
tpdavmgnpkvkahgkkvlgafsdglahldnlkgtfatlselhcdklhvd
penfrllgnvlvcvlahhfgkeftppvqaayqkvvagvanalahkyh
>HBA_HUMAN_2b Hemoglobin subunit alpha OS=Homo sapiens OX=9606 GN=HBA2 PE=1 SV=2
malspadktnvkaawgkvgahageygaealermflsfpttktyfphfdls
hgsaqvkghgkkvadaltnavahvddmpnalsalsdlhahklrvdpvnfk
llshcllvtlaahlpaeftpavhasldkflasvstvltskyr
>HBB_HUMAN_2b Hemoglobin subunit beta OS=Homo sapiens OX=9606 GN=HBB PE=1 SV=2
mahltpeeksavtalwgkvnvdevggealgrllvvypwtqrffesfgdls
tpdavmgnpkvkahgkkvlgafsdglahldnlkgtfatlselhcdklhvd
penfrllgnvlvcvlahhfgkeftppvqaayqkvvagvanalahkyh
```
stderr:
```
I1108 11:39:45.759545 140705398323008 utils.py:40] Finished hmmbuild query in 0.248 seconds
I1108 11:39:45.767869 140705398323008 hmmsearch.py:103] Launching sub-process ['/usr/bin/hmmsearch', '--noali', '--cpu', '8', '--F1', '0.1', '--F2', '0.1', '--F3', '0.1', '--incE', '100', '-E', '100', '--domE', '100', '--incdomE', '100', '-A', '/mnt/alphafold/tmp/tmpug569gkx/output.sto', '/mnt/alphafold/tmp/tmpug569gkx/query.hmm', '/db/pdb_seqres/pdb_seqres.txt']
I1108 11:39:45.830403 140705398323008 utils.py:36] Started hmmsearch (pdb_seqres.txt) query
I1108 11:39:51.148423 140705398323008 utils.py:40] Finished hmmsearch (pdb_seqres.txt) query in 5.318 seconds
I1108 11:39:51.492158 140705398323008 jackhmmer.py:133] Launching subprocess "/usr/bin/jackhmmer -o /dev/null -A /mnt/alphafold/tmp/tmpwtlriesd/output.sto --noali --F1 0.0005 --F2 5e-05 --F3 5e-07 --incE 0.0001 -E 0.0001 --cpu 8 -N 1 /tmp/tmpuj71bpee.fasta /db/small_bfd/bfd-first_non_consensus_sequences.fasta"
I1108 11:39:51.550291 140705398323008 utils.py:36] Started Jackhmmer (bfd-first_non_consensus_sequences.fasta) query
I1108 11:43:15.578521 140705398323008 utils.py:40] Finished Jackhmmer (bfd-first_non_consensus_sequences.fasta) query in 204.028 seconds
I1108 11:43:15.585719 140705398323008 templates.py:940] Searching for template for: malspadktnvkaawgkvgahageygaealermflsfpttktyfphfdlshgsaqvkghgkkvadaltnavahvddmpnalsalsdlhahklrvdpvnfkllshcllvtlaahlpaeftpavhasldkflasvstvltskyr
Traceback (most recent call last):
File "/app/alphafold/run_alphafold.py", line 422, in <module>
app.run(main)
File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 312, in run
_run_main(main, args)
File "/opt/conda/lib/python3.7/site-packages/absl/app.py", line 258, in _run_main
sys.exit(main(argv))
File "/app/alphafold/run_alphafold.py", line 406, in main
random_seed=random_seed)
File "/app/alphafold/run_alphafold.py", line 174, in predict_structure
msa_output_dir=msa_output_dir)
File "/app/alphafold/alphafold/data/pipeline_multimer.py", line 269, in process
is_homomer_or_monomer=is_homomer_or_monomer)
File "/app/alphafold/alphafold/data/pipeline_multimer.py", line 214, in _process_single_chain
msa_output_dir=chain_msa_output_dir)
File "/app/alphafold/alphafold/data/pipeline.py", line 225, in process
hits=pdb_template_hits)
File "/app/alphafold/alphafold/data/templates.py", line 968, in get_templates
kalign_binary_path=self._kalign_binary_path)
File "/app/alphafold/alphafold/data/templates.py", line 727, in _process_single_hit
query_sequence)
File "/app/alphafold/alphafold/data/templates.py", line 657, in _build_query_to_hit_index_mapping
min_idx = min(x for x in indices_query if x > -1)
ValueError: min() arg is an empty sequence
``` | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/624/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/624/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/623 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/623/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/623/comments | https://api.github.com/repos/deepmind/alphafold/issues/623/events | https://github.com/deepmind/alphafold/issues/623 | 1,435,259,241 | I_kwDOFoWQLM5VjFFp | 623 | Illegal character in pub_seqres.txt database file | {
"avatar_url": "https://avatars.githubusercontent.com/u/683829?v=4",
"events_url": "https://api.github.com/users/doricke/events{/privacy}",
"followers_url": "https://api.github.com/users/doricke/followers",
"following_url": "https://api.github.com/users/doricke/following{/other_user}",
"gists_url": "https://api.github.com/users/doricke/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/doricke",
"id": 683829,
"login": "doricke",
"node_id": "MDQ6VXNlcjY4MzgyOQ==",
"organizations_url": "https://api.github.com/users/doricke/orgs",
"received_events_url": "https://api.github.com/users/doricke/received_events",
"repos_url": "https://api.github.com/users/doricke/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/doricke/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/doricke/subscriptions",
"type": "User",
"url": "https://api.github.com/users/doricke"
} | [
{
"color": "E99695",
"default": false,
"description": "Something isn't working",
"id": 3094826889,
"name": "error report",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODg5",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/error%20report"
},
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"I also am getting a similar error during the hmmbuild query step :\r\n\r\n```\r\nI1110 08:13:51.837537 140292054591296 run_docker.py:255] Parse failed (sequence file /mnt/pdb_seqres_database_path/pdb_seqres.txt):\r\nI1110 08:13:51.837775 140292054591296 run_docker.py:255] Line 1366492: illegal character 0\r\n```\r\nLine 1366491-1366492 of the pdb_seqres.txt looks like this:\r\n\r\n```\r\n>7ooo_B mol:na length:11 DNA (5'-D(*CP*TP*(RWQ)P*TP*CP*TP*TP*TP*G)-3')\r\nCT05ATCTTTG\r\n```\r\n\r\n",
"You can fix this by filtering out the bad sequences from `pdb_seqres.txt`. We will submit a fix to the download script for this.\r\n\r\n\r\nTo fix for now, run the following 2 commands in the directory with `pdb_seqres.txt`:\r\n\r\n```sh\r\ngrep --after-context=1 --no-group-separator '>.* mol:protein' \"pdb_seqres.txt\" > \"pdb_seqres_filtered.txt\"\r\nmv \"pdb_seqres_filtered.txt\" \"pdb_seqres.txt\"\r\n```",
"Thank you. This worked for me. I am now able to run AF2 with `--model_preset=multimer`",
"Thanks, we fixed this in the pdb_seqres download script in v2.3.0."
] | "2022-11-03T21:07:43Z" | "2022-12-13T11:56:25Z" | "2022-12-13T11:56:24Z" | NONE | null | Parse failed (sequence file /db/pdb_seqres/pdb_seqres.txt):
Line 1366022: illegal character 0
Errors in file:
>7ooo_B mol:na length:11 DNA (5'-D(*CP*TP*(RWQ)P*TP*CP*TP*TP*TP*G)-3')
CT05ATCTTTG
>7ooo_E mol:na length:11 DNA (5'-D(*CP*TP*(RWQ)P*TP*CP*TP*TP*TP*G)-3')
CT05ATCTTTG
>7ozz_B mol:na length:11 DNA (5'-D(*CP*TP*(RWR)P*TP*CP*TP*TP*TP*G)-3')
CT05HTCTTTG
This is likely a PDB parsing error. For 7ozz, the 05H is likely coming from the HETATM entries:
ATOM 251 O4 DT B 12 -29.103 0.246 5.217 1.00 94.87 O
ATOM 252 C5 DT B 12 -28.572 1.806 6.922 1.00100.00 C
ATOM 253 C7 DT B 12 -29.343 2.919 6.285 1.00 97.49 C
ATOM 254 C6 DT B 12 -27.896 1.952 8.066 1.00102.37 C
HETATM 255 C1 05H B 13 -19.291 0.434 4.896 1.00100.82 C
HETATM 256 C1' 05H B 13 -20.815 -2.819 1.936 1.00 98.67 C
HETATM 257 C11 05H B 13 -25.832 1.973 4.789 1.00 82.44 C
HETATM 258 C2 05H B 13 -22.765 -2.247 0.431 1.00 92.33 C
HETATM 259 C2' 05H B 13 -19.788 -3.180 0.872 1.00 98.03 C
HETATM 260 C21 05H B 13 -23.920 -1.793 5.039 1.00 89.93 C
HETATM 261 C3 05H B 13 -20.565 -0.673 8.783 1.00 98.27 C
HETATM 262 C3' 05H B 13 -18.651 -2.241 1.179 1.00 97.27 C
HETATM 263 C4 05H B 13 -23.763 -0.151 0.487 1.00 89.17 C
HETATM 264 C4' 05H B 13 -18.700 -2.056 2.670 1.00 96.22 C
HETATM 265 C41 05H B 13 -25.360 -0.401 3.891 1.00 84.99 C
HETATM 266 C5 05H B 13 -22.810 0.257 1.532 1.00 92.64 C
HETATM 267 C5' 05H B 13 -18.184 -0.672 2.999 1.00 90.23 C
HETATM 268 C51 05H B 13 -25.154 0.642 4.907 1.00 85.30 C
HETATM 269 C6 05H B 13 -21.878 -0.660 1.967 1.00 90.50 C
HETATM 270 C61 05H B 13 -24.340 0.369 5.978 1.00 96.29 C
HETATM 271 C7 05H B 13 -22.853 1.639 2.117 1.00 86.19 C
HETATM 272 C71 05H B 13 -19.481 0.396 6.382 1.00107.13 C
HETATM 273 N1 05H B 13 -21.833 -1.885 1.435 1.00 88.64 N
HETATM 274 N11 05H B 13 -23.699 -0.813 6.045 1.00 93.76 N
HETATM 275 N3 05H B 13 -23.683 -1.382 -0.001 1.00 91.20 N
HETATM 276 N31 05H B 13 -24.720 -1.558 4.007 1.00 87.08 N
HETATM 277 N5' 05H B 13 -18.512 -0.505 4.395 1.00 99.47 N
HETATM 278 O2 05H B 13 -22.767 -3.380 -0.090 1.00 94.75 O
HETATM 279 O2' 05H B 13 -20.614 -1.724 7.810 1.00107.20 O
HETATM 280 O21 05H B 13 -23.387 -2.920 5.061 1.00102.39 O
HETATM 281 O3 05H B 13 -19.840 1.281 4.218 1.00 97.98 O
HETATM 282 O3' 05H B 13 -17.346 -2.673 0.724 1.00106.63 O
HETATM 283 O4 05H B 13 -24.624 0.643 0.067 1.00 90.61 O
HETATM 284 O4' 05H B 13 -20.087 -2.179 2.993 1.00108.67 O
HETATM 285 O41 05H B 13 -26.111 -0.183 2.926 1.00 91.45 O
HETATM 286 O5' 05H B 13 -21.611 2.197 9.740 1.00102.52 O
HETATM 287 OP1 05H B 13 -21.601 4.692 10.303 1.00137.19 O
HETATM 288 OP2 05H B 13 -22.877 3.934 8.214 1.00 90.96 O
HETATM 289 P 05H B 13 -22.373 3.591 9.699 1.00130.32 P
HETATM 290 C1'1 05H B 13 -22.792 -1.087 7.175 1.00 93.73 C
HETATM 291 C2'1 05H B 13 -21.353 -1.242 6.698 1.00105.90 C
HETATM 292 C3'1 05H B 13 -20.955 0.194 6.627 1.00110.52 C
HETATM 293 C4'1 05H B 13 -21.317 0.436 8.057 1.00108.99 C
HETATM 294 C5'1 05H B 13 -21.020 1.859 8.518 1.00104.57 C
HETATM 295 O4'1 05H B 13 -22.699 0.045 8.059 1.00110.59 O
ATOM 296 P DT B 14 -16.630 -1.995 -0.578 1.00 97.66 P
ATOM 297 OP1 DT B 14 -15.200 -2.365 -0.526 1.00104.42 O
ATOM 298 OP2 DT B 14 -17.060 -0.575 -0.633 1.00 98.23 O | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/623/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/623/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/622 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/622/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/622/comments | https://api.github.com/repos/deepmind/alphafold/issues/622/events | https://github.com/deepmind/alphafold/issues/622 | 1,433,377,670 | I_kwDOFoWQLM5Vb5uG | 622 | Locally install and run AlphaFold | {
"avatar_url": "https://avatars.githubusercontent.com/u/95124914?v=4",
"events_url": "https://api.github.com/users/galshuler/events{/privacy}",
"followers_url": "https://api.github.com/users/galshuler/followers",
"following_url": "https://api.github.com/users/galshuler/following{/other_user}",
"gists_url": "https://api.github.com/users/galshuler/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/galshuler",
"id": 95124914,
"login": "galshuler",
"node_id": "U_kgDOBat9sg",
"organizations_url": "https://api.github.com/users/galshuler/orgs",
"received_events_url": "https://api.github.com/users/galshuler/received_events",
"repos_url": "https://api.github.com/users/galshuler/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/galshuler/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/galshuler/subscriptions",
"type": "User",
"url": "https://api.github.com/users/galshuler"
} | [] | open | false | null | [] | null | [
"Based on those specs, you can probably run AlphaFold with reduced databases on your local computer, though if you do not have a dedicated graphics card it will probably be very slow.",
"Assuming you have a dedicated graphics card and do not need high speed results, you could also run with full databases on the HDD"
] | "2022-11-02T15:44:41Z" | "2022-11-02T16:31:25Z" | null | NONE | null | Hi,
It is possible to install and run Alpha Fold on my local computer (not on VM or cloud) with the given specification:
I9-12900 CPU
128gb ddr4-3200
Asus Prime Z690P 2tb NVME + 8TB HDD
Regards,
Gal.
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/622/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/622/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/621 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/621/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/621/comments | https://api.github.com/repos/deepmind/alphafold/issues/621/events | https://github.com/deepmind/alphafold/issues/621 | 1,432,256,746 | I_kwDOFoWQLM5VXoDq | 621 | compare the predicted and actual structure? | {
"avatar_url": "https://avatars.githubusercontent.com/u/105665325?v=4",
"events_url": "https://api.github.com/users/mukulem/events{/privacy}",
"followers_url": "https://api.github.com/users/mukulem/followers",
"following_url": "https://api.github.com/users/mukulem/following{/other_user}",
"gists_url": "https://api.github.com/users/mukulem/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/mukulem",
"id": 105665325,
"login": "mukulem",
"node_id": "U_kgDOBkxTLQ",
"organizations_url": "https://api.github.com/users/mukulem/orgs",
"received_events_url": "https://api.github.com/users/mukulem/received_events",
"repos_url": "https://api.github.com/users/mukulem/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/mukulem/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/mukulem/subscriptions",
"type": "User",
"url": "https://api.github.com/users/mukulem"
} | [] | open | false | null | [] | null | [] | "2022-11-01T23:52:29Z" | "2022-11-01T23:52:29Z" | null | NONE | null | Hi, I was wondering which python library did you use to compare the predicted and actual structure.
<img width="692" alt="image" src="https://user-images.githubusercontent.com/105665325/199346204-4843da4c-808a-4231-aed9-78cd723b25b3.png">
something similar to this.
Thanks | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/621/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/621/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/620 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/620/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/620/comments | https://api.github.com/repos/deepmind/alphafold/issues/620/events | https://github.com/deepmind/alphafold/issues/620 | 1,431,033,291 | I_kwDOFoWQLM5VS9XL | 620 | docker build -f docker/Dockerfile -t alphafold . Build failed | {
"avatar_url": "https://avatars.githubusercontent.com/u/49931482?v=4",
"events_url": "https://api.github.com/users/hezhiqiang8909/events{/privacy}",
"followers_url": "https://api.github.com/users/hezhiqiang8909/followers",
"following_url": "https://api.github.com/users/hezhiqiang8909/following{/other_user}",
"gists_url": "https://api.github.com/users/hezhiqiang8909/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/hezhiqiang8909",
"id": 49931482,
"login": "hezhiqiang8909",
"node_id": "MDQ6VXNlcjQ5OTMxNDgy",
"organizations_url": "https://api.github.com/users/hezhiqiang8909/orgs",
"received_events_url": "https://api.github.com/users/hezhiqiang8909/received_events",
"repos_url": "https://api.github.com/users/hezhiqiang8909/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/hezhiqiang8909/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/hezhiqiang8909/subscriptions",
"type": "User",
"url": "https://api.github.com/users/hezhiqiang8909"
} | [] | open | false | null | [] | null | [
"Hi thanks for raising this. Can you please provide more details e.g. about your machine, docker version etc?",
"> 嗨,谢谢你提出这个问题。您能否提供更多详细信息,例如关于您的机器、docker 版本等?\r\n\r\n\r\n\r\n",
"> 您好,谢谢您提出这个问题。您可以提供更多详细信息,例如关于的机器、docker 版本等?\r\n\r\n\r\n",
"> 您好,谢谢您提出这个问题。您可以提供更多详细信息,例如关于的机器、docker 版本等?\r\n\r\nThis is an Alibaba Cloud, Virginia public cloud machine",
"> 嗨,谢谢你提出这个问题。您能否提供更多详细信息,例如关于您的机器、docker 版本等?\r\n\r\nI'm very sorry, I already know the reason for this error. It is caused by compiling on a machine without a GPU driver. To use the Dockerfile in the docker directory to create a docker image, a machine with a GPU driver needs to be installed."
] | "2022-11-01T08:01:23Z" | "2022-11-03T10:08:59Z" | null | NONE | null | 
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/620/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/620/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/619 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/619/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/619/comments | https://api.github.com/repos/deepmind/alphafold/issues/619/events | https://github.com/deepmind/alphafold/issues/619 | 1,427,701,000 | I_kwDOFoWQLM5VGP0I | 619 | 1 | {
"avatar_url": "https://avatars.githubusercontent.com/u/97654261?v=4",
"events_url": "https://api.github.com/users/Pleyades12/events{/privacy}",
"followers_url": "https://api.github.com/users/Pleyades12/followers",
"following_url": "https://api.github.com/users/Pleyades12/following{/other_user}",
"gists_url": "https://api.github.com/users/Pleyades12/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Pleyades12",
"id": 97654261,
"login": "Pleyades12",
"node_id": "U_kgDOBdIV9Q",
"organizations_url": "https://api.github.com/users/Pleyades12/orgs",
"received_events_url": "https://api.github.com/users/Pleyades12/received_events",
"repos_url": "https://api.github.com/users/Pleyades12/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Pleyades12/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Pleyades12/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Pleyades12"
} | [] | closed | false | null | [] | null | [] | "2022-10-28T19:07:40Z" | "2022-10-31T17:14:30Z" | "2022-10-31T17:14:30Z" | NONE | null | null | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/619/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/619/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/618 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/618/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/618/comments | https://api.github.com/repos/deepmind/alphafold/issues/618/events | https://github.com/deepmind/alphafold/issues/618 | 1,426,800,293 | I_kwDOFoWQLM5VCz6l | 618 | Do I need to upgrade the database? | {
"avatar_url": "https://avatars.githubusercontent.com/u/30679826?v=4",
"events_url": "https://api.github.com/users/lonelymito/events{/privacy}",
"followers_url": "https://api.github.com/users/lonelymito/followers",
"following_url": "https://api.github.com/users/lonelymito/following{/other_user}",
"gists_url": "https://api.github.com/users/lonelymito/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/lonelymito",
"id": 30679826,
"login": "lonelymito",
"node_id": "MDQ6VXNlcjMwNjc5ODI2",
"organizations_url": "https://api.github.com/users/lonelymito/orgs",
"received_events_url": "https://api.github.com/users/lonelymito/received_events",
"repos_url": "https://api.github.com/users/lonelymito/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/lonelymito/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/lonelymito/subscriptions",
"type": "User",
"url": "https://api.github.com/users/lonelymito"
} | [] | open | false | null | [] | null | [
"It is not necessary, that is a different database. Database which AlphaFold uses for inference of a new structure hasn't changed, only database that contains already predicted structures is growing but that one is not used by AlphaFold for future predictions. ",
"I thought, that the --max_template_date corresponds to the deposition date in the pdb. And the local copy of the pdb database is used for compiling MSA, so maybe it would be useful to update this from time to time.\r\n"
] | "2022-10-28T07:13:41Z" | "2022-11-07T13:09:37Z" | null | NONE | null | Considering the more and more protein structures deposited in PDB, do I need to upgrede my AlphaFold database? and how to? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/618/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/618/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/617 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/617/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/617/comments | https://api.github.com/repos/deepmind/alphafold/issues/617/events | https://github.com/deepmind/alphafold/pull/617 | 1,426,261,367 | PR_kwDOFoWQLM5BswTS | 617 | Make database downloadble on macOS | {
"avatar_url": "https://avatars.githubusercontent.com/u/96356?v=4",
"events_url": "https://api.github.com/users/sparanoid/events{/privacy}",
"followers_url": "https://api.github.com/users/sparanoid/followers",
"following_url": "https://api.github.com/users/sparanoid/following{/other_user}",
"gists_url": "https://api.github.com/users/sparanoid/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/sparanoid",
"id": 96356,
"login": "sparanoid",
"node_id": "MDQ6VXNlcjk2MzU2",
"organizations_url": "https://api.github.com/users/sparanoid/orgs",
"received_events_url": "https://api.github.com/users/sparanoid/received_events",
"repos_url": "https://api.github.com/users/sparanoid/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/sparanoid/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/sparanoid/subscriptions",
"type": "User",
"url": "https://api.github.com/users/sparanoid"
} | [] | open | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/617/checks?check_run_id=9151370678) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request."
] | "2022-10-27T21:01:21Z" | "2022-10-30T21:15:38Z" | null | NONE | null | Use `-p` option to make download scripts work both on Linux and macOS. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/617/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/617/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/617.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/617",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/617.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/617"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/616 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/616/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/616/comments | https://api.github.com/repos/deepmind/alphafold/issues/616/events | https://github.com/deepmind/alphafold/issues/616 | 1,424,947,009 | I_kwDOFoWQLM5U7vdB | 616 | run time eorror | {
"avatar_url": "https://avatars.githubusercontent.com/u/89890896?v=4",
"events_url": "https://api.github.com/users/LAZY118/events{/privacy}",
"followers_url": "https://api.github.com/users/LAZY118/followers",
"following_url": "https://api.github.com/users/LAZY118/following{/other_user}",
"gists_url": "https://api.github.com/users/LAZY118/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/LAZY118",
"id": 89890896,
"login": "LAZY118",
"node_id": "MDQ6VXNlcjg5ODkwODk2",
"organizations_url": "https://api.github.com/users/LAZY118/orgs",
"received_events_url": "https://api.github.com/users/LAZY118/received_events",
"repos_url": "https://api.github.com/users/LAZY118/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/LAZY118/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/LAZY118/subscriptions",
"type": "User",
"url": "https://api.github.com/users/LAZY118"
} | [] | closed | false | null | [] | null | [
"Hi! This looks like you are using AlphaPullDown, so I would suggest raising this issue on their github. From the stacktrace it looks like you may need to update your CUDA driver. The recommended version for the Alphafold repo is 11.1.1 and we provide a Docker image with a working environment. Thanks!"
] | "2022-10-27T02:51:09Z" | "2022-10-27T13:39:50Z" | "2022-10-27T13:39:50Z" | NONE | null | Hello, I try to use the AlphafoldMultimerV2 to predict interactions between baits and candidates in pulldown mode, but every time it showed the same error, could anybody help me to deal with the error issues?
(AlphaPulldown) [LAZY@spgpu ~]$ run_multimer_jobs.py --mode=pulldown --num_cycle=3 --num_predictions_per_model=1 --output_path=/data/projects/pulldown_test/4/pridict_output/short1 --data_dir=/data/alphafold_database/alphafold2-data/ --protein_lists=/data/projects/pulldown_test/4/11.txt,/data/projects/pulldown_test/4/4_shorter.txt --monomer_objects_dir=/data/projects/pulldown_test/4/shorter1
2022-10-26 22:20:22.867691: I tensorflow/stream_executor/platform/default/dso_loader.cc:53] Successfully opened dynamic library libcudart.so.11.0
I1026 22:20:24.809013 140194327537472 utils.py:169] checking if output_dir exists /data/projects/pulldown_test/4/pridict_output/short1
I1026 22:20:26.530570 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P00720
I1026 22:20:28.374641 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P00971
I1026 22:20:33.116755 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P04415
I1026 22:20:34.247322 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P04418
I1026 22:20:35.527069 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P04524
I1026 22:20:38.995062 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P07071
I1026 22:20:40.817292 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P12726
I1026 22:20:41.742229 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P13304
I1026 22:20:42.988400 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P13342
I1026 22:20:45.438112 140194327537472 run_multimer_jobs.py:158] done creating multimer b11_and_P16009
I1026 22:20:46.188549 140194327537472 xla_bridge.py:264] Unable to initialize backend 'tpu_driver': NOT_FOUND: Unable to find driver in registry given worker:
I1026 22:20:47.116462 140194327537472 xla_bridge.py:264] Unable to initialize backend 'tpu': INVALID_ARGUMENT: TpuPlatform is not available.
I1026 22:20:54.713402 140194327537472 run_multimer_jobs.py:236] now running prediction on b11_and_P00720
I1026 22:20:54.713624 140194327537472 predict_structure.py:40] Checking for /data/projects/pulldown_test/4/pridict_output/short1/b11_and_P00720/ranking_debug.json
I1026 22:20:54.714354 140194327537472 predict_structure.py:48] Running model model_1_multimer_v2_pred_0 on b11_and_P00720
I1026 22:20:54.714848 140194327537472 model.py:166] Running predict with shape(feat) = {'aatype': (803,), 'residue_index': (803,), 'seq_length': (), 'msa': (3773, 803), 'num_alignments': (), 'template_aatype': (4, 803), 'template_all_atom_mask': (4, 803, 37), 'template_all_atom_positions': (4, 803, 37, 3), 'asym_id': (803,), 'sym_id': (803,), 'entity_id': (803,), 'deletion_matrix': (3773, 803), 'deletion_mean': (803,), 'all_atom_mask': (803, 37), 'all_atom_positions': (803, 37, 3), 'assembly_num_chains': (), 'entity_mask': (803,), 'num_templates': (), 'cluster_bias_mask': (3773,), 'bert_mask': (3773, 803), 'seq_mask': (803,), 'msa_mask': (3773, 803)}
2022-10-26 22:20:54.951156: E external/org_tensorflow/tensorflow/stream_executor/gpu/asm_compiler.cc:105] You are using ptxas 8.x, but TF requires ptxas 9.x (and strongly prefers >= 11.1). Compilation of XLA kernels below will likely fail.
You may not need to update CUDA; cherry-picking the ptxas binary is often sufficient.
2022-10-26 22:20:55.158621: W external/org_tensorflow/tensorflow/stream_executor/gpu/asm_compiler.cc:230] Falling back to the CUDA driver for PTX compilation; ptxas does not support CC 8.6
2022-10-26 22:20:55.158715: W external/org_tensorflow/tensorflow/stream_executor/gpu/asm_compiler.cc:233] Used ptxas at ptxas
2022-10-26 22:20:55.163476: E external/org_tensorflow/tensorflow/stream_executor/cuda/cuda_driver.cc:632] failed to get PTX kernel "shift_right_logical_3" from module: CUDA_ERROR_NOT_FOUND: named symbol not found
2022-10-26 22:20:55.163587: E external/org_tensorflow/tensorflow/compiler/xla/pjrt/pjrt_stream_executor_client.cc:2141] Execution of replica 0 failed: INTERNAL: Could not find the corresponding function
Traceback (most recent call last):
File "/spshared/apps/miniconda3/envs/AlphaPulldown/bin/run_multimer_jobs.py", line 328, in <module>
app.run(main)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/absl/app.py", line 312, in run
_run_main(main, args)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/absl/app.py", line 258, in _run_main
sys.exit(main(argv))
File "/spshared/apps/miniconda3/envs/AlphaPulldown/bin/run_multimer_jobs.py", line 324, in main
predict_multimers(multimers)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/bin/run_multimer_jobs.py", line 275, in predict_multimers
random_seed=random_seed,
File "/spshared/apps/miniconda3/envs/AlphaPulldown/bin/run_multimer_jobs.py", line 249, in predict_individual_jobs
seqs=multimer_object.input_seqs,
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/alphapulldown/predict_structure.py", line 58, in predict
processed_feature_dict, random_seed=model_random_seed
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/alphafold/model/model.py", line 167, in predict
result = self.apply(self.params, jax.random.PRNGKey(random_seed), feat)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/_src/random.py", line 125, in PRNGKey
key = prng.seed_with_impl(impl, seed)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/_src/prng.py", line 232, in seed_with_impl
return PRNGKeyArray(impl, impl.seed(seed))
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/_src/prng.py", line 272, in threefry_seed
lax.shift_right_logical(seed_arr, lax_internal._const(seed_arr, 32)))
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/_src/lax/lax.py", line 444, in shift_right_logical
return shift_right_logical_p.bind(x, y)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/core.py", line 325, in bind
return self.bind_with_trace(find_top_trace(args), args, params)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/core.py", line 328, in bind_with_trace
out = trace.process_primitive(self, map(trace.full_raise, args), params)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/core.py", line 678, in process_primitive
return primitive.impl(*tracers, **params)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/_src/dispatch.py", line 98, in apply_primitive
return compiled_fun(*args)
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/_src/dispatch.py", line 118, in <lambda>
return lambda *args, **kw: compiled(*args, **kw)[0]
File "/spshared/apps/miniconda3/envs/AlphaPulldown/lib/python3.7/site-packages/jax/_src/dispatch.py", line 556, in _execute_compiled
out_bufs_flat = compiled.execute(input_bufs_flat)
RuntimeError: INTERNAL: Could not find the corresponding function
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/616/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/616/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/615 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/615/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/615/comments | https://api.github.com/repos/deepmind/alphafold/issues/615/events | https://github.com/deepmind/alphafold/issues/615 | 1,424,914,022 | I_kwDOFoWQLM5U7nZm | 615 | predicting structure of octamer | {
"avatar_url": "https://avatars.githubusercontent.com/u/24600576?v=4",
"events_url": "https://api.github.com/users/TerenceChen95/events{/privacy}",
"followers_url": "https://api.github.com/users/TerenceChen95/followers",
"following_url": "https://api.github.com/users/TerenceChen95/following{/other_user}",
"gists_url": "https://api.github.com/users/TerenceChen95/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/TerenceChen95",
"id": 24600576,
"login": "TerenceChen95",
"node_id": "MDQ6VXNlcjI0NjAwNTc2",
"organizations_url": "https://api.github.com/users/TerenceChen95/orgs",
"received_events_url": "https://api.github.com/users/TerenceChen95/received_events",
"repos_url": "https://api.github.com/users/TerenceChen95/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/TerenceChen95/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/TerenceChen95/subscriptions",
"type": "User",
"url": "https://api.github.com/users/TerenceChen95"
} | [] | closed | false | null | [] | null | [] | "2022-10-27T02:02:47Z" | "2022-10-27T03:18:13Z" | "2022-10-27T03:18:13Z" | NONE | null | Hi,
I was trying to predict predict the structure of an octamer, and got the following error:
alphafold.data.templates.SequenceNotInTemplateError: Could not find the template sequence in 7a0l_A
Traceback (most recent call last):
File "/mnt/nas/tools/alphafold_non_docker_v220/alphafold/data/templates.py", line 362, in _realign_pdb_template_to_query
aligner.align([old_template_sequence, new_template_sequence]))
File "/mnt/nas/tools/alphafold_non_docker_v220/alphafold/data/tools/kalign.py", line 98, in align
raise RuntimeError('Kalign failed\nstdout:\n%s\n\nstderr:\n%s\n'
RuntimeError: Kalign failed
Traceback (most recent call last):
File "/mnt/nas/tools/alphafold_non_docker_v220/run_alphafold.py", line 442, in <module>
app.run(main)
File "/mnt/nas/tools/anaconda3/envs/alphafold_non_docker/lib/python3.8/site-packages/absl/app.py", line 312, in run
_run_main(main, args)
File "/mnt/nas/tools/anaconda3/envs/alphafold_non_docker/lib/python3.8/site-packages/absl/app.py", line 258, in _run_main
sys.exit(main(argv))
File "/mnt/nas/tools/alphafold_non_docker_v220/run_alphafold.py", line 415, in main
predict_structure(
File "/mnt/nas/tools/alphafold_non_docker_v220/run_alphafold.py", line 185, in predict_structure
feature_dict = data_pipeline.process(
File "/mnt/nas/tools/alphafold_non_docker_v220/alphafold/data/pipeline_multimer.py", line 264, in process
chain_features = self._process_single_chain(
File "/mnt/nas/tools/alphafold_non_docker_v220/alphafold/data/pipeline_multimer.py", line 212, in _process_single_chain
chain_features = self._monomer_data_pipeline.process(
File "/mnt/nas/tools/alphafold_non_docker_v220/alphafold/data/pipeline.py", line 223, in process
templates_result = self.template_featurizer.get_templates(
File "/mnt/nas/tools/alphafold_non_docker_v220/alphafold/data/templates.py", line 960, in get_templates
result = _process_single_hit(
File "/mnt/nas/tools/alphafold_non_docker_v220/alphafold/data/templates.py", line 786, in _process_single_hit
error = ('%s_%s (sum_probs: %.2f, rank: %d): feature extracting errors: '
TypeError: must be real number, not NoneType
Strangely, I can run the code successfullly in tetramer setting. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/615/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/615/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/614 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/614/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/614/comments | https://api.github.com/repos/deepmind/alphafold/issues/614/events | https://github.com/deepmind/alphafold/pull/614 | 1,419,948,224 | PR_kwDOFoWQLM5BXlFo | 614 | added a test file | {
"avatar_url": "https://avatars.githubusercontent.com/u/36188891?v=4",
"events_url": "https://api.github.com/users/mzelling/events{/privacy}",
"followers_url": "https://api.github.com/users/mzelling/followers",
"following_url": "https://api.github.com/users/mzelling/following{/other_user}",
"gists_url": "https://api.github.com/users/mzelling/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/mzelling",
"id": 36188891,
"login": "mzelling",
"node_id": "MDQ6VXNlcjM2MTg4ODkx",
"organizations_url": "https://api.github.com/users/mzelling/orgs",
"received_events_url": "https://api.github.com/users/mzelling/received_events",
"repos_url": "https://api.github.com/users/mzelling/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/mzelling/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/mzelling/subscriptions",
"type": "User",
"url": "https://api.github.com/users/mzelling"
} | [] | closed | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/614/checks?check_run_id=9058137969) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request.",
"This PR was meant to take place within my fork. Please disregard-- apologies."
] | "2022-10-23T21:48:45Z" | "2022-10-23T21:56:35Z" | "2022-10-23T21:49:18Z" | NONE | null | This pull request is a test. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/614/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/614/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/614.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/614",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/614.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/614"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/613 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/613/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/613/comments | https://api.github.com/repos/deepmind/alphafold/issues/613/events | https://github.com/deepmind/alphafold/issues/613 | 1,418,245,597 | I_kwDOFoWQLM5UiLXd | 613 | Problem with docker build -f docker/Dockerfile -t alphafold . | {
"avatar_url": "https://avatars.githubusercontent.com/u/54129071?v=4",
"events_url": "https://api.github.com/users/fengsidu/events{/privacy}",
"followers_url": "https://api.github.com/users/fengsidu/followers",
"following_url": "https://api.github.com/users/fengsidu/following{/other_user}",
"gists_url": "https://api.github.com/users/fengsidu/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/fengsidu",
"id": 54129071,
"login": "fengsidu",
"node_id": "MDQ6VXNlcjU0MTI5MDcx",
"organizations_url": "https://api.github.com/users/fengsidu/orgs",
"received_events_url": "https://api.github.com/users/fengsidu/received_events",
"repos_url": "https://api.github.com/users/fengsidu/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/fengsidu/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/fengsidu/subscriptions",
"type": "User",
"url": "https://api.github.com/users/fengsidu"
} | [] | open | false | null | [] | null | [
"Hi thanks for the question. I haven't been able to reproduce this error. Are you using the same Dockerfile in the latest version release or have you made some changes?",
"> Hi thanks for the question. I haven't been able to reproduce this error. Are you using the same Dockerfile in the latest version release or have you made some changes?\r\n\r\nHello, I did not make any changes to the Dockerfile during the initial installation, and this problem also occurred. This problem also occurred when I added a mirror source to the Dockerfile to speed up the download.",
"> 嗨,谢谢你的问题。我无法重现此错误。您是在最新版本中使用相同的 Dockerfile 还是进行了一些更改?\r\n\r\nMy machine is in Virginia, the network is very good, the dockerfile file is not modified, and the latest code pulled, the above problem also occurs",
"Hi thanks for raising. What version of docker are you using and on what type of machine?",
"> 您好,感谢您提出。您使用的是什么版本的 docker 以及在什么类型的机器上?\r\n\r\nI'm very sorry, I already know the reason for this error. It is caused by compiling on a machine without a GPU driver. To use the Dockerfile in the docker directory to create a docker image, a machine with a GPU driver needs to be installed.",
"Hi @hezhiqiang8909 thank you for the update. @fengsidu does your machine have a CUDA driver?"
] | "2022-10-21T12:11:29Z" | "2022-11-03T12:37:21Z" | null | NONE | null | The code always reports the following error, I don't know how to solve it
```
The following packages will be UPDATED:
ca-certificates 2022.3.29-h06a4308_1 --> 2022.07.19-h06a4308_0
certifi 2021.10.8-py39h06a4308_2 --> 2022.9.24-py39h06a4308_0
conda 4.12.0-py39h06a4308_0 --> 4.13.0-py39h06a4308_0
openssl 1.1.1n-h7f8727e_0 --> 1.1.1q-h7f8727e_0
Preparing transaction: ...working... done
Verifying transaction: ...working... done
Executing transaction: ...working... done
# >>>>>>>>>>>>>>>>>>>>>> ERROR REPORT <<<<<<<<<<<<<<<<<<<<<<
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1082, in __call__
print(_format_exc(exc_val, exc_tb), file=sys.stderr)
File "/opt/conda/lib/python3.9/site-packages/conda/cli/main.py", line 87, in _main
if isinstance(exit_code, int):
File "/opt/conda/lib/python3.9/site-packages/conda/cli/conda_argparse.py", line 84, in do_call
def do_call(args, parser):
File "/opt/conda/lib/python3.9/site-packages/conda/cli/main_install.py", line 20, in execute
install(args, parser, 'install')
File "/opt/conda/lib/python3.9/site-packages/conda/cli/install.py", line 316, in install
# of fns. That way, we fall to the next fn.
File "/opt/conda/lib/python3.9/site-packages/conda/cli/install.py", line 345, in handle_txn
File "/opt/conda/lib/python3.9/site-packages/conda/core/link.py", line 281, in execute
self._execute(tuple(concat(interleave(self.prefix_action_groups.values()))))
File "/opt/conda/lib/python3.9/site-packages/conda/core/link.py", line 753, in _execute
action.cleanup()
File "/opt/conda/lib/python3.9/site-packages/conda/core/path_actions.py", line 1015, in cleanup
if not isdir(self.holding_full_path):
File "/opt/conda/lib/python3.9/site-packages/conda/gateways/disk/delete.py", line 179, in rm_rf
remove_empty_parent_paths(path)
File "/opt/conda/lib/python3.9/site-packages/conda/gateways/disk/delete.py", line 152, in remove_empty_parent_paths
rmdir(parent_path)
OSError: [Errno 22] Invalid argument: '/opt/conda/lib/python3.9/site-packages/conda-4.12.0-py3.9.egg-info'
`$ /opt/conda/bin/conda install -qy conda==4.13.0`
environment variables:
CIO_TEST=<not set>
CONDA_ROOT=/opt/conda
CURL_CA_BUNDLE=<not set>
LD_LIBRARY_PATH=/usr/local/nvidia/lib:/usr/local/nvidia/lib64
PATH=/opt/conda/bin:/usr/local/nvidia/bin:/usr/local/cuda/bin:/usr/local/sb
in:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin
REQUESTS_CA_BUNDLE=<not set>
SSL_CERT_FILE=<not set>
active environment : None
user config file : /root/.condarc
populated config files :
conda version : 4.12.0
conda-build version : not installed
python version : 3.9.12.final.0
virtual packages : __linux=3.10.0=0
__glibc=2.27=0
__unix=0=0
__archspec=1=x86_64
base environment : /opt/conda (writable)
conda av data dir : /opt/conda/etc/conda
conda av metadata url : None
channel URLs : https://repo.anaconda.com/pkgs/main/linux-64
https://repo.anaconda.com/pkgs/main/noarch
https://repo.anaconda.com/pkgs/r/linux-64
https://repo.anaconda.com/pkgs/r/noarch
package cache : /opt/conda/pkgs
/root/.conda/pkgs
envs directories : /opt/conda/envs
/root/.conda/envs
platform : linux-64
user-agent : conda/4.12.0 requests/2.27.1 CPython/3.9.12 Linux/3.10.0-327.el7.x86_64 ubuntu/18.04.6 glibc/2.27
UID:GID : 0:0
netrc file : None
offline mode : False
An unexpected error has occurred. Conda has prepared the above report.
Upload successful.
The command '/bin/bash -o pipefail -c conda install -qy conda==4.13.0 && conda install -y -c conda-forge openmm=7.5.1 cudatoolkit==${CUDA_VERSION} pdbfixer pip python=3.7 && conda clean --all --force-pkgs-dirs --yes' returned a non-zero code: 1
```
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/613/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/613/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/612 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/612/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/612/comments | https://api.github.com/repos/deepmind/alphafold/issues/612/events | https://github.com/deepmind/alphafold/issues/612 | 1,414,927,024 | I_kwDOFoWQLM5UVhKw | 612 | GPU Memory Resource Exhaustion issue | {
"avatar_url": "https://avatars.githubusercontent.com/u/83024487?v=4",
"events_url": "https://api.github.com/users/bijuissacch224610/events{/privacy}",
"followers_url": "https://api.github.com/users/bijuissacch224610/followers",
"following_url": "https://api.github.com/users/bijuissacch224610/following{/other_user}",
"gists_url": "https://api.github.com/users/bijuissacch224610/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/bijuissacch224610",
"id": 83024487,
"login": "bijuissacch224610",
"node_id": "MDQ6VXNlcjgzMDI0NDg3",
"organizations_url": "https://api.github.com/users/bijuissacch224610/orgs",
"received_events_url": "https://api.github.com/users/bijuissacch224610/received_events",
"repos_url": "https://api.github.com/users/bijuissacch224610/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/bijuissacch224610/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/bijuissacch224610/subscriptions",
"type": "User",
"url": "https://api.github.com/users/bijuissacch224610"
} | [
{
"color": "A65D8A",
"default": false,
"description": "Out of memory error message",
"id": 3938926450,
"name": "out of memory",
"node_id": "LA_kwDOFoWQLM7qxz9y",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/out%20of%20memory"
}
] | closed | false | null | [] | null | [
"We improved the GPU usage for AlphaFold-Multimer in [v2.3.0](https://github.com/deepmind/alphafold/releases/tag/v2.3.0) - hopefully this addresses your issue. Closing this issue now, feel free to reopen if you still encounter this in v2.3.0."
] | "2022-10-19T12:56:30Z" | "2022-12-13T11:58:50Z" | "2022-12-13T11:58:49Z" | NONE | null | I am running a multimer analysis using local AlphaFold 2.2.2 installed on our server and have successfully predicted structures for monomer and multimer analyses. Recently I need to predict a protein complex using 3 protein sequences, one of which is 2482 amino acids long while other two are shorter.
I have provided adequate memory as seen here
<img width="569" alt="Screen Shot 2022-10-18 at 4 08 25 PM" src="https://user-images.githubusercontent.com/83024487/196695740-c47553c0-e986-48a2-9159-f7be5b484773.png">
I have tried running variations of various solutions provided here on Github issues but keep getting same error about resource exhaustion. When I look at log file, the GPU RAM is not being fully utilized.
<img width="931" alt="Screen Shot 2022-10-18 at 4 05 54 PM" src="https://user-images.githubusercontent.com/83024487/196696465-da305b12-3cb6-4197-8910-5446c9cbbb42.png">
My configurations are as below
<img width="514" alt="Screen Shot 2022-10-19 at 8 52 30 AM" src="https://user-images.githubusercontent.com/83024487/196696500-a4f76297-52af-4593-8dd7-b6c593b5fa36.png">
<img width="332" alt="Screen Shot 2022-10-19 at 8 52 45 AM" src="https://user-images.githubusercontent.com/83024487/196696523-00da029b-7be5-42ab-b7a4-2e0e790ebe7e.png">
Can you help me with this? The error seems to be from AlphaFold is analyzing the longest protein sequence in the complex. To reduce the load on memory, I am running my multimer analysis using "-db_preset=reduced_dbs" parameter
Any help from the developers or the community is deeply appreciated
Thanks
-B. Issac | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/612/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/612/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/611 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/611/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/611/comments | https://api.github.com/repos/deepmind/alphafold/issues/611/events | https://github.com/deepmind/alphafold/issues/611 | 1,410,820,954 | I_kwDOFoWQLM5UF2ta | 611 | Is there any solution that we can run AF2 by using GCP TPU vm? | {
"avatar_url": "https://avatars.githubusercontent.com/u/12453629?v=4",
"events_url": "https://api.github.com/users/edwardlo12/events{/privacy}",
"followers_url": "https://api.github.com/users/edwardlo12/followers",
"following_url": "https://api.github.com/users/edwardlo12/following{/other_user}",
"gists_url": "https://api.github.com/users/edwardlo12/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/edwardlo12",
"id": 12453629,
"login": "edwardlo12",
"node_id": "MDQ6VXNlcjEyNDUzNjI5",
"organizations_url": "https://api.github.com/users/edwardlo12/orgs",
"received_events_url": "https://api.github.com/users/edwardlo12/received_events",
"repos_url": "https://api.github.com/users/edwardlo12/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/edwardlo12/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/edwardlo12/subscriptions",
"type": "User",
"url": "https://api.github.com/users/edwardlo12"
} | [] | open | false | null | [] | null | [] | "2022-10-17T03:19:46Z" | "2022-10-17T03:19:46Z" | null | NONE | null | We are trying to infer a sequence with 5000 amino acids. But AF2 can only support single GPU, so we think we can try to use TPU. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/611/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/611/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/610 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/610/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/610/comments | https://api.github.com/repos/deepmind/alphafold/issues/610/events | https://github.com/deepmind/alphafold/issues/610 | 1,410,112,012 | I_kwDOFoWQLM5UDJoM | 610 | No such file or directory: '.../pdb_mmcif/mmcif_files/XXXX.cif' | {
"avatar_url": "https://avatars.githubusercontent.com/u/34373061?v=4",
"events_url": "https://api.github.com/users/biomolecule/events{/privacy}",
"followers_url": "https://api.github.com/users/biomolecule/followers",
"following_url": "https://api.github.com/users/biomolecule/following{/other_user}",
"gists_url": "https://api.github.com/users/biomolecule/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/biomolecule",
"id": 34373061,
"login": "biomolecule",
"node_id": "MDQ6VXNlcjM0MzczMDYx",
"organizations_url": "https://api.github.com/users/biomolecule/orgs",
"received_events_url": "https://api.github.com/users/biomolecule/received_events",
"repos_url": "https://api.github.com/users/biomolecule/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/biomolecule/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/biomolecule/subscriptions",
"type": "User",
"url": "https://api.github.com/users/biomolecule"
} | [] | open | false | null | [] | null | [
"I am expierencing the same issue with another protein not being found (7nvm) even though max_template_date is set.",
"Hi thanks for raising this. Have you updated the pdb_seqres file more recently than the PDB mmcif files? This error is likely to happen when they are out of sync. In the future we can add a more informative error message.",
"I am experiencing the same issue and first tried changing the max_template_date, but that didn't help. \r\nThen I re-ran both download_pdb_mmcif.sh and download_pdb_seqres.sh yesterday. But the error persists, mmcif_files/6ek0.cif is missing. \r\nWhat I found is that 6ek0 appears in the obsolete.dat. Any idea how to proceed?\r\nThanks and best!\r\nPhilipp",
"> I am experiencing the same issue and first tried changing the max_template_date, but that didn't help. Then I re-ran both download_pdb_mmcif.sh and download_pdb_seqres.sh yesterday. But the error persists, mmcif_files/6ek0.cif is missing. What I found is that 6ek0 appears in the obsolete.dat. Any idea how to proceed? Thanks and best! Philipp\r\n\r\nhttps://files.wwpdb.org/pub/pdb/data/structures/divided/mmCIF/ek/6eko.cif.gz",
"> I am experiencing the same issue and first tried changing the max_template_date, but that didn't help. Then I re-ran both download_pdb_mmcif.sh and download_pdb_seqres.sh yesterday. But the error persists, mmcif_files/6ek0.cif is missing. What I found is that 6ek0 appears in the obsolete.dat. Any idea how to proceed? Thanks and best! Philipp\r\n\r\nor sync the PDB mmcif files",
"Hej @Maiya19724, \r\nThanks a lot for our help! Sadly, this is not the missing file - 6ek0 (with a zero) is not in the pdb ek-folder. \r\n\r\n> or sync the PDB mmcif files\r\nThis is what download_pdb_mmcif.sh is doing, right? I did this and 6ek0 was not in there.\r\n\r\nThanks and best!\r\nPhilipp\r\n",
"> Hej @Maiya19724, Thanks a lot for our help! Sadly, this is not the missing file - 6ek0 (with a zero) is not in the pdb ek-folder.\r\n> \r\n> > or sync the PDB mmcif files\r\n> > This is what download_pdb_mmcif.sh is doing, right? I did this and 6ek0 was not in there.\r\n> \r\n> Thanks and best! Philipp\r\n\r\nsorry for that."
] | "2022-10-15T09:47:08Z" | "2022-12-01T05:47:14Z" | null | NONE | null | Now l'm running AF2 multimer with "run_alphafold.py" . Both MSAs' generation for two chains are success, as well as the template search for the first chain. BUT python report an error during that of the second:
`FileNotFoundError: [Errno 2] No such file or directory: '.../pdb_mmcif/mmcif_files/7xiv.cif'`
The publication date of the 7xiv is 2022-06, but the download date of pdb_mmcif is earlier, so, as my concerned, the most simple solution is to set the AF2's paramater "max_template_date" even earlier than database download, such as
`--max_template_date=2020-05-14`
But the error occurs again. Update pdb_mmcif might be another solution, but it is unacceptable to upgrade it everytime running AF2. It seems the " --max_template_date" does not work.
What should l do to avoid such error?
Regards

| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/610/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/610/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/609 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/609/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/609/comments | https://api.github.com/repos/deepmind/alphafold/issues/609/events | https://github.com/deepmind/alphafold/issues/609 | 1,409,145,531 | I_kwDOFoWQLM5T_dq7 | 609 | Geometric algebra | {
"avatar_url": "https://avatars.githubusercontent.com/u/1799429?v=4",
"events_url": "https://api.github.com/users/EelcoHoogendoorn/events{/privacy}",
"followers_url": "https://api.github.com/users/EelcoHoogendoorn/followers",
"following_url": "https://api.github.com/users/EelcoHoogendoorn/following{/other_user}",
"gists_url": "https://api.github.com/users/EelcoHoogendoorn/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/EelcoHoogendoorn",
"id": 1799429,
"login": "EelcoHoogendoorn",
"node_id": "MDQ6VXNlcjE3OTk0Mjk=",
"organizations_url": "https://api.github.com/users/EelcoHoogendoorn/orgs",
"received_events_url": "https://api.github.com/users/EelcoHoogendoorn/received_events",
"repos_url": "https://api.github.com/users/EelcoHoogendoorn/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/EelcoHoogendoorn/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/EelcoHoogendoorn/subscriptions",
"type": "User",
"url": "https://api.github.com/users/EelcoHoogendoorn"
} | [] | open | false | null | [] | null | [
"Just replacing the /geometry/, r3.py and quat_affine.py math functions with GA equivalents should not have to be a lot of work, come to think of it, as long as the state representations are kept the same..",
"Note that the current JAX backend only implements array-of-struct memory layout multivectors. Adding support for struct-of-array should not be hard at all; the thing im still trying to wrap my head around is what should happen if SOA and AOS multivectors interact? Is there a general rule for this; or would it make most sense to implement a user-overridable lookup table to specify what type you are supposed to allocate when you multiply a SOA-quat and a AOS-vector, and so on?\r\n\r\nI do not quite understand the mix of SOA and OAS in alphafold in the first place. My first guess would be that SOA is always preferred, when working on GPU/TPU, with large batches of identical objects; as is the case for alphafold. Why mix in AOS layouts in the first place? Is that a deliberate choice; or more a matter of it being too annoying to rewrite all code to SOA?",
"@Augustin-Zidek I see youve been working on a lot of this part of the codebase; would you be able to comment on the above questions concerning SOA-vs-AOS?"
] | "2022-10-14T10:37:34Z" | "2022-10-17T15:06:50Z" | null | NONE | null | Ive recently authored a [geometric algebra](https://github.com/EelcoHoogendoorn/numga) package for JAX, and it occurred to me that a project like alphafold might benefit quite a bit from adopting it.
If nothing else, one feature of using numga I have found very useful in my own work, is to easily swap out the execution strategy for your operations. In my experience, this can matter up to 10x, both in compilation and runtime performance. Right now alphafold appears to use a mix of dense and sparse product execution styles; eg ([quat_multiply](https://github.com/deepmind/alphafold/blob/5cb2f8c480aa8314c02a93c6fbfc3f48f0ce8af0/alphafold/model/quat_affine.py#L153)) and ([vecs_cross_vecs](https://github.com/deepmind/alphafold/blob/5cb2f8c480aa8314c02a93c6fbfc3f48f0ce8af0/alphafold/model/r3.py#L265)).
Numga completely seperates the mathematical logic from the execution logic, and it is easy to swap out execution styles. If you toggle [this](https://github.com/EelcoHoogendoorn/numga/blob/31de8b96d2f2a60690cc511ad2436a6d177da855/numga/examples/physics/run_chain.py#L24) line you can get an impression of how much it matters on your hardware, on a fairly representative piece of code being executed. In my benchmarking, dense products can compile up to 10x faster, while sparse ones tend to execute 10x faster (at least on CPU).
While it is super elegant, it is not obvious to me that going all-in on the GA 'motor' paradigm is the right call for a performance sensitive application like allphafold; and i have some open questions with respect to numerical integration errors myself. But one can perfectly well use a geometric algebra library in conjunction with a 'classical' translation + quaternion state encoding. For my own work, my preferred approach is to start prototyping my code in the most mathematically elegant manner, and to drop down to lower level constructs only where I can show it provides a benefit; and to keep both implementations around, so I can unit test them against one another. But maybe encoding all states with motors would work out really well for alphafold; I wouldnt dare say without having tried.
I dont have the bandwidth to dive into alphafold to try and make a branch myself, but id be happy to see where I can help if anyone else is interested in giving it a go. If nothing else, playing around a little with geometric algebra is a lot of fun, I can promise that much. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/609/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/609/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/608 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/608/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/608/comments | https://api.github.com/repos/deepmind/alphafold/issues/608/events | https://github.com/deepmind/alphafold/issues/608 | 1,401,430,241 | I_kwDOFoWQLM5TiCDh | 608 | Carriage returns in pdb_seqres.txt | {
"avatar_url": "https://avatars.githubusercontent.com/u/63256188?v=4",
"events_url": "https://api.github.com/users/therealchrisneale/events{/privacy}",
"followers_url": "https://api.github.com/users/therealchrisneale/followers",
"following_url": "https://api.github.com/users/therealchrisneale/following{/other_user}",
"gists_url": "https://api.github.com/users/therealchrisneale/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/therealchrisneale",
"id": 63256188,
"login": "therealchrisneale",
"node_id": "MDQ6VXNlcjYzMjU2MTg4",
"organizations_url": "https://api.github.com/users/therealchrisneale/orgs",
"received_events_url": "https://api.github.com/users/therealchrisneale/received_events",
"repos_url": "https://api.github.com/users/therealchrisneale/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/therealchrisneale/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/therealchrisneale/subscriptions",
"type": "User",
"url": "https://api.github.com/users/therealchrisneale"
} | [
{
"color": "D4C5F9",
"default": false,
"description": "Further information is requested",
"id": 3094826897,
"name": "usage question",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODk3",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/usage%20question"
}
] | closed | false | null | [] | null | [
"Carriage returns should be handled ok by the MSA search tools, fine to leave as is."
] | "2022-10-07T16:37:09Z" | "2022-12-13T11:59:49Z" | "2022-12-13T11:59:49Z" | NONE | null | Hello,
I see carriage returns in some database input files:
$ ./alphafold/scripts/download_pdb_seqres.sh TEST
$ grep -r $'\r' TEST/pdb_seqres/pdb_seqres.txt | wc -l
1476032
Should I remove them (e.g., sed -i "s/\r$//" TEST/pdb_seqres/pdb_seqres.txt) or is it not expected to represent any problem?
Thank you,
Chris. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/608/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/608/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/607 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/607/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/607/comments | https://api.github.com/repos/deepmind/alphafold/issues/607/events | https://github.com/deepmind/alphafold/issues/607 | 1,399,876,021 | I_kwDOFoWQLM5TcGm1 | 607 | Running "run_docker.py" says ModuleNotFoundError: No module named 'asbl' | {
"avatar_url": "https://avatars.githubusercontent.com/u/115172389?v=4",
"events_url": "https://api.github.com/users/Aaronro16/events{/privacy}",
"followers_url": "https://api.github.com/users/Aaronro16/followers",
"following_url": "https://api.github.com/users/Aaronro16/following{/other_user}",
"gists_url": "https://api.github.com/users/Aaronro16/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Aaronro16",
"id": 115172389,
"login": "Aaronro16",
"node_id": "U_kgDOBt1kJQ",
"organizations_url": "https://api.github.com/users/Aaronro16/orgs",
"received_events_url": "https://api.github.com/users/Aaronro16/received_events",
"repos_url": "https://api.github.com/users/Aaronro16/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Aaronro16/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Aaronro16/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Aaronro16"
} | [] | closed | false | null | [] | null | [
"I created this thread for those that are having the same issue"
] | "2022-10-06T15:32:55Z" | "2022-10-06T15:33:45Z" | "2022-10-06T15:33:45Z" | NONE | null | I just installed AlphaFold and for the first time that I tried to run it gived to me the next error:
File "alphafold/docker/run_docker.py", line 22, in <module>
from absl import app
ModuleNotFoundError: No module named 'absl'
I used:
pip uninstall absl-py
pip install absl-py
pip3 uninstall absl-py
pip3 install absl-py
and didn't work, later I tried with the next command and was solved:
python3 -m pip install absl-py
The same issue happened for the docker library:
File "docker/run_docker.py", line 25, in <module>
import docker
ModuleNotFoundError: No module named 'docker'
And again solved with the same command but specifying the library docker:
python3 -m pip install docker | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/607/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/607/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/606 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/606/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/606/comments | https://api.github.com/repos/deepmind/alphafold/issues/606/events | https://github.com/deepmind/alphafold/issues/606 | 1,398,735,817 | I_kwDOFoWQLM5TXwPJ | 606 | Link format for downloading pdb files from a third party application | {
"avatar_url": "https://avatars.githubusercontent.com/u/72611578?v=4",
"events_url": "https://api.github.com/users/alisa667/events{/privacy}",
"followers_url": "https://api.github.com/users/alisa667/followers",
"following_url": "https://api.github.com/users/alisa667/following{/other_user}",
"gists_url": "https://api.github.com/users/alisa667/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/alisa667",
"id": 72611578,
"login": "alisa667",
"node_id": "MDQ6VXNlcjcyNjExNTc4",
"organizations_url": "https://api.github.com/users/alisa667/orgs",
"received_events_url": "https://api.github.com/users/alisa667/received_events",
"repos_url": "https://api.github.com/users/alisa667/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/alisa667/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/alisa667/subscriptions",
"type": "User",
"url": "https://api.github.com/users/alisa667"
} | [] | open | false | null | [] | null | [] | "2022-10-06T03:50:46Z" | "2022-10-06T03:50:46Z" | null | NONE | null | Hello,
I would like to access Alphafold DB from a third party application. As I understand the link format is **https://alphafold.ebi.ac.uk/files/AF-[a UniProt accession]-F[a fragment number]-model_v3.pdb**. Can I use this format in my application to download PDB files? I would like to know whether it is guaranteed to be the same for all UniProt accession numbers and whether there is a possibility of it being changed.
Thank you,
Alisa | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/606/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/606/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/605 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/605/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/605/comments | https://api.github.com/repos/deepmind/alphafold/issues/605/events | https://github.com/deepmind/alphafold/pull/605 | 1,397,689,819 | PR_kwDOFoWQLM5ANYc0 | 605 | Add conda lib path to LD_LIBRARY_PATH | {
"avatar_url": "https://avatars.githubusercontent.com/u/25644865?v=4",
"events_url": "https://api.github.com/users/gabrielctn/events{/privacy}",
"followers_url": "https://api.github.com/users/gabrielctn/followers",
"following_url": "https://api.github.com/users/gabrielctn/following{/other_user}",
"gists_url": "https://api.github.com/users/gabrielctn/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/gabrielctn",
"id": 25644865,
"login": "gabrielctn",
"node_id": "MDQ6VXNlcjI1NjQ0ODY1",
"organizations_url": "https://api.github.com/users/gabrielctn/orgs",
"received_events_url": "https://api.github.com/users/gabrielctn/received_events",
"repos_url": "https://api.github.com/users/gabrielctn/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/gabrielctn/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/gabrielctn/subscriptions",
"type": "User",
"url": "https://api.github.com/users/gabrielctn"
} | [] | open | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/605/checks?check_run_id=8721030002) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request."
] | "2022-10-05T12:08:22Z" | "2022-10-05T12:08:27Z" | null | NONE | null | Fix the following errors which occured when running the singularity transformed docker image of alphafold 2.2.4 Dockerfile:
```
Could not load dynamic library 'libcusolver.so.11'
```
Which generates the error:
```
RuntimeError: jaxlib/cuda/cusolver_kernels.cc:44: operation cusolverDnCreate(&handle) failed: cuSolver internal error
```
Steps to generate the Singularity image by converting the docker image:
```
$ docker build -f docker/Dockerfile -t alphafold:2.2.4 .
$ docker run -v /var/run/docker.sock:/var/run/docker.sock -v /tmp:/output --privileged -t --rm quay.io/singularity/docker2singularity -n alphafold_2.2.4.sif alphafold:2.2.4
$ singularity build /tmp/alphafold_2.2.4.sif docker-daemon://alphafold:2.2.4
``` | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/605/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/605/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/605.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/605",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/605.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/605"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/604 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/604/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/604/comments | https://api.github.com/repos/deepmind/alphafold/issues/604/events | https://github.com/deepmind/alphafold/issues/604 | 1,395,514,280 | I_kwDOFoWQLM5TLduo | 604 | Port to Metal and macOS | {
"avatar_url": "https://avatars.githubusercontent.com/u/71743241?v=4",
"events_url": "https://api.github.com/users/philipturner/events{/privacy}",
"followers_url": "https://api.github.com/users/philipturner/followers",
"following_url": "https://api.github.com/users/philipturner/following{/other_user}",
"gists_url": "https://api.github.com/users/philipturner/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/philipturner",
"id": 71743241,
"login": "philipturner",
"node_id": "MDQ6VXNlcjcxNzQzMjQx",
"organizations_url": "https://api.github.com/users/philipturner/orgs",
"received_events_url": "https://api.github.com/users/philipturner/received_events",
"repos_url": "https://api.github.com/users/philipturner/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/philipturner/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/philipturner/subscriptions",
"type": "User",
"url": "https://api.github.com/users/philipturner"
} | [] | open | false | null | [] | null | [] | "2022-10-04T00:39:32Z" | "2022-10-04T00:39:32Z" | null | NONE | null | I've been taking a shot at porting this from NVIDIA/CUDA to M1/Metal, so that I can run it on my personal MacBook Pro (1 TB disk). I anticipated being able to port the entire framework in a single day, but faced significant challenges regarding disk space. It would have been much less challenging if the total database collection were even just halved in size.
I wish to engineer proteins that never existed before in biology, and may have no evolutionary history. AlphaFold would let me rapidly search the solution space of amino acid sequences, until encountering proteins that suit my needs. I fear that AlphaFold, heavily tuned for MSAs, would perform poorly on proteins foreign to biological evolution. Nevertheless, this would be a great tool in my toolbox.
I'm thinking of either (1) purchasing an external HDD or (2) seeing whether you reduce the collective database size from 600 (actually 700) GB to something like 200 GB. When/if I finish the port, would you consider merging these contributions into the main branch? I don't think that's very likely, but "never say never".
Fork and documentation of my porting efforts: https://github.com/philipturner/alphafold-metal | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/604/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/604/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/603 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/603/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/603/comments | https://api.github.com/repos/deepmind/alphafold/issues/603/events | https://github.com/deepmind/alphafold/issues/603 | 1,393,197,056 | I_kwDOFoWQLM5TCoAA | 603 | GPU is not fully used. | {
"avatar_url": "https://avatars.githubusercontent.com/u/114789491?v=4",
"events_url": "https://api.github.com/users/daishuyan/events{/privacy}",
"followers_url": "https://api.github.com/users/daishuyan/followers",
"following_url": "https://api.github.com/users/daishuyan/following{/other_user}",
"gists_url": "https://api.github.com/users/daishuyan/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/daishuyan",
"id": 114789491,
"login": "daishuyan",
"node_id": "U_kgDOBteMcw",
"organizations_url": "https://api.github.com/users/daishuyan/orgs",
"received_events_url": "https://api.github.com/users/daishuyan/received_events",
"repos_url": "https://api.github.com/users/daishuyan/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/daishuyan/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/daishuyan/subscriptions",
"type": "User",
"url": "https://api.github.com/users/daishuyan"
} | [] | open | false | null | [] | null | [] | "2022-10-01T02:51:12Z" | "2022-10-01T02:51:12Z" | null | NONE | null | Hi, I've installed AF2 on my own linux computer. And tried used it to calculate structures of proteins with different lengths (~100-2000 aa). I found a problem, that the GPU is almost not use, only ~250MB memory is used by AF2 in all predictions.
I tried to find a solution, but there is no one report the same issu**e as me**.
**My system is:**
OS: Ubuntu 20.04.1
CPU: Intel Core i9-9900K 3.60 GHz x 16
GPU: NVIDIA RTX 3080Ti 12 GB
Driver version: 495.29.05
CUDA version: 11.5
RAM: 32 GB
**Output of nvidia-smi when AF2 running:**
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 495.29.05 Driver Version: 495.29.05 CUDA Version: 11.5 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 NVIDIA GeForce ... On | 00000000:01:00.0 On | N/A |
| 0% 50C P8 34W / 350W | 2368MiB / 12045MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=============================================================================|
| 0 N/A N/A 1188 G /usr/lib/xorg/Xorg 29MiB |
| 0 N/A N/A 28400 G /usr/lib/xorg/Xorg 83MiB |
| 0 N/A N/A 28527 G /usr/bin/gnome-shell 46MiB |
| 0 N/A N/A 29313 G /usr/lib/firefox/firefox 150MiB |
| 0 N/A N/A 55039 C python 247MiB |
+-----------------------------------------------------------------------------+
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/603/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/603/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/602 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/602/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/602/comments | https://api.github.com/repos/deepmind/alphafold/issues/602/events | https://github.com/deepmind/alphafold/issues/602 | 1,391,467,178 | I_kwDOFoWQLM5S8Bqq | 602 | Can I use cudatoolkit==11.7.0 instead of cudatoolkit==11.1.1 ? | {
"avatar_url": "https://avatars.githubusercontent.com/u/63256188?v=4",
"events_url": "https://api.github.com/users/therealchrisneale/events{/privacy}",
"followers_url": "https://api.github.com/users/therealchrisneale/followers",
"following_url": "https://api.github.com/users/therealchrisneale/following{/other_user}",
"gists_url": "https://api.github.com/users/therealchrisneale/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/therealchrisneale",
"id": 63256188,
"login": "therealchrisneale",
"node_id": "MDQ6VXNlcjYzMjU2MTg4",
"organizations_url": "https://api.github.com/users/therealchrisneale/orgs",
"received_events_url": "https://api.github.com/users/therealchrisneale/received_events",
"repos_url": "https://api.github.com/users/therealchrisneale/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/therealchrisneale/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/therealchrisneale/subscriptions",
"type": "User",
"url": "https://api.github.com/users/therealchrisneale"
} | [] | closed | false | null | [] | null | [
"Hi Chris thanks for reaching out. We haven't tested AlphaFold with this version of Cuda. I would recommend running some test structures and checking that the metrics match what is expected. We have recently seen some variability depending on hardware https://github.com/deepmind/alphafold/issues/597 so its worth checking this."
] | "2022-09-29T20:35:38Z" | "2022-09-30T12:51:52Z" | "2022-09-30T12:51:52Z" | NONE | null | Hello,
our cluster has nvidia kernel driver version 510.85.02. I can build the container with cudatoolkit 11.1.1, but when I run I get the errors listed below and the GPUs are not found/used. When I modify the Dockerfile to use cuda 11.7.0, it builds and runs just fine and the output structure is sensible. Is there any reason to be cautious of using cudatoolkit 11.7.0 instead of 11.1.1 ?
In case it matters:
(1) I am building this on charliecloud instead of docker
(2) I modified the LD_LIBRARY_PATH a bit because it listed /usr/local/nvidia/lib:/usr/local/nvidia/lib64 but out of conda the libcuda.so.1 was actually in /usr/local/cuda/compat
### Here's the issue I was having when using cudatoolkit 11.1.1 inside the container:
I0929 20:05:58.839441 140412502734656 xla_bridge.py:350] Unable to initialize backend 'tpu_driver': NOT_FOUND: Unable to find driver in registry given worker:
2022-09-29 20:05:59.142936: E external/org_tensorflow/tensorflow/stream_executor/cuda/cuda_driver.cc:265] failed call to cuInit: CUDA_ERROR_SYSTEM_DRIVER_MISMATCH: system has unsupported display driver / cuda driver combination
2022-09-29 20:05:59.146190: E external/org_tensorflow/tensorflow/stream_executor/cuda/cuda_diagnostics.cc:313] kernel version 510.85.2 does not match DSO version 455.45.1 -- cannot find working devices in this configuration
I0929 20:05:59.189327 140412502734656 xla_bridge.py:350] Unable to initialize backend 'cuda': FAILED_PRECONDITION: No visible GPU devices.
I0929 20:05:59.189445 140412502734656 xla_bridge.py:350] Unable to initialize backend 'rocm': NOT_FOUND: Could not find registered platform with name: "rocm". Available platform names are: CUDA Host Interpreter
I0929 20:05:59.189768 140412502734656 xla_bridge.py:350] Unable to initialize backend 'tpu': module 'jaxlib.xla_extension' has no attribute 'get_tpu_client'
W0929 20:05:59.189845 140412502734656 xla_bridge.py:357] No GPU/TPU found, falling back to CPU. (Set TF_CPP_MIN_LOG_LEVEL=0 and rerun for more info.)
### When I build the container with cudatoolkit 11.7.0 and run it, the GPUs are found and the lack of TPUs is expected on this system:
I0929 13:37:31.731364 140268510418752 xla_bridge.py:356] Unable to initialize backend 'tpu_driver': NOT_FOUND: Unable to find driver in registry given worker:
I0929 13:37:33.668347 140268510418752 xla_bridge.py:356] Unable to initialize backend 'rocm': NOT_FOUND: Could not find registered platform with name: "rocm". Available platform names are: Interpreter Host CUDA
I0929 13:37:33.668632 140268510418752 xla_bridge.py:356] Unable to initialize backend 'tpu': module 'jaxlib.xla_extension' has no attribute 'get_tpu_client'
I0929 13:37:33.668713 140268510418752 xla_bridge.py:356] Unable to initialize backend 'plugin': xla_extension has no attributes named get_plugin_device_client. Compile TensorFlow with //tensorflow/compiler/xla/python:enable_plugin_device set to true (defaults to false) to enable this.
Thank you for your advice,
Chris. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/602/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/602/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/601 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/601/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/601/comments | https://api.github.com/repos/deepmind/alphafold/issues/601/events | https://github.com/deepmind/alphafold/pull/601 | 1,387,987,781 | PR_kwDOFoWQLM4_tPTx | 601 | Add docker image build test. | {
"avatar_url": "https://avatars.githubusercontent.com/u/69149348?v=4",
"events_url": "https://api.github.com/users/jkwmoore/events{/privacy}",
"followers_url": "https://api.github.com/users/jkwmoore/followers",
"following_url": "https://api.github.com/users/jkwmoore/following{/other_user}",
"gists_url": "https://api.github.com/users/jkwmoore/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/jkwmoore",
"id": 69149348,
"login": "jkwmoore",
"node_id": "MDQ6VXNlcjY5MTQ5MzQ4",
"organizations_url": "https://api.github.com/users/jkwmoore/orgs",
"received_events_url": "https://api.github.com/users/jkwmoore/received_events",
"repos_url": "https://api.github.com/users/jkwmoore/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/jkwmoore/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/jkwmoore/subscriptions",
"type": "User",
"url": "https://api.github.com/users/jkwmoore"
} | [] | closed | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/601/checks?check_run_id=8576728881) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request.",
"That was meant to go to my own main >_<"
] | "2022-09-27T15:50:17Z" | "2022-09-27T15:50:48Z" | "2022-09-27T15:50:35Z" | NONE | null | null | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/601/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/601/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/601.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/601",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/601.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/601"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/600 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/600/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/600/comments | https://api.github.com/repos/deepmind/alphafold/issues/600/events | https://github.com/deepmind/alphafold/issues/600 | 1,385,298,093 | I_kwDOFoWQLM5Skfit | 600 | difference of results between locally run alphafold and alphafold colab | {
"avatar_url": "https://avatars.githubusercontent.com/u/80506994?v=4",
"events_url": "https://api.github.com/users/danny2551515/events{/privacy}",
"followers_url": "https://api.github.com/users/danny2551515/followers",
"following_url": "https://api.github.com/users/danny2551515/following{/other_user}",
"gists_url": "https://api.github.com/users/danny2551515/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/danny2551515",
"id": 80506994,
"login": "danny2551515",
"node_id": "MDQ6VXNlcjgwNTA2OTk0",
"organizations_url": "https://api.github.com/users/danny2551515/orgs",
"received_events_url": "https://api.github.com/users/danny2551515/received_events",
"repos_url": "https://api.github.com/users/danny2551515/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/danny2551515/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/danny2551515/subscriptions",
"type": "User",
"url": "https://api.github.com/users/danny2551515"
} | [] | open | false | null | [] | null | [
"Hi yes its possible to get different results as they are different environments and the model is stochastic. Could you please provide full reproducibility details about your run? Also, have you updated to the latest version?",
"Hello Htomlinson14\r\n\r\nSorry about the late response.\r\n\r\nI normally use AlphaFold V.2.2.2, and after I checked your comment, I used V.2.2.4, the latest version, for structural prediction.\r\n\r\nI carried out the prediction under these four conditions:\r\n\r\n \r\n1. use full DB and templates\r\n \r\npython3 /data/AF/alphafold/docker/run_docker.py \\\r\n--fasta_paths=${fasta_file_path} \\\r\n--max_template_date=3000-01-01 \\\r\n--model_preset=monomer_ptm \\\r\n--data_dir=/data/AF/AFDB/ \\\r\n--docker_user=0 \\\r\n--gpu_devices=0 \\\r\n--output_dir=/data/result/\r\n \r\n2. use reduced DB and templates\r\n \r\npython3 /data/AF/alphafold/docker/run_docker.py \\\r\n--fasta_paths=${fasta_file_path} \\\r\n--max_template_date=3000-01-01 \\\r\n--model_preset=monomer_ptm \\\r\n--data_dir=/data/AF/AFDB/ \\\r\n--docker_user=0 \\\r\n--gpu_devices=0 \\\r\n--output_dir=/data/result/ \\\r\n--db_preset=reduced_dbs\r\n \r\n3. use full DB and disable templates\r\n \r\npython3 /data/AF/alphafold/docker/run_docker.py \\\r\n--fasta_paths=${fasta_file_path} \\\r\n--max_template_date=1000-01-01 \\\r\n--model_preset=monomer_ptm \\\r\n--data_dir=/data/AF/AFDB/ \\\r\n--docker_user=0 \\\r\n--gpu_devices=0 \\\r\n--output_dir=/data/result/\r\n \r\n4. use reduced DB and disable templates\r\n \r\npython3 /data/AF/alphafold/docker/run_docker.py \\\r\n--fasta_paths=${fasta_file_path} \\\r\n--max_template_date=1000-01-01 \\\r\n--model_preset=monomer_ptm \\\r\n--data_dir=/data/AF/AFDB/ \\\r\n--docker_user=0 \\\r\n--gpu_devices=0 \\\r\n--output_dir=/data/result/ \\\r\n--db_preset=reduced_dbs\r\n\r\nUnder these four conditions, the five predictions I obtained respectively from both V.2.2.2 and V.2.2.4 were very different from the predictions from AlphaFold colab.\r\n\r\nAre you aware of the factors behind these differences occuring?\r\n\r\nLike I asked earlier, how can I obtain similar predictions from local AlphaFold and AlphaFold Colab?\r\n",
"Hi can you please report the machine (GPU) you are using and also the iptm+ptm results for each of these runs?",
"You may also wish to follow the discussions on https://github.com/deepmind/alphafold/issues/597",
"The average plddt value from the AlphaFold colab was as follows:\r\n\r\nver.alphafold colab\r\n\r\navg_plddt = 67.1059722222222\r\n\r\nThe machine that has AlphaFold installed uses Quadro RTX 8000. In AlphaFold V.2.2.2 and V.2.2.4, the predictions we obtained under the following four conditions are as follows:\r\n\r\n\r\n1.full db & templates = 3000-01-01\r\nver.2.2.4\r\n \"plddts\": {\r\n \"model_1_ptm_pred_0\": 97.25193926550712,\r\n \"model_2_ptm_pred_0\": 97.18683996607362,\r\n \"model_3_ptm_pred_0\": 54.927574842232,\r\n \"model_4_ptm_pred_0\": 58.54845161102359,\r\n \"model_5_ptm_pred_0\": 43.06532542208311\r\n },\r\n\r\nver.2.2.2\r\n \"plddts\": {\r\n \"model_1_ptm_pred_0\": 97.36867929339586,\r\n \"model_2_ptm_pred_0\": 97.21677934497643,\r\n \"model_3_ptm_pred_0\": 50.224661763080945,\r\n \"model_4_ptm_pred_0\": 39.979508179484505,\r\n \"model_5_ptm_pred_0\": 44.549697156675734\r\n }\r\n\r\n2.full db & templates = 1000-01-01\r\nver.2.2.4\r\n \"plddts\": {\r\n \"model_1_ptm_pred_0\": 43.664334482582525,\r\n \"model_2_ptm_pred_0\": 48.08251340941516,\r\n \"model_3_ptm_pred_0\": 61.68443432122087,\r\n \"model_4_ptm_pred_0\": 48.93973191788411,\r\n \"model_5_ptm_pred_0\": 39.43275963009237\r\n }\r\n\r\nver.2.2.2\r\n \"plddts\": {\r\n \"model_1_ptm_pred_0\": 58.981938345566,\r\n \"model_2_ptm_pred_0\": 46.08298427146891,\r\n \"model_3_ptm_pred_0\": 58.97027318035094,\r\n \"model_4_ptm_pred_0\": 55.71557161934546,\r\n \"model_5_ptm_pred_0\": 44.21769917365889\r\n }\r\n\r\n\r\n3.reduced db & templates = 3000-01-01\r\nver.2.2.4\r\n \"plddts\": {\r\n \"model_1_ptm_pred_0\": 97.26357937406594,\r\n \"model_2_ptm_pred_0\": 97.13925092424562,\r\n \"model_3_ptm_pred_0\": 40.45697902340831,\r\n \"model_4_ptm_pred_0\": 41.859432024373355,\r\n \"model_5_ptm_pred_0\": 40.82802636714861\r\n },\r\n\r\nver.2.2.2\r\n \"plddts\": {\r\n \"model_1_ptm_pred_0\": 97.46789556103246,\r\n \"model_2_ptm_pred_0\": 97.30054276924895,\r\n \"model_3_ptm_pred_0\": 53.98386122906149,\r\n \"model_4_ptm_pred_0\": 45.08604272171022,\r\n \"model_5_ptm_pred_0\": 38.54566059335381\r\n }\r\n\r\n\r\n4.reduced db & templates = 1000-01-01\r\nver.2.2.4\r\n \"plddts\": {\r\n \"model_1_ptm_pred_0\": 48.53682881088287,\r\n \"model_2_ptm_pred_0\": 49.97295933519744,\r\n \"model_3_ptm_pred_0\": 55.95563452058329,\r\n \"model_4_ptm_pred_0\": 54.111995563263015,\r\n \"model_5_ptm_pred_0\": 42.91102593340323\r\n }\r\n\r\nver.2.2.2\r\n \"plddts\": {\r\n \"model_1_ptm_pred_0\": 43.48648526660625,\r\n \"model_2_ptm_pred_0\": 52.457455662168826,\r\n \"model_3_ptm_pred_0\": 48.408318636112455,\r\n \"model_4_ptm_pred_0\": 61.21301963988726,\r\n \"model_5_ptm_pred_0\": 52.85276750489996\r\n }\r\n",
"Hi - thanks very much for providing these values. I can't say exactly what is happening here but a few things to consider:\r\n* The dictionary keys say e.g. \"model_1_ptm_pred_0\" but are listed as plddt. Are these definitely pLDDT and not pTM?\r\n* It seems that a newer template DB is being used than what is searched against in the colab. This could be why the results for local runs with templates are much higher, as there could be near exact hits. We don't recommend turning off templates to achieve reproducibility\r\n* Predictions with low confidence (40-60 pLDDT is fairly low) are often highly variable, so its hard to compare results with high precision in this range.",
"Yes, when I checked the Ranking_debug.json file, it was marked as plddt. Should I check the ptm value in the pkl file and tell you?",
"Ok cool. I think in this case the second and third bullets above are most relevant, particularly the comments on the potential impact of newer templates. Thanks!",
"Hi @Htomlinson14 .\r\n\r\nI repeated attempts to match the results of local version and colab version.\r\n\r\nI confirmed advice from Issue #126 to disable HHBlits on UniClust in addition to db-preset and templates.\r\n\r\nCould you tell me how to disable HHBlits?"
] | "2022-09-26T02:07:14Z" | "2022-10-10T23:02:59Z" | null | NONE | null | Hello. I am making structural predictions using the colab version and the local version.
I have confirmed that there is a big difference between the two versions.
To solve this difference, I run alphafold locally with -db_preset=reduced_dbs and max_template_date=1000-01-01.
But there is still a big difference.
Is it possible to get similar results from the alphafold to the alphafold colab version? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/600/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/600/timeline | null | reopened | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/599 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/599/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/599/comments | https://api.github.com/repos/deepmind/alphafold/issues/599/events | https://github.com/deepmind/alphafold/issues/599 | 1,384,912,924 | I_kwDOFoWQLM5SjBgc | 599 | A more general fasta file parser | {
"avatar_url": "https://avatars.githubusercontent.com/u/4644611?v=4",
"events_url": "https://api.github.com/users/ichxw/events{/privacy}",
"followers_url": "https://api.github.com/users/ichxw/followers",
"following_url": "https://api.github.com/users/ichxw/following{/other_user}",
"gists_url": "https://api.github.com/users/ichxw/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ichxw",
"id": 4644611,
"login": "ichxw",
"node_id": "MDQ6VXNlcjQ2NDQ2MTE=",
"organizations_url": "https://api.github.com/users/ichxw/orgs",
"received_events_url": "https://api.github.com/users/ichxw/received_events",
"repos_url": "https://api.github.com/users/ichxw/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ichxw/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ichxw/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ichxw"
} | [] | open | false | null | [] | null | [] | "2022-09-25T06:20:13Z" | "2022-09-25T06:20:13Z" | null | NONE | null | The fasta parser currently doesn't accept ; at the beginning of one line as a comment symbol. Could the fasta parser be more general? like the reference here: https://en.wikipedia.org/wiki/FASTA_format | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/599/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/599/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/598 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/598/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/598/comments | https://api.github.com/repos/deepmind/alphafold/issues/598/events | https://github.com/deepmind/alphafold/issues/598 | 1,383,122,403 | I_kwDOFoWQLM5ScMXj | 598 | How does AlphaFold2 determine the protonation state/hydrogen | {
"avatar_url": "https://avatars.githubusercontent.com/u/36098720?v=4",
"events_url": "https://api.github.com/users/shaoqx/events{/privacy}",
"followers_url": "https://api.github.com/users/shaoqx/followers",
"following_url": "https://api.github.com/users/shaoqx/following{/other_user}",
"gists_url": "https://api.github.com/users/shaoqx/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/shaoqx",
"id": 36098720,
"login": "shaoqx",
"node_id": "MDQ6VXNlcjM2MDk4NzIw",
"organizations_url": "https://api.github.com/users/shaoqx/orgs",
"received_events_url": "https://api.github.com/users/shaoqx/received_events",
"repos_url": "https://api.github.com/users/shaoqx/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/shaoqx/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/shaoqx/subscriptions",
"type": "User",
"url": "https://api.github.com/users/shaoqx"
} | [] | open | false | null | [] | null | [] | "2022-09-22T23:28:13Z" | "2022-09-22T23:29:11Z" | null | NONE | null | Hi! I'm working on projects related to protonating the protein under a specific pH. I found outputs of AlphaFold2 prediction contain hydrogens in the model and I wonder how AlphaFold2 determines the protonation state of residues?
I traced back to the feature of AlphaFold2 used for training and found in the Nature paper it says `"one is atom37 where each heavy atom ... The other representation we employ is called atom14 ... 14 is chosen because it is the maximum number of heavy atoms for any standard amino acid."` But I found in the feature map from intermediate files that there are features for hydrogen atoms.
So I wonder if AlphaFold2 predicts positions of hydrogen atoms by training on datasets with existing hydrogens? If so, how are those hydrogens determined in the training set? If not, how is positions of hydrogens determined.
(I also tested some predictions for systems like 1NVG, the protonation state around the Zn center is not correct.) | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/598/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/598/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/597 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/597/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/597/comments | https://api.github.com/repos/deepmind/alphafold/issues/597/events | https://github.com/deepmind/alphafold/issues/597 | 1,382,696,313 | I_kwDOFoWQLM5SakV5 | 597 | Multimer predictions degraded after updating from 2.2.3 to 2.2.4? | {
"avatar_url": "https://avatars.githubusercontent.com/u/13498154?v=4",
"events_url": "https://api.github.com/users/lucajovine/events{/privacy}",
"followers_url": "https://api.github.com/users/lucajovine/followers",
"following_url": "https://api.github.com/users/lucajovine/following{/other_user}",
"gists_url": "https://api.github.com/users/lucajovine/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/lucajovine",
"id": 13498154,
"login": "lucajovine",
"node_id": "MDQ6VXNlcjEzNDk4MTU0",
"organizations_url": "https://api.github.com/users/lucajovine/orgs",
"received_events_url": "https://api.github.com/users/lucajovine/received_events",
"repos_url": "https://api.github.com/users/lucajovine/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/lucajovine/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/lucajovine/subscriptions",
"type": "User",
"url": "https://api.github.com/users/lucajovine"
} | [] | open | false | null | [] | null | [
"Hi thanks for raising this. Can you please provide more details of the test\nrun so this can be reproduced?\n\nOn Thu, 22 Sept 2022 at 17:12, Luca Jovine ***@***.***> wrote:\n\n> Hello, this is ranking_debug.json from a test multimer run that I had\n> performed with version 2.2.3:\n>\n> { \"iptm+ptm\": { \"model_1_multimer_v2_pred_0\": 0.8584136076107545,\n> \"model_2_multimer_v2_pred_0\": 0.8055674020453231,\n> \"model_3_multimer_v2_pred_0\": 0.6845710872128711,\n> \"model_4_multimer_v2_pred_0\": 0.8423123043127994,\n> \"model_5_multimer_v2_pred_0\": 0.8676285262204777 }, \"order\": [\n> \"model_5_multimer_v2_pred_0\", \"model_1_multimer_v2_pred_0\",\n> \"model_4_multimer_v2_pred_0\", \"model_2_multimer_v2_pred_0\",\n> \"model_3_multimer_v2_pred_0\" ] }\n>\n> and this is from an equivalent job, carried out using the same exact\n> input, after updating to 2.2.4:\n>\n> { \"iptm+ptm\": { \"model_1_multimer_v2_pred_0\": 0.40774006137839586,\n> \"model_2_multimer_v2_pred_0\": 0.44277279108467366,\n> \"model_3_multimer_v2_pred_0\": 0.442618376771567,\n> \"model_4_multimer_v2_pred_0\": 0.5631646773883024,\n> \"model_5_multimer_v2_pred_0\": 0.48998768122405234 }, \"order\": [\n> \"model_4_multimer_v2_pred_0\", \"model_5_multimer_v2_pred_0\",\n> \"model_2_multimer_v2_pred_0\", \"model_3_multimer_v2_pred_0\",\n> \"model_1_multimer_v2_pred_0\" ] }\n>\n> Is anyone else experiencing the same? I am not getting any obvious error\n> upon running 2.2.4, so it's not clear to me why there should be such a\n> significant difference...\n>\n> —\n> Reply to this email directly, view it on GitHub\n> <https://github.com/deepmind/alphafold/issues/597>, or unsubscribe\n> <https://github.com/notifications/unsubscribe-auth/ADKS2BZQTT7WLUCNIVLWTRLV7SAOLANCNFSM6AAAAAAQTGX6SM>\n> .\n> You are receiving this because you are subscribed to this thread.Message\n> ID: ***@***.***>\n>\n",
"Sure, of course. This was my input FASTA:\r\n\r\n```\r\n>protein1\r\nIDWDVYCSQDESIPAKFISRLVTSKDQALEKTEINCSNGLVPITQEFGINMMLIQYTRNELLDSPGMCVFWGPYSVPKNDTVVLYTVTARLKWSEGPPTNLSIQCYMPK\r\n>protein2\r\nRSWHYVEPKFLNKAFEVALKVQIIAGFDRGLVKWLRVHGRTLSTVQKKALYFVNRRYMQTHWANYMLWINKKIDALGRTPVVGDYTRLGAEIGRRIDMAYFYDFLKDKNMIPKYLPYMEEINRMRPADVPVKYM\r\n```\r\n(which basically corresponds to our PDB deposition 5IIB)\r\n\r\nFlags were --db_preset=full_dbs --model_preset=multimer --num_multimer_predictions_per_model=1 --gpu_devices=\"0\" --max_template_date=3000-01-01",
"Also...\r\n\r\n```\r\n> python run_alphafold.py --helpshort\r\n/usr/local/conda/miniconda3/envs/af2/lib/python3.7/site-packages/haiku/_src/data_structures.py:37: FutureWarning: jax.tree_structure is deprecated, and will be removed in a future release. Use jax.tree_util.tree_structure instead.\r\n PyTreeDef = type(jax.tree_structure(None))\r\nTraceback (most recent call last):\r\n File \"run_alphafold.py\", line 39, in <module>\r\n from alphafold.relax import relax\r\n File \"/media/3p5TBssd1/usr/local/alphafold/alphafold/alphafold/relax/relax.py\", line 18, in <module>\r\n from alphafold.relax import amber_minimize\r\n File \"/media/3p5TBssd1/usr/local/alphafold/alphafold/alphafold/relax/amber_minimize.py\", line 25, in <module>\r\n from alphafold.relax import cleanup\r\n File \"/media/3p5TBssd1/usr/local/alphafold/alphafold/alphafold/relax/cleanup.py\", line 22, in <module>\r\n import pdbfixer\r\nModuleNotFoundError: No module named 'pdbfixer'\r\n```\r\n\r\nI guess this could be automatically installed via conda?",
"just my 2 cents, I don't have the issue with \r\n``` \r\n --use_gpu_relax=true \\\r\n --model_preset=multimer \\\r\n --max_template_date=3000-01-01 \\\r\n --db_preset=full_dbs\r\n ```\r\n \r\n pasted sorted values: (2.2.4 on the left and 2.2.3 on the right)\r\n ```\r\n \"model_5_multimer_v2_pred_1\": 0.8600176497731321, \"model_2_multimer_v2_pred_4\": 0.8423162898031145,\r\n \"model_4_multimer_v2_pred_1\": 0.8355726960868132, \"model_2_multimer_v2_pred_2\": 0.8345795690267899,\r\n \"model_5_multimer_v2_pred_0\": 0.8106497232061523, \"model_4_multimer_v2_pred_4\": 0.8274770532116257,\r\n \"model_5_multimer_v2_pred_4\": 0.7618542529543955 \"model_5_multimer_v2_pred_1\": 0.8100580241139267,\r\n \"model_1_multimer_v2_pred_2\": 0.7462422015606857, \"model_1_multimer_v2_pred_1\": 0.7556708441527138,\r\n \"model_5_multimer_v2_pred_3\": 0.7262914654950798, \"model_1_multimer_v2_pred_2\": 0.7530240249541638,\r\n \"model_4_multimer_v2_pred_2\": 0.6850626261632934, \"model_4_multimer_v2_pred_0\": 0.7453822555113954,\r\n \"model_1_multimer_v2_pred_4\": 0.6504379081135182, \"model_1_multimer_v2_pred_3\": 0.7074747369853926,\r\n \"model_4_multimer_v2_pred_3\": 0.6282972043309893, \"model_4_multimer_v2_pred_2\": 0.6217676770784255,\r\n \"model_4_multimer_v2_pred_0\": 0.5659582021061171, \"model_5_multimer_v2_pred_2\": 0.5892705180579751,\r\n \"model_1_multimer_v2_pred_3\": 0.5530272925711416, \"model_1_multimer_v2_pred_4\": 0.5886967664945151,\r\n \"model_1_multimer_v2_pred_1\": 0.550091036479859, \"model_4_multimer_v2_pred_1\": 0.5816013102240326,\r\n \"model_2_multimer_v2_pred_0\": 0.537335888385569, \"model_2_multimer_v2_pred_0\": 0.517386942967343,\r\n \"model_2_multimer_v2_pred_2\": 0.534451564803337, \"model_1_multimer_v2_pred_0\": 0.5026680309406077,\r\n \"model_4_multimer_v2_pred_4\": 0.5258449296466584, \"model_4_multimer_v2_pred_3\": 0.48747237933686044,\r\n \"model_2_multimer_v2_pred_3\": 0.515407234197668, \"model_5_multimer_v2_pred_3\": 0.46593239818276644,\r\n \"model_3_multimer_v2_pred_1\": 0.499675159211949, \"model_3_multimer_v2_pred_2\": 0.43583198796278766,\r\n \"model_2_multimer_v2_pred_4\": 0.4734043448923909, \"model_2_multimer_v2_pred_1\": 0.42842107067582047,\r\n \"model_3_multimer_v2_pred_0\": 0.46022060251713326, \"model_5_multimer_v2_pred_0\": 0.4262880199002601,\r\n \"model_1_multimer_v2_pred_0\": 0.4401072457868955, \"model_3_multimer_v2_pred_1\": 0.42030398029562555,\r\n \"model_5_multimer_v2_pred_2\": 0.4233857428348928, \"model_3_multimer_v2_pred_4\": 0.41386853973521476,\r\n \"model_3_multimer_v2_pred_4\": 0.4185971424788965, \"model_3_multimer_v2_pred_0\": 0.4004228507119986,\r\n \"model_3_multimer_v2_pred_3\": 0.4112920469041982, \"model_2_multimer_v2_pred_3\": 0.381738114315064,\r\n \"model_3_multimer_v2_pred_2\": 0.3990563873655274, \"model_3_multimer_v2_pred_3\": 0.3696506974209459,\r\n \"model_2_multimer_v2_pred_1\": 0.3888554107330705, \"model_5_multimer_v2_pred_4\": 0.35258544121857827\r\n```",
"Thanks @lucajovine. Are you running this with the docker image? It should have all of the prerequisites installed. pdbfixer isn't available on pypi, so needs to be installed with conda.\r\n\r\nThanks @truatpasteurdotfr we also didn't see any issues in internal testing. ",
"Hi, thank you - am re-installing 2.2.4 from scratch rather than pulling it into my existing installation (although this always worked fine in the past, maybe something odd happened this time). Will re-run the test after that and let you know what happened!\r\n@Htomlinson14 yes this was from the docker image\r\n",
"Hello again, so I reinstalled from scratch 2.2.3 and 2.2.4 on two different machines and re-run 4 test jobs on each (two different monomers and two different multimers, with multimer_1 being the same as above).\r\nI am afraid that the problem persists, even though (1) it does not seem to happen with monomers and (2) it is much more evident for one multimer than the other:\r\n\r\n```\r\nmonomer_1\r\n\t2.2.3\r\n\t\t\"model_1_pred_0\": 92.51882397715578,\r\n\t\t\"model_2_pred_0\": 92.2056299645637,\r\n\t\t\"model_3_pred_0\": 92.03138974894895,\r\n\t\t\"model_4_pred_0\": 93.1682804560119,\r\n\t\t\"model_5_pred_0\": 93.65241384700947\r\n\t2.2.4\r\n\t\t\"model_1_pred_0\": 92.53758910826187,\r\n\t\t\"model_2_pred_0\": 92.94215515721487,\r\n\t\t\"model_3_pred_0\": 93.52598782948384,\r\n\t\t\"model_4_pred_0\": 93.42160017797602,\r\n\t\t\"model_5_pred_0\": 93.70445135155603\r\n\r\nmonomer_2\r\n\t2.2.3\r\n\t\t\"model_1_pred_0\": 85.85987502475369,\r\n\t\t\"model_2_pred_0\": 84.90289928449474,\r\n\t\t\"model_3_pred_0\": 86.95139195928073,\r\n\t\t\"model_4_pred_0\": 86.64831516433784,\r\n\t\t\"model_5_pred_0\": 86.92184064848513\r\n\t2.2.4\r\n\t\t\"model_1_pred_0\": 85.94278137586902,\r\n\t\t\"model_2_pred_0\": 84.20654974770405,\r\n\t\t\"model_3_pred_0\": 86.83553028330901,\r\n\t\t\"model_4_pred_0\": 87.14120498958022,\r\n\t\t\"model_5_pred_0\": 87.1268406084284\r\n\r\nmultimer_1\r\n\t2.2.3\r\n\t\t\"model_1_multimer_v2_pred_0\": 0.8501568830419893,\r\n\t\t\"model_2_multimer_v2_pred_0\": 0.8486699468672132,\r\n\t\t\"model_3_multimer_v2_pred_0\": 0.3229293496900477,\r\n\t\t\"model_4_multimer_v2_pred_0\": 0.8455477128109672,\r\n\t\t\"model_5_multimer_v2_pred_0\": 0.852226493932774\t\r\n\t2.2.4\r\n\t\t\"model_1_multimer_v2_pred_0\": 0.5117391870226631,\r\n\t\t\"model_2_multimer_v2_pred_0\": 0.4098071256975262,\r\n\t\t\"model_3_multimer_v2_pred_0\": 0.43044599005298734,\r\n\t\t\"model_4_multimer_v2_pred_0\": 0.5445095546638219,\r\n\t\t\"model_5_multimer_v2_pred_0\": 0.5859474538776686\r\n\r\nmultimer_2\r\n\t2.2.3\r\n\t\t\"model_1_multimer_v2_pred_0\": 0.8336408586490794,\r\n\t\t\"model_2_multimer_v2_pred_0\": 0.8327994809055634,\r\n\t\t\"model_3_multimer_v2_pred_0\": 0.8024798134822844,\r\n\t\t\"model_4_multimer_v2_pred_0\": 0.811666152078712,\r\n\t\t\"model_5_multimer_v2_pred_0\": 0.8072626282045929\r\n\t2.2.4\r\n\t\t\"model_1_multimer_v2_pred_0\": 0.7638937624852511,\r\n\t\t\"model_2_multimer_v2_pred_0\": 0.7730047759497934,\r\n\t\t\"model_3_multimer_v2_pred_0\": 0.8002452325904037,\r\n\t\t\"model_4_multimer_v2_pred_0\": 0.7938463939736111,\r\n\t\t\"model_5_multimer_v2_pred_0\": 0.7926498563280233\r\n```\r\n\r\nI have not done such careful comparisons before, but I always ran a couple of tests after updating to new versions and - although of course some minor variability is normal - I do not recall seeing differences of the kind observed for multimer_1...",
"Hi thanks very much for this. I will investigate and get back to you.",
"Hi again, I'm not seeing the same performance issues on the fasta sequence you provided. Below are the results I have obtained:\r\n\r\n```\r\n2.2.3\r\n \"model_1_multimer_v2_pred_0\": 0.7026557510461339,\r\n \"model_2_multimer_v2_pred_0\": 0.4509017402688641,\r\n \"model_3_multimer_v2_pred_0\": 0.380578570853769,\r\n \"model_4_multimer_v2_pred_0\": 0.6910764110918439,\r\n \"model_5_multimer_v2_pred_0\": 0.7656100030107779\r\n\r\n2.2.4\r\n \"model_1_multimer_v2_pred_0\": 0.6930976486128585,\r\n \"model_2_multimer_v2_pred_0\": 0.840286483893667,\r\n \"model_3_multimer_v2_pred_0\": 0.36765421415043464,\r\n \"model_4_multimer_v2_pred_0\": 0.6591293362123893,\r\n \"model_5_multimer_v2_pred_0\": 0.8106014168732179\r\n```\r\n\r\nWhat hardware are you using?",
"Hi, both jobs were run on PCs running Ubuntu 20.04, one with a Quadro RTX 5000 (2.2.3) and the other with a GeForce RTX 2070 (2.2.4).",
"Hi thanks for this. Would it be possible to reverse the experiment?\r\n\r\nAlso, what cuda versions are on these machines? (run nvidia-smi)",
"2.2.4 (2022-09-22) with data freshly re-downloaded on DGX A100 (driver 515.65.01) singularity image from docker `docker://ghcr.io/truatpasteurdotfr/alphafold:main` and ealier version with 2.2.3 (2022-09-13)\r\n```\r\n}tru@myrdal:~/alphafold$ head -n 8 alphafold-2022-09-13-1905-data-2.2.4-20220923-num_multimer_predictions_per_model_1-IALsG/issue-587/ranking_debug.json \r\n{\r\n \"iptm+ptm\": {\r\n \"model_1_multimer_v2_pred_0\": 0.8568935778701908,\r\n \"model_2_multimer_v2_pred_0\": 0.8499162052690372,\r\n \"model_3_multimer_v2_pred_0\": 0.2961016070831388,\r\n \"model_4_multimer_v2_pred_0\": 0.8552748738237211,\r\n \"model_5_multimer_v2_pred_0\": 0.851201296704003\r\n },\r\ntru@myrdal:~/alphafold$ head -n 8 alphafold-2022-09-22-2049-data-2.2.4-20220923-num_multimer_predictions_per_model_1-OCW7T/issue-587/ranking_debug.json \r\n{\r\n \"iptm+ptm\": {\r\n \"model_1_multimer_v2_pred_0\": 0.8550884037892792,\r\n \"model_2_multimer_v2_pred_0\": 0.39117327804768387,\r\n \"model_3_multimer_v2_pred_0\": 0.6731524796823938,\r\n \"model_4_multimer_v2_pred_0\": 0.8525504678600079,\r\n \"model_5_multimer_v2_pred_0\": 0.8554723246526093\r\n },\r\n```",
"On ColabFold, we've been using the latest version of jax. One user reported getting different results depending on which GPU they got. They report that A100 gives different result compared to V100 or T4.",
"...good point @sokrypton: my first comment would've exactly been that I was not aware that the type of GPU that one used made a difference to the actual quality of an inference - other than of course by having enough VRAM to run the corresponding job to start with. But if it does, then this would be a quite crucial piece of information.\r\n\r\n@Htomlinson14: yes of course I can reverse the experiment. But since one of the machines is currently running other jobs using 2.2.3, is it fine if on that one I just create a new conda environment to build the 2.2.4 docker image and then run the reverse test, or could the slightly different setup introduce further issues?\r\nAs for CUDA, both machines have 11.5.",
"Thanks for raising this. For now, if you are using an affected GPU we recommend pinning to previous versions of the repo. We will investigate the issue further.",
"OK sure, looking forward to hearing what you find...",
"Sorry I couldn't follow. Which GPUs are affected now?",
"I'm beginning to suspect the old jax library was more consistent across GPUs, while the new jax library is not.\r\nUnfortunately, I don't have access to any high-end GPUs (like A100), so I can't do any tests on my side.\r\n\r\nI think one informative test to do would be: run on 2.2.3 with A100 and non-A100, then repeat the same experiment with 2.2.4 on A100 and non-A100. ",
"Same here, but if DeepMind/Alphabet wants to indefinitely lend us one to test out things I will not say no! ;-)",
"I think DeepMind/Alphabet only uses TPUs :P ",
"It's OK, I'll take anything!!",
"I will be trying on a dual A40 server where I have enough free space to download data, not sure I can do that on my workstation with an RTX 2080...\r\n",
"A40 (48GB), Driver Version: 510.54\r\n```\r\n \"iptm+ptm\": {\r\n \"model_1_multimer_v2_pred_0\": 0.8501128109383987,\r\n \"model_2_multimer_v2_pred_0\": 0.8540356829322757,\r\n \"model_3_multimer_v2_pred_0\": 0.6771353362331958,\r\n \"model_4_multimer_v2_pred_0\": 0.8452873344090073,\r\n \"model_5_multimer_v2_pred_0\": 0.8578954696460785\r\n },\r\n```\r\nRTX-2080ti (11GB), Driver Version: 515.65.01\r\n```\r\n \"iptm+ptm\": {\r\n \"model_1_multimer_v2_pred_0\": 0.8535170484093717,\r\n \"model_2_multimer_v2_pred_0\": 0.8481280255454735,\r\n \"model_3_multimer_v2_pred_0\": 0.7481300076762746,\r\n \"model_4_multimer_v2_pred_0\": 0.8371862945478382,\r\n \"model_5_multimer_v2_pred_0\": 0.8328140753338644\r\n },\r\n```\r\nI will try to get access on a V100 and rtx1080 machine.",
"@truatpasteurdotfr interesting! can you repeat this but with v2.2.3 (to see if the outputs are more consistent between GPUs with the original pinned version of jax).",
"In case it is useful, here's results for the test posted in this thread from v2.2.4 + cray + A100 + nvidia kernel 510.85.02 + cudatoolkit 11.7.0 + charliecloud in place of docker + databases downloaded on Sept 23, 2022 + removed the 4 offending entries from pdb_seqres/pdb_seqres.txt that match ^CT05\r\n\r\n{\r\n \"iptm+ptm\": {\r\n \"model_1_multimer_v2_pred_0\": 0.8440023053565382,\r\n \"model_2_multimer_v2_pred_0\": 0.8485773361074703,\r\n \"model_3_multimer_v2_pred_0\": 0.26827771102715653,\r\n \"model_4_multimer_v2_pred_0\": 0.8483706916239296,\r\n \"model_5_multimer_v2_pred_0\": 0.8648632996723908\r\n },\r\n \"order\": [\r\n \"model_5_multimer_v2_pred_0\",\r\n \"model_2_multimer_v2_pred_0\",\r\n \"model_4_multimer_v2_pred_0\",\r\n \"model_1_multimer_v2_pred_0\",\r\n \"model_3_multimer_v2_pred_0\"\r\n ]\r\n}\r\n\r\nVisualizing them, I see that model 3 has a chain B Arg56 that adopts a different chi3 rotamer, which would sterically clash with chain A (probably at Glu46) if chain A adopted the same relative orientation that it does in the other 4 models. I didn't read up on the methods used to do multimers, but if the side chain rotamers get locked in before the packing is evaluated, or there are other similar things happening that are sensitive to the order of events, then this system may be subject to substantial statistical noise.",
"rtx 1080ti (Driver Version: 510.54 )out of memory, as it can not expand into the cpu RAM,,,\r\n```\r\n2022-10-01 07:07:58.693069: W external/org_tensorflow/tensorflow/compiler/xla/service/platform_util.cc:190] unable to create StreamExecutor for CUDA:0: failed initializing StreamExecutor for CUDA device ordinal 0: INTERNAL: failed call to cuDevicePrimaryCtxRetain: CUDA_ERROR_OUT_OF_MEMORY: out of memory; total memory reported: 11721179136\r\n```",
"Hi all. We have run AF multimer on several different GPUs and are not seeing any significant differences between predictions. @lucajovine would you mind checking to see if there are any differences in the MSAs or templates, on the two machines you are using? Thanks!",
"@Htomlinson14 There should not have been, because the databases used by one of them were a clone of those of the other...",
"Thanks -- sounds like its not a data issue. Its been difficult to reproduce internally and we have tried on several different GPUs. We will continue to look into this internally but until then we would recommend using v2.2.3, which has no difference in features. It seems likely that jax numerics have changed between jax versions on your hardware. One way to check this would be to change the jax and jaxlib versions, rebuild the docker and rerun the model. One could do this at various jax versions to diagnose whether this is the issue.",
"OK thanks, will stick to 2.2.3 for now and - compatibly with other people's work on the same machine - keep in mind the check you suggested."
] | "2022-09-22T16:12:05Z" | "2022-10-19T04:34:59Z" | null | NONE | null | Hello, this is ranking_debug.json from a test multimer run that I had performed with version 2.2.3:
`{
"iptm+ptm": {
"model_1_multimer_v2_pred_0": 0.8584136076107545,
"model_2_multimer_v2_pred_0": 0.8055674020453231,
"model_3_multimer_v2_pred_0": 0.6845710872128711,
"model_4_multimer_v2_pred_0": 0.8423123043127994,
"model_5_multimer_v2_pred_0": 0.8676285262204777
},
"order": [
"model_5_multimer_v2_pred_0",
"model_1_multimer_v2_pred_0",
"model_4_multimer_v2_pred_0",
"model_2_multimer_v2_pred_0",
"model_3_multimer_v2_pred_0"
]
}`
and this is from an equivalent job, carried out using the same exact input, after updating to 2.2.4:
`{
"iptm+ptm": {
"model_1_multimer_v2_pred_0": 0.40774006137839586,
"model_2_multimer_v2_pred_0": 0.44277279108467366,
"model_3_multimer_v2_pred_0": 0.442618376771567,
"model_4_multimer_v2_pred_0": 0.5631646773883024,
"model_5_multimer_v2_pred_0": 0.48998768122405234
},
"order": [
"model_4_multimer_v2_pred_0",
"model_5_multimer_v2_pred_0",
"model_2_multimer_v2_pred_0",
"model_3_multimer_v2_pred_0",
"model_1_multimer_v2_pred_0"
]
}`
Is anyone else experiencing the same? I am not getting any obvious error upon running 2.2.4, so it's not clear to me why there should be such a significant difference... | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/597/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/597/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/596 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/596/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/596/comments | https://api.github.com/repos/deepmind/alphafold/issues/596/events | https://github.com/deepmind/alphafold/pull/596 | 1,382,260,979 | PR_kwDOFoWQLM4_a3sn | 596 | fix bibtex in readme | {
"avatar_url": "https://avatars.githubusercontent.com/u/24830662?v=4",
"events_url": "https://api.github.com/users/mathe42/events{/privacy}",
"followers_url": "https://api.github.com/users/mathe42/followers",
"following_url": "https://api.github.com/users/mathe42/following{/other_user}",
"gists_url": "https://api.github.com/users/mathe42/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/mathe42",
"id": 24830662,
"login": "mathe42",
"node_id": "MDQ6VXNlcjI0ODMwNjYy",
"organizations_url": "https://api.github.com/users/mathe42/orgs",
"received_events_url": "https://api.github.com/users/mathe42/received_events",
"repos_url": "https://api.github.com/users/mathe42/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/mathe42/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/mathe42/subscriptions",
"type": "User",
"url": "https://api.github.com/users/mathe42"
} | [] | closed | false | null | [] | null | [
"Thanks for your pull request! It looks like this may be your first contribution to a Google open source project. Before we can look at your pull request, you'll need to sign a Contributor License Agreement (CLA).\n\nView this [failed invocation](https://github.com/deepmind/alphafold/pull/596/checks?check_run_id=8492253152) of the CLA check for more information.\n\nFor the most up to date status, view the checks section at the bottom of the pull request.",
"Fixed in https://github.com/deepmind/alphafold/commit/7b87da537498245b861e7831175c777ba89b3828, thanks for catching this!"
] | "2022-09-22T11:05:41Z" | "2022-11-01T14:18:28Z" | "2022-11-01T14:18:27Z" | NONE | null | null | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/596/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/596/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/596.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/596",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/596.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/596"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/595 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/595/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/595/comments | https://api.github.com/repos/deepmind/alphafold/issues/595/events | https://github.com/deepmind/alphafold/issues/595 | 1,378,505,618 | I_kwDOFoWQLM5SKlOS | 595 | AttributeError: module 'jax' has no attribute 'tree_multimap' | {
"avatar_url": "https://avatars.githubusercontent.com/u/20445196?v=4",
"events_url": "https://api.github.com/users/mnaidoo01/events{/privacy}",
"followers_url": "https://api.github.com/users/mnaidoo01/followers",
"following_url": "https://api.github.com/users/mnaidoo01/following{/other_user}",
"gists_url": "https://api.github.com/users/mnaidoo01/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/mnaidoo01",
"id": 20445196,
"login": "mnaidoo01",
"node_id": "MDQ6VXNlcjIwNDQ1MTk2",
"organizations_url": "https://api.github.com/users/mnaidoo01/orgs",
"received_events_url": "https://api.github.com/users/mnaidoo01/received_events",
"repos_url": "https://api.github.com/users/mnaidoo01/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/mnaidoo01/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/mnaidoo01/subscriptions",
"type": "User",
"url": "https://api.github.com/users/mnaidoo01"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"Hi thanks for raising this! This duplicates https://github.com/deepmind/alphafold/issues/589 so will close this issue",
"Thank you so much for getting back."
] | "2022-09-19T20:49:26Z" | "2022-09-20T14:43:31Z" | "2022-09-20T11:17:25Z" | NONE | null | This is related to #[456 ](https://github.com/deepmind/alphafold/issues/456).
"5. Run AlphaFold and download prediction" (in Colab notebook [example](https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb)), started throwing an error recently and now expects 'jax.tree_multimap' to be changed to 'jax.tree_map' (see [stackflow](https://stackoverflow.com/questions/73693738/module-jax-has-no-attribute-tree-multimap-in-alphafold2-colab))
[tfgg](https://github.com/tfgg) noted this back in May but maybe we should update this to accommodate the latest jax module in Colab. (jax 0.3.17 jaxlib 0.3.15+cuda11.cudnn805) | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/595/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/595/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/593 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/593/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/593/comments | https://api.github.com/repos/deepmind/alphafold/issues/593/events | https://github.com/deepmind/alphafold/issues/593 | 1,375,230,940 | I_kwDOFoWQLM5R-Fvc | 593 | Too many command-line arguments : No space in the command | {
"avatar_url": "https://avatars.githubusercontent.com/u/109017995?v=4",
"events_url": "https://api.github.com/users/Azjer2014/events{/privacy}",
"followers_url": "https://api.github.com/users/Azjer2014/followers",
"following_url": "https://api.github.com/users/Azjer2014/following{/other_user}",
"gists_url": "https://api.github.com/users/Azjer2014/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Azjer2014",
"id": 109017995,
"login": "Azjer2014",
"node_id": "U_kgDOBn97iw",
"organizations_url": "https://api.github.com/users/Azjer2014/orgs",
"received_events_url": "https://api.github.com/users/Azjer2014/received_events",
"repos_url": "https://api.github.com/users/Azjer2014/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Azjer2014/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Azjer2014/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Azjer2014"
} | [] | open | false | null | [] | null | [
"`--model_names=monomer` may be `--model_preset=monomer`.",
"Hi, \r\n\r\nI'm running into the same issue and below is my command:\r\n\r\n```\r\nsingularity run --home /scratch/users/sbusi/af2/alphafold/alphafold -B /scratch/users/sbusi/af2/alphafold/alphafold/data:/data -B .:/etc --pwd /scratch/users/sbusi/af2 --nv /scratch/users/sbusi/af2/af2-container/alphafold_2.2.3.sif run_alphafold.py \\\r\n--fasta_paths=${ALPHAFOLD_DATADIR}/h1.fa \\\r\n--output_dir=${ALPHAFOLD_DIR}/output_af2 \\\r\n--data_dir=${ALPHAFOLD_DATADIR} \\\r\n--uniref90_database_path=${ALPHAFOLD_DIR}/uniref90/uniref90.fasta \\\r\n--mgnify_database_path=${ALPHAFOLD_DIR}/mgnify/mgy_clusters_2018_12.fa \\\r\n--bfd_database_path=${ALPHAFOLD_DIR}/bfd \\\r\n--pdb70_database_path=${ALPHAFOLD_DIR}/pdb70 \\\r\n--template_mmcif_dir=${ALPHAFOLD_DIR}/pdb_mmcif/mmcif_files \\\r\n--obsolete_pdbs_path=${ALPHAFOLD_DIR}/pdb_mmcif/obsolete.dat \\\r\n--model_preset=monomer \\\r\n--max_template_date=2021-11-01 \\\r\n--use_gpu_relax=True \\\r\n--db_preset=full_dbs\r\n```\r\n\r\nThanks for your input on this.",
"> `--model_names=monomer` may be `--model_preset=monomer`.\r\n\r\nUsing your suggested parameter gives a different error about using a wrong parameter. The parameters in my command are correct. ",
">I have not troubleshoot this error yet but I can see your command is pointing to the actual files in the databases (mine direct to folders) so I'm going to try your way to see if it works. \r\n\r\n",
"> Using your suggested parameter gives a different error about using a wrong parameter. The parameters in my command are correct.\r\n\r\nHow about `--model_names=model_1` then?"
] | "2022-09-16T00:12:06Z" | "2022-12-04T15:12:00Z" | null | NONE | null | Hello, I'm trying to run alphafold using the command indicated below. I'm getting the error "Too many command-line arguments". I made sure to remove all the spaces. I'm not sure why I still get this error. I would appreciate any help to figure out the problem.
```
singularity run --home /Alphafold/alphafold-2.2.0 -e alphafold.sif run_alphafold.py \
--data_dir=/Alphafold/alphafold-2.2.0/data/ \
--output_dir=/Alphafold/alphafold-2.2.0/my-files/output/ \
--fasta_paths=/Alphafold/alphafold-2.2.0/my-files/input/wild-type.fasta \
--pdb70_database_path=/Alphafold/alphafold-2.2.0/data/pdb70/ \
--mgnify_database_path=/Alphafold/alphafold-2.2.0/data/mgnify/ \
--uniref90_database_path=/Alphafold/alphafold-2.2.0/data/uniref90/ \
--uniclust30_database_path=/Alphafold/alphafold-2.2.0/data/uniclust30/ \
--bfd_database_path=/Alphafold/alphafold-2.2.0/data/bfd/ \
--template_mmcif_dir=/Alphafold/alphafold-2.2.0/data/mmCIF/ \
--obsolete_pdbs_path=/Alphafold/alphafold-2.2.0/data/pdb_mmcif/ \
--max_template_date=2020-05-14 \
--model_names=monomer
```
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/593/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/593/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/592 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/592/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/592/comments | https://api.github.com/repos/deepmind/alphafold/issues/592/events | https://github.com/deepmind/alphafold/issues/592 | 1,373,926,292 | I_kwDOFoWQLM5R5HOU | 592 | one error about alphafold,ask for help online | {
"avatar_url": "https://avatars.githubusercontent.com/u/72640716?v=4",
"events_url": "https://api.github.com/users/285880830/events{/privacy}",
"followers_url": "https://api.github.com/users/285880830/followers",
"following_url": "https://api.github.com/users/285880830/following{/other_user}",
"gists_url": "https://api.github.com/users/285880830/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/285880830",
"id": 72640716,
"login": "285880830",
"node_id": "MDQ6VXNlcjcyNjQwNzE2",
"organizations_url": "https://api.github.com/users/285880830/orgs",
"received_events_url": "https://api.github.com/users/285880830/received_events",
"repos_url": "https://api.github.com/users/285880830/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/285880830/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/285880830/subscriptions",
"type": "User",
"url": "https://api.github.com/users/285880830"
} | [] | open | false | null | [] | null | [] | "2022-09-15T05:06:10Z" | "2022-09-15T05:06:10Z" | null | NONE | null | CE8eSpRY.fasta
报错:I0910 11:42:53.106589 139985805367104 hhsearch.py:76] Launching subprocess "hhsearch -i /tmp/tmpd42j2lo8/query.a3m -o /tmp/tmpd42j2lo8/output.hhr -maxseq 1000000 -d /mnt/pdb70/pdb70"
I0910 11:42:53.259523 139985805367104 utils.py:36] Started HHsearch query
I0910 11:42:53.617978 139985805367104 utils.py:40] Finished HHsearch query in 0.358 seconds
Traceback (most recent call last):
File "run_alphafold.py", line 338, in <module>
app.run(main)
File "/home/alphafold/miniconda3/envs/alphafold/lib/python3.8/site-packages/absl/app.py", line 312, in run
_run_main(main, args)
File "/home/alphafold/miniconda3/envs/alphafold/lib/python3.8/site-packages/absl/app.py", line 258, in _run_main
sys.exit(main(argv))
File "run_alphafold.py", line 310, in main
predict_structure(
File "run_alphafold.py", line 170, in predict_structure
feature_dict = data_pipeline.process(
File "/home/alphafold/alphafold/alphafold/data/pipeline.py", line 166, in process
hhsearch_result = self.hhsearch_pdb70_runner.query(uniref90_msa_as_a3m)
File "/home/alphafold/alphafold/alphafold/data/tools/hhsearch.py", line 85, in query
raise RuntimeError(
RuntimeError: HHSearch failed:
stdout:
stderr:
- 11:42:53.447 INFO: /tmp/tmpd42j2lo8/query.a3m is in A2M, A3M or FASTA format
- 11:42:53.447 WARNING: Ignoring invalid symbol '*' at pos. 1623 in line 2 of /tmp/tmpd42j2lo8/query.a3m
- 11:42:53.610 ERROR: [subseq from] CRISPR-associated endonuclease Cas9/Csn1 n=212 Tax=root TaxID=1 RepID=CAS9_STRP1
- 11:42:53.610 ERROR: Error in /opt/conda/conda-bld/hhsuite_1616660820288/work/src/hhalignment.cpp:1244: Compress:
- 11:42:53.610 ERROR: sequences in /tmp/tmpd42j2lo8/query.a3m do not all have the same number of columns,
- 11:42:53.610 ERROR:
e.g. first sequence and sequence UniRef90_Q99ZW2/2-1048.
- 11:42:53.610 ERROR: Check input format for '-M a2m' option and consider using '-M first' or '-M 50'
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/592/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/592/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/591 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/591/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/591/comments | https://api.github.com/repos/deepmind/alphafold/issues/591/events | https://github.com/deepmind/alphafold/issues/591 | 1,372,632,542 | I_kwDOFoWQLM5R0LXe | 591 | AlphaFold (or hmmsearch) is not able to parse some pdb_seqres.txt due to unusual residue naming | {
"avatar_url": "https://avatars.githubusercontent.com/u/547146?v=4",
"events_url": "https://api.github.com/users/TeletcheaLab/events{/privacy}",
"followers_url": "https://api.github.com/users/TeletcheaLab/followers",
"following_url": "https://api.github.com/users/TeletcheaLab/following{/other_user}",
"gists_url": "https://api.github.com/users/TeletcheaLab/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/TeletcheaLab",
"id": 547146,
"login": "TeletcheaLab",
"node_id": "MDQ6VXNlcjU0NzE0Ng==",
"organizations_url": "https://api.github.com/users/TeletcheaLab/orgs",
"received_events_url": "https://api.github.com/users/TeletcheaLab/received_events",
"repos_url": "https://api.github.com/users/TeletcheaLab/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/TeletcheaLab/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/TeletcheaLab/subscriptions",
"type": "User",
"url": "https://api.github.com/users/TeletcheaLab"
} | [] | closed | false | null | [] | null | [
"I've also encountered this issue. I hope `pdb_seqres.txt` itself or the AlphaFold pipeline will be improved.",
"Duplicate of #569 - please follow updates on that issue."
] | "2022-09-14T09:10:51Z" | "2022-09-19T15:13:10Z" | "2022-09-19T15:13:09Z" | NONE | null | Dear all,
I had trouble running a prediction with updated pdb_seqres.txt files since some entries contain unusual DNA residue names, PDB code 7ooo, 7oos and 7ozz. These nucleic acids are modified residues but do not follow DNA alphabet, so the parser fails with an error on the letter "0" (zero)
Traceback here and details below:
Traceback (most recent call last):
File "/app/alphafold/run_alphafold.py", line 422, in <module>
app.run(main)
File "/opt/alphafoldenv/lib/python3.8/site-packages/absl/app.py", line 312, in run
_run_main(main, args)
File "/opt/alphafoldenv/lib/python3.8/site-packages/absl/app.py", line 258, in _run_main
sys.exit(main(argv))
File "/app/alphafold/run_alphafold.py", line 398, in main
predict_structure(
File "/app/alphafold/run_alphafold.py", line 172, in predict_structure
feature_dict = data_pipeline.process(
File "/app/alphafold/alphafold/data/pipeline_multimer.py", line 264, in process
chain_features = self._process_single_chain(
File "/app/alphafold/alphafold/data/pipeline_multimer.py", line 212, in _process_single_chain
chain_features = self._monomer_data_pipeline.process(
File "/app/alphafold/alphafold/data/pipeline.py", line 185, in process
pdb_templates_result = self.template_searcher.query(msa_for_templates)
File "/app/alphafold/alphafold/data/tools/hmmsearch.py", line 79, in query
return self.query_with_hmm(hmm)
File "/app/alphafold/alphafold/data/tools/hmmsearch.py", line 112, in query_with_hmm
raise RuntimeError(
RuntimeError: hmmsearch failed:
stdout:
# hmmsearch :: search profile(s) against a sequence database
# HMMER 3.3.2 (Nov 2020); http://hmmer.org/
# Copyright (C) 2020 Howard Hughes Medical Institute.
# Freely distributed under the BSD open source license.
# - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
# query HMM file: /tmp/tmp2i0w1r3m/query.hmm
# target sequence database: /scratch/shared/dataset/alphafold_data/pdb_seqres/pdb_seqres.txt
# MSA of all hits saved to file: /tmp/tmp2i0w1r3m/output.sto
# show alignments in output: no
# sequence reporting threshold: E-value <= 100
# domain reporting threshold: E-value <= 100
# sequence inclusion threshold: E-value <= 100
# domain inclusion threshold: E-value <= 100
# MSV filter P threshold: <= 0.1
# Vit filter P threshold: <= 0.1
# Fwd filter P threshold: <= 0.1
# number of worker threads: 8
# - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Query: query [M=242]
stderr:
Parse failed (sequence file /scratch/shared/dataset/alphafold_data/pdb_seqres/pdb_seqres.txt):
Line 1364756: illegal character 0
After manually editing the file to remove the "05H" character (the modified DNA nucleotide) the error is gone. Here is a full diff:
diff -Naup pdb_seqres/pdb_seqres.txt-orig pdb_seqres/pdb_seqres.txt
--- pdb_seqres/pdb_seqres.txt-orig 2022-09-13 00:19:53.000000000 +0200
+++ pdb_seqres/pdb_seqres.txt 2022-09-13 00:36:37.000000000 +0200
@@ -1360655,9 +1360655,9 @@ CAAAGAAAAG
>7ooo_D mol:na length:10 RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3')
CAAAGAAAAG
>7ooo_B mol:na length:11 DNA (5'-D(*CP*TP*(RWQ)P*TP*CP*TP*TP*TP*G)-3')
-CT05ATCTTTG
+CTATCTTTG
>7ooo_E mol:na length:11 DNA (5'-D(*CP*TP*(RWQ)P*TP*CP*TP*TP*TP*G)-3')
-CT05ATCTTTG
+CTATCTTTG
>7oop_A mol:protein length:1970 DNA-directed RNA polymerase II subunit RPB1
MHGGGPPSGDSACPLRTIKRVQFGVLSPDELKRMSVTEGGIKYPETTEGGRPKLGGLMDPRQGVIERTGRCQTCAGNMTECPGHFGHIELAKPVFHVGFLVKTMKVLRCVCFFCSKLLVDSNNPKIKDILAKSKGQPKKRLTHVYDLCKGKNICEGGEEMDNKFGVEQPEGDEDLTKEKGHGGCGRYQPRIRRSGLELYAEWKHVNEDSQEKKILLSPERVHEIFKRISDEECFVLGMEPRYARPEWMIVTVLPVPPLSVRPAVVMQGSARNQDDLTHKLADIVKINNQLRRNEQNGAAAHVIAEDVKLLQFHVATMVDNELPGLPRAMQKSGRPLKSLKQRLKGKEGRVRGNLMGKRVDFSARTVITPDPNLSIDQVGVPRSIAANMTFAEIVTPFNIDRLQELVRRGNSQYPGAKYIIRDNGDRIDLRFHPKPSDLHLQTGYKVERHMCDGDIVIFNRQPTLHKMSMMGHRVRILPWSTFRLNLSVTTPYNADFDGDEMNLHLPQSLETRAEIQELAMVPRMIVTPQSNRPVMGIVQDTLTAVRKFTKRDVFLERGEVMNLLMFLSTWDGKVPQPAILKPRPLWTGKQIFSLIIPGHINCIRTHSTHPDDEDSGPYKHISPGDTKVVVENGELIMGILCKKSLGTSAGSLVHISYLEMGHDITRLFYSNIQTVINNWLLIEGHTIGIGDSIADSKTYQDIQNTIKKAKQDVIEVIEKAHNNELEPTPGNTLRQTFENQVNRILNDARDKTGSSAQKSLSEYNNFKSMVVSGAKGSKINISQVIAVVGQQNVEGKRIPFGFKHRTLPHFIKDDYGPESRGFVENSYLAGLTPTEFFFHAMGGREGLIDTAVKTAETGYIQRRLIKSMESVMVKYDATVRNSINQVVQLRYGEDGLAGESVEFQNLATLKPSNKAFEKKFRFDYTNERALRRTLQEDLVKDVLSNAHIQNELEREFERMREDREVLRVIFPTGDSKVVLPCNLLRMIWNAQKIFHINPRLPSDLHPIKVVEGVKELSKKLVIVNGDDPLSRQAQENATLLFNIHLRSTLCSRRMAEEFRLSGEAFDWLLGEIESKFNQAIAHPGEMVGALAAQSLGEPATQMTLNTFHYAGVSAKNVTLGVPRLKELINISKKPKTPSLTVFLLGQSARDAERAKDILCRLEHTTLRKVTANTAIYYDPNPQSTVVAEDQEWVNVYYEMPDFDVARISPWLLRVELDRKHMTDRKLTMEQIAEKINAGFGDDLNCIFNDDNAEKLVLRIRIMNSDENKMQEEEEVVDKMDDDVFLRCIESNMLTDMTLQGIEQISKVYMHLPQTDNKKKIIITEDGEFKALQEWILETDGVSLMRVLSEKDVDPVRTTSNDIVEIFTVLGIEAVRKALERELYHVISFDGSYVNYRHLALLCDTMTCRGHLMAITRHGVNRQDTGPLMKCSFEETVDVLMEAAAHGESDPMKGVSENIMLGQLAPAGTGCFDLLLDAEKCKYGMEIPTNIPGLGAAGPTGMFFGSAPSPMGGISPAMTPWNQGATPAYGAWSPSVGSGMTPGAAGFSPSAASDASGFSPGYSPAWSPTPGSPGSPGPSSPYIPSPGGAMSPSYSPTSPAYEPRSPGGYTPQSPSYSPTSPSYSPTSPSYSPTSPNYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPNYSPTSPNYTPTSPSYSPTSPSYSPTSPNYTPTSPNYSPTSPSYSPTSPSYSPTSPSYSPSSPRYTPQSPTYTPSSPSYSPSSPSYSPTSPKYTPTSPSYSPSSPEYTPTSPKYSPTSPKYSPTSPKYSPTSPTYSPTTPKYSPTSPTYSPTSPVYTPTSPKYSPTSPTYSPTSPKYSPTSPTYSPTSPKGSTYSPTSPGYSPTSPTYSLTSPAISPDDSDEEN
>7oop_J mol:protein length:67 DNA-directed RNA polymerases I, II, and III subunit RPABC5
@@ -1360717,7 +1360717,7 @@ MWKDKEFQVLFVLTILTLISGTIFYSTVEGLRPIDALYFS
>7oos_A mol:na length:10 RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3')
CAAAGAAAAG
>7oos_B mol:na length:11 DNA (5'-D(*CP*TP*(RWT)P*TP*CP*TP*TP*TP*G)-3')
-CT05KTCTTTG
+CTTCTTTG
>7oot_A mol:protein length:141 Interferon regulatory factor 4
MGSHHHHHHSAALEVLFQGPGGNGKLRQWLIDQIDSGKYPGLVWENEEKSIFRIPWKHAGKQDYNREEDAALFKAWALFKGKFREGIDKPDPPTWKTRLRCALNKSNDFEELVERSQLDISDPYKVYRIVPEGAKKGAKQL
>7oot_B mol:protein length:141 Interferon regulatory factor 4
@@ -1364753,7 +1364753,7 @@ GSHMEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEA
>7ozz_A mol:na length:10 RNA (5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3')
CAAAGAAAAG
>7ozz_B mol:na length:11 DNA (5'-D(*CP*TP*(RWR)P*TP*CP*TP*TP*TP*G)-3')
-CT05HTCTTTG
+CTTCTTTG
>7p00_H mol:protein length:298 Antibody fragment scFv16
MKFLVNVALVFMVVYISYIYADYKDDDDKHHHHHHHHHHLEVLFQGPDVQLVESGGGLVQPGGSRKLSCSASGFAFSSFGMHWVRQAPEKGLEWVAYISSGSGTIYYADTVKGRFTISRDDPKNTLFLQMTSLRSEDTAMYYCVRSIYYYGSSPFDFWGQGTTLTVSSGGGGSGGGGSGGGGSDIVMTQATSSVPVTPGESVSISCRSSKSLLHSNGNTYLYWFLQRPGQSPQLLIYRMSNLASGVPDRFSGSGSGTAFTLTISRLEAEDVGVYYCMQHLEYPLTFGAGTKLELKAAA
>7p00_B mol:protein length:354 Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
I do not think this error belongs to HHMsearch (the parse failed error), but to AlphaFold.
May be an exception should be triggered, but not halt the whole process ?
Thanks a lot to your time, I'll report to HMMsearch too (linking this issue). | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/591/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/591/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/590 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/590/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/590/comments | https://api.github.com/repos/deepmind/alphafold/issues/590/events | https://github.com/deepmind/alphafold/issues/590 | 1,367,467,458 | I_kwDOFoWQLM5RgeXC | 590 | How to get pdb structure files in bulk download from Google Cloud | {
"avatar_url": "https://avatars.githubusercontent.com/u/97283999?v=4",
"events_url": "https://api.github.com/users/TrineAalborg/events{/privacy}",
"followers_url": "https://api.github.com/users/TrineAalborg/followers",
"following_url": "https://api.github.com/users/TrineAalborg/following{/other_user}",
"gists_url": "https://api.github.com/users/TrineAalborg/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/TrineAalborg",
"id": 97283999,
"login": "TrineAalborg",
"node_id": "U_kgDOBcxvnw",
"organizations_url": "https://api.github.com/users/TrineAalborg/orgs",
"received_events_url": "https://api.github.com/users/TrineAalborg/received_events",
"repos_url": "https://api.github.com/users/TrineAalborg/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/TrineAalborg/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/TrineAalborg/subscriptions",
"type": "User",
"url": "https://api.github.com/users/TrineAalborg"
} | [] | closed | false | null | [] | null | [
"Hi Trine! Thanks for the question. Since PDBx/mmCIF is now the default file format, we don't plan to support pdb formats in general and they aren't available in the cloud dataset. They are, however, available on the EMBL FTP site for SwissProt and various proteomes. There are also open source converters between the two formats that can be found. Thanks!"
] | "2022-09-09T08:21:27Z" | "2022-09-12T12:44:04Z" | "2022-09-12T12:44:04Z" | NONE | null | Using the described method for whole-proteome structure file download from the Google Cloud database produces the structure files in mmCIF format. Are the structure files available in pdb format for bulk download, and how do I specify this in the download command? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/590/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/590/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/589 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/589/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/589/comments | https://api.github.com/repos/deepmind/alphafold/issues/589/events | https://github.com/deepmind/alphafold/issues/589 | 1,364,378,783 | I_kwDOFoWQLM5RUsSf | 589 | New Jax version broke Alphafold on Colab? | {
"avatar_url": "https://avatars.githubusercontent.com/u/18224663?v=4",
"events_url": "https://api.github.com/users/danjaco/events{/privacy}",
"followers_url": "https://api.github.com/users/danjaco/followers",
"following_url": "https://api.github.com/users/danjaco/following{/other_user}",
"gists_url": "https://api.github.com/users/danjaco/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/danjaco",
"id": 18224663,
"login": "danjaco",
"node_id": "MDQ6VXNlcjE4MjI0NjYz",
"organizations_url": "https://api.github.com/users/danjaco/orgs",
"received_events_url": "https://api.github.com/users/danjaco/received_events",
"repos_url": "https://api.github.com/users/danjaco/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/danjaco/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/danjaco/subscriptions",
"type": "User",
"url": "https://api.github.com/users/danjaco"
} | [] | closed | false | null | [] | null | [
"I am getting the same error message. Just in case it might be related to this problem, I was able to obtain protein predictions before the relax_use_gpu option was included in the notebook. \r\n\r\nError message:\r\n\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:195: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.\r\n leaves, structure = jax.tree_flatten(mapping)\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/data_structures.py:203: FutureWarning: jax.tree_unflatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_unflatten instead.\r\n self._mapping = jax.tree_unflatten(self._structure, self._leaves)\r\n/opt/conda/lib/python3.7/site-packages/alphafold/model/mapping.py:50: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.\r\n values_tree_def = jax.tree_flatten(values)[1]\r\n/opt/conda/lib/python3.7/site-packages/alphafold/model/mapping.py:54: FutureWarning: jax.tree_unflatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_unflatten instead.\r\n return jax.tree_unflatten(values_tree_def, flat_axes)\r\n/opt/conda/lib/python3.7/site-packages/alphafold/model/mapping.py:129: FutureWarning: jax.tree_flatten is deprecated, and will be removed in a future release. Use jax.tree_util.tree_flatten instead.\r\n flat_sizes = jax.tree_flatten(in_sizes)[0]\r\n/opt/conda/lib/python3.7/site-packages/haiku/_src/stateful.py:457: FutureWarning: jax.tree_leaves is deprecated, and will be removed in a future release. Use jax.tree_util.tree_leaves instead.\r\n length = jax.tree_leaves(xs)[0].shape[0]\r\n\r\n---------------------------------------------------------------------------\r\n\r\nUnfilteredStackTrace Traceback (most recent call last)\r\n\r\n[<ipython-input-5-420c51c385fa>](https://localhost:8080/#) in <module>\r\n 44 processed_feature_dict = model_runner.process_features(np_example, random_seed=0)\r\n---> 45 prediction = model_runner.predict(processed_feature_dict, random_seed=random.randrange(sys.maxsize))\r\n 46 \r\n\r\n87 frames\r\n\r\nUnfilteredStackTrace: AttributeError: module 'jax' has no attribute 'tree_multimap'\r\n\r\nThe stack trace below excludes JAX-internal frames.\r\nThe preceding is the original exception that occurred, unmodified.\r\n\r\n--------------------\r\n\r\nThe above exception was the direct cause of the following exception:\r\n\r\nAttributeError Traceback (most recent call last)\r\n\r\n[/opt/conda/lib/python3.7/site-packages/haiku/_src/stateful.py](https://localhost:8080/#) in difference(before, after)\r\n 310 params_before, params_after = box_and_fill_missing(before.params,\r\n 311 after.params)\r\n--> 312 params_after = jax.tree_multimap(functools.partial(if_changed, is_new_param),\r\n 313 params_before, params_after)\r\n 314 \r\n\r\nAttributeError: module 'jax' has no attribute 'tree_multimap'",
"only work with multiple sequences,if I run with only one sequence, \"AttributeError: module 'jax' has no attribute 'tree_multimap'\" comes out",
"I am having this issue as well when trying to run single sequences in the colab notebook. Has anyone found a workaround that allows notebooks to run to completion?",
"It doesn't look as if someone has suggested a workaround. I tried again today using a single sequence and I am still getting the same error message reported a week ago. Hopefully someone will look into this issue as soon as possible, as it has completely broken the Colab notebook (at least when running single sequences).",
"I just successfully used a different Colab notebook: \r\nhttps://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb",
"Thank you very much for letting us know @ruth-hanna! Hopefully this issue will be solved soon and we will be able to use this notebook as well.",
"HI! Thanks for raising this issue. We're currently testing a fix and should hopefully be able to push an update shortly. Thanks!",
"Is there any update on the 'jax' issue for https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb ? I also have this issue for predictions from one protein sequence. Thank you for trying to fix it!",
"Hi thanks for this! This has now been fixed in https://github.com/deepmind/alphafold/releases/tag/v2.2.4.",
"I'm getting another error now, at the database search stage:\r\n\r\n\"NameError: name 'model_type_to_use' is not defined\" (line 80)\r\n\r\n\r\n",
"Did you execute cell 3 before cell 4? This is where model_type_to_use is defined. Thanks!",
"Doh thanks, yes I was a a bit too hurried."
] | "2022-09-07T09:36:08Z" | "2022-09-22T11:23:15Z" | "2022-09-21T16:59:52Z" | NONE | null | Was getting deprecation warnings for a while, now appears not to work at all.

| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/589/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/589/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/588 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/588/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/588/comments | https://api.github.com/repos/deepmind/alphafold/issues/588/events | https://github.com/deepmind/alphafold/issues/588 | 1,362,859,732 | I_kwDOFoWQLM5RO5bU | 588 | Low GPU memory-usage and 0 GPU-Util | {
"avatar_url": "https://avatars.githubusercontent.com/u/69333850?v=4",
"events_url": "https://api.github.com/users/xuxiaochen1209/events{/privacy}",
"followers_url": "https://api.github.com/users/xuxiaochen1209/followers",
"following_url": "https://api.github.com/users/xuxiaochen1209/following{/other_user}",
"gists_url": "https://api.github.com/users/xuxiaochen1209/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/xuxiaochen1209",
"id": 69333850,
"login": "xuxiaochen1209",
"node_id": "MDQ6VXNlcjY5MzMzODUw",
"organizations_url": "https://api.github.com/users/xuxiaochen1209/orgs",
"received_events_url": "https://api.github.com/users/xuxiaochen1209/received_events",
"repos_url": "https://api.github.com/users/xuxiaochen1209/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/xuxiaochen1209/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/xuxiaochen1209/subscriptions",
"type": "User",
"url": "https://api.github.com/users/xuxiaochen1209"
} | [] | open | false | null | [] | null | [
"And I found there is only one result_model_1_multimer_v2_pred_0.pkl, one unrelaxed_model_1_multimer_v2_pred_0.pdb and one features.pkl file in the output file, not 5 pdb files and 5 unrelaxed pdb files as usual. "
] | "2022-09-06T07:54:11Z" | "2022-09-07T09:28:11Z" | null | NONE | null | Hello,
I had a problem with running alphafold. The first two hours are very smooth, and I think the MSA part is finished in these two hours. However, when it showd:
I0905 13:06:56.466166 140453353674560 model.py:175] Output shape was {'distogram': {'bin_edges': (63,), 'logits': (691, 691, 64)}, 'experimentally_resolved': {'logits': (691, 37)}, 'masked_msa': {'logits': (252, 691, 22)}, 'predicted_aligned_error': (691, 691), 'predicted_lddt': {'logits': (691, 50)}, 'structure_module': {'final_atom_mask': (691, 37), 'final_atom_positions': (691, 37, 3)}, 'plddt': (691,), 'aligned_confidence_probs': (691, 691, 64), 'max_predicted_aligned_error': (), 'ptm': (), 'iptm': (), 'ranking_confidence': ()}
I0905 13:06:56.467109 140453353674560 run_alphafold.py:202] Total JAX model model_1_multimer_v2_pred_0 on VHVL predict time (includes compilation time, see --benchmark): 246.2s
This step takes forever. I checked the CPU usage, memory usage, and the GPU usage and they are:
PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND
35488 dell 20 0 69.9g 4.8g 594148 R 100.0 3.8 1591:11 python /h+
total used free shared buff/cache available
Mem: 128357 6557 1730 106 120069 121081
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 515.43.04 Driver Version: 515.43.04 CUDA Version: 11.7 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 NVIDIA GeForce ... Off | 00000000:3B:00.0 Off | N/A |
| 30% 33C P2 101W / 320W | 5886MiB / 10240MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
| 1 NVIDIA GeForce ... Off | 00000000:5E:00.0 Off | N/A |
| 30% 25C P0 88W / 320W | 0MiB / 10240MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
| 2 NVIDIA GeForce ... Off | 00000000:B1:00.0 Off | N/A |
| 30% 25C P0 89W / 320W | 0MiB / 10240MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
| 3 NVIDIA GeForce ... Off | 00000000:D9:00.0 Off | N/A |
| 30% 25C P0 94W / 320W | 0MiB / 10240MiB | 0% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=============================================================================|
| 0 N/A N/A 35488 C python 1020MiB |
+-----------------------------------------------------------------------------+
The GPU memory is not very high since I saw some people's A100 had a menory usage with over 20000MiB. What's more, the GPU-Util is only 0-1%. I'm not sure whether it's because the graphic driver/CUDA/CUDNN/JAX versions are not matched (driver version: 515.43.04, CUDA version: 11.7, CUDNN version: 8.4.1.50, jaxlib version: 0.3.15+cuda11.cudnn82, python version: 3.8). I didn't see any error log, but it just didn't move on for over 30 hours. I also used 'conda activate alphafold' and tested in python3:
>>> import torch
>>> print(torch.cuda.is_available())
True
>>> from torch.backends import cudnn
>>> print(cudnn.is_available())
True
It seems that the CUDA and CUDNN works. So I'm confused and did anyone have this problem before and could you please kindly teach me how to solve it? Thanks a lot for your kind guide. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/588/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/588/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/587 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/587/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/587/comments | https://api.github.com/repos/deepmind/alphafold/issues/587/events | https://github.com/deepmind/alphafold/issues/587 | 1,359,670,383 | I_kwDOFoWQLM5RCuxv | 587 | Alphafold2 Modulenotfound | {
"avatar_url": "https://avatars.githubusercontent.com/u/112677174?v=4",
"events_url": "https://api.github.com/users/SolkenSlibs/events{/privacy}",
"followers_url": "https://api.github.com/users/SolkenSlibs/followers",
"following_url": "https://api.github.com/users/SolkenSlibs/following{/other_user}",
"gists_url": "https://api.github.com/users/SolkenSlibs/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/SolkenSlibs",
"id": 112677174,
"login": "SolkenSlibs",
"node_id": "U_kgDOBrdRNg",
"organizations_url": "https://api.github.com/users/SolkenSlibs/orgs",
"received_events_url": "https://api.github.com/users/SolkenSlibs/received_events",
"repos_url": "https://api.github.com/users/SolkenSlibs/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/SolkenSlibs/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/SolkenSlibs/subscriptions",
"type": "User",
"url": "https://api.github.com/users/SolkenSlibs"
} | [] | closed | false | null | [] | null | [
"Hi! Is it possible that your colab runtime restarted? When this happens, you will need to rerun steps 1 and 2 to make sure that the relevant packages have been installed, as these do not persist between sessions. Thanks!",
"> Hi! Is it possible that your colab runtime restarted? When this happens, you will need to rerun steps 1 and 2 to make sure that the relevant packages have been installed, as these do not persist between sessions. Thanks!\r\n\r\nI would rerun steps 1 and 2 but unfortunately would get a separate error for the second one. Unfortunately, I am now receiving a different error and am unable to get past download alphafold.\r\n\r\nRuntimeError Traceback (most recent call last)\r\nin\r\n46 raise RuntimeError('Colab TPU runtime not supported. Change it to GPU via Runtime -> Change Runtime Type -> Hardware accelerator -> GPU.')\r\n47 elif jax.local_devices()[0].platform == 'cpu':\r\n---> 48 raise RuntimeError('Colab CPU runtime not supported. Change it to GPU via Runtime -> Change Runtime Type -> Hardware accelerator -> GPU.')\r\n49 else:\r\n50 print(f'Running with {jax.local_devices()[0].device_kind} GPU')\r\n\r\nRuntimeError: Colab CPU runtime not supported. Change it to GPU via Runtime -> Change Runtime Type -> Hardware accelerator -> GPU.\r\n\r\nWhen I do go into runtime type it is already set to GPU and I haven't changed anything in there previously I am not sure what I am doing wrong as I really only found out about this site recently.",
"Hi! I haven't been able to reproduce your issue. Are you still having this problem? Thanks!",
"> Hi! I haven't been able to reproduce your issue. Are you still having this problem? Thanks!\r\n\r\nNot sure what changed but it seems to be working fine right now. I am currently running 3 structures and I am currently at 6% hopefully nothing happens.",
"Thats great thanks! I will close the issue but feel free to reopen if it comes back."
] | "2022-09-02T03:49:38Z" | "2022-10-03T09:25:39Z" | "2022-10-03T09:25:39Z" | NONE | null | Hi, I'm new to using Alphafold2 and I have been looking for the solution for this problem but haven't found it so sorry in advance if it has already been posted. I am running 3 sequences and around halfway through the second or third sequence I keep getting this error when I do the whole thing just stops working for a couple of hours and I am not sure what to do.
ModuleNotFoundError Traceback (most recent call last)
[<ipython-input-1-c47351bef025>](https://localhost:8080/#) in <module>
19 import matplotlib.pyplot as plt
20 import numpy as np
---> 21 import py3Dmol
22
23 from alphafold.model import model
ModuleNotFoundError: No module named 'py3Dmol'
---------------------------------------------------------------------------
NOTE: If your import is failing due to a missing package, you can
manually install dependencies using either !pip or !apt.
To view examples of installing some common dependencies, click the
"Open Examples" button below. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/587/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/587/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/586 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/586/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/586/comments | https://api.github.com/repos/deepmind/alphafold/issues/586/events | https://github.com/deepmind/alphafold/issues/586 | 1,358,808,191 | I_kwDOFoWQLM5Q_cR_ | 586 | Rescoring and Constraints in multimer modeling. | {
"avatar_url": "https://avatars.githubusercontent.com/u/10496600?v=4",
"events_url": "https://api.github.com/users/entropybit/events{/privacy}",
"followers_url": "https://api.github.com/users/entropybit/followers",
"following_url": "https://api.github.com/users/entropybit/following{/other_user}",
"gists_url": "https://api.github.com/users/entropybit/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/entropybit",
"id": 10496600,
"login": "entropybit",
"node_id": "MDQ6VXNlcjEwNDk2NjAw",
"organizations_url": "https://api.github.com/users/entropybit/orgs",
"received_events_url": "https://api.github.com/users/entropybit/received_events",
"repos_url": "https://api.github.com/users/entropybit/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/entropybit/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/entropybit/subscriptions",
"type": "User",
"url": "https://api.github.com/users/entropybit"
} | [] | open | false | null | [] | null | [] | "2022-09-01T12:58:41Z" | "2022-09-01T13:00:17Z" | null | NONE | null | Hi,
since I am working on complexes I was very happy about the multimer modeling.
However, if one has available distance constraints from the lab saying which residues interact with
each other, it would be highly useful if those constraints could be considered somehow.
With a normal docking approach I would just generate a big ensemble and select the models based on my constraints from the ensemble. Of course that leaves me with many models to choose from instead of the few ranked models produced
by AF2. I was also thinking if I could just rescore those models by using them as AF2 templates but that would still mean a few hundred to thousand AF2 runs depending on the protein complex ^^'.
And I am also not sure if that really is a "rescoring" of that complex with AF2.
So in summary, considering constraints would be really great, and also some kind of rescoring mechanism to
gauge the quality of an arbitrary input structure with the AF2 model.
Regards and Thanks in advance,
Benjamin Mayer,
TUD, Germany
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/586/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/586/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/585 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/585/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/585/comments | https://api.github.com/repos/deepmind/alphafold/issues/585/events | https://github.com/deepmind/alphafold/issues/585 | 1,358,635,890 | I_kwDOFoWQLM5Q-yNy | 585 | models variability for AF multimer | {
"avatar_url": "https://avatars.githubusercontent.com/u/10754760?v=4",
"events_url": "https://api.github.com/users/fglaser/events{/privacy}",
"followers_url": "https://api.github.com/users/fglaser/followers",
"following_url": "https://api.github.com/users/fglaser/following{/other_user}",
"gists_url": "https://api.github.com/users/fglaser/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/fglaser",
"id": 10754760,
"login": "fglaser",
"node_id": "MDQ6VXNlcjEwNzU0NzYw",
"organizations_url": "https://api.github.com/users/fglaser/orgs",
"received_events_url": "https://api.github.com/users/fglaser/received_events",
"repos_url": "https://api.github.com/users/fglaser/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/fglaser/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/fglaser/subscriptions",
"type": "User",
"url": "https://api.github.com/users/fglaser"
} | [] | open | false | null | [] | null | [] | "2022-09-01T10:45:38Z" | "2022-09-01T10:45:38Z" | null | NONE | null | Dear all,
I am thinking for a while about alphafold model variability for multimer models. Even if use 20 multimer predictions (100 models) the resulting complex prediction are very very similar, but in reality there should be be some variability in the models as the complex is accommodating its potential energy surface.
Is there a parameter in AF that would allow me to produce more "variability in the resutls of AF multimer?
Thanks a lot,
Fabian Glaser
Technion, Israel | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/585/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/585/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/584 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/584/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/584/comments | https://api.github.com/repos/deepmind/alphafold/issues/584/events | https://github.com/deepmind/alphafold/issues/584 | 1,358,140,710 | I_kwDOFoWQLM5Q85Um | 584 | inquiry regarding input format | {
"avatar_url": "https://avatars.githubusercontent.com/u/101581813?v=4",
"events_url": "https://api.github.com/users/zjq1011/events{/privacy}",
"followers_url": "https://api.github.com/users/zjq1011/followers",
"following_url": "https://api.github.com/users/zjq1011/following{/other_user}",
"gists_url": "https://api.github.com/users/zjq1011/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/zjq1011",
"id": 101581813,
"login": "zjq1011",
"node_id": "U_kgDOBg4D9Q",
"organizations_url": "https://api.github.com/users/zjq1011/orgs",
"received_events_url": "https://api.github.com/users/zjq1011/received_events",
"repos_url": "https://api.github.com/users/zjq1011/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/zjq1011/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/zjq1011/subscriptions",
"type": "User",
"url": "https://api.github.com/users/zjq1011"
} | [] | open | false | null | [] | null | [] | "2022-09-01T02:03:13Z" | "2022-09-01T02:03:13Z" | null | NONE | null | Hello,
just in case, I want to confirm if the up-to-date Alphafold2 only accepts amino acids sequence as input.
Is it possible to add some restrictions before the predictions?
Also, there seem to be no options to specify whether the protein is from prokaryotes and eukaryotes,
will it influence the results?
Thanks. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/584/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/584/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/583 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/583/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/583/comments | https://api.github.com/repos/deepmind/alphafold/issues/583/events | https://github.com/deepmind/alphafold/issues/583 | 1,355,894,493 | I_kwDOFoWQLM5Q0U7d | 583 | Different output from the same sequence | {
"avatar_url": "https://avatars.githubusercontent.com/u/8283969?v=4",
"events_url": "https://api.github.com/users/barbarashih/events{/privacy}",
"followers_url": "https://api.github.com/users/barbarashih/followers",
"following_url": "https://api.github.com/users/barbarashih/following{/other_user}",
"gists_url": "https://api.github.com/users/barbarashih/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/barbarashih",
"id": 8283969,
"login": "barbarashih",
"node_id": "MDQ6VXNlcjgyODM5Njk=",
"organizations_url": "https://api.github.com/users/barbarashih/orgs",
"received_events_url": "https://api.github.com/users/barbarashih/received_events",
"repos_url": "https://api.github.com/users/barbarashih/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/barbarashih/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/barbarashih/subscriptions",
"type": "User",
"url": "https://api.github.com/users/barbarashih"
} | [
{
"color": "D4C5F9",
"default": false,
"description": "Further information is requested",
"id": 3094826897,
"name": "usage question",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODk3",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/usage%20question"
}
] | closed | false | null | [] | null | [
"Lines 105-109 in run_alphafold.py seem very relevant, and the README also has a note on reproducibility that is important. As interpreted by me, the gist of it is that runs are consistently similar, but not identical. You can make the runs even more consistent by setting the random seed used, but GPU inference is nondeterministic so very slight differences will still likely occur. In my experience, after aligning the structures in PyMOL, most structures with decent confidence scores are extremely similar even without setting the seed.",
"Thank you very much for your answer @tcoates5 !"
] | "2022-08-30T14:51:39Z" | "2022-09-13T19:57:18Z" | "2022-09-13T19:57:18Z" | NONE | null | Hi,
Thank you for sharing this great tool. I noticed that when I run Alphafold2 on the same sequence, I would get slightly different result each time. Is there any way we can ensure getting identical output from identical sequence, or is that something that's unavoidable?
Thank you
Barbara | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/583/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/583/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/582 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/582/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/582/comments | https://api.github.com/repos/deepmind/alphafold/issues/582/events | https://github.com/deepmind/alphafold/issues/582 | 1,354,445,704 | I_kwDOFoWQLM5QuzOI | 582 | protein protein interaction Indexeror | {
"avatar_url": "https://avatars.githubusercontent.com/u/19805255?v=4",
"events_url": "https://api.github.com/users/hellorp1990/events{/privacy}",
"followers_url": "https://api.github.com/users/hellorp1990/followers",
"following_url": "https://api.github.com/users/hellorp1990/following{/other_user}",
"gists_url": "https://api.github.com/users/hellorp1990/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/hellorp1990",
"id": 19805255,
"login": "hellorp1990",
"node_id": "MDQ6VXNlcjE5ODA1MjU1",
"organizations_url": "https://api.github.com/users/hellorp1990/orgs",
"received_events_url": "https://api.github.com/users/hellorp1990/received_events",
"repos_url": "https://api.github.com/users/hellorp1990/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/hellorp1990/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/hellorp1990/subscriptions",
"type": "User",
"url": "https://api.github.com/users/hellorp1990"
} | [
{
"color": "D4C5F9",
"default": false,
"description": "Further information is requested",
"id": 3094826897,
"name": "usage question",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODk3",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/usage%20question"
}
] | open | false | null | [] | null | [
"Are `EUXXXX` and `12H` sequence names? If so, the [FASTA format](https://en.wikipedia.org/wiki/FASTA_format) requires the following formatting:\r\n\r\n```\r\n>EUXXXX\r\nMKMASNDATPSDGSTANLVPEVNNEVMALEPVVGAAIAAPVAGQQNVIDPWIRNNFVQAPGGEFTVSPRNAPGEILWSAPLGPDLNPYLSHLARMYNGYAGGFEVQVILAGNAFTAGKIIFAAVPPNFPTEGLSPSQVTMFPHIIVDVRQLEPVLIPLPDVRNNFYHYNQSNDPTIKLIAMLYTPLRANNAGDDVFTVSCRVLTRPSPDFDFIFLVPPTVESRTKPFSVPILTVEEMTNSRFPIPLEKLFTGPSSAFVVQPQNGRCTTDGVL\r\n>12H\r\nMEWNWVVLFLLSLTAGVYAQGQMQQSGAELVKPKLSCKTSGF\r\n```\r\n\r\nSee https://github.com/deepmind/alphafold#examples for more details.",
"@Augustin-Zidek Yes they are started with > (i added them here as well, but for some reason > is removed from the comment)",
"I see, `>` is a special character in Markdown so it didn't render properly -- I fixed that in your comment. Could you try without spaces before the `>` characters?",
"Could you also post the full command you are using to launch this and the full error?",
"@Augustin-Zidek When i ran the first sequence ( > EUXXXX) alone -monomer pipeline. And it ran fine but when i tried to ran multimer or protein protein interaction pipeline its showed error. \r\n\r\nFull error:\r\n\r\n\r\n",
"@Augustin-Zidek input command:\r\n\r\n--fasta_paths=input.fasta \\\r\n --max_template_date=2022-08-29 \\\r\n --model_preset=multimer \\",
"The error is clearly coming from the FASTA parser, so there must be something wrong with the FASTA format. \r\n\r\n* Is the listing you provided above exactly what is in the FASTA file (including blank lines, leading/trailing spaces, etc.)? \r\n* Is the FASTA encoded as ASCII or UTF-8?"
] | "2022-08-29T14:58:00Z" | "2022-08-29T15:28:39Z" | null | NONE | null | Hi, I am new to protein folding area and I am having a little issue with running protein protein interaction with Alphafold.
My inputs:
```
>EUXXXX
MKMASNDATPSDGSTANLVPEVNNEVMALEPVVGAAIAAPVAGQQNVIDPWIRNNFVQAPGGEFTVSPRNAPGEILWSAPLGPDLNPYLSHLARMYNGYAGGFEVQVILAGNAFTAGKIIFAAVPPNFPTEGLSPSQVTMFPHIIVDVRQLEPVLIPLPDVRNNFYHYNQSNDPTIKLIAMLYTPLRANNAGDDVFTVSCRVLTRPSPDFDFIFLVPPTVESRTKPFSVPILTVEEMTNSRFPIPLEKLFTGPSSAFVVQPQNGRCTTDGVL
>12H
MEWNWVVLFLLSLTAGVYAQGQMQQSGAELVKPKLSCKTSGF
```
I am having an error: IndexError: list index out of range (i believe this error is related to input file formatting).
Can anyone help with this issue?
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/582/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/582/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/579 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/579/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/579/comments | https://api.github.com/repos/deepmind/alphafold/issues/579/events | https://github.com/deepmind/alphafold/issues/579 | 1,351,327,790 | I_kwDOFoWQLM5Qi6Au | 579 | Can we limit the depth of the MSA? | {
"avatar_url": "https://avatars.githubusercontent.com/u/45075374?v=4",
"events_url": "https://api.github.com/users/many-states/events{/privacy}",
"followers_url": "https://api.github.com/users/many-states/followers",
"following_url": "https://api.github.com/users/many-states/following{/other_user}",
"gists_url": "https://api.github.com/users/many-states/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/many-states",
"id": 45075374,
"login": "many-states",
"node_id": "MDQ6VXNlcjQ1MDc1Mzc0",
"organizations_url": "https://api.github.com/users/many-states/orgs",
"received_events_url": "https://api.github.com/users/many-states/received_events",
"repos_url": "https://api.github.com/users/many-states/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/many-states/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/many-states/subscriptions",
"type": "User",
"url": "https://api.github.com/users/many-states"
} | [
{
"color": "C5DEF5",
"default": false,
"description": "New feature or request",
"id": 3094826893,
"name": "feature request",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODkz",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/feature%20request"
},
{
"color": "D4C5F9",
"default": false,
"description": "Further information is requested",
"id": 3094826897,
"name": "usage question",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODk3",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/usage%20question"
}
] | open | false | null | [] | null | [
"It looks like the functionality is there, based on the existence of [truncate_stockholm_msa(stockholm_msa_path: str, max_sequences: int)](https://github.com/deepmind/alphafold/blob/86a0b8ec7a39698a7c2974420c4696ea4cb5743a/alphafold/data/parsers.py#L277) and the places it is called, like in the [Jackhmmer wrapper](https://github.com/deepmind/alphafold/blob/86a0b8ec7a39698a7c2974420c4696ea4cb5743a/alphafold/data/tools/jackhmmer.py#L155). Default values for `max_sequences` are defined for the different search databases in the [class init of DataPipeline](https://github.com/deepmind/alphafold/blob/86a0b8ec7a39698a7c2974420c4696ea4cb5743a/alphafold/data/pipeline.py#L125), but the upstream instantiations of pipelines leave them as default."
] | "2022-08-25T18:59:13Z" | "2022-08-26T16:35:43Z" | null | NONE | null | We have some users at our site folding large protein complexes using the multimer workflow, and have run into issues with memory exhausted on the GPUs even using the reduced databases. In the log file we notice it reporting what looks like a deep MSA:
`I0822 12:49:34.498847 140441650169664 pipeline.py:238] Final (deduplicated) MSA size: 11437 sequences.`
In the prediction stage, it doesn't look like the 11k sequences are used, as the msa dimensions are smaller:
`I0822 12:59:51.185315 140441650169664 model.py:166] Running predict with shape(feat) = {'aatype': (4205,), 'residue_index': (4205,), 'seq_length': (), 'msa': (3072, 4205), ...`
But this still looks larger than the MSA depth of 30 to 100 that I believe was identified as optimal in the original publication. Does this MSA depth appear abnormal, and if so is there anything we can do to restrict the MSA to a smaller number of sequences?
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/579/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/579/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/578 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/578/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/578/comments | https://api.github.com/repos/deepmind/alphafold/issues/578/events | https://github.com/deepmind/alphafold/issues/578 | 1,350,405,987 | I_kwDOFoWQLM5QfY9j | 578 | I am using alphafold to predict protein structure. while running the first step, i found the error int he comment. Can anyone help me with this error? | {
"avatar_url": "https://avatars.githubusercontent.com/u/112063013?v=4",
"events_url": "https://api.github.com/users/jeid94/events{/privacy}",
"followers_url": "https://api.github.com/users/jeid94/followers",
"following_url": "https://api.github.com/users/jeid94/following{/other_user}",
"gists_url": "https://api.github.com/users/jeid94/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/jeid94",
"id": 112063013,
"login": "jeid94",
"node_id": "U_kgDOBq3yJQ",
"organizations_url": "https://api.github.com/users/jeid94/orgs",
"received_events_url": "https://api.github.com/users/jeid94/received_events",
"repos_url": "https://api.github.com/users/jeid94/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/jeid94/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/jeid94/subscriptions",
"type": "User",
"url": "https://api.github.com/users/jeid94"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
},
{
"color": "FEF2C0",
"default": false,
"description": "AlphaFold colab issue",
"id": 3313914804,
"name": "colab",
"node_id": "MDU6TGFiZWwzMzEzOTE0ODA0",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/colab"
}
] | closed | false | null | [] | null | [
"Sorry about this! See https://github.com/deepmind/alphafold/issues/573 for a workaround, we are working on a fix.",
"> Sorry about this! See #573 for a workaround, we are working on a fix.\r\n\r\nthank you !!",
"Fixed in https://github.com/deepmind/alphafold/commit/dad1f286aeaa158702a98aeee0f5661f30de6da8"
] | "2022-08-25T06:46:35Z" | "2022-08-25T09:26:47Z" | "2022-08-25T09:26:47Z" | NONE | null | Hello everyone,
I am using alphafold to predict protein structure. while running the first step, i found the following error. Can anyone helps me please.
CalledProcessError Traceback (most recent call last)
[<ipython-input-1-0c215f21152d>](https://localhost:8080/#) in <module>
35 PATH=get_ipython().run_line_magic('env', 'PATH')
36 get_ipython().run_line_magic('env', 'PATH=/opt/conda/bin:{PATH}')
---> 37 get_ipython().run_line_magic('shell', 'conda update -qy conda && conda install -qy -c conda-forge python=3.7 openmm=7.5.1 pdbfixer')
38 pbar.update(80)
39
2 frames
[/usr/local/lib/python3.7/dist-packages/google/colab/_system_commands.py](https://localhost:8080/#) in check_returncode(self)
133 if self.returncode:
134 raise subprocess.CalledProcessError(
--> 135 returncode=self.returncode, cmd=self.args, output=self.output)
136
137 def _repr_pretty_(self, p, cycle): # pylint:disable=unused-argument
CalledProcessError: Command 'conda update -qy conda && conda install -qy -c conda-forge python=3.7 openmm=7.5.1 pdbfixer' returned non-zero exit status 1. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/578/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/578/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/577 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/577/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/577/comments | https://api.github.com/repos/deepmind/alphafold/issues/577/events | https://github.com/deepmind/alphafold/issues/577 | 1,350,260,504 | I_kwDOFoWQLM5Qe1cY | 577 | 2.2.2 build TLS issues | {
"avatar_url": "https://avatars.githubusercontent.com/u/27769885?v=4",
"events_url": "https://api.github.com/users/MrDotOne/events{/privacy}",
"followers_url": "https://api.github.com/users/MrDotOne/followers",
"following_url": "https://api.github.com/users/MrDotOne/following{/other_user}",
"gists_url": "https://api.github.com/users/MrDotOne/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/MrDotOne",
"id": 27769885,
"login": "MrDotOne",
"node_id": "MDQ6VXNlcjI3NzY5ODg1",
"organizations_url": "https://api.github.com/users/MrDotOne/orgs",
"received_events_url": "https://api.github.com/users/MrDotOne/received_events",
"repos_url": "https://api.github.com/users/MrDotOne/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/MrDotOne/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/MrDotOne/subscriptions",
"type": "User",
"url": "https://api.github.com/users/MrDotOne"
} | [
{
"color": "cfd3d7",
"default": true,
"description": "This issue or pull request already exists",
"id": 3094826892,
"name": "duplicate",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODky",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/duplicate"
}
] | closed | false | null | [] | null | [
"Sorry about this! See https://github.com/deepmind/alphafold/issues/573 for a workaround, we are working on a fix.",
"Fixed in https://github.com/deepmind/alphafold/commit/dad1f286aeaa158702a98aeee0f5661f30de6da8."
] | "2022-08-25T03:16:57Z" | "2022-08-25T09:27:00Z" | "2022-08-25T09:27:00Z" | NONE | null | Trying to build a recent clone of the repo on our HPC and i am receiving the following error:
cryptography-37.0.2 | 1.5 MB | ########## | 100%
ocl-icd-system-1.0.0 | 4 KB | ########## | 100%
Preparing transaction: ...working... done
Verifying transaction: ...working... done
Executing transaction: ...working... By downloading and using the CUDA Toolkit conda packages, you accept the terms and conditions of the CUDA End User License Agreement (EULA): https://docs.nvidia.com/cuda/eula/index.html
done
Retrieving notices: ...working... failed
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1125, in __call__
File "/opt/conda/lib/python3.9/site-packages/conda/cli/main.py", line 86, in main_subshell
File "/opt/conda/lib/python3.9/site-packages/conda/cli/conda_argparse.py", line 93, in do_call
File "/opt/conda/lib/python3.9/site-packages/conda/notices/core.py", line 75, in wrapper
File "/opt/conda/lib/python3.9/site-packages/conda/notices/core.py", line 39, in display_notices
File "/opt/conda/lib/python3.9/site-packages/conda/notices/http.py", line 36, in get_notice_responses
File "/opt/conda/lib/python3.9/site-packages/conda/notices/http.py", line 39, in <genexpr>
File "/opt/conda/lib/python3.9/concurrent/futures/_base.py", line 609, in result_iterator
File "/opt/conda/lib/python3.9/concurrent/futures/_base.py", line 446, in result
File "/opt/conda/lib/python3.9/concurrent/futures/_base.py", line 391, in __get_result
File "/opt/conda/lib/python3.9/concurrent/futures/thread.py", line 58, in run
File "/opt/conda/lib/python3.9/site-packages/conda/notices/http.py", line 42, in <lambda>
File "/opt/conda/lib/python3.9/site-packages/conda/notices/cache.py", line 37, in wrapper
File "/opt/conda/lib/python3.9/site-packages/conda/notices/http.py", line 58, in get_channel_notice_response
File "/opt/conda/lib/python3.9/site-packages/requests/sessions.py", line 600, in get
File "/opt/conda/lib/python3.9/site-packages/requests/sessions.py", line 587, in request
File "/opt/conda/lib/python3.9/site-packages/requests/sessions.py", line 701, in send
File "/opt/conda/lib/python3.9/site-packages/requests/adapters.py", line 460, in send
File "/opt/conda/lib/python3.9/site-packages/requests/adapters.py", line 263, in cert_verify
OSError: Could not find a suitable TLS CA certificate bundle, invalid path: /opt/conda/lib/python3.9/site-packages/certifi/cacert.pem
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/bin/conda", line 13, in <module>
sys.exit(main())
File "/opt/conda/lib/python3.9/site-packages/conda/cli/main.py", line 129, in main
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1413, in conda_exception_handler
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1128, in __call__
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1170, in handle_exception
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1181, in handle_unexpected_exception
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1251, in print_unexpected_error_report
ModuleNotFoundError: No module named 'conda.cli.main_info'
The command '/bin/bash -c conda update -qy conda && conda install -y -c conda-forge openmm=7.5.1 cudatoolkit==${CUDA_VERSION} pdbfixer pip python=3.7' returned a non-zero code: 1
Has anyone else seen this, and if so, how does one get around it?
Thank you. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/577/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/577/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/576 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/576/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/576/comments | https://api.github.com/repos/deepmind/alphafold/issues/576/events | https://github.com/deepmind/alphafold/pull/576 | 1,349,726,109 | PR_kwDOFoWQLM49vACm | 576 | workaround for issue with latest conda version | {
"avatar_url": "https://avatars.githubusercontent.com/u/40922889?v=4",
"events_url": "https://api.github.com/users/Meghpal/events{/privacy}",
"followers_url": "https://api.github.com/users/Meghpal/followers",
"following_url": "https://api.github.com/users/Meghpal/following{/other_user}",
"gists_url": "https://api.github.com/users/Meghpal/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Meghpal",
"id": 40922889,
"login": "Meghpal",
"node_id": "MDQ6VXNlcjQwOTIyODg5",
"organizations_url": "https://api.github.com/users/Meghpal/orgs",
"received_events_url": "https://api.github.com/users/Meghpal/received_events",
"repos_url": "https://api.github.com/users/Meghpal/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Meghpal/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Meghpal/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Meghpal"
} | [] | closed | false | null | [] | null | [
"Thank you! I submitted the fix separately."
] | "2022-08-24T16:26:12Z" | "2022-08-25T09:28:14Z" | "2022-08-25T09:28:14Z" | NONE | null | Solves #573 by not updating to latest conda version | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/576/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/576/timeline | null | null | 0 | {
"diff_url": "https://github.com/deepmind/alphafold/pull/576.diff",
"html_url": "https://github.com/deepmind/alphafold/pull/576",
"merged_at": null,
"patch_url": "https://github.com/deepmind/alphafold/pull/576.patch",
"url": "https://api.github.com/repos/deepmind/alphafold/pulls/576"
} | true |
https://api.github.com/repos/deepmind/alphafold/issues/575 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/575/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/575/comments | https://api.github.com/repos/deepmind/alphafold/issues/575/events | https://github.com/deepmind/alphafold/issues/575 | 1,349,650,559 | I_kwDOFoWQLM5Qcgh_ | 575 | gzip: database//mgnify/mgy_clusters_2018_12.fa.gz: No such file or directory | {
"avatar_url": "https://avatars.githubusercontent.com/u/56931786?v=4",
"events_url": "https://api.github.com/users/tianrenmaogithub/events{/privacy}",
"followers_url": "https://api.github.com/users/tianrenmaogithub/followers",
"following_url": "https://api.github.com/users/tianrenmaogithub/following{/other_user}",
"gists_url": "https://api.github.com/users/tianrenmaogithub/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/tianrenmaogithub",
"id": 56931786,
"login": "tianrenmaogithub",
"node_id": "MDQ6VXNlcjU2OTMxNzg2",
"organizations_url": "https://api.github.com/users/tianrenmaogithub/orgs",
"received_events_url": "https://api.github.com/users/tianrenmaogithub/received_events",
"repos_url": "https://api.github.com/users/tianrenmaogithub/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/tianrenmaogithub/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/tianrenmaogithub/subscriptions",
"type": "User",
"url": "https://api.github.com/users/tianrenmaogithub"
} | [
{
"color": "F9D0C4",
"default": false,
"description": "Issue setting up AlphaFold",
"id": 3317500161,
"name": "setup",
"node_id": "MDU6TGFiZWwzMzE3NTAwMTYx",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/setup"
}
] | open | false | null | [] | null | [
"Did you run the command line. I had the same error but it worked when I assigned a directory.\r\n`/home/usrn/alphafold/scripts/download_pdb70.sh directory_path`\r\n\r\nIf you are in the alphafold directory then it should look like this\r\n`scripts/download_pdb70.sh directory_path`\r\n\r\nOne thing also is that the directory_path should have a lot of storage. I got an error when the directory I chose was sub 2TB, I had to get a 8TB SSD to not run across the same issue using the full database.",
"> Did you run the command line. I had the same error but it worked when I assigned a directory. `/home/usrn/alphafold/scripts/download_pdb70.sh directory_path`\r\n> \r\n> If you are in the alphafold directory then it should look like this `scripts/download_pdb70.sh directory_path`\r\n> \r\n> One thing also is that the directory_path should have a lot of storage. I got an error when the directory I chose was sub 2TB, I had to get a 8TB SSD to not run across the same issue using the full database.\r\n\r\nSorry for the typo. I meant download_mgnify.sh should be edited. \r\n",
"Oh it should still work just edit the .sh file and your directory_path. Hopefully it works for you.",
"Did you use full path or relative path? You should use the full directory path for this to work.",
"> Did you use full path or relative path? You should use the full directory path for this to work.\r\n\r\nI see. I used a relative path. Thank you!\r\nSince an absolute path is required, the pushd and popd statements are actually removable because gunzip decompresses a zip file in the same place and you don't need to go to that directory.\r\nBecause many people prefer to feed a relative path, I think it is better to make it accept relative path by removing the \"${ROOT_DIR}/\" from the gunzip \"${ROOT_DIR}/${BASENAME}\" while keeping the pusd and popd. It will work for both absolute and relative paths.\r\n"
] | "2022-08-24T15:40:05Z" | "2022-08-25T14:58:49Z" | null | NONE | null | I got this error when running download_all_data.sh, which runs download_mgnify.sh.
I looked into the latter and these lines were causing the error.
pushd "${ROOT_DIR}"
gunzip "${ROOT_DIR}/${BASENAME}"
popd
It went to the directory to unzip the file, but the path was wrong. The ${ROOT_DIR}/ should be removed. | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/575/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/575/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/574 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/574/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/574/comments | https://api.github.com/repos/deepmind/alphafold/issues/574/events | https://github.com/deepmind/alphafold/issues/574 | 1,348,959,049 | I_kwDOFoWQLM5QZ3tJ | 574 | Can you reduce the GPU memory occupation, improve the computing speed and save the running time? | {
"avatar_url": "https://avatars.githubusercontent.com/u/68001792?v=4",
"events_url": "https://api.github.com/users/ZihaoXingUP/events{/privacy}",
"followers_url": "https://api.github.com/users/ZihaoXingUP/followers",
"following_url": "https://api.github.com/users/ZihaoXingUP/following{/other_user}",
"gists_url": "https://api.github.com/users/ZihaoXingUP/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ZihaoXingUP",
"id": 68001792,
"login": "ZihaoXingUP",
"node_id": "MDQ6VXNlcjY4MDAxNzky",
"organizations_url": "https://api.github.com/users/ZihaoXingUP/orgs",
"received_events_url": "https://api.github.com/users/ZihaoXingUP/received_events",
"repos_url": "https://api.github.com/users/ZihaoXingUP/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ZihaoXingUP/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ZihaoXingUP/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ZihaoXingUP"
} | [] | closed | false | null | [] | null | [
"We improved the GPU usage for AlphaFold-Multimer in [v2.3.0](https://github.com/deepmind/alphafold/releases/tag/v2.3.0) - hopefully this addresses your issue. Closing this issue now, feel free to reopen if you still encounter this in v2.3.0."
] | "2022-08-24T06:56:18Z" | "2022-12-13T12:02:12Z" | "2022-12-13T12:02:12Z" | NONE | null | Can you reduce the GPU memory occupation, improve the computing speed and save the running time? | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/574/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/574/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/573 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/573/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/573/comments | https://api.github.com/repos/deepmind/alphafold/issues/573/events | https://github.com/deepmind/alphafold/issues/573 | 1,348,447,888 | I_kwDOFoWQLM5QX66Q | 573 | DeepMind AlphaFold2 colab notebook already crashes during 3rd party software installation cell | {
"avatar_url": "https://avatars.githubusercontent.com/u/80322296?v=4",
"events_url": "https://api.github.com/users/MWNautilus/events{/privacy}",
"followers_url": "https://api.github.com/users/MWNautilus/followers",
"following_url": "https://api.github.com/users/MWNautilus/following{/other_user}",
"gists_url": "https://api.github.com/users/MWNautilus/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/MWNautilus",
"id": 80322296,
"login": "MWNautilus",
"node_id": "MDQ6VXNlcjgwMzIyMjk2",
"organizations_url": "https://api.github.com/users/MWNautilus/orgs",
"received_events_url": "https://api.github.com/users/MWNautilus/received_events",
"repos_url": "https://api.github.com/users/MWNautilus/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/MWNautilus/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/MWNautilus/subscriptions",
"type": "User",
"url": "https://api.github.com/users/MWNautilus"
} | [
{
"color": "E99695",
"default": false,
"description": "Something isn't working",
"id": 3094826889,
"name": "error report",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODg5",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/error%20report"
},
{
"color": "ffffff",
"default": false,
"description": "An issue with Docker",
"id": 3094826898,
"name": "docker",
"node_id": "MDU6TGFiZWwzMDk0ODI2ODk4",
"url": "https://api.github.com/repos/deepmind/alphafold/labels/docker"
}
] | closed | false | null | [] | null | [
"The latest version (4.14.0) of conda is in some way causing this. This is auto-installed when we run `conda update -qy conda` in the \"Install 3rd part software\" section. There is a workaround:\r\n\r\nIn the section you want to locate this piece of code:\r\n\r\n```python\r\n%shell conda update -qy conda \\\r\n && conda install -qy -c conda-forge \\\r\n python=3.7 \\\r\n openmm=7.5.1 \\\r\n pdbfixer\r\n```\r\n\r\nand change it with \r\n\r\n```python\r\n%shell conda install -qy conda=4.13.0 \\\r\n && conda install -qy -c conda-forge \\\r\n python=3.7 \\\r\n openmm=7.5.1 \\\r\n pdbfixer\r\n```",
"Note: the workaround code should have a space between \"install\" and \"-qy\"!",
"Thanks Michael! I've corrected the snippet for everyone's comfort.",
"Fixed in https://github.com/deepmind/alphafold/commit/dad1f286aeaa158702a98aeee0f5661f30de6da8. Many thanks @Meghpal.",
"my !gget setup alphafold on google colab invariably fails with these comments\r\n\r\nThu Oct 6 12:51:12 2022 ERROR \r\n Please install AlphaFold third-party dependency openmm v7.5.1 by running the following command from the command line: \r\n 'conda install -qy conda==4.13.0 && conda install -qy -c conda-forge openmm=7.5.1' \r\n (Recommendation: Follow with 'conda update -qy conda' to update conda to the latest version afterwards.)\r\n\r\nInspite of me doing \r\n\r\n!conda install -qy conda==4.13.0 && conda install -qy -c conda-forge openmm=7.5.1\r\nfollowed by !conda update -qy conda\r\n\r\nAny suggestions as what might begoing wrong? My runs on \r\n\r\n!conda install -qy conda==4.13.0 && conda install -qy -c conda-forge openmm=7.5.1\r\n!conda update -qy conda \r\n\r\nalways runs fine without having any brush of error.\r\n\r\nThanks in advance\r\n\r\nSucheta"
] | "2022-08-23T19:28:05Z" | "2022-10-06T12:56:13Z" | "2022-08-25T09:27:36Z" | NONE | null | Here is the log:
`17%
17/100 [elapsed: 05:10 remaining: 02:43]
Found existing installation: tensorflow 2.8.2+zzzcolab20220719082949
Uninstalling tensorflow-2.8.2+zzzcolab20220719082949:
Successfully uninstalled tensorflow-2.8.2+zzzcolab20220719082949
Reading package lists...
Building dependency tree...
Reading state information...
The following package was automatically installed and is no longer required:
libnvidia-common-460
Use 'sudo apt autoremove' to remove it.
The following additional packages will be installed:
libdivsufsort3
Suggested packages:
hmmer-doc
The following NEW packages will be installed:
hmmer libdivsufsort3
0 upgraded, 2 newly installed, 0 to remove and 20 not upgraded.
Need to get 1,164 kB of archives.
After this operation, 11.9 MB of additional disk space will be used.
Get:1 http://archive.ubuntu.com/ubuntu bionic/universe amd64 libdivsufsort3 amd64 2.0.1-3 [44.4 kB]
Get:2 http://archive.ubuntu.com/ubuntu bionic/universe amd64 hmmer amd64 3.1b2+dfsg-5ubuntu1 [1,119 kB]
Fetched 1,164 kB in 0s (9,405 kB/s)
debconf: unable to initialize frontend: Dialog
debconf: (No usable dialog-like program is installed, so the dialog based frontend cannot be used. at /usr/share/perl5/Debconf/FrontEnd/Dialog.pm line 76, <> line 2.)
debconf: falling back to frontend: Readline
debconf: unable to initialize frontend: Readline
debconf: (This frontend requires a controlling tty.)
debconf: falling back to frontend: Teletype
dpkg-preconfigure: unable to re-open stdin:
Selecting previously unselected package libdivsufsort3:amd64.
(Reading database ... 155676 files and directories currently installed.)
Preparing to unpack .../libdivsufsort3_2.0.1-3_amd64.deb ...
Unpacking libdivsufsort3:amd64 (2.0.1-3) ...
Selecting previously unselected package hmmer.
Preparing to unpack .../hmmer_3.1b2+dfsg-5ubuntu1_amd64.deb ...
Unpacking hmmer (3.1b2+dfsg-5ubuntu1) ...
Setting up libdivsufsort3:amd64 (2.0.1-3) ...
Setting up hmmer (3.1b2+dfsg-5ubuntu1) ...
Processing triggers for man-db (2.8.3-2ubuntu0.1) ...
Processing triggers for libc-bin (2.27-3ubuntu1.5) ...
Looking in indexes: https://pypi.org/simple, https://us-python.pkg.dev/colab-wheels/public/simple/
Collecting py3dmol
Downloading py3Dmol-1.8.1-py2.py3-none-any.whl (6.5 kB)
Installing collected packages: py3dmol
Successfully installed py3dmol-1.8.1
PREFIX=/opt/conda
Unpacking payload ...
Collecting package metadata (current_repodata.json): done
Solving environment: done
## Package Plan ##
environment location: /opt/conda
added / updated specs:
- _libgcc_mutex==0.1=main
- _openmp_mutex==4.5=1_gnu
- brotlipy==0.7.0=py39h27cfd23_1003
- ca-certificates==2022.3.29=h06a4308_1
- certifi==2021.10.8=py39h06a4308_2
- cffi==1.15.0=py39hd667e15_1
- charset-normalizer==2.0.4=pyhd3eb1b0_0
- colorama==0.4.4=pyhd3eb1b0_0
- conda-content-trust==0.1.1=pyhd3eb1b0_0
- conda-package-handling==1.8.1=py39h7f8727e_0
- conda==4.12.0=py39h06a4308_0
- cryptography==36.0.0=py39h9ce1e76_0
- idna==3.3=pyhd3eb1b0_0
- ld_impl_linux-64==2.35.1=h7274673_9
- libffi==3.3=he6710b0_2
- libgcc-ng==9.3.0=h5101ec6_17
- libgomp==9.3.0=h5101ec6_17
- libstdcxx-ng==9.3.0=hd4cf53a_17
- ncurses==6.3=h7f8727e_2
- openssl==1.1.1n=h7f8727e_0
- pip==21.2.4=py39h06a4308_0
- pycosat==0.6.3=py39h27cfd23_0
- pycparser==2.21=pyhd3eb1b0_0
- pyopenssl==22.0.0=pyhd3eb1b0_0
- pysocks==1.7.1=py39h06a4308_0
- python==3.9.12=h12debd9_0
- readline==8.1.2=h7f8727e_1
- requests==2.27.1=pyhd3eb1b0_0
- ruamel_yaml==0.15.100=py39h27cfd23_0
- setuptools==61.2.0=py39h06a4308_0
- six==1.16.0=pyhd3eb1b0_1
- sqlite==3.38.2=hc218d9a_0
- tk==8.6.11=h1ccaba5_0
- tqdm==4.63.0=pyhd3eb1b0_0
- tzdata==2022a=hda174b7_0
- urllib3==1.26.8=pyhd3eb1b0_0
- wheel==0.37.1=pyhd3eb1b0_0
- xz==5.2.5=h7b6447c_0
- yaml==0.2.5=h7b6447c_0
- zlib==1.2.12=h7f8727e_1
The following NEW packages will be INSTALLED:
_libgcc_mutex pkgs/main/linux-64::_libgcc_mutex-0.1-main
_openmp_mutex pkgs/main/linux-64::_openmp_mutex-4.5-1_gnu
brotlipy pkgs/main/linux-64::brotlipy-0.7.0-py39h27cfd23_1003
ca-certificates pkgs/main/linux-64::ca-certificates-2022.3.29-h06a4308_1
certifi pkgs/main/linux-64::certifi-2021.10.8-py39h06a4308_2
cffi pkgs/main/linux-64::cffi-1.15.0-py39hd667e15_1
charset-normalizer pkgs/main/noarch::charset-normalizer-2.0.4-pyhd3eb1b0_0
colorama pkgs/main/noarch::colorama-0.4.4-pyhd3eb1b0_0
conda pkgs/main/linux-64::conda-4.12.0-py39h06a4308_0
conda-content-tru~ pkgs/main/noarch::conda-content-trust-0.1.1-pyhd3eb1b0_0
conda-package-han~ pkgs/main/linux-64::conda-package-handling-1.8.1-py39h7f8727e_0
cryptography pkgs/main/linux-64::cryptography-36.0.0-py39h9ce1e76_0
idna pkgs/main/noarch::idna-3.3-pyhd3eb1b0_0
ld_impl_linux-64 pkgs/main/linux-64::ld_impl_linux-64-2.35.1-h7274673_9
libffi pkgs/main/linux-64::libffi-3.3-he6710b0_2
libgcc-ng pkgs/main/linux-64::libgcc-ng-9.3.0-h5101ec6_17
libgomp pkgs/main/linux-64::libgomp-9.3.0-h5101ec6_17
libstdcxx-ng pkgs/main/linux-64::libstdcxx-ng-9.3.0-hd4cf53a_17
ncurses pkgs/main/linux-64::ncurses-6.3-h7f8727e_2
openssl pkgs/main/linux-64::openssl-1.1.1n-h7f8727e_0
pip pkgs/main/linux-64::pip-21.2.4-py39h06a4308_0
pycosat pkgs/main/linux-64::pycosat-0.6.3-py39h27cfd23_0
pycparser pkgs/main/noarch::pycparser-2.21-pyhd3eb1b0_0
pyopenssl pkgs/main/noarch::pyopenssl-22.0.0-pyhd3eb1b0_0
pysocks pkgs/main/linux-64::pysocks-1.7.1-py39h06a4308_0
python pkgs/main/linux-64::python-3.9.12-h12debd9_0
readline pkgs/main/linux-64::readline-8.1.2-h7f8727e_1
requests pkgs/main/noarch::requests-2.27.1-pyhd3eb1b0_0
ruamel_yaml pkgs/main/linux-64::ruamel_yaml-0.15.100-py39h27cfd23_0
setuptools pkgs/main/linux-64::setuptools-61.2.0-py39h06a4308_0
six pkgs/main/noarch::six-1.16.0-pyhd3eb1b0_1
sqlite pkgs/main/linux-64::sqlite-3.38.2-hc218d9a_0
tk pkgs/main/linux-64::tk-8.6.11-h1ccaba5_0
tqdm pkgs/main/noarch::tqdm-4.63.0-pyhd3eb1b0_0
tzdata pkgs/main/noarch::tzdata-2022a-hda174b7_0
urllib3 pkgs/main/noarch::urllib3-1.26.8-pyhd3eb1b0_0
wheel pkgs/main/noarch::wheel-0.37.1-pyhd3eb1b0_0
xz pkgs/main/linux-64::xz-5.2.5-h7b6447c_0
yaml pkgs/main/linux-64::yaml-0.2.5-h7b6447c_0
zlib pkgs/main/linux-64::zlib-1.2.12-h7f8727e_1
Preparing transaction: done
Executing transaction: done
installation finished.
WARNING:
You currently have a PYTHONPATH environment variable set. This may cause
unexpected behavior when running the Python interpreter in Miniconda3.
For best results, please verify that your PYTHONPATH only points to
directories of packages that are compatible with the Python interpreter
in Miniconda3: /opt/conda
env: PATH=/opt/conda/bin:/opt/bin:/usr/local/nvidia/bin:/usr/local/cuda/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/tools/node/bin:/tools/google-cloud-sdk/bin
Collecting package metadata (current_repodata.json): ...working... done
Solving environment: ...working... done
## Package Plan ##
environment location: /opt/conda
added / updated specs:
- conda
The following packages will be downloaded:
package | build
---------------------------|-----------------
_openmp_mutex-5.1 | 1_gnu 21 KB
ca-certificates-2022.07.19 | h06a4308_0 124 KB
certifi-2022.6.15 | py39h06a4308_0 153 KB
cffi-1.15.1 | py39h74dc2b5_0 228 KB
conda-4.14.0 | py39h06a4308_0 915 KB
cryptography-37.0.1 | py39h9ce1e76_0 1.3 MB
cytoolz-0.11.0 | py39h27cfd23_0 350 KB
ld_impl_linux-64-2.38 | h1181459_1 654 KB
libgcc-ng-11.2.0 | h1234567_1 5.3 MB
libgomp-11.2.0 | h1234567_1 474 KB
libstdcxx-ng-11.2.0 | h1234567_1 4.7 MB
ncurses-6.3 | h5eee18b_3 781 KB
openssl-1.1.1q | h7f8727e_0 2.5 MB
pip-22.1.2 | py39h06a4308_0 2.5 MB
requests-2.28.1 | py39h06a4308_0 92 KB
setuptools-63.4.1 | py39h06a4308_0 1.1 MB
sqlite-3.39.2 | h5082296_0 1.1 MB
tk-8.6.12 | h1ccaba5_0 3.0 MB
toolz-0.11.2 | pyhd3eb1b0_0 49 KB
tqdm-4.64.0 | py39h06a4308_0 126 KB
urllib3-1.26.11 | py39h06a4308_0 182 KB
xz-5.2.5 | h7f8727e_1 339 KB
zlib-1.2.12 | h7f8727e_2 106 KB
------------------------------------------------------------
Total: 25.9 MB
The following NEW packages will be INSTALLED:
cytoolz pkgs/main/linux-64::cytoolz-0.11.0-py39h27cfd23_0
toolz pkgs/main/noarch::toolz-0.11.2-pyhd3eb1b0_0
The following packages will be REMOVED:
colorama-0.4.4-pyhd3eb1b0_0
conda-content-trust-0.1.1-pyhd3eb1b0_0
six-1.16.0-pyhd3eb1b0_1
The following packages will be UPDATED:
_openmp_mutex 4.5-1_gnu --> 5.1-1_gnu
ca-certificates 2022.3.29-h06a4308_1 --> 2022.07.19-h06a4308_0
certifi 2021.10.8-py39h06a4308_2 --> 2022.6.15-py39h06a4308_0
cffi 1.15.0-py39hd667e15_1 --> 1.15.1-py39h74dc2b5_0
conda 4.12.0-py39h06a4308_0 --> 4.14.0-py39h06a4308_0
cryptography 36.0.0-py39h9ce1e76_0 --> 37.0.1-py39h9ce1e76_0
ld_impl_linux-64 2.35.1-h7274673_9 --> 2.38-h1181459_1
libgcc-ng 9.3.0-h5101ec6_17 --> 11.2.0-h1234567_1
libgomp 9.3.0-h5101ec6_17 --> 11.2.0-h1234567_1
libstdcxx-ng 9.3.0-hd4cf53a_17 --> 11.2.0-h1234567_1
ncurses 6.3-h7f8727e_2 --> 6.3-h5eee18b_3
openssl 1.1.1n-h7f8727e_0 --> 1.1.1q-h7f8727e_0
pip 21.2.4-py39h06a4308_0 --> 22.1.2-py39h06a4308_0
requests pkgs/main/noarch::requests-2.27.1-pyh~ --> pkgs/main/linux-64::requests-2.28.1-py39h06a4308_0
setuptools 61.2.0-py39h06a4308_0 --> 63.4.1-py39h06a4308_0
sqlite 3.38.2-hc218d9a_0 --> 3.39.2-h5082296_0
tk 8.6.11-h1ccaba5_0 --> 8.6.12-h1ccaba5_0
tqdm pkgs/main/noarch::tqdm-4.63.0-pyhd3eb~ --> pkgs/main/linux-64::tqdm-4.64.0-py39h06a4308_0
urllib3 pkgs/main/noarch::urllib3-1.26.8-pyhd~ --> pkgs/main/linux-64::urllib3-1.26.11-py39h06a4308_0
xz 5.2.5-h7b6447c_0 --> 5.2.5-h7f8727e_1
zlib 1.2.12-h7f8727e_1 --> 1.2.12-h7f8727e_2
Preparing transaction: ...working... done
Verifying transaction: ...working... done
Executing transaction: ...working... done
Collecting package metadata (current_repodata.json): ...working... done
Solving environment: ...working... failed with initial frozen solve. Retrying with flexible solve.
Solving environment: ...working... failed with repodata from current_repodata.json, will retry with next repodata source.
Collecting package metadata (repodata.json): ...working... done
Solving environment: ...working... done
## Package Plan ##
environment location: /opt/conda
added / updated specs:
- openmm=7.5.1
- pdbfixer
- python=3.7
The following packages will be downloaded:
package | build
---------------------------|-----------------
ca-certificates-2022.6.15 | ha878542_0 149 KB conda-forge
certifi-2022.6.15 | py37h89c1867_0 155 KB conda-forge
cffi-1.14.6 | py37hc58025e_0 225 KB conda-forge
colorama-0.4.5 | pyhd8ed1ab_0 18 KB conda-forge
conda-4.14.0 | py37h89c1867_0 1010 KB conda-forge
conda-package-handling-1.8.1| py37h540881e_1 1.0 MB conda-forge
cryptography-37.0.2 | py37h38fbfac_0 1.5 MB conda-forge
cudatoolkit-11.7.0 | hd8887f6_10 831.6 MB conda-forge
fftw-3.3.10 |nompi_h77c792f_102 6.4 MB conda-forge
libblas-3.9.0 |15_linux64_openblas 12 KB conda-forge
libcblas-3.9.0 |15_linux64_openblas 12 KB conda-forge
libgfortran-ng-12.1.0 | h69a702a_16 23 KB conda-forge
libgfortran5-12.1.0 | hdcd56e2_16 1.8 MB conda-forge
liblapack-3.9.0 |15_linux64_openblas 12 KB conda-forge
libopenblas-0.3.20 |pthreads_h78a6416_0 10.1 MB conda-forge
numpy-1.21.6 | py37h976b520_0 6.1 MB conda-forge
ocl-icd-2.3.1 | h7f98852_0 119 KB conda-forge
ocl-icd-system-1.0.0 | 1 4 KB conda-forge
openmm-7.5.1 | py37h96c4ddf_1 10.7 MB conda-forge
openssl-1.1.1o | h166bdaf_0 2.1 MB conda-forge
pdbfixer-1.7 | pyhd3deb0d_0 167 KB conda-forge
pip-22.2.2 | pyhd8ed1ab_0 1.5 MB conda-forge
pycosat-0.6.3 |py37h540881e_1010 107 KB conda-forge
pysocks-1.7.1 | py37h89c1867_5 28 KB conda-forge
python-3.7.10 |hffdb5ce_100_cpython 57.3 MB conda-forge
python_abi-3.7 | 2_cp37m 4 KB conda-forge
requests-2.28.1 | pyhd8ed1ab_0 53 KB conda-forge
ruamel_yaml-0.15.80 |py37h5e8e339_1006 270 KB conda-forge
setuptools-65.2.0 | py37h89c1867_0 1.4 MB conda-forge
six-1.16.0 | pyh6c4a22f_0 14 KB conda-forge
tqdm-4.64.0 | pyhd8ed1ab_0 81 KB conda-forge
urllib3-1.25.8 | py37hc8dfbb8_1 160 KB conda-forge
------------------------------------------------------------
Total: 934.1 MB
The following NEW packages will be INSTALLED:
colorama conda-forge/noarch::colorama-0.4.5-pyhd8ed1ab_0
cudatoolkit conda-forge/linux-64::cudatoolkit-11.7.0-hd8887f6_10
fftw conda-forge/linux-64::fftw-3.3.10-nompi_h77c792f_102
libblas conda-forge/linux-64::libblas-3.9.0-15_linux64_openblas
libcblas conda-forge/linux-64::libcblas-3.9.0-15_linux64_openblas
libgfortran-ng conda-forge/linux-64::libgfortran-ng-12.1.0-h69a702a_16
libgfortran5 conda-forge/linux-64::libgfortran5-12.1.0-hdcd56e2_16
liblapack conda-forge/linux-64::liblapack-3.9.0-15_linux64_openblas
libopenblas conda-forge/linux-64::libopenblas-0.3.20-pthreads_h78a6416_0
numpy conda-forge/linux-64::numpy-1.21.6-py37h976b520_0
ocl-icd conda-forge/linux-64::ocl-icd-2.3.1-h7f98852_0
ocl-icd-system conda-forge/linux-64::ocl-icd-system-1.0.0-1
openmm conda-forge/linux-64::openmm-7.5.1-py37h96c4ddf_1
pdbfixer conda-forge/noarch::pdbfixer-1.7-pyhd3deb0d_0
python_abi conda-forge/linux-64::python_abi-3.7-2_cp37m
six conda-forge/noarch::six-1.16.0-pyh6c4a22f_0
The following packages will be REMOVED:
brotlipy-0.7.0-py39h27cfd23_1003
cytoolz-0.11.0-py39h27cfd23_0
The following packages will be UPDATED:
conda-package-han~ pkgs/main::conda-package-handling-1.8~ --> conda-forge::conda-package-handling-1.8.1-py37h540881e_1
cryptography pkgs/main::cryptography-37.0.1-py39h9~ --> conda-forge::cryptography-37.0.2-py37h38fbfac_0
pip pkgs/main/linux-64::pip-22.1.2-py39h0~ --> conda-forge/noarch::pip-22.2.2-pyhd8ed1ab_0
pycosat pkgs/main::pycosat-0.6.3-py39h27cfd23~ --> conda-forge::pycosat-0.6.3-py37h540881e_1010
pysocks pkgs/main::pysocks-1.7.1-py39h06a4308~ --> conda-forge::pysocks-1.7.1-py37h89c1867_5
setuptools pkgs/main::setuptools-63.4.1-py39h06a~ --> conda-forge::setuptools-65.2.0-py37h89c1867_0
The following packages will be SUPERSEDED by a higher-priority channel:
ca-certificates pkgs/main::ca-certificates-2022.07.19~ --> conda-forge::ca-certificates-2022.6.15-ha878542_0
certifi pkgs/main::certifi-2022.6.15-py39h06a~ --> conda-forge::certifi-2022.6.15-py37h89c1867_0
cffi pkgs/main::cffi-1.15.1-py39h74dc2b5_0 --> conda-forge::cffi-1.14.6-py37hc58025e_0
conda pkgs/main::conda-4.14.0-py39h06a4308_0 --> conda-forge::conda-4.14.0-py37h89c1867_0
openssl pkgs/main::openssl-1.1.1q-h7f8727e_0 --> conda-forge::openssl-1.1.1o-h166bdaf_0
python pkgs/main::python-3.9.12-h12debd9_0 --> conda-forge::python-3.7.10-hffdb5ce_100_cpython
requests pkgs/main/linux-64::requests-2.28.1-p~ --> conda-forge/noarch::requests-2.28.1-pyhd8ed1ab_0
ruamel_yaml pkgs/main::ruamel_yaml-0.15.100-py39h~ --> conda-forge::ruamel_yaml-0.15.80-py37h5e8e339_1006
tqdm pkgs/main/linux-64::tqdm-4.64.0-py39h~ --> conda-forge/noarch::tqdm-4.64.0-pyhd8ed1ab_0
urllib3 pkgs/main::urllib3-1.26.11-py39h06a43~ --> conda-forge::urllib3-1.25.8-py37hc8dfbb8_1
Preparing transaction: ...working... done
Verifying transaction: ...working... done
Executing transaction: ...working... By downloading and using the CUDA Toolkit conda packages, you accept the terms and conditions of the CUDA End User License Agreement (EULA): https://docs.nvidia.com/cuda/eula/index.html
done
Retrieving notices: ...working... failed
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1125, in __call__
File "/opt/conda/lib/python3.9/site-packages/conda/cli/main.py", line 86, in main_subshell
File "/opt/conda/lib/python3.9/site-packages/conda/cli/conda_argparse.py", line 93, in do_call
File "/opt/conda/lib/python3.9/site-packages/conda/notices/core.py", line 75, in wrapper
File "/opt/conda/lib/python3.9/site-packages/conda/notices/core.py", line 39, in display_notices
File "/opt/conda/lib/python3.9/site-packages/conda/notices/http.py", line 36, in get_notice_responses
File "/opt/conda/lib/python3.9/site-packages/conda/notices/http.py", line 39, in <genexpr>
File "/opt/conda/lib/python3.9/concurrent/futures/_base.py", line 609, in result_iterator
File "/opt/conda/lib/python3.9/concurrent/futures/_base.py", line 446, in result
File "/opt/conda/lib/python3.9/concurrent/futures/_base.py", line 391, in __get_result
File "/opt/conda/lib/python3.9/concurrent/futures/thread.py", line 58, in run
File "/opt/conda/lib/python3.9/site-packages/conda/notices/http.py", line 42, in <lambda>
File "/opt/conda/lib/python3.9/site-packages/conda/notices/cache.py", line 37, in wrapper
File "/opt/conda/lib/python3.9/site-packages/conda/notices/http.py", line 58, in get_channel_notice_response
File "/opt/conda/lib/python3.9/site-packages/requests/sessions.py", line 600, in get
File "/opt/conda/lib/python3.9/site-packages/requests/sessions.py", line 587, in request
File "/opt/conda/lib/python3.9/site-packages/requests/sessions.py", line 701, in send
File "/opt/conda/lib/python3.9/site-packages/requests/adapters.py", line 460, in send
File "/opt/conda/lib/python3.9/site-packages/requests/adapters.py", line 263, in cert_verify
OSError: Could not find a suitable TLS CA certificate bundle, invalid path: /opt/conda/lib/python3.9/site-packages/certifi/cacert.pem
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/bin/conda", line 13, in <module>
sys.exit(main())
File "/opt/conda/lib/python3.9/site-packages/conda/cli/main.py", line 129, in main
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1413, in conda_exception_handler
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1128, in __call__
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1170, in handle_exception
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1181, in handle_unexpected_exception
File "/opt/conda/lib/python3.9/site-packages/conda/exceptions.py", line 1251, in print_unexpected_error_report
ModuleNotFoundError: No module named 'conda.cli.main_info'
---------------------------------------------------------------------------
CalledProcessError Traceback (most recent call last)
[<ipython-input-1-0c215f21152d>](https://localhost:8080/#) in <module>
35 PATH=get_ipython().run_line_magic('env', 'PATH')
36 get_ipython().run_line_magic('env', 'PATH=/opt/conda/bin:{PATH}')
---> 37 get_ipython().run_line_magic('shell', 'conda update -qy conda && conda install -qy -c conda-forge python=3.7 openmm=7.5.1 pdbfixer')
38 pbar.update(80)
39
2 frames
[/usr/local/lib/python3.7/dist-packages/google/colab/_system_commands.py](https://localhost:8080/#) in check_returncode(self)
133 if self.returncode:
134 raise subprocess.CalledProcessError(
--> 135 returncode=self.returncode, cmd=self.args, output=self.output)
136
137 def _repr_pretty_(self, p, cycle): # pylint:disable=unused-argument
CalledProcessError: Command 'conda update -qy conda && conda install -qy -c conda-forge python=3.7 openmm=7.5.1 pdbfixer' returned non-zero exit status 1.`
Michael. | {
"+1": 2,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 2,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/573/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/573/timeline | null | completed | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/572 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/572/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/572/comments | https://api.github.com/repos/deepmind/alphafold/issues/572/events | https://github.com/deepmind/alphafold/issues/572 | 1,348,150,048 | I_kwDOFoWQLM5QWyMg | 572 | AlphaFold Multimer v2.2.2 Incorrectly Predicts Homo-oligomer Despite PDB Structure | {
"avatar_url": "https://avatars.githubusercontent.com/u/107201368?v=4",
"events_url": "https://api.github.com/users/Aroids/events{/privacy}",
"followers_url": "https://api.github.com/users/Aroids/followers",
"following_url": "https://api.github.com/users/Aroids/following{/other_user}",
"gists_url": "https://api.github.com/users/Aroids/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/Aroids",
"id": 107201368,
"login": "Aroids",
"node_id": "U_kgDOBmPDWA",
"organizations_url": "https://api.github.com/users/Aroids/orgs",
"received_events_url": "https://api.github.com/users/Aroids/received_events",
"repos_url": "https://api.github.com/users/Aroids/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/Aroids/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/Aroids/subscriptions",
"type": "User",
"url": "https://api.github.com/users/Aroids"
} | [] | open | false | null | [] | null | [] | "2022-08-23T15:35:49Z" | "2022-08-23T15:36:12Z" | null | NONE | null | An interesting case of ~accurate protomer prediction but poor multimer prediction:
I predicted a homo-oligomer complex (ring) for a well-characterized protein with AlphaFold Multimer v2.2.2 (using the full database). The protein has a nearly complete PDB structure which AlphaFold uses as a template. The predicted structure has the expected general topology; however, the alpha-carbons of the predicted and PDB structures align very poorly (RMSD 26), and it is visually apparent that there is excessive space between the chains of the predicted structure resulting in an abnormally large ring diameter. The alignment object suggests that the chains of the predicted structure should be in a different order.
The individual chains of the predicted structure align to the individual chains of the PDB structure fairly well (RMSD ~2). When the chains of the predicted structure are aligned individually, the resulting 'reassembled' predicted structure globally aligns to the PDB structure with an RMSD <2.
| {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/572/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/572/timeline | null | null | null | null | false |
https://api.github.com/repos/deepmind/alphafold/issues/571 | https://api.github.com/repos/deepmind/alphafold | https://api.github.com/repos/deepmind/alphafold/issues/571/labels{/name} | https://api.github.com/repos/deepmind/alphafold/issues/571/comments | https://api.github.com/repos/deepmind/alphafold/issues/571/events | https://github.com/deepmind/alphafold/issues/571 | 1,346,670,446 | I_kwDOFoWQLM5QRI9u | 571 | Jackhmmer/UniRef90 Issue | {
"avatar_url": "https://avatars.githubusercontent.com/u/106316626?v=4",
"events_url": "https://api.github.com/users/ena2016/events{/privacy}",
"followers_url": "https://api.github.com/users/ena2016/followers",
"following_url": "https://api.github.com/users/ena2016/following{/other_user}",
"gists_url": "https://api.github.com/users/ena2016/gists{/gist_id}",
"gravatar_id": "",
"html_url": "https://github.com/ena2016",
"id": 106316626,
"login": "ena2016",
"node_id": "U_kgDOBlZDUg",
"organizations_url": "https://api.github.com/users/ena2016/orgs",
"received_events_url": "https://api.github.com/users/ena2016/received_events",
"repos_url": "https://api.github.com/users/ena2016/repos",
"site_admin": false,
"starred_url": "https://api.github.com/users/ena2016/starred{/owner}{/repo}",
"subscriptions_url": "https://api.github.com/users/ena2016/subscriptions",
"type": "User",
"url": "https://api.github.com/users/ena2016"
} | [] | closed | false | null | [] | null | [
"Okay, I'm running the idea that this runtime error could be a lack of vRAM. Going to plug in my dinosaur GPU acc and see if it does the trick. I'm praying it doesn't short or explode while running.\r\n\r\nUpdate:\r\nIt didn't work, I got the same error message.",
"Updated to v2.2.3 and attempted to truncate the TR/pMHCII as much as possible. it gets stuck on the HLA alpha chain and stops there. I'm switching the order to HLA beta first. Probably wont do anything but trying everything here.",
"Figured out my issue, my TRpMHCII complex sequences had to be cleaned up using one of the scripts above. Everything is working now. \r\n\r\nIt could be an interesting feature to have AF validate fasta files before running the MSAs, there are open source Fasta validators on GitHub too, maybe a collab, then create a script to clean the files before the run. \r\n\r\nScript flow\r\nCheck/validate fasta file -> clean up fasta file script (while depositing cleaned up fasta file in the output path, to create reproducibility folder) -> run AF\r\n\r\nThis was a headache but definitely made me go through all the issues on AF related to jackhmmer, will leave this issue here in case people run into same error."
] | "2022-08-22T16:20:36Z" | "2022-08-27T00:11:44Z" | "2022-08-27T00:11:44Z" | NONE | null | Hi all, I ran into this issue running .fasta for a DRA/B pMHC TRA/B complex model.
### Input-
`python3 docker/run_docker.py \
--data_dir=/media/eric/NewCupcake \
--output_dir=/media/eric/OldCupcake/tmp/alphafold \
--fasta_paths=/media/eric/OldCupcake/FastaFiles/TRpep79_HLA–DQB10201DQA10201.fasta\
--max_template_date=2020-08-20 \
--model_preset=multimer
`
### Output-
`I0822 10:24:30.640870 139730341103424 run_docker.py:255] max_sto_sequences=self.uniref_max_hits)
I0822 10:24:30.640907 139730341103424 run_docker.py:255] File "/app/alphafold/alphafold/data/pipeline.py", line 94, in run_msa_tool
I0822 10:24:30.640944 139730341103424 run_docker.py:255] result = msa_runner.query(input_fasta_path, max_sto_sequences)[0] # pytype: disable=wrong-arg-count
I0822 10:24:30.640981 139730341103424 run_docker.py:255] File "/app/alphafold/alphafold/data/tools/jackhmmer.py", line 172, in query
I0822 10:24:30.641017 139730341103424 run_docker.py:255] input_fasta_path, self.database_path, max_sequences)
I0822 10:24:30.641054 139730341103424 run_docker.py:255] File "/app/alphafold/alphafold/data/tools/jackhmmer.py", line 143, in _query_chunk
I0822 10:24:30.641093 139730341103424 run_docker.py:255] 'Jackhmmer failed\nstderr:\n%s\n' % stderr.decode('utf-8'))
I0822 10:24:30.641130 139730341103424 run_docker.py:255] RuntimeError: Jackhmmer failed
I0822 10:24:30.641168 139730341103424 run_docker.py:255] stderr:
I0822 10:24:30.641204 139730341103424 run_docker.py:255] Fatal exception (source file p7_alidisplay.c, line 728):
I0822 10:24:30.641240 139730341103424 run_docker.py:255] backconverted trace didn't end at expected place on model (UniRef90_S9X7U2/chain_D)
I0822 10:24:30.641277 139730341103424 run_docker.py:255]
I0822 10:24:30.641314 139730341103424 run_docker.py:255]
`
I'm running source code from v2.2.2
I tried opening UniRef90 and deleting the sequences but it did not work.
I tried redownloading from UnitProt and that didn't solve the issue.
I redownloaded the whole database and this did not work either.
I'm stuck on what to try next. Any help is appreciated.
UPDATE 8/23/2022
The culprit is the MHCII molecules these are the sequences:
>DQA102:01
`EDIVADHVASYGVNLYQSYGPSGQFTHEFDGDEEFYVDLERKETVWKLPLFHRLR.FDPQFALTNIAVLKHNLNILIKRSNSTAATNEVPEVTVFSKSPVTLGQPNTLICLVDNIFPPVVNITWLSNGHSVTEGVSETSFLSKSDHSFFKISYLTFLPSADEIYDCKVEHWGLDEPLLKHWEPEIPAPMSEVDGGGGGAQLEKELQALEKENAQLEWELQALEKELAQ`
>DQB102:01
`RDSPEDFVYQFKGMCYFTNGTERVRLVSRSIYNREEIVRFDSDVGEFRAVTLLGLPAAEYWNSQKDILERKRAAVDRVCRHNYQLELRTTLQRRVEPTVTISPSRTEALNHHNLLVCSVTDFYPAQIKVRWFRNDQEETAGVVSTPLIRNGDWTFQILVMLEMTPQRGDVYTCHVEHPSLQSPITVEWRAQSESAQSKVDGGGGGAQLKKKLQALKKKNAQLKWKLQALKKKLAQ`
Is there a way to have Jackhmmer skip sequences that have utf-8 errors?
Update 8/24/2022
I was reading HMMER Userguide and I'm going to try --noali flag, will update to see if it worked.
Update 8/24/2022
It didn't work. :/
Gonna try these next..
Find any utf-8 in fasta file
`grep -axv '.*' /media/eric/NewCupcake/uniref90/uniref90.fasta`
Remove any utf-8 in fasta file
`iconv -f utf-8 -t utf-8 -c /media/eric/NewCupcake/uniref90/uniref90.fasta -o /media/eric/NewCupcake/uniref90/uniref90v2.fasta`
Update:
It also did not work | {
"+1": 0,
"-1": 0,
"confused": 0,
"eyes": 0,
"heart": 0,
"hooray": 0,
"laugh": 0,
"rocket": 0,
"total_count": 0,
"url": "https://api.github.com/repos/deepmind/alphafold/issues/571/reactions"
} | https://api.github.com/repos/deepmind/alphafold/issues/571/timeline | null | completed | null | null | false |